Abstract
Recognition of acetylated chromatin by the bromodomains and extra-terminal domain (BET) family proteins is a hallmark for transcriptional activation and anchoring viral genomes to mitotic chromosomes of the host. One of the BET family proteins BRD2 interacts with acetylated chromatin during mitosis and leads to transcriptional activation in culture cells. Here, we report the crystal structures of the N-terminal bromodomain of human BRD2 (BRD2-BD1; residues 74–194) in complex with each of three different Lys-12-acetylated H4 peptides. The BRD2-BD1 recognizes the H4 tail acetylated at Lys-12 (H4K12ac), whereas the side chain of hypoacetylated Lys-8 of H4 binds at the cavity of the dimer interface of BRD2-BD1. From binding studies, we identified the BRD2-BD1 residues that are responsible for recognition of the Lys-12-acetylated H4 tail. In addition, mutation to Lys-8 in the Lys-12-acetylated H4 tail decreased the binding to BRD2-BD1, implicating the critical role of Lys-8 in the Lys-12-acetylated H4 tail for the recognition by BRD2-BD1. Our findings provide a structural basis for deciphering the histone code by the BET bromodomain through the binding with a long segment of the histone H4 tail, which presumably prevents erasure of the histone code during the cell cycle.
Keywords: Chromatin/Regulation, Methods/X-ray Crystallography, Protein/Structure, Crystal Structure, Protein Domains, Acetylation, BET Family, BRD2 BD1, Bromodomain
Introduction
In eukaryotes, genomic DNA is complexed with core histones, consisting of two H2A-H2B dimers and one H3-H4 tetramer, to form a nucleoprotein architecture called the nucleosome (1, 2). These four core histones include a central core domain, and N- and C-terminal tail regions. The N-terminal histone tails, especially that of histone H4, are rich in lysine residues, onto which several different acetyltransferases and methyltransferases covalently add post-translational modifications (3, 4). The histone codes defined by combinations of such a histone tail modification are considered as a key regulatory mechanism for DNA metabolisms that trigger alteration of the chromatin structure and/or association of several different trans-acting factors (5–7). Among these post-translational modifications, acetylation of the histone tails and its recognition are typical hallmarks for activation of chromatin DNA (4, 7, 8, 10). The acetylated N-terminal tails of the histones are selectively recognized by the bromodomain, an ∼110-amino acid-long domain found in several chromatin-associated factors, including nuclear histone acetyltransferases, ATP-dependent chromatin-remodeling factors, and the bromodomain and extra-terminal domain (BET)6 family of nuclear proteins (11, 12). The human BET family, including BRD2, BRD3, BRD4, and BRDT, has a unique architecture with two tandem bromodomains and a conserved extra-terminal domain (13–15). In the intact nuclei, the BET protein BRD2 associates through its bromodomains mainly with the acetylated lysine 12 (K12ac) of H4, one of the active marks of chromatin, and activates transcription (16). Another study indicates that BRD2 binds chromatin containing Lys-12- or Lys-5-acetylated histone H4, although it scarcely bound to those containing Lys-16-acetylated H4 or Lys-9-acetylated H3 (17). Interestingly, the BET proteins BRD2 and BRD4 associate with such acetylated chromatins throughout the cell cycle (16, 18, 19), whereas other non-BET bromodomain proteins dissociate from the chromosomes during mitosis (20–22). This tight retention on chromosomes is a unique feature of the BET proteins, which is utilized by papilloma virus and presumably by Kaposi sarcoma-associated herpesvirus, for tethering their genomes to the mitotic chromosome of the host and for propagating them during cell division of the host (23, 24). Furthermore, disruption of Brd2 in mice causes severe obesity without type 2 diabetes (25), suggesting that BRD2 is a potential therapeutic target.
Recently, we showed from structural and biochemical studies that the N-terminal bromodomain of BRD2 forms a dimer (26). In this study, to understand the molecular mechanism of BRD2 in the histone code recognition, we determined the crystal structures of the N-terminal bromodomain (BD1; see Fig. 1A) of BRD2 in complex with the acetylated histone H4 tails. We found that the BRD2-BD1 bromodomain recognized a long segment of the histone H4 tail that is acetylated at Lys-12 (H4K12ac). From structural and biochemical analyses, we also suggest that not only K12ac but also Lys-8 are important for the recognition of the Lys-12-acetylated histone H4 tail by BRD2-BD1.
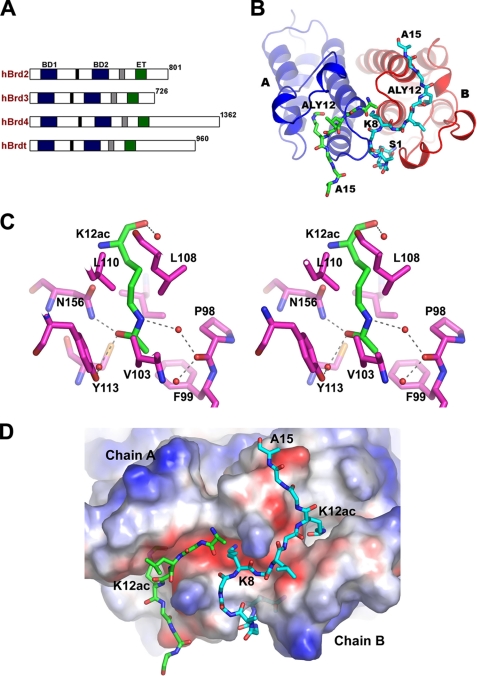
FIGURE 1.
Crystal structure of the BRD2-BD1 bromodomain in complex with Lys-12-acetylated histone H4 tails. A, schematic representation of human BET proteins. B, structure of the BD1 dimer complex. The BD1 molecules and the H4K12ac tails are shown as ribbon and sticks, respectively. C, recognition mode of K12ac. The hydrophobic environment around K12ac and water molecules (red balls) is indicated by the stick model. Molecular interactions involving K12ac are shown with broken lines. D, electrostatic surface potential of the BD1 dimer in the complex. The H4K12ac tails bound to the protein dimer are depicted as sticks. The residues Ser-1 to Gly-6 of the H4K12ac tail, which associate with molecule B, run through the dimeric interface. Lys-8 side chain interacts with the negatively charged cavity of BD2, which is produced by the dimer association. E, stereo view of the close-up view of the BD1 complex structure in the binding site region. The BD1 molecules A and B are shown as a ribbon diagram, and the H4K12ac tails are shown as sticks. F, electrostatic interaction of Lys-8 in the H4K12ac tail at the BD1 dimeric interface. Structural figures were generated with PyMol.
EXPERIMENTAL PROCEDURES
Expression and Purification of BRD2-BD1
The BRD2-BD1 protein used for crystallization and binding analysis was expressed and purified as described previously (26). Several BRD2-BD1 mutants were directly introduced into the prepared pET15b-BRD2-BD1 constructs by PCRs, using a QuikChange II XL site-directed mutagenesis kit (Stratagene). The wild-type and the BD1 mutants (Y113A, I154A, N156A, P158D, D160A, P111D/D112A, D112A/I116E, K157A/D160A, and P156D/D160A) were expressed in Escherichia coli Rosetta (DE3) and were purified by chromatography on a HisTrap HP column and a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare). The purity of the proteins was confirmed by SDS-PAGE.
Crystallization
All crystallization trials were carried out using the hanging-drop vapor-diffusion method by mixing 1 μl of protein solution (5 mg ml−1 in 20 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl and 2 mm dithiothreitol) with 1 μl of various reservoir solutions and equilibrating the mixtures against 500 μl of reservoir solution at 20 °C. Single crystals of size ∼0.5 × 0.3 × 0.03 mm3 were obtained in a drop containing 25–30% polyethylene glycol monomethyl ether 5000, 0.2 m ammonium sulfate, 0.1 m MES buffer (pH 6.7). For the protein complex with the peptide P2 (Table 1), the soaking method was employed by including the peptide (2 mm) in a drop containing the native protein crystals and incubating the mixture for 24 h before starting the data collection. In the cases of the P1 and P3 peptide complexes, the co-crystallization procedure was carried out by mixing the respective peptide and protein solutions (10:1 molar ratio) and incubating them at 4 °C for about 4 h before setting up the crystallization experiment. In this case, the polyethylene glycol monomethyl ether 5000 concentration and the pH in the reservoir solution were slightly different from the conditions used for obtaining the native crystals. All of the crystals were soaked briefly in a cryoprotectant containing 10% (w/v) glycerol and were flash-frozen in liquid nitrogen prior to the start of data collection.
TABLE 1.
Histone H4 peptides used in this study
| H4 peptide | Primary sequencea | Acetylated lysine position | N-terminal region |
|---|---|---|---|
| P1 | SGRGKGGKGLGK(ac)GGA | 12 | 1–15 |
| P2 | SGRGK(ac)GGKGLGK(ac)GGA | 5, 12 | 1–15 |
| P3 | LGK(ac)GGAKRHRKV | 12 | 10–21 |
a K(ac) indicates an ϵ-acetylated lysine.
Structure Determination
Complete diffraction data sets for each of the protein complex crystals with the acetylated histone H4 peptides (P1, P2, and P3) were collected at beamline BL44B2 at SPring-8, Harima, Japan. The crystal-to-detector distance was set to 190 mm. The oscillation range per image was 1.0°, with no overlap between two contiguous images. The data were processed and scaled with the HKL2000 program suite (27). The structures of BD1-P1, BD1-P2, and BD1-P3 complexes, respectively, were solved by the molecular replacement procedure using the apo-form of BD1 as a model (26). The crystals of BD1 complexes belonged to the space group C2, with three BD1 molecules, and contained two H4 tail peptide chains in the asymmetric unit. After maximum likelihood refinement with REFMAC5, from the CCP4 suite (28), the 2mFo − DFc and mFo − DFc maps were calculated to identify and to build the H4 tail in the complex structure. The graphics program O (29) was used to build the model of the complexes. The crystallographic data and refinement statistics of the BD1-P1, BD1-P2, and BD1-P3 complexes are summarized in Table 2. The stereochemistry of the protein and the H4 tail peptides is good, as checked with PROCHECK (30).
TABLE 2.
Summary of data collection and refinement statistics
| BD1 + P1 | BD1 + P2 | BD1 + P3 | |
|---|---|---|---|
| Data collection | |||
| Wavelength | 0.9791 Å | 0.9791 Å | 0.9791 Å |
| Resolution | 20 to 2.04 Å | 20 to 2.04 Å | 20 to 2.04 Å |
| Redundancya | 3.5 (2.8) | 3.6 (3.0) | 3.6 (3.6) |
| Unique reflections | 25,699 | 26,362 | 26,788 |
| Completeness | 96.1% (78.1%) | 98.4% (88.1%) | 99.5% (98.9%) |
| Rmergeb | 8.9% (30.5%) | 8.4% (36.7%) | 5.7% (21.2%) |
| Refinement statistics | |||
| Resolution | 20 to 2.04 Å | 20 to 2.3 Å | 20 to 2.04 Å |
| σ cutoff | 0 | 0 | 0 |
| Reflections | 24,375 | 17,759 | 25,421 |
| No. of protein residues | 333 | 333 | 333 |
| No. of peptide residues | 25 | 20 | 2 |
| No. of water molecules | 250 | 250 | 250 |
| Rcrystc | 18.8% | 17.9% | 19.2% |
| Rfreed | 24.8% | 24.3% | 23.1% |
| Average B factors | 31.86 | 34.63 | 31.48 |
| Root mean square deviations | |||
| Bond lengths | 0.017 Å | 0.022 Å | 0.017 Å |
| Bond angles | 1.55° | 1.69° | 1.41° |
a Numbers in parentheses are values in the highest resolution shell.
b Rmerge = Σ|Iobs − 〈I〉|/Σ〈I〉 summed over all observations and reflections.
c Rcryst = Σ|Fobs − Fcalc|/ΣFobs.
d Rfree calculated with 5% of data omitted from refinement.
Surface Plasmon Resonance Binding Assays
For binding assays of BRD2-BD1 mutants, the biotinylated H4 tail peptide acetylated at Lys-12 (K12ac), corresponding to the sequence of the N-terminal 15 residues of histone H4, was purchased from NeoMPS Inc. The peptide was immobilized on flow cell 2 and d-biotin was immobilized on flow cell 1, as a reference, of a sensor chip SA (Biacore, GE Healthcare). The binding affinities of the BRD2-BD1 proteins against the H4 tail peptide were examined on a Biacore 3000 (Biacore, GE Healthcare), with PBS (pH 7.4) as the running buffer. The reference value from the blank flow cell 1 was subtracted from the value of the functionalized flow cell 2 to obtain the actual response units. For binding assays to the H4 peptides with different acetylation patterns, biotinylated H4 tail peptides corresponding to the sequence of the N-terminal 20 residues of histone H4 were purchased from Toray Research Center Inc. Binding assays were carried out on a Biacore 3000 (Biacore, GE Healthcare). The BRD2-BD1 protein was loaded for 1 min with a 20 μl/min flow rate onto the peptide-immobilized sensor chip SA, and the flow was further continued for 1 min to detect the dissociation of the protein from the immobilized peptides in the PBS buffer. The results were analyzed using the BIAevaluation software version 4.1 (Biacore, GE Healthcare).
RESULTS AND DISCUSSION
BRD2-BD1 Complexed with a Histone H4 Tail Peptide 1
As the BRD2 bromodomains prefer to interact with N-acetyl-lysine either at positions 5 or 12 of histone H4 (H4K5ac or H4K12ac) (16, 17), we have attempted to crystallize BRD2-BD1 with various acetylated H4 tails (Table 1), and finally we determined the complex structure of BRD2-BD1 with the Lys-12-acetylated histone H4 tail peptide (peptide, P1; residues 1–15) (Fig. 1, A and B). The BD1-P1 complex crystal was obtained by co-crystallization, and it diffracted beyond 2.2 Å resolution. The tertiary structure of BD1 in the complex is very similar to the native structure of BD1 (26) and BD2 (31, 32) as well as to other known bromodomain structures (33–38). As observed in the native structure of BD1, the noncrystallography symmetry arrangement of the three molecules is unique, as compared with those found in other protein structures. The molecules are not related by 3-fold symmetry. Molecule A is transformed into molecule B by 2-fold symmetry, whereas the third noncrystallography symmetry molecule C is related by a rotation of 110° to the molecule B. Molecule C forms a dimer with a molecule generated by crystallographic symmetry (supplemental Fig. 1).
The difference Fourier map clearly showed the electron density for the acetylated histone H4 tail in the cleft region of BD1 (Fig. 1B and supplemental Fig. 2). The acetylated H4 tail bound to molecules A and B, but it was absent from molecule C (data not shown). The absence of the H4 tail in the binding site of molecule C is most likely due to the extended regions (i.e. N and C termini) of molecule B that blocked the entry of the H4 tail to molecule C. The electron density was missing for residues 1–7 in molecule A, as the residues in this region are flanking away from the protein (supplemental Fig. 2A). However, to our surprise, the full-length N-terminal H4 tail from residues 1 to 15 was readily traceable for molecule B, except for the side chains of Arg-3 and Lys-5 (supplemental Fig. 2B). As the H4 tail bound to molecule B was most well defined, the explanations below are based on the results for molecule B, unless otherwise stated.
In the complex structure, the acetylated H4 tail binds in the narrow cleft region with an extended, nonspecific conformation. The main chain atom N of K12ac produces a bifurcated electrostatic interaction with the side chains of Asn-156 and Asp-160. The side chain of K12ac sits in the deep hydrophobic pocket, formed by the residues Pro-98, Phe-99, Val-103, Leu-108, Lys-110, Cys-152, and Ile-162 (Fig. 1C). The acetyl group of K12ac is positioned in the cavity in such a way that the OH of the carbonyl group electrostatically interacts with Nδ2 of Asn-156, as well as indirectly with the Tyr-113 side chain through a water molecule (Fig. 1C), which is conserved in other bromodomain structures (13). A series of hydrogen bond networks between the water molecules exist on one side of the K12ac side chain. The side chain atom Nζ of K12ac electrostatically interacts with a water molecule (Fig. 1C). The other water molecule acts as a bridge between Nζ of K12ac and the carbonyl group of Pro-98. Moreover, the hydrophobic residues Phe-99, Val-103, and Ile-162 surround the methyl (CH3) group of K12ac (Fig. 1C). The charge distribution on the cavity surface, based on the electrostatic surface analysis, is complementary to that of the aliphatic moiety (Fig. 1D).
Recognition of Histone H4 Tail Peptide 1 by BRD2-BD1
Based on the BD1-P1 complex structure, the H4K12ac tail interacting BD1 residues in the peptide-binding site are highlighted in Fig. 1E. The main chain nitrogen and carbonyl oxygen atoms of Gly-13 (the +1 position to K12ac) form hydrogen bonds with Oδ1 of Asp-160. The carbonyl oxygen atom of Gly-13 contributes a main chain hydrogen bond to the amino group of Asp-161. Gly-14 (+2 position) does not interact with the protein. Ala-15 (+3 position), which is positioned at the end of the cleft, forms two hydrogen bonds, through its main chain N and O atoms, with the carbonyl group of Thr-159 and the carbonyl group of Thr-168 of the dimer-related molecule A, respectively.
The residues Lys-8 (−4 position) and Leu-10 (−2 position), which bind at the other side of the shallow pocket, have numerous interactions with the protein. An electrostatic interaction is formed between the carbonyl group of Leu-10 (−2 position) and Nζ of Lys-157 (Fig. 1E). However, the other residue Lys-8 (−4 position) forms many hydrophilic interactions with the main chain atoms of the protein, through its positively charged side chain (Fig. 1, D and F). The Nζ of Lys-8 interacts with the carbonyl groups of Tyr-153, Ile-154, and Asn-156, which are located in helix αB, and with Tyr-153 and Ile-154 of the neighboring dimeric molecule A (Fig. 1F). In addition, it interacts with a tightly bound water molecule. In the case of the H4 tail bound to molecule A, no such interactions exist, because Lys-8 of the H4 tail cannot be accommodated in the same orientation as that observed on molecule B. The electron density for the side chain of Lys-8 of the H4 tail bound to molecule A is weak (supplemental Fig. 2A), because it is exposed to the solvent region.
Gly-6 and Gly-7 of the H4 tail are located away from molecule B, and thus lack interactions with the protein. Due to a kink at Gly-7, a turn comprising residues 5–7 is located near the BC loop (156–158) of the adjacent molecule A. Unexpectedly, the first seven residues (Ser-1 to Gly-7) are sandwiched in a pocket formed between the dimeric interface (Fig. 1D), which possesses an electronegative surface. Although the main chain was traceable from Ser-1 to Lys-5, no electron density for the side chains of Arg-3 and Lys-5 was visible (supplemental Fig. 2B). These results imply that the formation of such a dimeric interface minimizes the flexibility of the N-terminal tail of histone H4 on BRD2-BD1.
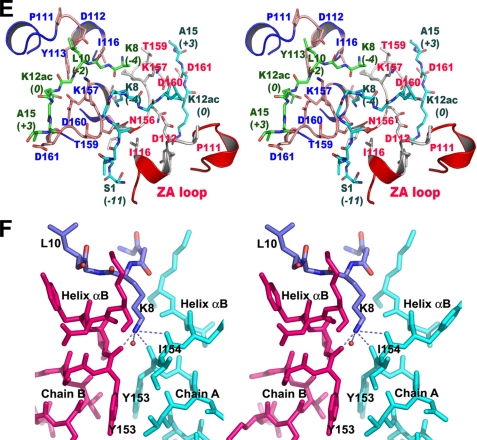
Overall, we observed six electrostatic interactions between the main chain of the protein and the side chains of the H4 tail. Intriguingly, the BD1-P1 complex structure revealed that at least 10 residues of the H4 tail interact with BRD2-BD1, as summarized in Fig. 2A. The sequence preference of BRD2-BD1 deduced from the BD1-P1 complex structure is XXKXLXKacXXA, where X should be a flexible residue such as glycine (Fig. 2A). Among all of the core histone sequences, this sequence is only found around K12ac of H4 (Fig. 2B). These results presumably explain a mechanism for highly selective association between BRD2 and H4K12ac (16).
FIGURE 2.
Schematic diagram for the BD1 residues interacting with Lys-12-acetylated histone H4 tail. A, interactions between BD1 and the H4 tail. The residues of BD1 that interact with the H4K12ac side chain are not indicated for clarity. The interacting residues corresponding to molecules A and B are labeled in blue and red, respectively. B, superposition of residues 1–15 are from the N-terminal histone tails. The K12ac residue and the sequence recognized by BD1, including the hypoacetylated Lys-8 in H4, are colored red and blue, respectively. The lysine residues that can be acetylated in the histone tails are also colored red.
Structure of the BD1 Complex with Histone H4 Peptide 2
Although the aforementioned results showed that the bromodomain BD1 of BRD2 binds to the acetyl-lysine at position 12 of histone H4, we carried out an additional structural analysis of another complex with a peptide containing acetyl-lysines at positions 5 (H4K5ac) and 12 (H4K12ac) of the H4 peptide, P2 (Table 1), to determine whether this protein essentially binds to H4K12ac. The structure of this protein complex indeed revealed that the BRD2-BD1 specifically interacts with the H4K12ac residue (supplemental Fig. 3). The mode of peptide binding to the protein is similar to that found in the BD1-P1 complex. However, the electron density for the peptide residues up to and including H4K5ac was completely missing, for both molecules A and B. Notably, as also observed in the BD1-P1 complex, Lys-8 of peptide P2 contributes similar hydrophilic interactions with the main chain atoms of the protein residues in the secondary binding site region. The electrostatic surface potential study suggested that the side chain of Lys-8 could enter into the narrow negatively charged cavity formed by the αB helices of molecules A and B (Fig. 1, D and F). Thus, the results gleaned from the analyses of the BD1-P1 and -P2 complexes suggest that Lys-8, which is positioned in the secondary binding site, might be a potential candidate for stabilizing the recognition of K12ac for the BD1 interaction.
Structure of the BD1 Complex with Histone H4 Tail Peptide 3
To determine whether Lys-8 of histone H4 is essential for the acetyl lysine position selectivity for the BD1 interaction, we solved yet another complex structure for an acetylated histone H4 peptide, P3 (Table 1), lacking residues 1–9 altogether. Remarkably, the complex structure for peptide P3 revealed that the peptide binds very weakly to the protein (supplemental Fig. 4). For molecule A, no electron density was observed, except for a weak electron density in the H4K12ac side chain-binding region. For molecule B, the electron density was also absent, except for H4K12ac. The electron density did not improve for the other residues, even after several rounds of refinement. This result suggests that the instability of this peptide binding is due to the lack of the anchoring residue at the −4 position of H4K12ac (corresponding to Lys-8 of histone H4), which electrostatically interacts with the BRD2-BD1 surface at the secondary binding site. Because the co-crystallized crystals of the BD1-P3 complex did not show any additional electron density in the peptide-binding site, we also tried the soaking method for this protein complex, which had been incubated with the P3 peptide overnight. However, the soaked experiment also failed to detect the additional electron density (data not shown).
When analyzed, the crystal packing of the above described structures, the peptide molecules do not have any potential interactions with any of the symmetry-related molecules, and the long segment of the peptides in the dimer is exposed to the solvent region. The observations provide further evidence that the complex formation in the crystals is not due to any kind of artifact but mainly to the specific recognition between the H4 peptide and the bromodomain.
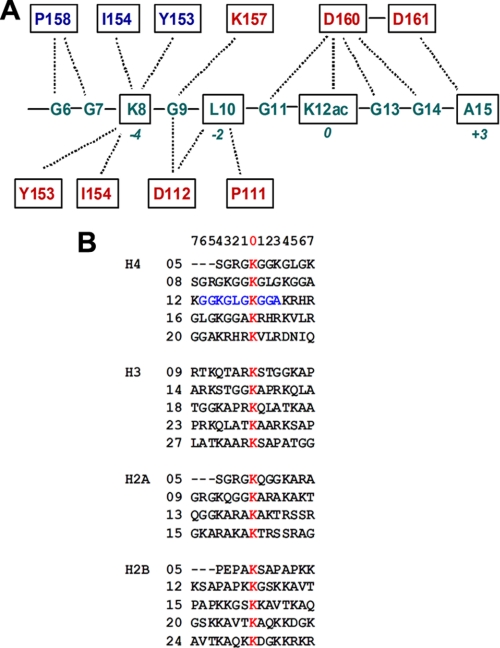
Binding Assay for BD1 and the Histone H4 Tail Interaction
To examine the specific recognition of the Lys-12-acetylated H4 tail by BRD2-BD1, we performed surface plasmon resonance analyses with a series of BD1 mutants (single mutants Y113A, I154A, N156A, P158D, and D160A and double mutants P111D/D112A, D112A/I116E, N156D/D160A, and K157A/D160A). The BD1 binding to the H4K12ac tail peptide was drastically reduced by all of the tested mutations (Fig. 3). As predicted (26), the weak binding of the I154A mutant, which positions at the dimeric interface and interacts with H4K8 through its carbonyl group (Fig. 1F), implies that the alteration of dimerization, and probably the reduction in Lys-8 binding, substantially decreased the H4K12ac binding activity of BRD2-BD1. Mutations to the H4-interacting residues such as Tyr-113, Asn-156, and Asp-160 further decreased the BD1 binding to the K12ac-containing H4 tail peptide, supporting that these residues are indeed responsible for the recognition of Lys-12-acetylated H4 tail in solution.
FIGURE 3.
Surface plasmon resonance binding between Lys-12-acetylated histone H4 tails and BRD2-BD1 mutants. The binding activity to the K12ac H4 tail peptide was compared between the wild-type BD1 (WT, red line) and the BD1 point mutants (Y113A, I154A, N156A, P158D, D160A, P111D/D112A, D112A/I116E, N156D/D160A, and K157A/D160A). The mutations are located in the H4 tail-interacting region as shown in Fig. 1E.
Interaction with the Histone H4 Tail with Different Patterns of Acetylation
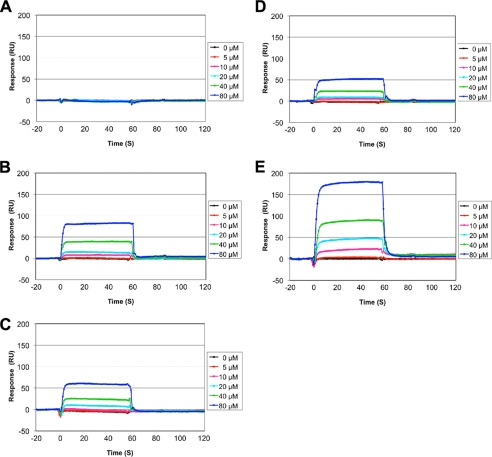
To address the preference of BRD2-BD1 for the acetylation status of the histone H4 tail, we analyzed the binding activity using histone H4 tail peptides with different patterns of acetylation (Fig. 4). As expected, BRD2-BD1 did not bind to nonacetylated H4 tail peptide (Fig. 4A). BD1 bound to the Lys-12-acetylated H4 tail peptide and to a less extent to the Lys-5-acetylated peptide (Fig. 4, B and C). The dissociation constants were approximately estimated to be 930 μm and 1.8 mm for K12ac and K5ac, respectively, as calculated by the steady-state affinity binding model. Based on the crystal structures and the in vitro binding assay, BRD2-BD1 and the H4K12ac tail are considered to bind each other in an equal ratio. To assess whether BRD2-BD1 binding to the Lys-12-acetylated H4 tail is affected by the side chain of Lys-8, we next performed the effect of K8A mutation on BD1 binding (Fig. 4D). As expected, the interaction between BD1 and the Lys-12-acetylated H4 tail peptide was significantly weakened by the K8A mutation. The dissociation constant was estimated to be ∼3.0 mm. These results suggest that the residues Lys-8 K12ac of H4 substantially contribute to the selective interaction between BRD2-BD1 and histone H4 in solution.
FIGURE 4.
BRD2-BD1 interaction to histone H4 tails with different patterns of acetylation. Dose-dependent binding results ranging from 5 to 80 μm of the protein concentration are shown. Acetylation patterns of the histone H4 (1–20) peptides immobilized to the sensor chip are as follows: A, no acetylation; B, Lys-12-acetylated; C, Lys-5-acetylated; D, Lys-12-acetylated with K8A mutation; E, di-acetylated at Lys-12 and Lys-5.
As BRD2 binds chromatin that is enriched in Lys-12- or Lys-5-acetylated histone H4 (17), we further tested the BD1 binding activity to di-acetylated H4 tail peptide at Lys-12 and Lys-5. To our surprise, BRD2-BD1 bound to the H4K5ac/K12ac peptide noticeably with the approximate dissociation constant of 360 μm (Fig. 4E). The observation of considerable interaction with BRD2-BD1, in an unknown manner, may be due to influence of additional acetylated lysine (K5ac) in the di-acetylated H4 peptide or may be due to the possibility of more than one mode of recognition by BRD2-BD1 for the multiacetylated H4 tail (see below).
Comparison between BET Family Bromodomains
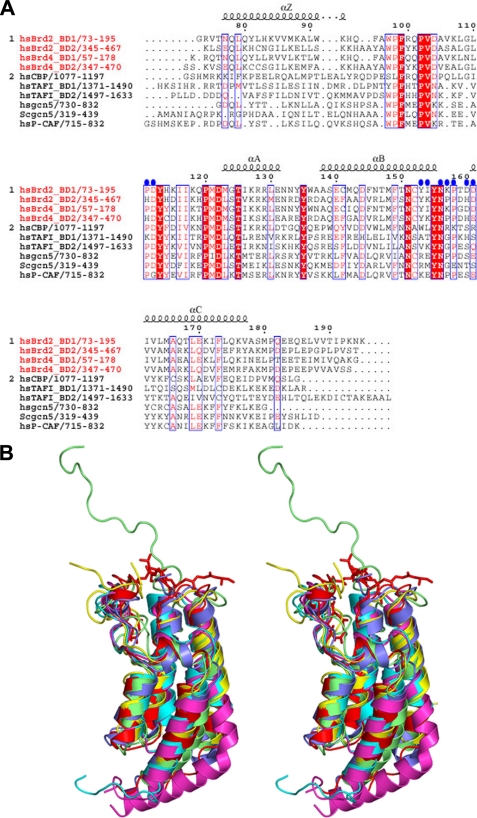
Sequence comparisons of the first and second bromodomain modules, BD1 and BD2 from the BRD2 and BRD4 proteins, revealed that the corresponding bromodomains in BRD2 and BRD4 (i.e. BRD2-BD1 versus BRD4-BD1; BRD2-BD2 versus BRD4-BD2) are highly homologous as compared with the nonequivalent bromodomains BD1 versus BD2 (Fig. 5A). The corresponding bromodomains share more than 75% sequence identity, whereas only about 44% sequence identity exists between BD1 and BD2. Sequence comparisons of other BET proteins also yielded similar results (data not shown). When the properties of the residues of BRD2-BD1 that interact with the acetylated H4 peptide were compared with those of BRD4-BD1, an intriguing result was observed; the acetylated H4 peptide-interacting residues are highly conserved between the first bromodomains of BRD2 and BRD4 (Fig. 5A). This suggests that the interaction mode of the first bromodomain with the acetylated histone H4 in BRD4, as well as in other BET proteins, might be similar to that observed in the BRD2-BD1 complexes.
FIGURE 5.
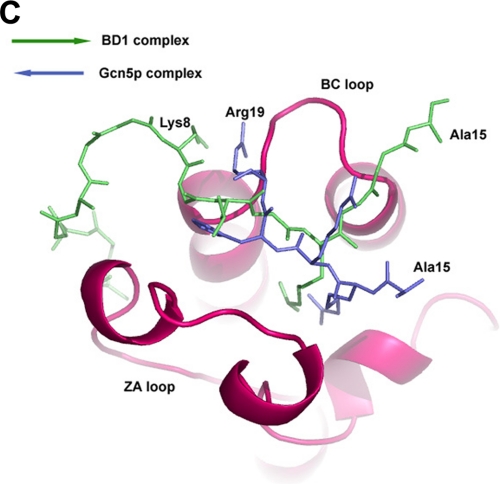
Comparison of BRD2-BD1 with other bromodomains. A, sequence comparison of the BET and non-BET bromodomains. The name and region of the BET family are indicated in red (i.e. BD1 and BD2 regions of human BRD2 and BRD4). The amino acid sequence alignment was produced by ClustalW (41) and was manually modified. Red characters indicate identical residues in the BET family (group 1) of BRD2 and BRD4 bromodomains. Characters on the red background indicate completely conserved amino acids among the BET (group 1) and non-BET (group 2) bromodomains. Blue circles denote residues that interact with the acetylated histone H4 tail residues (except for the K12ac side chain interactions, for clarity). The figure was generated by ESpript (9). B, stereo view of the superposition of BRD2-BD1-peptide complex with the other known bromodomain structures. The peptide in the BRD2-BD1 complex is shown as sticks model for clarity. Color code is as follows: red, BD1 complex; cyan, TAF1-bromodomain-1; magenta, TAF1-bromodomain-2; slate blue, Gcn5-acetylated histone H4 peptide complex; yellow, PCAF-tat-peptide complex; olive green, cAMP-response element-binding protein-binding protein-p53 peptide complex. C, orientation of the acetylated H4 peptides in BRD2-BD1 (green) and Gcn5 (slate blue) complexes.
While we were preparing this manuscript, the crystal structure of the BD1 domain of mouse BRDT (BRDT-BD1) in complex with acetylated histone H4 peptide was reported (39). Unexpectedly, BRDT-BD1 strongly associates with the histone H4 peptide di-acetylated at Lys-5 and Lys-8 (H4K5acK8ac) although it faintly associates with the H4 peptides mono-acetylated either at Lys-5, Lys-8, Lys-12, or Lys-16 (39). In addition, the side chains of K5ac and K8ac bind to BRDT-BD1 at the same acetylated lysine-binding pocket simultaneously. This feature is completely absent in the structure of the BRD2-BD1 complex (supplemental Fig. 5). Another recent report indicates that the bromodomains of BRDT specifically bind to histone H4 in which both Lys-5 and Lys-8 are acetylated, although it did not bind to those acetylated at either Lys-5, Lys-8, Lys-12, or Lys-16 (40). This preference of BRDT is quite different from that of BRD2 that favors acetylation of Lys-12 most among the four single acetylations (16). Thus, the N-terminal bromodomains of BET proteins may flexibly recognize different patterns of H4 acetylation, possibly utilizing at least the two different manners of H4 tail interactions. Consequently, multiple acetylation of H4 at Lys-5, Lys-8, and Lys-12 may contribute to tight and complex association of BET proteins with chromatin.
In case of the second bromodomain of BRD2/BRD4, it also recognize the N-terminal H4 tail acetylated at Lys-12 (H4K12ac) (16, 31, 32). By comparing the sequences between BD1 and BD2, the functional residues of BD1 involved in the acetylated histone H4 interactions are not highly conserved with their equivalent residues in BD2 (Fig. 5A). We have also observed that the mode of acetylated histone H4 recognition by BD2 is quite different from BD1, although both the bromodomains recognize H4K12ac.7
Comparison with Other Bromodomain Structures
The left-handed four-helical domain of BRD2-BD1 is conserved in the bromodomain family (33–38). The overall structures of these bromodomains are similar; however, the H4 tail-binding site possesses major structural variations, particularly in the ZA and BC loop regions (data not shown).
When we compared the BD1 complex structure with other known bromodomain complex structures, such as Gcn5 with an acetylated H4 tail peptide (35), PCAF with an acetylated tat peptide (38), and cAMP-response element-binding protein-binding protein with an acetylated p53 peptide (37), we observed from our structural studies that the H4 tail peptide in the BD1 complex form more interactions with their protein partners than those found in other bromodomain complexes (Fig. 5B). The complex structure of the Gcn5 bromodomain with the H4 tail peptide, corresponding to residues 15–29 (AK(ac)RHRKILRNSIQGI) (35), showed that this bromodomain preferentially binds to the Lys-16-acetylated H4 tail. Although they used a 15-amino acid-long peptide containing H4K16ac, just 4 residues (i.e. residues 16–19) bound to the protein, and only a limited number of contacts were formed with 2 residues, H4K16ac+2 (His-18) and H4K16ac+3 (Arg-19). When compared, the native structure of TAF1 double bromodomains (36) by superimposing monomer of the BD1 dimer with that of the TAF1 double bromodomain for Cα atoms, it showed that the packing is quite different between them (supplemental Fig. 6). The peptide binding regions of the TAF1 double bromodomain are quite close to each other, such that two acetyl-lysine residues from a single H4 tail could interact with them simultaneously. However, in the BD1 dimer case, the two bromodomains interact with two independent peptides, and hence BD1 may interact with two acetylated H4 tails through its dimerization.
TAF1 binds to the N-terminal tail of histone H4, which is mono-acetylated either at Lys-8, Lys-12, or Lys-16, as well as to that of histone H3 acetylated at Lys-14, suggesting that TAF1 possesses broader recognition specificity (16, 36). Similarly, the PCAF protein, which contains a single bromodomain, also has broader specificity for acetylated histone recognition (16, 33). This broader specificity, which may be due to weaker interactions, would occur between the bromodomains of TAF1/PCAF and the H3/H4 tails; thus, their association with or dissociation from the histones would depend on their respective recognitions.
On the other hand, the BET family proteins, BRD2 and BRD3, preferentially recognize the histone H4 N-terminal tail acetylated at Lys-12 (16, 17). This narrow and specific recognition directly correlates with the structural features observed in BRD2-BD1 where the long portion of the H4 peptide, spanning about 10 amino acids, binds with the bromodomain (Fig. 1, B and D, and Fig. 2).
Another noteworthy difference was observed in both the location and direction of the bound peptides in the two bromodomain complexes from BRD2 and Gcn5, and the backbones of the peptides in the two corresponding structures are oriented nearly antiparallel to each other (Fig. 5C). This suggests that the interaction modes of the BRD2 and Gcn5 bromodomains with histone H4 are completely different and that the two sequences in the vicinity of the acetylated Lys-12 and the acetylated Lys-16 are recognized in different manners by BRD2 and Gcn5, respectively, although they both bind to acetylated H4 peptides. In the case of the Gcn5 complex structure (35), only a few intermolecular contacts were observed compared with BRD2-BD1. Because the acetylated histone H4 interacting residues are highly conserved among the first bromodomains in the BET proteins, the orientation of the acetylated histone H4 tail may be the same for the first bromodomain in BET protein family members.
In summary, this study describes how BRD2-BD1 deciphers the histone code by means of recognizing H4K12ac, in which the large numbers of intermolecular interactions occur with the N-terminal 15 residues of the acetylated histone H4 tail. In addition, the BRD2-BD1 can recognize two H4K12ac tails simultaneously. We also suggest the role of Lys-8 in determining the recognition of a specific acetylated lysine position for BRD2. These results provide a starting point for further structural and functional analyses aimed at understanding how the BET family proteins interact with acetylated chromatin, and how they regulate the transcriptional machinery in a histone acetylation-dependent manner.
Supplementary Material
Acknowledgments
We thank Drs. N. Igarashi (BL6A, Photon Factory) and M. Yamamoto (BL44B2, SPring-8) for their help in data collection; C. Shang, M. Wakamori and E. Adachi for their technical assistance; T. Kanno for critical reading of the manuscript; and T. Nakayama and A. Ishii for their clerical assistance.
This work was authored, in whole or in part, by National Institutes of Health staff. This work was supported in part by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, by the RIKEN Structural Genomics/Proteomics Initiative, and the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
The atomic coordinates and structure factors (codes 2DVQ, 2DVR, and 2DVS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
T. Umehara, B. Padmanabhan, and S. Yokoyama, unpublished results.
- BET
- bromodomain and extra-terminal domain
- BRD2
- bromodomain-containing 2
- BD1
- bromodomain 1
- MES
- 4-morpholineethanesulfonic acid
- PCAF
- p300/CBP-associated factor.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Kornberg R. D., Lorch Y. (1999) Cell 98, 285–294 [DOI] [PubMed] [Google Scholar]
- 3.Rice J. C., Allis C. D. (2001) Curr. Opin. Cell Biol. 13, 263–273 [DOI] [PubMed] [Google Scholar]
- 4.Roth S. Y., Denu J. M., Allis C. D. (2001) Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 5.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 6.Fischle W., Wang Y., Allis C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 7.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownell J. E., Allis C. D. (1996) Curr. Opin. Genet. Dev. 6, 176–184 [DOI] [PubMed] [Google Scholar]
- 9.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 10.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes S. R., Dollard C., Winston F., Beck S., Trowsdale J., Dawid I. B. (1992) Nucleic Acids Res. 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denis G. V. (2001) Front. Biosci. 6, D1065–D1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng L., Zhou M. M. (2002) FEBS Lett. 513, 124–128 [DOI] [PubMed] [Google Scholar]
- 14.Wu S. Y., Chiang C. M. (2007) J. Biol. Chem. 282, 13141–13145 [DOI] [PubMed] [Google Scholar]
- 15.Mujtaba S., Zeng L., Zhou M. M. (2007) Oncogene 26, 5521–5527 [DOI] [PubMed] [Google Scholar]
- 16.Kanno T., Kanno Y., Siegel R. M., Jang M. K., Lenardo M. J., Ozato K. (2004) Mol. Cell 13, 33–43 [DOI] [PubMed] [Google Scholar]
- 17.LeRoy G., Rickards B., Flint S. J. (2008) Mol. Cell 30, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey A., Ellenberg J., Farina A., Coleman A. E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. (2000) Mol. Cell. Biol. 20, 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Balbás M. A., Dey A., Rabindran S. K., Ozato K., Wu C. (1995) Cell 83, 29–38 [DOI] [PubMed] [Google Scholar]
- 21.Muchardt C., Reyes J. C., Bourachot B., Leguoy E., Yaniv M. (1996) EMBO J. 15, 3394–3402 [PMC free article] [PubMed] [Google Scholar]
- 22.Kruhlak M. J., Hendzel M. J., Fischle W., Bertos N. R., Hameed S., Yang X. J., Verdin E., Bazett-Jones D. P. (2001) J. Biol. Chem. 276, 38307–38319 [DOI] [PubMed] [Google Scholar]
- 23.You J., Croyle J. L., Nishimura A., Ozato K., Howley P. M. (2004) Cell 117, 349–360 [DOI] [PubMed] [Google Scholar]
- 24.Viejo-Borbolla A., Ottinger M., Brüning E., Bürger A., König R., Kati E., Sheldon J. A., Schulz T. F. (2005) J. Virol. 79, 13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F., Liu H., Blanton W. P., Belkina A., Lebrasseur N. K., Denis G. V. (2010) Biochem. J. 425, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura Y., Umehara T., Nakano K., Jang M. K., Shirouzu M., Morita S., Uda-Tochio H., Hamana H., Terada T., Adachi N., Matsumoto T., Tanaka A., Horikoshi M., Ozato K., Padmanabhan B., Yokoyama S. (2007) J. Biol. Chem. 282, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 29.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallgr. 26, 283–291 [Google Scholar]
- 31.Huang H., Zhang J., Shen W., Wang X., Wu J., Wu J., Shi Y. (2007) BMC Struct. Biol. 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Wang X., Zhang J., Huang H., Ding B., Wu J., Shi Y. (2008) Biochemistry 47, 6403–6417 [DOI] [PubMed] [Google Scholar]
- 33.Dhalluin C., Carlson J. E., Zeng L., He C., Aggarwal A. K., Zhou M. M. (1999) Nature 399, 491–496 [DOI] [PubMed] [Google Scholar]
- 34.Hudson B. P., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2000) J. Mol. Biol. 304, 355–370 [DOI] [PubMed] [Google Scholar]
- 35.Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., Travers A. A. (2000) EMBO J. 19, 6141–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobson R. H., Ladurner A. G., King D. S., Tjian R. (2000) Science 288, 1422–1425 [DOI] [PubMed] [Google Scholar]
- 37.Mujtaba S., He Y., Zeng L., Yan S., Plotnikova O., Sachchidanand, Sanchez R., Zeleznik-Le N. J., Ronai Z., Zhou M. M. (2004) Mol. Cell 13, 251–263 [DOI] [PubMed] [Google Scholar]
- 38.Mujtaba S., He Y., Zeng L., Farooq A., Carlson J. E., Ott M., Verdin E., Zhou M. M. (2002) Mol. Cell 9, 575–586 [DOI] [PubMed] [Google Scholar]
- 39.Morinière J., Rousseaux S., Steuerwald U., Soler-López M., Curtet S., Vitte A. L., Govin J., Gaucher J., Sadoul K., Hart D. J., Krijgsveld J., Khochbin S., Müller C. W., Petosa C. (2009) Nature 461, 664–668 [DOI] [PubMed] [Google Scholar]
- 40.Sasaki K., Ito T., Nishino N., Khochbin S., Yoshida M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16257–16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgins D. G., Bleasby A. J., Fuchs R. (1992) Comput. Appl. Biosci. 8, 189–191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.