Abstract
The sequence specificity of the recombination activating gene (RAG) complex during V(D)J recombination has been well studied. RAGs can also act as structure-specific nuclease; however, little is known about the mechanism of its action. Here, we show that in addition to DNA structure, sequence dictates the pattern and efficiency of RAG cleavage on altered DNA structures. Cytosine nucleotides are preferentially nicked by RAGs when present at single-stranded regions of heteroduplex DNA. Although unpaired thymine nucleotides are also nicked, the efficiency is many fold weaker. Induction of single- or double-strand breaks by RAGs depends on the position of cytosines and whether it is present on one or both of the strands. Interestingly, RAGs are unable to induce breaks when adenine or guanine nucleotides are present at single-strand regions. The nucleotide present immediately next to the bubble sequence could also affect RAG cleavage. Hence, we propose “C(d)C(S)C(S)” (d, double-stranded; s, single-stranded) as a consensus sequence for RAG-induced breaks at single-/double-strand DNA transitions. Such a consensus sequence motif is useful for explaining RAG cleavage on other types of DNA structures described in the literature. Therefore, the mechanism of RAG cleavage described here could explain facets of chromosomal rearrangements specific to lymphoid tissues leading to genomic instability.
Keywords: DNA Damage, DNA Enzymes, DNA Recombination, Immunology, Purine, Chromosomal Translocation, Double-strand Break, Genomic Instability, RAGs, V(D)J Recombination
Introduction
The recombination activating gene (RAG)3 complex, consisting of RAG1 and RAG2, is the nuclease responsible for V(D)J recombination, a physiological process by which immunoglobulin and T-cell receptor diversity is generated. RAGs are normally expressed in B-cells and T-cells (1). During V(D)J recombination, the variable (V), diversity (D), and joining (J) subexons are rearranged. Specific sequences present at the ends of the subexon, called recombination signal sequences (RSS) are recognized by RAGs. Each RSS consists of a conserved heptamer and nonamer, separated by a nonconserved spacer, the length of which designates RSS as a 12-signal or 23-signal. Normally during V(D)J recombination, a 12-signal pairs with a 23-signal with the help of proteins like HMGB1 (high mobility group box 1). The nick induced by RAGs during V(D)J recombination is consistently 5′ of the heptamers (2–14). The nicked strand is then converted to a hairpin in each V, D, and J coding end by a transesterification reaction, leaving each of the signal ends blunt (15). The hairpins are then opened by the Artemis-DNAPKcs complex (16). After cleavage, the RAG complex remains tightly bound to the two signal ends and less tightly bound to the coding end, in a postcleavage complex (17, 18). Finally, the complete exon coding for antibody or TCR (T-cell receptor) is generated by joining of the broken subexons by nonhomologous DNA endjoining (NHEJ) (19, 20).
In the recent past, studies have shown that cryptic RSS sites present elsewhere in the genome can also act as off-target sites for RAG misrecognition, leading to chromosomal translocations in lymphoid cancers such as leukemia (21–24). In addition to its sequence-specific endonuclease activities, recent studies have shown that the RAG complex can act as a structure-specific nuclease (22). We previously showed that a non-B DNA structure formed at the BCL2 major breakpoint region (MBR) on chromosome 18 involved in t(14;18) translocation in follicular lymphoma can be cleaved by RAGs (25–29). Further, it was also shown that in addition to the BCL2 MBR structure, other non-B DNA structures, such as heteroduplex DNA and heterologous loops are also targets for the RAG complex (30, 31). The RAG complex can also cleave 3′ overhangs, flap DNA, and gap structures (32). All of these studies were done at physiological concentrations of divalent cation, Mg2+. Two independent groups have also shown that RAGs are able to cleave hairpin structures in Mn2+-containing buffer, and this was observed to a much lesser extent in Mg2+ (33, 34). In a recent study, we found that the most common translocations in early human B cells occur at CpG sites. We proposed that deamination of methylated cytosines at CpG can lead to small unpaired bubble-like regions in the genome, which RAGs can cleave to generate breaks (35).
The above studies, therefore, suggest that changes in the normal B-DNA structure could make the region vulnerable to RAGs when present in lymphoid tissues. This could explain why lymphoid cells possess elevated levels of chromosomal translocation and other rearrangements compared with non-lymphoid tissues. Such elevated levels of pathological chromosomal rearrangement leads to the altered expression of critical genes resulting in human lymphoid malignancies such as leukemia and lymphoma (28, 36). Because such structural changes in the DNA could lead to elevated levels of single- and double-strand breaks, those could also account for the increased genomic instability.
Our previous studies have shown that when acting as a structure-specific nuclease, RAGs recognize and bind to the single-stranded region of heteroduplex DNA, and the efficiency of the cleavage depends on the length of the single-stranded region (28, 30, 31). However, there are many important questions unanswered. What dictates the specificity of RAG cleavage when it acts as a structure-specific nuclease? Because, mostly such cleavage on altered DNA structures is pathologic, it is important to know the mechanism which determines whether a given region may be cleaved by RAGs or not. This will also help to understand the mechanism of genomic instability in lymphoid cells. We previously reported the occurrence of strand bias, when RAGs cleave on non-B DNA structures (30). Because double-strand break formation needs two independent nicks in close proximity on the DNA, it was not understood how such nicks can lead to DSBs during chromosomal rearrangements. It has been shown that during sequence-specific cleavage of RSS by RAGs, the coding sequence influences the nicking (37–39). Thus, the role of neighboring sequences, when RAGs act as a structure-specific nuclease, deserves examination.
In the present study we have attempted to understand the mechanism by which RAGs cleave altered DNA structures. Here we report that RAG cleavage on heteroduplex DNA is sequence-dependent. RAGs can cleave heteroduplex DNA only when pyrimidines are present at the single-stranded region. Cytosines are preferred, and when present on both strands, RAG cleavage leads to a double-strand break. We further show that the sequence dependence is also applicable to all other structural alterations studied.
EXPERIMENTAL PROCEDURES
Enzymes, Chemicals, and Reagents
Chemical reagents were obtained from Sigma, Amresco, and SRL. DNA-modifying enzymes were purchased from New England Biolabs (Beverly, MA) and Fermentas (Glen Burnie, MD). Radioisotope-labeled nucleotides were purchased from BRIT (Hyderabad, India). Culture media were from Sera Laboratory International Limited (West Sussex, RH17 5PB UK), and fetal bovine serum and PenStrep were from Invitrogen.
Oligomers
Oligomers were from Sigma. The sequences of oligomers are shown in supplemental Table S1. The oligomers were purified using 8–15% denaturing polyacrylamide gel electrophoresis. The complementary oligomers were annealed in 100 mm NaCl and 1 mm EDTA by boiling for 10 min, followed by slow cooling. Hairpin loop-forming oligomers were incubated without salt in boiling water and were immediately kept on ice for 30 min before use.
5′ End Labeling of Oligomers
The 5′ end labeling of the oligomeric DNA was done using T4 polynucleotide kinase in a buffer containing 20 mm Tris acetate (pH 7.9), 10 mm magnesium acetate, 50 mm potassium acetate,1 mm DTT, and [γ-32P]ATP at 37 °C for 1 h. The labeled substrates were purified using Qiagen quick nucleotide removal kit and stored at −80 °C until used.
RAG Expression and Purification
Both core RAG1 (cRAG1) and core RAG2 (cRAG2) are cloned in vector pEBG in the BamHI/NotI site, and they are expressed as N-terminal GST fusion proteins under transcriptional control of elongation factor 1α promoter as described earlier (31, 39). Expression vectors for mouse cRAG1 (amino acids 384–1008) and mouse cRAG2 (amino acids 1–383) were transiently transfected into 293T cells (human embryonic kidney epithelial cells expressing simian virus 40 large tumor (T) antigen) by the calcium phosphate precipitation method. Cells were harvested after 48 h, and proteins were purified as described earlier (31, 39). Purity was tested on SDS-PAGE and by Western blotting (supplemental Fig. S1A). MBP cRAGs (RAG1, amino acids 384–1040; RAG2, amino acids 1–383) and full-length RAGs (FLRAG1, 1–1040 amino acids; FLRAG2, 1–527 amino acids) were purified using a mild method as described (40). Briefly, 14 plates of 293T cells were transfected with 10 μg of plasmids each for MBP cRAG1/cRAG2 or cRAG1/FLRAG2 by the calcium phosphate method. For MBP FLRAG1/cRAG2 purification, 20 μg of FLRAG1 and 10 μg of cRAG2 plasmids were used. After 48 h of transfection, cells were harvested, and proteins were purified using amylose resin column (New England Biolabs). Eluted fractions of MBP-RAG proteins were checked by CBB staining (supplemental Fig. S1B). The activity was checked by site-specific nicking on standard RSS. However, because of poor solubility, our efforts to purify FLRAG1 and FLRAG2 complex were unsuccessful.
Copurified MBP cRAG1 and cRAG2 were fractionated on a Biogel P-100 (Bio-Rad) column equilibrated with 25 mm HEPES (pH 7.4), 150 mm KCl, 10 mm MgCl2, 10% glycerol, and 2 mm DTT. Fractions (100 μl) were collected and tested for the presence of cRAG1 and cRAG2 proteins by silver staining (supplemental Fig. S2A). The activity of the fractions was checked by RAG cleavage assay on standard 12-RSS and 6-nt bubble substrates with the (C/C)6 sequence.
RAG Cleavage on Oligomeric DNA
Appropriate oligomeric substrates were incubated with RAG proteins for 1 h at 37 °C in a buffer containing 25 mm MOPS, (pH 7.0), 30 mm KCl, 30 mm potassium glutamate, and 5 mm MgCl2 as described earlier (30, 31). In the control, RAG reaction buffer alone was used. Reactions were terminated by adding the loading dye containing formamide, and products were resolved on 15% denaturing polyacrylamide gels. The gels were dried and exposed to a PhosphorImager screen, and signal was detected using a Fuji PhosphorImager FLA9000 (Fuji, Japan). Incubation times used for time-course experiments are indicated in the respective figure legends. When RAG cleavage reactions were performed to study DNA double-strand breaks, native dye with glycerol was added to the sample following the cleavage reaction, were loaded onto a 15% native polyacrylamide gel, and signals were detected as described above. Each experiment described in the present study was done a minimum of two independent times (independent reaction incubations) with complete agreement.
For quantification of RAG cleavage, MultiGauge software (v3.0) was used. We first selected a rectangular area covering the substrate DNA band in the lane containing no RAG and quantified the intensity. Then we placed the same size rectangle on all the cleaved bands resulting from RAG activity and quantified. An equal area from elsewhere in the gel where there was no specific band was used as background and was subtracted. We considered the no-RAG control substrate as 100% and compared with the cleavage product intensities. For example, we got “x” as the substrate band intensity and “y” as the intensity of cleavage products of equal area after background subtraction; the % of cleavage was calculated as (y/x)100.
Electrophoretic Mobility Shift Assay
The [γ-32P]ATP-labeled bubble substrates were incubated with RAGs in a buffer containing 25 mm MOPS (pH 7.0), 30 mm KCl, 30 mm potassium glutamate, 1 mm MnCl2, and 5 mm MgCl2. In the no-RAG control reactions, buffer alone was used. A 45-bp double-stranded oligomer (0.1 μm) was used as nonspecific DNA. Reaction mixtures were incubated at 37 °C for 10 min. The products were resolved on a 6% native polyacrylamide gel, and signals were detected using a PhosphorImager.
P1 Nuclease Cleavage Assay
The substrate DNA containing different types of bubble sequences was incubated with P1 nuclease as described earlier (41). In each experiment, a 5′ end-labeled substrate DNA was tested for P1 nuclease sensitivity by incubating with increasing concentrations (0.001, 0.01, and 0.1 units) of P1 nuclease in a buffer containing 10 mm Tris-HCl (pH 7.9), 10 mm MgCl2, 50 mm NaCl, and 1 mm DTT at 37 °C for 30 min. Reaction products were then resolved on a 12–15% denaturing PAGE and analyzed.
RESULTS
Previously we have shown that the non-B DNA structure is a target for RAG cleavage under physiological conditions (22). This was shown in the context of a non-B DNA structure formed at BCL2 MBR (29). Subsequently, we found that a symmetrical bubble or heterologous loop present on an oligomeric DNA could also be cleaved by RAGs (31). During these studies, we noted a clear strand bias in the RAG cleavage efficiency between the top and bottom strands of bubble structures (30), which was inexplicable.
RAG Cleavage Efficiency of the DNA Strands Changes with Sequence of the Bubbles
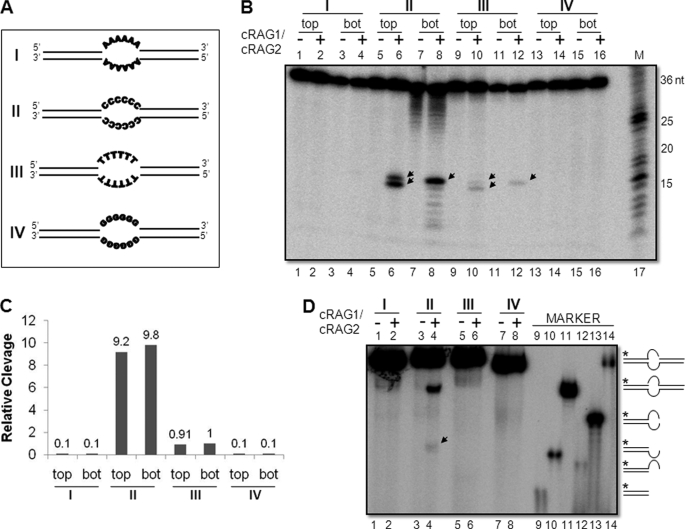
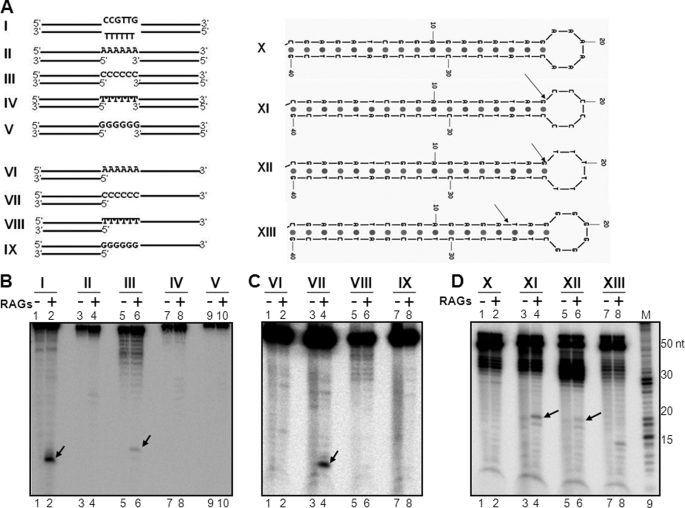
To understand the mechanistic aspects of the strand bias during RAG cleavage on heteroduplex DNA, we synthesized oligomers with different bubble sequences. Preliminary results showed that the efficiency of RAG cleavage on heteroduplex DNA is dependent on the sequence composition of the bubble (data not shown). Based on this, we tested which would be the most favored nucleotide for optimal RAG cleavage of non-B DNA structures. To address this question, we generated oligomeric DNA substrates containing bubble sequences with (A/A)6, (C/C)6, (T/T)6, or (G/G)6 (Fig. 1A). In all cases, the length of the double-stranded arms was 15-bp each. Oligomeric DNA, with either top or bottom strands radiolabeled, was incubated with RAGs at 37 °C for 1 h, and the products were resolved on a 15% PAGE gel. Results showed efficient RAG cleavage on both top and bottom strands when the heteroduplex DNA with cytosine bubbles were used (Fig. 1, B, lanes 5–8 and C). Distinct RAG cleavage was also seen when thymine bubbles were used (Fig. 1B, lanes 9–12), though the efficiency of the cleavage was many fold lower (Fig. 1C). To our surprise, we could not detect any RAG cleavage on heteroduplex DNA, when adenines and guanines were present as bubble sequences (Fig. 1B, lanes 1–4, 13–16, and C). Consistent with this, we found that RAG binding also occurs preferentially to cytosine or thymine bubble-containing substrates (supplemental Fig. S3). Further, we confirmed the presence of 6-nt bubbles in all four heteroduplex DNA substrates by P1 nuclease cleavage assay (data not shown). Hence, our results show that the presence of altered DNA structures alone is not sufficient for RAG cleavage, and cytosines are the favored nucleotides for its cleavage on heteroduplex DNA.
FIGURE 1.
Comparison of efficiency of RAG cleavage with respect to composition of sequences in the bubble region of heteroduplex DNA. A, diagrammatic representation of oligomeric substrates containing 6-nt bubbles. The bubble sequences are shown in bold letters. The double-stranded arms are of 15-bp length each and indicated with double lines. Sequence of the bubble region are (A/A)6, (C/C)6, (T/T)6, or (G/G)6 and are denoted as I, II, III, and IV, respectively. B, polyacrylamide gel profile showing RAG cleavage on heteroduplex DNA, I, II, III, and IV described in A. RAG cleavage reactions were done in the buffer containing 5 mm MgCl2 using DNA substrates, which were [γ32P]ATP-labeled either on the top or bottom strand for 1 h at 37 °C and were resolved on a 15% denaturing PAGE gel. top indicates that radiolabeled strand is the top strand. bot indicates that the bottom strand is radiolabeled. The Klenow partial-digested 1-nt ladder was used as marker. RAG-specific cleavage products are indicated by an arrow. C, bar diagram showing quantification of RAG cleavage efficiency of the top and bottom strands of heteroduplex DNA structures described in B. The RAG cleavage products were quantified using MultiGauge software. The substrate amount in the respective no-RAG lane was taken as 100%, and the relative cleavage of products was calculated and indicated as %. In all cases, background was subtracted. The calculated percentage is shown on top of the respective columns in the bar diagram. D, detection of RAG-induced DSBs on bubble substrates shown in A. RAG cleavage products were resolved on 15% native PAGE gels. The markers for possible nicked or double-stranded breaks are shown on the right hand side. The bands due to DSBs are indicated by an arrow.
Comparable results were also seen when the length of the double-stranded arms was increased to 25 from 15 bp while maintaining bubble sequences as (A/A)6, (C/C)6, (T/T)6, or (G/G)6 (supplemental Fig. S4, A–C). These results suggest that irrespective of the length of the flanking region, RAGs can nick heteroduplex DNA structures, when appropriate sequences are present.
Because we noticed that efficient RAG cleavage on heteroduplex DNA structures was seen only when stretches of cytosines were present, we wondered what would be the status of RAG cleavage when the same sequences are present on a duplex DNA. Results showed that RAGs do not cleave cytosines when present on a duplex DNA (C/G)6, even if they are present in stretches (data not shown). Therefore, the observed cytosine specificity of RAGs is restricted to heteroduplex DNA.
RAG Cleavage on Heteroduplex DNA with Cytosine Bubbles Leads to Induction of Double-stranded Breaks
DNA double-strand breaks are prerequisites for formation of chromosomal translocations and other chromosomal rearrangements. Hence, we wondered whether the RAG-induced nicks in the top and bottom strands could contribute to formation of DSBs. To test this, the heteroduplex DNA substrates (Fig. 1A) were incubated with RAGs, and cleavage products were resolved on a native polyacrylamide gel. Results showed that in the case of bubbles with cytosines, RAG cleavage led to the formation of double-strand breaks (Fig. 1D, lanes 3 and 4) suggesting that both strands of the same molecule were cleaved. However, we could not find RAG cleavage on any other substrates including bubbles with thymines (Fig. 1D). Further the identity of the band due to DSB was studied using specific markers as indicated (Fig. 1D, lanes 9–14). Results showed that the DSB observed was due to independent cuts at two different single/double-strand transitions positioned diagonally (Fig. 1D, lanes 4 and 10). Hence, it is evident that that when cytosines are present at the single-stranded region on both top and bottom strands of heteroduplex DNA it can lead to DSBs. Further, we found that RAG cleavage on cytosine bubble could induce DSBs, irrespective of the length of the side arms (supplemental Fig. S4D). However, in none of the other substrates DSBs could be induced (supplemental Fig. S4D). Therefore, our study shows that pyrimidines, particularly cytosines are the most favored nucleotides for DSB formation.
MBP-tagged Core and Full-length RAGs Show Cytosine Specificity on Heteroduplex DNA
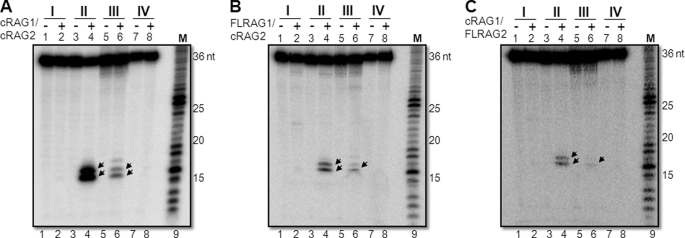
Because the above experiments were performed using GST-tagged cRAGs, we were interested in testing whether the observed sequence specificity of RAGs holds true when MBP-tagged cRAGs or FLRAGs were used (supplemental Fig. S1B). This is particularly important based on the report that GST may induce dimerization of the target protein and can have an effect on its properties. To test this, MBP core RAGs were incubated with bubble substrates with (A/A)6, (C/C)6, (T/T)6, or (G/G)6 single-stranded regions (Fig. 1A). Results showed that like GST cRAGs, the cleavage efficiency of MBP cRAGs was also many fold higher when cytosines were present at the bubble region (Fig. 2A, lanes 3 and 4). Besides, fractionation of MBP cRAGs on a size exclusion chromatography column indicated that the nuclease activity exhibited by RAGs indeed comigrated with RAG proteins (supplemental Fig. S2). The cleavage at the thymine-containing bubble was many fold weaker (Fig. 2A, lanes 5 and 6). Bubbles with adenine or guanine did not show any cleavage even with MBP cRAGs. Comparable results were obtained when full-length RAGs (FLRAG1/cRAG2 or cRAG1/FLRAG2) were used (Fig. 2, B and C). However, in both combinations of full-length RAGs, the overall efficiency of RAG cleavage was weaker. This suggests that the cytosine preference when RAGs act as a structure-specific nuclease is an inherent property of RAGs and that tags did not affect the cleavage property.
FIGURE 2.
Comparison of cleavage efficiency when core RAGs or full-length RAGs were used on different heteroduplex DNA. The bubble sequences containing (A/A)6, (C/C)6, (T/T)6, and (G/G)6 denoted as I, II, III, and IV, respectively, were incubated in a buffer containing 5 mm MgCl2 with either MBP cRAG1/cRAG2 (A) or MBP FLRAG1/cRAG2 (B) or MBP cRAG1/FLRAG2 (C). The RAG cleavage products were resolved on a denaturing PAGE gel and are indicated by arrows. M is a 1-nt molecular weight ladder. For other details refer to the Fig. 1 legend.
Prolonged Incubation Does Not Alter the Sequence Preference Exhibited by RAGs on Heteroduplex DNA
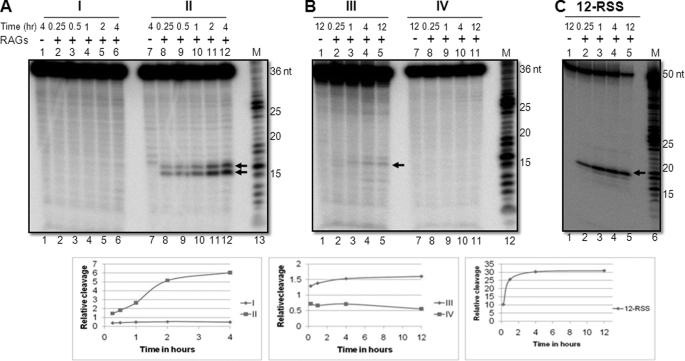
The above experiments were performed using an incubation time of 1 h. Therefore, we wondered whether increasing the RAG reaction time could change the sequence specificity. Besides, the kinetics of RAG cleavage when it acts as a structure-specific nuclease has never been studied. Time course experiments were performed on different heteroduplex DNA substrates containing (A/A)6, (C/C)6, (T/T)6, or (G/G)6 single-stranded regions (Fig. 1A). Results showed that the observed sequence preference on RAG cleavage remained unaltered irrespective of time of incubation (Fig. 3, A and B). The cytosine-containing bubble was getting preferentially cleaved while thymine cleavage remained weak (Fig. 3A, lanes 7–12; B, lanes 1–5). Adenine and guanine bubble cleavage was undetectable even with prolonged incubation time (Fig. 3A, lanes 1–6; B, lanes 7–11). Interestingly, we noted an increase in the cleavage efficiency of cytosine with an increase in the incubation time (Fig. 3A, lanes 7–12). This was true even in the case of thymine (Fig. 3B, lanes 1–5). However, as shown previously, RAG cleavage efficiency on RSS remained the same after 60 min (Fig. 3C).
FIGURE 3.
Comparison of time kinetics of RAG cleavage reaction of different heteroduplex DNA substrates. The bubble sequences containing (A/A)6, (C/C)6, (T/T)6, and (G/G)6 denoted as I, II, III, and IV, respectively, or oligomer containing 12 RSS were incubated for different time periods as indicated with either cRAG1/cRAG2 (GST-tagged) and resolved on a 15% denaturing PAGE gel. A, RAG cleavage kinetics of poly A/A and C/C heteroduplexes. B, RAG cleavage kinetics of poly T/T and G/G heteroduplexes. C, RAG cleavage kinetics of 12-RSS. RAG cleavage products are indicated by an arrow. M is the 1-nt molecular weight ladder. In each panel, RAG cleavage products resulted from respective gels were quantified and presented.
Two Cytosines Present at the Double-strand/Single-strand Junctions Are Critical for RAG Cleavage
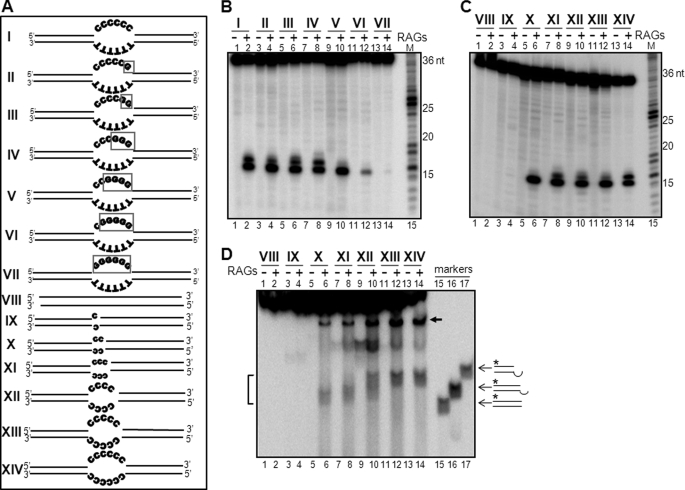
Our studies thus far showed that bubbles with six cytosines are efficiently cleaved by RAGs, compared with thymines, adenines, or guanines. However, it may be possible that all cytosines may not be important for RAG cleavage. To investigate the minimum number of cytosines required for RAG cleavage, we synthesized new oligomeric bubble substrates with decreasing numbers of cytosines at the bubble region by replacing them with guanines (Fig. 4A, I–VII). The bubble region in the antiparallel strand was TTTTTT in all cases. RAG cleavage studies showed that there was no significant difference in the efficiency of RAG cleavage when the number of cytosines was between 3 and 6 (Fig. 4B, lanes 1–8). In these cases, we could see two cleaved products, one at the 15-nt position at the junction of the single-strand/double-strand transition (Fig. 4B). The second product, which was weaker in intensity, was due to a cleavage at the first cytosine of the bubble. When the number of cytosines was reduced to 2, the efficiency of RAG cleavage at the junction remained the same; however, the cleavage at the first internal cytosine disappeared (Fig. 4B, lanes 9 and 10). Interestingly, when the number of cytosines was reduced to 1, cleavage efficiency reduced dramatically (Fig. 4B, lanes 11 and 12). When all cytosines were replaced with guanines, RAG cleavage was almost undetectable (Fig. 4B, lanes 13 and 14). P1 nuclease analysis confirmed the presence of a 6-nt bubble region in all the substrates (supplemental Fig. S5, A and B). Thus, our results showed that only two cytosines are critical for RAG cleavage on heteroduplex DNA even when a bubble of 6-nt length was present.
FIGURE 4.
Two cytosines within the bubble region are sufficient for optimal RAG cleavage on heteroduplex DNA structures. A, diagrammatic representation of oligomeric substrates containing either 6-nt bubbles with varying number of cytosines (denoted as I–VII) or with varying lengths of bubble region, 0–6 (denoted as VIII–XIV). The changes from Cs to Gs are indicated using a box (see I-VII). In all cases, the length of upstream and downstream sequences are the same (15 bp). The duplex DNA substrate (0-nt bubble) comprises the top strand of the 6-nt bubble (denoted as VIII) paired with the corresponding complementary sequence. In each case, the top strand oligomer was radiolabeled with [γ32P]ATP and used. B, polyacrylamide gel profile showing RAG cleavage on heteroduplex substrates I-VII. M is the 1-nt ladder. C, polyacrylamide gel profile showing RAG cleavage on heteroduplex substrates VIII-XIV shown in panel A. M is the 1-nt ladder. D, RAG induced double-strand breaks when DNA contains a bubble as small as 2 nt. Heteroduplex DNA substrates VIII-XIV were subjected to RAG cleavage, and the products were resolved on a 15% native PAGE gel to detect DSBs. The bands due to DSBs (bracketed) and single-strand breaks (arrow) are indicated. Molecular markers used for identifying the positions of DSBs are shown.
To check the minimum length of the bubble that can be cleaved by RAGs, when sequences at both strands of the bubble are cytosines, we generated substrates containing bubbles with 1–6 nucleotides of cytosines (Fig. 4A, IX–XIV). Results showed that RAGs could cleave the bubble substrates efficiently when the lengths of the bubbles were 2–6 nt (Fig. 4C, lanes 5–14). Interestingly, cleavage was weak when the length of the bubble was 1 nt (Fig. 4C, lanes 3 and 4). P1 nuclease analysis confirmed the bubble region in respective substrates (supplemental Fig. S5, A and C). When a single nucleotide mismatch of C/A was used in the context of CCG/GAC in an oligomeric DNA substrate of 31-bp length, we could detect specific RAG cleavage with low efficiency in one strand (supplemental Fig. S6A). The cleavage efficiency was better when a single nucleotide mismatch of C/C was used in the context of CCG/GCC in an oligomeric DNA substrate of 31-bp length (supplemental Fig. S6B, lanes 3 and 4). More importantly, a C/C mismatch in this case also led to detectable RAG nicking on both top and bottom strands (supplemental Fig. S6B, lanes 3–6). These results suggest that the immediate flanking sequence of the mismatch region also affects the efficiency of RAG cleavage. Further, we also tested whether the observed cleavage at the 1-nt mismatch could be influenced by the length of the double-stranded arms. To test this, we generated a 70-nt oligomer with either CCG/GAC or CCG/GCC 1-nt mismatches. Results showed detectable RAG cleavage on both substrates, although the efficiency was weak (supplemental Fig. S6C) suggesting that the length of the double-stranded arms did not affect the RAG cleavage efficiency even when a 1-nt mismatch is present.
Based on the above results, we tested whether the RAG cleavage at 2-nt cytosine bubbles could lead to DSB formation. Following RAG cleavage of the above substrates (Fig. 4A, VIII–XIV), products were analyzed on a native PAGE. Interestingly, we found that RAGs could induce DSBs when cytosines were present on bubble sequences except in the case of the 1-nt bubble (Fig. 4D). The strong band seen below substrates, following treatment with RAGs (Fig. 4D, lanes 6, 8, 10, 12, and 14) were identified as the product caused by RAG nicking resulting in a single-strand break (supplemental Fig. S7). In the case of a 1-nt bubble, the DSB formation was undetectable (Fig. 4D, lanes 3 and 4). Therefore, our results confirm that as low as a 2-nt bubble with cytosine could generate DSBs upon cleavage with RAGs.
Cytosine Preference Is Seen for RAG Cleavage on 3′ Overhangs, Gaps, and Hairpin Loops
Because we find that RAG cleavage is preferred when cytosines are present on bubble structures, we tested whether a similar rule applies for other DNA structures studied in the literature (32). To experimentally evaluate the hypothesis, we have generated oligomeric substrates containing gaps, 3′ overhang, or stem loop structures (Fig. 5A). In the case of gap structures, the region corresponding to the gap was synthesized with AAAAAA, CCCCCC, TTTTTT, or GGGGGG (Fig. 5A, II-V). In the case of overhangs, the 6 nt at the overhang region next to double-stranded DNA was replaced with AAAAAA, CCCCCC, TTTTTT, or GGGGGG (Fig. 5A, VI-IX). In the case of stem loops, the loop region was synthesized with AAAAAA, CCCCCC, TTTTTT, or GGGGGG sequences (Fig. 5A, X-XIII). A 6-nt bubble substrate containing two cytosines was used as positive control (Fig. 5A, I). In all cases, radiolabeled substrate DNA was incubated in RAG reaction buffer containing 5 mm MgCl2 as described earlier. Results showed that efficient RAG cleavage at gap or overhang DNA structures was seen only when CCCCCC was present within the single-stranded regions (Fig. 5, B, lanes 5 and 6 and C, lanes 3 and 4). In the case of other sequences, we could not find any detectable RAG cleavage under the physiological concentrations of MgCl2 used (Fig. 5, B and C). When similar studies were performed using stem loops, we noticed that RAG cleavage was preferred when CCCCCC was present at the single-strand/double-strand transition (Fig. 5D, lanes 3 and 4). The cleavage at the thymine loop was weaker (Fig. 5D, lanes 5 and 6). Although we did find a band in the stem loop containing GGGGGG, it did not match with the normal cleavage position. These results suggest that the observed RAG cleavage preference in the bubble structures is a general characteristic and is applicable to other types of DNA structures as well.
FIGURE 5.
RAG cleavage on gap DNA structures, protruding overhangs, and hairpin structures. A, diagrammatic representation of oligomeric substrates containing gap DNA structures (denoted as I–V), 3′ overhangs (denoted as VI–IX), and hairpin loops (denoted as X–XIII) with poly A, C, T, or G at the single-stranded region as indicated. In the case of gap substrates, the length of the double-stranded arm is 15 bp toward one side and 10 bp toward the other side. In the case of overhangs, the length of the double-stranded region is 15 bp, whereas the length of the single-stranded region was also 15 nt. In the case of hairpin loop structures, the length of the loop region was 6 nt whereas the length of the double-stranded arm was 18 bp. B–D, polyacrylamide gel profile showing RAG cleavage on gap structures (B), protruding overhangs (C), and hairpin structures (D). In all panels, RAG cleavage reactions were done in a buffer containing 5 mm MgCl2 for 1 h at 37 °C and were resolved on a 15% denaturing PAGE gel. For other details refer to the Fig. 1 legend.
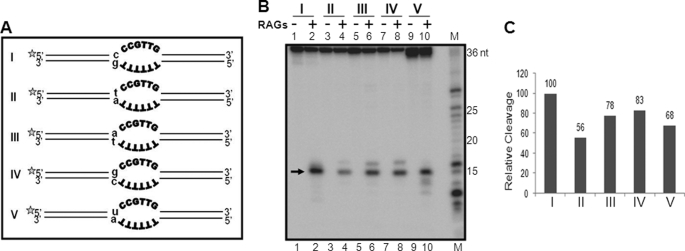
An Immediate Single Nucleotide Mutation Alters the Cleavage Efficiency at Bubble Sequences
We have tested the role of neighboring sequences on RAG cleavage in non-B DNA structures two different ways. In one of the experiments, RAG cleavage was performed following the swapping of the duplex arms of the heteroduplex DNA, and results showed no difference in the cleavage pattern and efficiency (data not shown). Next we tested whether a single nucleotide mutation immediately next to the bubble region can affect the cleavage efficiency as seen in the case of the 1-nt bubble described above. A 36-bp oligomeric substrate containing a 6-nt bubble with 2 cytosines immediately next to the single-stranded region was used for the study (Fig. 6A). Five different single nucleotide mutations at the double-stranded region immediately flanking the bubble sequence was created by changing C/G of the wild-type oligomer to T/A, A/T, G/C, or U/A (Fig. 6A). To our surprise, we found that the C → T mutation led to ∼50% reduction in the RAG cleavage efficiency (Fig. 6, B, lanes 1–4 and C). It was also observed that C → A, C → G, or C → U conversion led to a reduction in the RAG cleavage efficiency, although it was limited (Fig. 6, B and C). We also observed a comparable reduction in RAG binding (data not shown). Hence, our results suggest that the nucleotide at double-stranded DNA, immediately upstream of the bubble region, can affect the RAG cleavage, and it is most efficient when cytosine is present.
FIGURE 6.
Effect of neighboring sequences on RAG cleavage on heteroduplex DNA structures. A, diagrammatic representation of oligomeric substrates containing 6-nt bubbles with different flanking single nucleotide mutations (indicated in small letters). In each case, the length of the arm is 15 bp. The asterisk indicates the strand that is radiolabeled with [γ32P]ATP. B, polyacrylamide gel profile showing the effect of a single nucleotide change on RAG cleavage at heteroduplex DNA structures. RAG-specific cleavage products are indicated by an arrow. C, bar diagram showing quantification of RAG cleavage efficiency at the top strand of heteroduplex DNA described in A and B. For other details refer to the Fig. 1 legend.
DISCUSSION
In the present study, we identified novel recognition sequences for the RAG complex when acting as a structure-specific nuclease. Further, we showed that both structure and sequence features of the heteroduplex DNA are important in determining the pattern and efficiency of RAG cleavage.
Sequences of the Single-stranded Region and Immediate Neighboring Sequences Affect RAG Cleavage on Heteroduplex DNA
RAGs are well-studied as a sequence-specific nuclease for their role in V(D)J recombination. Its specificity at recombination signal sequences are extensively characterized by different groups (37–39, 42–44). Earlier studies by us and others (22) have shown that RAGs can also act as a structure-specific nuclease. In the present study, we find that although having an altered DNA structure is important for RAG recognition of the bubble-containing sequences, structure alone is not sufficient for its reactivity. Instead we find that the sequence composition of the bubbles dictate the pattern and efficiency of RAG cleavage. We find that cytosines are preferred over thymine for RAG-induced single- and double-strand breaks. Although overall cleavage efficiency was comparable between the top and bottom strands in respective cases, we found that cleavage at the top strand resulted in two bands, whereas it was only one in the case of the bottom strands. Such a difference in RAG cleavage could be due to differences in the neighboring sequences. However, more studies are required to identify the exact mechanism. The most interesting finding was that when the sequences of the bubbles were purines (adenine or guanine), there was no cleavage at the heteroduplex DNA at all. We also found that when cytosines were present, as small as 2-nt bubbles were sufficient for robust RAG cleavage. However, when nucleotides other than cytosines were present, the number of nucleotides required for optimal cleavage was 6 (30). Furthermore, the presence of a cytosine in the double-stranded region just upstream of the bubble sequence resulted in the highest RAG cleavage efficiency. Changing the C to A, or T or G significantly reduced the RAG cleavage efficiency. However, it is important to point out that the change of C to U did not change the cleavage efficiency, whereas a C to T conversion dramatically reduced RAG cleavage efficiency. This is understandable as both cytosine and uracil do not have the methyl group that thymine possesses.
Based on the above studies, it appears that for optimal RAG cleavage at altered DNA structures, two cytosines close to the 5′-end of the heteroduplex region are important. The presence of a cytosine in the duplex DNA next to the bubble is also preferred. Thus, we propose that “C(d)C(S)C(S)” (the subscript “d” denotes double-stranded, while “s” stands for single-stranded DNA) could be a consensus sequence for RAGs to induce single-strand breaks. A consensus sequence for inducing double-strand breaks could be C(d)C(S)C(S)/C(S)C(S). The presence of A or G in place of C in the single-stranded DNA could abolish the RAG cleavage completely. However, a replacement with T could still be cleaved, but with a much lower efficiency. In an earlier study, it has been shown that GC-rich sequences are the most fragile sites in the genome, though no clear consensus sequence was discernable (45).
Sequence Composition Specificity of RAGs on Bubble Structures Reflects a More General Property of RAGs
The RAG cleavage at sequences other than RSS sites was first reported for 3′ overhangs, flap DNA structures, and gap DNAs, and the authors showed that RAGs could cleave at single-strand/double-strand transitions even in buffers containing Mg2+ (32). In earlier studies, it was also shown that when the heptamer of the RSS was used for RAG cleavage in a single-stranded DNA context, RAGs could cleave at single-strand/double-strand transitions (38). It has also been shown that during V(D)J recombination, the intermediates containing flaps and overhangs could be cleaved by RAGs (46). Later, our own studies have shown that a non-B DNA structure present at the BCL2 MBR sequence could be cleaved by RAGs, which was also extended to other types of DNA structures, like heteroduplexes and heterologous loops (25–31, 35). All those experiments were performed in buffers that were close to physiologic conditions in which 5 mm MgCl2 was used. Hairpins, which are V(D)J recombination intermediates, were also used for testing RAG activity and in two independent studies, it was reported that in the presence Mn2+, RAGs were able to cleave hairpin intermediates (33, 34).
Because our studies have shown that RAG cleavage on altered DNA structures is preferred when cytosines but not purines are present on the bubble region, we wondered whether that could be true in other DNA structures studied in the literature. By using different overhang, gap, and stem loop substrates containing Cs, As, Ts, or Gs, we found that in all cases, cytosines were preferred for RAG cleavage. As in the bubble substrates, adenines, and guanines did not contribute toward specific RAG cleavage. It is more important to point out that in all cases Mg2+ was used for RAG cleavage rather than Mn2+. These data suggest that the observed sequence preference noticed during RAG cleavage on heteroduplex DNA structures is a more general property of RAGs when it acts as a structure-specific nuclease. Therefore, the sequence motif identified by us can be used to explain the published studies from the literature. It is interesting to point out that all the overhang substrates used in the earlier studies had cytosines at the single-strand/double-strand junctions (32, 47).
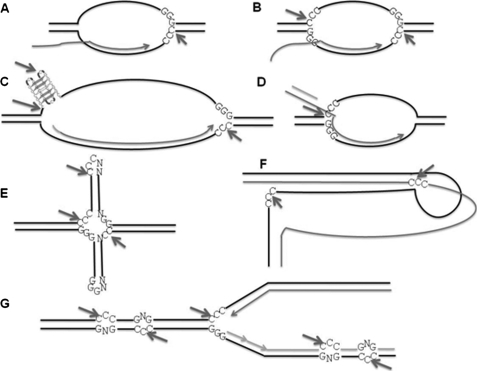
How Often Would One Expect Heteroduplex DNA Structures in the Human Genome in Lymphoid Tissues?
Normally DNA in our genome is expected to be in the B-form duplex conformation. However, when RAGs act as a structure-specific nuclease, it always recognizes the single-stranded region present in the altered DNA structure. Therefore, one of the major questions that arises is how often one would see such a type of structure in the human genome? Also what is the mechanism by which a B-DNA may be converted to a heteroduplex DNA? For duplex DNA to get converted into altered DNA, first it needs to get unpaired. This could be due to breathing of DNA, melting due to supercoiling during replication or transcription (48) (Fig. 7). Moreover, for each type of structure formation, specific types of sequences are required (28, 48, 49). For example, the presence of inverted repeats or palindromic sequences could lead to cruciform structures (50). The presence of direct repeats could lead to misaligned sequences. Homopurine:homopyrimidine stretches with mirror repeat symmetry could lead to formation of triplex DNA (49). G-quartets may be formed in stretches of Gs when appropriate conditions are provided (28, 48). G/C repeats during transcription could also lead to formation of RNA/DNA hybrids (51). In addition to these, even spontaneous deamination of cytosine could lead to mismatches. Deamination of a methylated cytosine could lead to a T:G mismatch (35). Any of these structures, when present in lymphoid tissues could be a target for RAGs, when appropriate sequences are present.
FIGURE 7.
Plausible RAG cleavage targets within the cells when it acts as a structure specific nuclease. A and B, bubble structures formed during transcription. Depending on the availability of Cs at the junction and size of the bubble, the RAG cleavage could lead to either SSB or DSB. C, transcription through a cytosine-rich template could lead to an RNA-DNA hybrid formation. The resulting G-rich single-stranded region may fold into G-quartet structures. D, D-loops formed during replication and recombination also could be a target for RAGs. E, cruciform structures with Cs at single-strand-double-strand transitions could be cleaved by RAGs. F, triple helical DNA formed at homopurine:pyrimidine tracts with mirror repeat symmetry could be a target for RAGs. G, mismatches generated during DNA replication could be a target for RAGs if those contain cytosine. In addition, tips of replication forks with cytosines could also be a target for RAGs.
RAG-induced Breaks in Heteroduplex DNA Structures: Relevance in Cancer and Genomic Instability
The observed RAG-induced breaks on bubble structures suggests that when such altered structures or mismatch regions are present in the cells, it could lead to different types of genomic rearrangements. Given that the sequence preference shown by core RAGs hold true for full-length RAGs as well, it suggests that this is an inherent property of RAGs and such a structural specificity is physiological. However, the rearrangements could be dependent on how often such structures may be present in the genome. Because RAGs are present only in lymphoid tissues, any type of rearrangement in other cell types is ruled out. As shown by our results, RAG-induced genomic instability could be controlled at the sequence level. Therefore, depending on the sequence of the single-stranded region of the heteroduplex DNA, either a single-strand break or double-strand break could be induced, which in turn could culminate in genomic instability and cancer (Fig. 7). Cancers like leukemia and lymphoma are restricted to lymphoid tissues and are characterized by presence of chromosomal abnormalities such as chromosomal translocations, deletions, inversions, and other mutations. Because such abnormalities might require multiple DSBs, it is possible that depending on the sequence, a DSB could be generated by RAGs by inducing two independent nicks (Fig. 7) (22, 52, 53). The probability to have such cytosines near single-strand/double-strand transitions is quite high, when structures like triplex DNA or G-quartets are formed in GC-rich sequences (28, 48, 49, 54) (Fig. 7). In cases where SSBs are generated, replication across a nick or cleavage by single-strand-specific enzymes such as Artemis could convert them into DSBs (55, 56).
Because we show that the C(d)C(S)C(S) motif can be the most favored sequence for RAG nicking and DSB formation on altered DNA structures including DNA mismatches and gaps in physiological conditions, this novel sequence motif could be a new recognition sequence for RAGs (Fig. 7), just like the RSS on a standard duplex B-DNA. This further suggests that the RAG cleavage pattern on altered DNA structures is context-specific, and both sequence and structural determinants act together to limit the RAG-induced genomic instability.
Supplementary Material
Acknowledgments
We thank Dr. Patrick Swanson for providing MBP RAG constructs and Mridula Nambiar for critical reading of the manuscript and help in cell culture. We thank Dr. Bibha Choudhary and members of the SCR laboratory for discussions and help. We also thank Dr. Raghavan Varadarajan for financial assistance.
This work was supported in part by grants from the Department of Science and Technology, India (SR/SO/BB-051/2007), and an Indian Institute of Science startup grant (to S. C. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
- RAG
- recombination-activating gene
- NHEJ
- nonhomologous DNA endjoining
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- MOPS
- 4-morpholinepropanesulfonic acid
- DSB
- double-stranded break
- SSB
- single-stranded break
- RSS
- recombination signal sequence
- MBR
- major breakpoint region
- MBP
- maltose-binding protein.
REFERENCES
- 1.Schatz D. G., Baltimore D. (2004) Cell 116, S103–S106 [DOI] [PubMed] [Google Scholar]
- 2.Lewis S. M. (1994) Adv. Immunol. 56, 27–150 [DOI] [PubMed] [Google Scholar]
- 3.Hiom K., Gellert M. (1998) Mol. Cell 1, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 4.Fugmann S. D., Lee A. I., Shockett P. E., Villey I. J., Schatz D. G. (2000) Annu. Rev. Immunol. 18, 495–527 [DOI] [PubMed] [Google Scholar]
- 5.Gellert M. (2002) Annu. Rev. Biochem. 71, 101–132 [DOI] [PubMed] [Google Scholar]
- 6.Roth D. B. (2003) Nat. Rev. Immunol. 3, 656–666 [DOI] [PubMed] [Google Scholar]
- 7.Schlissel M. S. (2003) Nat. Rev. Immunol. 3, 890–899 [DOI] [PubMed] [Google Scholar]
- 8.Sadofsky M. J. (2004) Immunol. Rev. 200, 83–89 [DOI] [PubMed] [Google Scholar]
- 9.Swanson P. C. (2004) Immunol. Rev. 200, 90–114 [DOI] [PubMed] [Google Scholar]
- 10.Oettinger M. A. (2004) Immunol. Rev. 200, 165–181 [DOI] [PubMed] [Google Scholar]
- 11.Jung D., Alt F. W. (2004) Cell 116, 299–311 [DOI] [PubMed] [Google Scholar]
- 12.Schatz D. G., Spanopoulou E. (2005) Curr. Top Microbiol. Immunol. 290, 49–85 [DOI] [PubMed] [Google Scholar]
- 13.Lieber M. R., Yu K., Raghavan S. C. (2006) DNA Repair 5, 1234–1245 [DOI] [PubMed] [Google Scholar]
- 14.Jung D., Giallourakis C., Mostoslavsky R., Alt F. W. (2006) Annu. Rev. Immunol. 24, 541–570 [DOI] [PubMed] [Google Scholar]
- 15.Roth D. B., Menetski J. P., Nakajima P. B., Bosma M. J., Gellert M. (1992) Cell 70, 983–991 [DOI] [PubMed] [Google Scholar]
- 16.Ma Y., Pannicke U., Schwarz K., Lieber M. R. (2002) Cell 108, 781–794 [DOI] [PubMed] [Google Scholar]
- 17.Agrawal A., Schatz D. G. (1997) Cell 89, 43–53 [DOI] [PubMed] [Google Scholar]
- 18.Jones J. M., Gellert M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12926–12931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieber M. R., Ma Y., Pannicke U., Schwarz K. (2003) Nat. Rev. Mol. Cell Biol. 4, 712–720 [DOI] [PubMed] [Google Scholar]
- 20.Rooney S., Chaudhuri J., Alt F. W. (2004) Immunol. Rev. 200, 115–131 [DOI] [PubMed] [Google Scholar]
- 21.Raghavan S. C., Kirsch I. R., Lieber M. R. (2001) J. Biol. Chem. 276, 29126–29133 [DOI] [PubMed] [Google Scholar]
- 22.Raghavan S. C., Lieber M. R. (2006) Bioessays 28, 480–494 [DOI] [PubMed] [Google Scholar]
- 23.Marculescu R., Le T., Simon P., Jaeger U., Nadel B. (2002) J. Exp. Med. 195, 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis S. M., Agard E., Suh S., Czyzyk L. (1997) Mol. Cell. Biol. 17, 3125–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghavan S. C., Chastain P., Lee J. S., Hegde B. G., Houston S., Langen R., Hsieh C. L., Haworth I. S., Lieber M. R. (2005) J. Biol. Chem. 280, 22749–22760 [DOI] [PubMed] [Google Scholar]
- 26.Raghavan S. C., Hsieh C. L., Lieber M. R. (2005) Mol. Cell. Biol. 25, 6475–6484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghavan S. C., Lieber M. R. (2004) Cell Cycle 3, 762–768 [PubMed] [Google Scholar]
- 28.Raghavan S. C., Lieber M. R. (2007) Front Biosci. 12, 4402–4408 [DOI] [PubMed] [Google Scholar]
- 29.Raghavan S. C., Swanson P. C., Wu X., Hsieh C. L., Lieber M. R. (2004) Nature 428, 88–93 [DOI] [PubMed] [Google Scholar]
- 30.Raghavan S. C., Gu J., Swanson P. C., Lieber M. R. (2007) DNA Repair 6, 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghavan S. C., Swanson P. C., Ma Y., Lieber M. R. (2005) Mol. Cell. Biol. 25, 5904–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santagata S., Besmer E., Villa A., Bozzi F., Allingham J. S., Sobacchi C., Haniford D. B., Vezzoni P., Nussenzweig M. C., Pan Z. Q., Cortes P. (1999) Mol. Cell 4, 935–947 [DOI] [PubMed] [Google Scholar]
- 33.Besmer E., Mansilla-Soto J., Cassard S., Sawchuk D. J., Brown G., Sadofsky M., Lewis S. M., Nussenzweig M. C., Cortes P. (1998) Mol. Cell 2, 817–828 [DOI] [PubMed] [Google Scholar]
- 34.Shockett P. E., Schatz D. G. (1999) Mol. Cell. Biol. 19, 4159–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai A. G., Lu H., Raghavan S. C., Muschen M., Hsieh C. L., Lieber M. R. (2008) Cell 135, 1130–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nambiar M., Kari V., Raghavan S. C. (2008) Biochim. Biophys. Acta 1786, 139–152 [DOI] [PubMed] [Google Scholar]
- 37.Cuomo C. A., Mundy C. L., Oettinger M. A. (1996) Mol. Cell. Biol. 16, 5683–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsden D. A., McBlane J. F., van Gent D. C., Gellert M. (1996) EMBO J. 15, 3197–3206 [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K., Lieber M. R. (1999) Mol. Cell. Biol. 19, 8094–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raval P., Kriatchko A. N., Kumar S., Swanson P. C. (2008) Nucleic Acids Res. 36, 2060–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naik A. K., Raghavan S. C. (2008) DNA Repair 7, 1384–1391 [DOI] [PubMed] [Google Scholar]
- 42.Lieber M. R., Hesse J. E., Mizuuchi K., Gellert M. (1987) Genes Dev. 1, 751–761 [DOI] [PubMed] [Google Scholar]
- 43.Roth D. B., Zhu C., Gellert M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10788–10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBlane J. F., van Gent D. C., Ramsden D. A., Romeo C., Cuomo C. A., Gellert M., Oettinger M. A. (1995) Cell 83, 387–395 [DOI] [PubMed] [Google Scholar]
- 45.Greenbaum J. A., Parker S. C., Tullius T. D. (2007) Genome Res 17, 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grawunder U., Lieber M. R. (1997) Nucleic Acids Res. 25, 1375–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishihara T., Nagawa F., Nishizumi H., Kodama M., Hirose S., Hayashi R., Sakano H. (2004) Mol. Cell. Biol. 24, 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinden R. R. (ed) (1994) DNA Structure and Function, Academic Press, San Diego, CA [Google Scholar]
- 49.Mirkin S. M. (2007) Nature 447, 932–940 [DOI] [PubMed] [Google Scholar]
- 50.Bacolla A., Wells R. D. (2004) J. Biol. Chem. 279, 47411–47414 [DOI] [PubMed] [Google Scholar]
- 51.Yu K., Chedin F., Hsieh C. L., Wilson T. E., Lieber M. R. (2003) Nat. Immunol. 4, 442–451 [DOI] [PubMed] [Google Scholar]
- 52.Franco S., Alt F. W., Manis J. P. (2006) DNA Repair 5, 1030–1041 [DOI] [PubMed] [Google Scholar]
- 53.Mills K. D., Ferguson D. O., Alt F. W. (2003) Immunol. Rev. 194, 77–95 [DOI] [PubMed] [Google Scholar]
- 54.Wells R. D. (2007) Trends Biochem. Sci. 32, 271–278 [DOI] [PubMed] [Google Scholar]
- 55.Kuzminov A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8241–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma Y., Schwarz K., Lieber M. R. (2005) DNA Repair 4, 845–851 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.