Abstract
Synaptic degeneration, including impairment of synaptic plasticity and loss of synapses, is an important feature of Alzheimer disease pathogenesis. Increasing evidence suggests that these degenerative synaptic changes are associated with an accumulation of soluble oligomeric assemblies of amyloid β (Aβ) known as ADDLs. In primary hippocampal cultures ADDLs bind to a subpopulation of neurons. However the molecular basis of this cell type-selective interaction is not understood. Here, using siRNA screening technology, we identified α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunits and calcineurin as candidate genes potentially involved in ADDL-neuron interactions. Immunocolocalization experiments confirmed that ADDL binding occurs in dendritic spines that express surface AMPA receptors, particularly the calcium-impermeable type II AMPA receptor subunit (GluR2). Pharmacological removal of the surface AMPA receptors or inhibition of AMPA receptors with antagonists reduces ADDL binding. Furthermore, using co-immunoprecipitation and photoreactive amino acid cross-linking, we found that ADDLs interact preferentially with GluR2-containing complexes. We demonstrate that calcineurin mediates an endocytotic process that is responsible for the rapid internalization of bound ADDLs along with surface AMPA receptor subunits, which then both colocalize with cpg2, a molecule localized specifically at the postsynaptic endocytic zone of excitatory synapses that plays an important role in activity-dependent glutamate receptor endocytosis. Both AMPA receptor and calcineurin inhibitors prevent oligomer-induced surface AMPAR and spine loss. These results support a model of disease pathogenesis in which Aβ oligomers interact selectively with neurotransmission pathways at excitatory synapses, resulting in synaptic loss via facilitated endocytosis. Validation of this model in human disease would identify therapeutic targets for Alzheimer disease.
Keywords: Receptors/Endocytosis, ADDL Binding, Excitatory Synapses, GYKI52466, GluR2, Photoreactive Cross-link, cpg2, Receptor Loss
Introduction
Alzheimer disease (AD)2 likely begins with deficits in synaptic transmission in brain regions that are critical for higher cognitive function (1), as stereological analyses of post-mortem samples show that synaptic loss correlates with cognitive dysfunction better than amyloid plaque or neurofibrillary tangle load (2–5). One of the triggers for such synaptic impairments is thought to be soluble oligomers of amyloid β (Aβ) peptides that accumulate in the brain and are potently toxic to synapses (6–8). Aβ synaptotoxicity is consistently observed with Aβ oligomers from synthetic preparations (9–11), cell-derived forms (7, 8, 12), and the brains of AD patients (13) and APPswe transgenic mice (14), such that these oligomers disrupt synaptic structure (9, 10, 15–18), inhibit long term potentiation (LTP) (7, 8, 19), and induce memory deficits (13, 20).
Although the mechanisms by which Aβ oligomers disrupt synaptic function are not known, Aβ oligomers bind to the dendritic processes of specific subtypes of hippocampal neurons with high affinity (9, 15, 21), leading to the hypothesis that there are synaptic binding targets for Aβ oligomers. Several studies have reported that Aβ and/or Aβ oligomers bind to excitatory neurotransmitter receptors, such as NMDA receptor subunits and α7nAchR (10, 17, 22), and/or signaling proteins expressed at postsynaptic membranes such as the insulin receptor, EphB2 receptor, and the prion protein (10, 21, 23, 24). In contrast, the AMPA receptor (AMPAR), the glutamate receptor subtype that mediates the majority of the fast excitatory synaptic transmission (25) essential for memory processing (26–28), has not been implicated as a binding partner for Aβ.
Despite a lack of data demonstrating direct Aβ binding to AMPARs, detrimental changes in AMPARs are a molecular correlate of synaptic losses in the AD brain. For example, the downscaling of AMPAR subunits GluR1 and GluR2/3 occurs selectively in the entorhinal cortex and CA1 of the hippocampus, regions that are most vulnerable to AD pathogenesis (29–33). This AMPAR reduction precedes neurofibrillary tangles (34), suggesting that it is an early molecular alteration in AD. Similar results are observed in AD transgenic mice (18, 35–38). Given the prominent role of AMPAR in excitatory synaptic transmission and the positive linear correlation of AMPAR expression with synaptic weight and size (39), the loss of AMPAR in AD raises the possibility that disruption of this receptor class might be a primary factor involved in the pathogenesis of AD.
In this study we sought to investigate synaptotoxic changes resulting from Aβ oligomer-neuron interaction. Employing an unbiased functional genomics screen in murine neuroblastoma N2A cells followed by verification in primary hippocampal neuronal cultures, we showed that the surface expression and function of AMPAR (preferentially GluR2) are required for ADDL-synaptic interaction, which in turn results in the surface AMPA receptor loss. This detrimental effect is mediated by an endocytosis depending on calcineurin activity. Our results suggest that the AMPAR-containing excitatory synapses and the related trafficking pathway are key targets of Aβ oligomers in AD.
EXPERIMENTAL PROCEDURES
Materials
Biotin-labeled and unlabeled synthetic Aβ42 peptides were from American Peptides (Sunnyvale, CA). Antibodies against Aβ1–16 (6E10) and Aβ42 (12F4) were from Covance. Antibodies against the N termini of GluR2 and GluR3 were purchased from Zymed Laboratories Inc. (South San Francisco, CA). Anti C-terminal GluR1, GluR2/3, and GluR4, anti-spinophilin, and anti-cpg2 antibodies were from Millipore (Billerica, MA) or Thermo Fisher Scientific (Fremont, CA). Alexa Fluor-streptavidin, anti-mouse, and anti-rabbit IgGs were from Invitrogen. Calcineurin, the surface biotinylation kit, and photoreactive amino acids were purchased from Fisher Scientific (Waltham, MA). All cell culture, SDS-PAGE, and Western blotting reagents and materials were from Invitrogen. GYKI52466 was purchased from Sigma; CNQX and IEM1406 were purchased from Tocris Bioscience (Charnwood, UK). siRNA transfection reagents were purchased from Dharmacon, Inc. (Chicago, IL).
Preparation of Aβ Oligomers
ADDLs and biotin-labeled ADDLs (bADDLs) were prepared from synthetic Aβ42 using a standardized protocol (11, 15).
siRNA Screen
siRNA pools (three sequence-independent siRNAs targeting the same transcript) were transfected at a final concentration of 25 nm to N2A cells using Dharmafect reagents in 384-well plates via a robotic protocol. After 3 days of transfection, the medium was removed and cells were treated with 20 μm bADDLs at 37 °C for 60 min before immunostaining. bADDL binding was visualized with streptavidin conjugated to fluorescein isothiocyanate (1:2000) followed by image acquisition and analysis with an IN Cell high content imaging station. bADDL binding intensity from siRNA-treated cells was measured and normalized against the total number of cells indicated by staining with 4′,6-diamidino-2-phenylindole. Data quality was assured through implementing quality control procedures such as adjustment of positional effects as described previously (40). To reduce the impact of outliers on hit selection, we used statistical methods to identify them (41) and excluded them in subsequent analyses. We calculated the strictly standardized mean difference (SSMD) for all siRNAs including outliers. SSMD is the ratio of the mean and standard deviation of a difference (42). We then used SSMD-based methods to select hits.
Primary Hippocampal Neuronal Cultures and Pharmacological Treatments
Primary neuronal cultures were prepared from the brains of embryonic day 18 Sprague-Dawley rats (Taconic Farms) maintained in neurobasal/B27 medium for 21 days as described (15). ADDLs or bADDLs at 500 nm were added for the indicated lengths of time. For pharmacological treatments, neurons were treated at designated concentrations for 30 min prior to the addition of ADDLs.
Cell-derived Aβ Oligomers
Cell-derived Aβ oligomers were generated from mouse primary cortical cultures prepared from APPswe and APP-YAC transgenic mice and CHO cells expressing mutant V717F (Val717 → Phe) human APP (7PA2 cells). Mouse primary cortical cultures were prepared from cortices at postnatal day 1 and maintained in neurobasal/B27 medium. Aβ42 levels from media were measured with ELISA on a weekly basis. 7PA2 cells were a kind gift from the Selkoe laboratory (Brigham and Women's Hospital). Cells were cultured and medium conditioned according to a published protocol (7, 8). To test the synaptic toxicity of cell-derived Aβ oligomers, rat hippocampal culture medium was replaced with media freshly collected from the above cells for the indicated times.
Aβ42 ELISAs
The total levels of Aβ42 were measured in a sandwich ELISA in which 6E10 was immobilized on a 96-well plate (5 μg/ml) in 0.2 m carbonate-bicarbonate buffer, pH 9.4, and blocked with 3% bovine serum albumin. Conditioned media from different cells were added to the plate and incubated at 4 °C overnight. The Aβ42 levels were detected with an anti-Aβ42 antibody (12F4) conjugated with alkaline phosphate followed by the CDP-Star substrate reaction. The data were read with an EnVision instrument (PerkinElmer Life Sciences).
Immunocytochemical Assays
Treated hippocampal neurons on coverslips were fixed with 4% formaldehyde. To minimize surface receptor internalization, 4% sucrose was added to the fixation solution. Neurons were blocked and permeabilized with 20% normal goat serum containing 0.1% Triton X-100 and incubated at 4 °C overnight with a primary antibody recognizing an AMPA receptor subunit or a synaptic marker. This treatment was followed by simultaneous incubation with a secondary antibody (1:500) and streptavidin (1:800), each conjugated with a different Alexa Fluor, in the dark at room temperature for 1 h. The coverslips were then mounted on a glass slide and visualized by a Nikon TE2000 confocal microscope with PerkinElmer rapid confocal imager UltraVIEW ERS software or by a Nikon ECLIPSE Ti epifluorescent microscope.
Surface Biotinylation
Surface biotinylation was performed using a surface labeling kit (Pierce). To minimize the internalization of the labeled surface protein, labeling was performed at 4 °C for 30 min according to the manufacturer's manual. The proteins following cell lysis were resolved on 4–20% SDS-PAGE and transferred to nitrocellulose membranes. AMPARs were detected with antibodies against GluR1, GluR2/3, and GluR4, respectively. Changes in the amount of surface receptor were measured after normalization with the total amount (surface plus intracellular pools) of receptors.
Internalization Assay
The internalization of bADDLs and AMPAR was performed by acid stripping based on a procedure for ligand and receptor internalization (43–46). For ADDL internalization, neurons were incubated with 500 nm bADDLs for various times. The neurons were rinsed with ice-cold PBS followed by incubation on ice for 3 min with 0.2 n acetic acid and 0.5 m NaCl. For AMPA receptor internalization, neurons were incubated with N-terminal anti-GluR2 (1:15) or anti-GluR3 (1:15) for 15 min at 37 °C. Neurons were then treated with ADDLs for 30 min followed by stripping. Cells were rinsed three times with ice-cold PBS, fixed, and permeabilized. The internalized bADDLs and AMPARs were detected with streptavidin and anti-mouse IgG, each labeled with a different Alexa Fluor fluorophore.
Co-immunoprecipitation
ADDL- or endogenous Aβ-treated neurons were lysed in a lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% protease inhibitor mixture, and 1 mg/ml cellular proteins. Samples were subjected to immunoprecipitation by 6E10. The precipitated complex was resolved on 4–20% SDS-PAGE and transferred to nitrocellulose membrane. The co-precipitated AMPA receptors were detected with antibodies raised against each GluR subunit. To detect GluR-cpg2 interaction, anti-GluR antibodies were immobilized covalently on to Sephadex resin using a cross-link kit from Pierce Biotechnology. The control and ADDL-treated hippocampal neurons were incubated with the beads. After being washed, the precipitated samples were separated on SDS-PAGE, and the co-precipitated cpg2 was detected with Western blots.
Photoreactive Cross-linking
Photoreactive amino acid cross-linking was performed according to the manufacturer's instructions (Pierce Biotechnology). 7PA2 and control CHO cells were cultured in leucine- and methionine-free Dulbecco's modified Eagle's limiting medium containing 10% PBS-dialyzed serum. l-Photo-leucine and l-photo-methionine were added to the medium to final concentrations of 4 and 2 mm, respectively. Media were replaced once during culture. Upon confluence of cells the media from both 7PA2 and control CHO cells were collected into a 50-ml test tube. The rat primary hippocampal neurons were prepared and cultured in a 24-well plate for 21 days. The neuronal culture medium was removed and replaced with medium containing photo-leucine and photo-methionine from 7PA2 or control CHO cells. Neurons were irradiated with a UV lamp at 350 nm for 15 min at room temperature. The media were removed, and neurons were washed with PBS, pH 7.4, and then lysed with a mixture of 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.8 m EDTA, 0.5 m EGTA, 1% Triton X-100, 0.5% Nonidet P-40, and 1% protease. The cell lysates were immunoprecipitated with the anti-Aβ antibody 6E10 followed by detection of co-precipitation of AMPARs with different anti-GluR subunit antibodies.
Data Analyses
Image and Western blots analyses were analyzed using ImageJ software. For bADDL binding, the streptavidin staining was quantified under the appropriate threshold. The values of integrated intensity were normalized with the number of neurons in each image plane. Data from more than 10 image planes were collected. For changes in AMPA receptors and synaptic spine markers, 3–5 dendrites from each neuron and 5–10 neurons from each treatment were measured. Both the number of receptors or drebrin puncta per length of dendrites and the intensity of the signals were measured under the appropriate thresholds. For changes in surface AMPA receptors, the intensity of the biotinylated and total receptors, both on Western blots, was measured, and the ratios of surface/total intensity were calculated. All data were analyzed with one-way analysis of variance. All animals used in this study were handled in full compliance with The United States Public Health Service Policy on Humane Care and Use of Laboratory Animals and the Guide for the Care and Use of Laboratory Animals.
RESULTS
AMPA Receptor Subunit and Calcineurin Implicated in bADDL Binding via High Throughput siRNA Screen
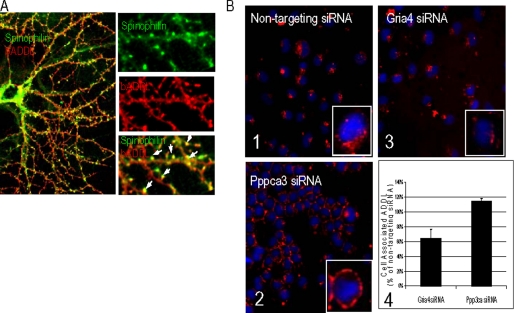
To generate neuroactive oligomeric Aβ assemblies, monomeric Aβ was aggregated into ADDLs and bADDLs. The oligomerization process was monitored by electrophoresis (species patterns are shown in supplemental Fig. 1A). When resolved on SDS-PAGE and stained with SYPRO Ruby, both the ADDL and bADDL preparations contained similar SDS-resistant species including apparent monomer, trimer, tetramer, and dodecamer. A small amount of a slower migrating species with an approximate size of 12 and 24 mers was seen in the bADDL preparation. These patterns are consistent with those in previous reports (15, 47). When bADDLs (500 nm) were administered to primary rat hippocampal neurons that had been cultured for 21 days in vitro, streptavidin-reactive puncta were observed on the dendrites of a subset of neurons at 10 min following treatment. This apparent bADDL binding is saturable, can be prevented by anti-Aβ antibodies, and is only visible when Aβ42 oligomers but not Aβ40 or monomeric or fibrillar preparations of Aβ42 are used (9, 10, 15, 47, 48). We assessed the subcellular site of neuronal interaction by adding antibodies to spinophilin, a postsynaptic marker for dendritic spines harboring excitatory synapses (49). Extensive colocalization between spinophilin and bADDLs was observed (Fig. 1A). Selective binding of bADDLs to spines implies that synaptic proteins present on dendritic spines are likely to regulate the interactions between oligomeric Aβ and neurons.
FIGURE 1.
Cell selectivity of bADDL binding. A, cultured hippocampal neurons (21 days in vitro) were treated with 500 nm bADDLs at 37 °C for 10 min. Cells were immunostained with an anti-spinophilin antibody followed by a secondary antibody-Alexa 488 and streptavidin-Alexa 555. bADDL staining shows a high degree of colocalization with spinophilin. B, bADDL biding in siRNA-transfected N2A cells: Panels 1–3, representative images of cells transfected with siRNA silencing Gria4 and Ppp3ca and control stained to visualize cell-associated ADDL (red) and nuclei (blue; stained by 4,6-diamidino-2-phenylindole). Gria4 and Ppp3ca were the two top hits from a screen aimed at identifying key proteins involved in the binding and processing of ADDL to neuronal cells. Images were acquired using an automated confocal imager (IN Cell 3000, GE Healthcare) equipped with a ×40 air objective. Image analysis and quantitation of the ADDL-associated fluorescent signal was performed using the imager proprietary software (Raven, GE Healthcare) and its “translocation” algorithm. Approximately 400 cells/well were imaged and analyzed. Images are reproduced using the same intensity scale. Panel 1, bADDL binding under non-target siRNA condition; panel 2, Pppca3 siRNA reduced ADDL internalization; panel 3, inhibited ADDL binding by Gria4 siRNA; panel 4, bar graph showing quantitation of cell-associated ADDL in Gria4 and Ppp3ca siRNA-treated samples (n = 3) obtained during the confirmation screen. Data are expressed as the percentage of cell-associated ADDL measured in samples treated with non-targeting siRNA. Both Gria4 and Ppp3ca siRNAs were the highest ranking genes among those that inhibited binding and blocked internalization of ADDL, respectively.
To screen for such molecules, we sought to model this bADDL-spine interaction in a cellular system amenable to high throughput siRNA transfection. N2A cells treated with bADDLs produced bADDL cellular staining that was readily quantified using high content imaging (Fig. 1B, panel 1), whereas many other cell lines tested did not, indicating that the binding was for a specific receptor(s). After a 60-min incubation with N2A cells, the majority of the bADDL staining was in the cytosolic compartment of the cell (Fig. 1B, panel 1, inset (enlarged)), suggesting internalization of bound bADDLs into N2A cells. We tested the effects of 6500 siRNAs targeting a variety of neuronal receptors and signaling proteins on bADDL/N2A cell interaction (see “Experimental Procedures”; detailed results of the screen will be published elsewhere). Approximately 20 siRNAs met the statistical criteria for having a reproducible effect on binding, among which two positive siRNAs were selected for further study. siRNAs targeting Pppca3, the gene encoding the catalytic subunit of calcineurin, resulted in a significant retainment of ADDL signal to the cell membrane (Fig. 1B, panel 2), in contrast to the cytosolic distribution of bADDLs in the non-siRNA-treated cells (Fig. 1B, panel 1). This result suggests that Pppca3 siRNA blocks ADDL internalization. In addition, siRNAs targeting Gria4, a gene encoding the type 4 AMPAR subunit (GluR4), showed a reproducible statistically significant reduction in bADDL binding to N2A cells (Fig. 1B, panel 3). The quantification of the changes in bADDL binding by Pppca3 and Gria4 siRNAs are summarized in Fig. 1B, panel 4.
We then assessed the expression of AMPAR subunits in the N2A cells used for the screen. Compared with primary hippocampal neurons, our N2A cells expressed low but detectable levels of AMPAR subunit RNA (data not shown). Western blotting with multiple antibodies revealed that the N2A cells expressed proteins that cross-reacted with GluR1, GluR2/3, and Glur4 antibodies (supplemental Fig. 1B). However, for each protein the banding pattern from hippocampal neurons and N2A cells was different. Given that the physiological relevance of bADDL binding with N2A cells is unclear, and particularly that those cells lack dendritic spines and express low levels of the neuronal AMPAR proteins, we focused on primary hippocampal neurons to assess the involvement of these two molecules in the Aβ oligomeric-synaptic interaction without further characterization of the N2A findings.
bADDLs Bind to GluR2-expressing Neurons
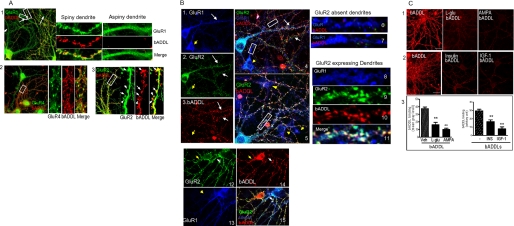
As a first step toward validating the findings from the siRNA screen, we examined whether bADDLs colocalize with AMPAR-immunoreactive spines in rat primary hippocampal neurons. We expanded our investigations to include all four AMPAR subunits, GluR1–GluR4, to determine whether all four regulate bADDL binding. In immunofluorescent colocalization experiments with AMPAR subtype-selective antibodies, GluR expression was detected on the soma and dendrites of both spiny and aspiny neurons. bADDL immunoreactivity was restricted largely to spiny structures and generally observed in spines containing GluR subunits, whereas neurons with aspiny dendrites showed little bADDL binding (Fig. 2A, panels 1 and 2). Although GluR1 (Fig. 2A, panel 1) and GluR4 (panel 2) are localized in the spines, they showed only partial colocalization with bound bADDLs. In comparison, GluR2 displayed a greater degree of colocalization with bADDL binding (Fig. 2A, panel 3). Neurons that did not extensively bind bADDLs (bADDL−) typically expressed either GluR1 or GluR4 in the soma and dendrites (Fig. 2A, panels 1 and 2) but generally showed low GluR2 immunoreactivity (Fig. 2B, panels 1–7, yellow arrows). In contrast, bADDL binding occurred mostly on dendrites that express abundant spines and GluR2 (Fig. 2B, panels 1–10). Dendrites lacking GluR1 (Fig. 2B, panels 1, 3, and 13) but expressing GluR2 showed the strongest bADDL binding. Together these data suggest that bADDLs interact with GluR-expressing spines, particularly when GluR2 is present.
FIGURE 2.
AMPAR-dependent bADDL binding. A, bADDL bindings are observed in AMPAR-containing dendritic spines. Panel 1, bADDL binding was observed only in those dendrites with spines. The bADDL binding shows partial colocalization with GluR1 immunoreactivity. Panel 2, bADDL binding shows a low degree of colocalization with GluR4. Panel 3, bADDL binding colocalizes well with GluR2. B, bADDLs bind to GluR2-expressing neurons. Triple immunostaining for GluR1, GluR2, and bADDL binding was applied to bADDL-treated neurons. Panels 1–3 show representative GluR1 (blue) and GluR2 (green) immunostaining and bADDL binding (red), respectively, acquired from the same microscopy plane under different filters. bADDL binding was observed largely in GluR2-expressing neurons (white arrows), whereas neurons expressing GluR1 but not GluR2 show negative bADDL binding (yellow arrows). Panel 4 is a merged image of panels 1–3. Panels 6 and 7 show enlarged images of dendritic fragments (rectangular boxes in panel 4) in which only GluR1 but not GluR2 is expressed. These dendrites display negative bADDL binding. Panel 5 is another merged image showing similar results. Panels 8–11 show an enlarged image of a dendritic fragment taken from panel 5 (rectangular box) with abundant GluR2 and GluR1. These dendrites show strong bADDL binding. Panels 12–15 show that although neurons expressing both GluR2 and GluR1 bind bADDLs (white arrows), neurons expressing only GluR2 (panels 12, yellow arrow) but not GluR1 (panels 13, yellow arrow) still bind bADDLs (panels 14 and 15, red color, yellow and white arrows). Together the results suggest that GluR2, but not GluR1, is required for bADDL binding. C, treating hippocampal neurons with different reagents that internalize AMPAR cause marked reductions in bADDL binding. l-Glu (50 μm), AMPA (100 μm), insulin (1 μm), and IGF-1 (1 μm) were preincubated with hippocampal neurons for 30 min at 37 °C followed by incubation with bADDL for 10 min at 37 °C. One-way analysis of variance showed significant differences in bADDL binding between neurons with and without pharmacological pretreatment. **, p < 0.01.
One way to test whether AMPARs are required for ADDL binding was to remove the receptors from the membrane surface via either siRNA or pharmacological treatments. siRNA transfection proved toxic to neurons cultured for longer than 4 days, well before bADDL binding could be detected. Thus we used pharmacological reagents to induce the internalization of GluR2/3. AMPA and glutamate have been known to stimulate internalization of GluR2/3 (50). Insulin and IGF-1 also cause internalization of AMPARs by distinct mechanisms (43, 43, 51–53). As shown in Fig. 2C, pretreatment of neurons for 30 min with each of these reagents significantly reduced bADDL binding (Fig. 2C, panels 1 and 2). These pharmacological data suggest that the presence of a surface AMPA receptor complex is essential for ADDL binding.
Rapid Neuronal Uptake of ADDLs Requires Calcineurin Activity
The observation that siRNAs targeting calcineurin led to peripheral membrane localization of bADDLs in N2A cells suggested that neurons might internalize bADDLs in a specific, regulated process. To first determine whether bADDLs are also internalized in primary neurons, we used an acid stripping method to selectively remove bADDLs bound to the cell surface (45, 46). Rat primary hippocampal neurons were treated with bADDLs to allow binding before stripping with a high salt, acidic solution that disrupts protein-protein interactions. Under this condition, bADDLs on the membrane surface are removed, and only those internalized and thereby protected from exposure to the extracellular medium would be retained and detectable with fluorescently labeled streptavidin after membrane permeabilization. ADDLs bound rapidly to the dendritic processes of neurons (Fig. 3A, panel 1). The acid solution removed the majority of bADDL staining after 1 or 5 min of incubation (Fig. 3A, panel 2), suggesting that bADDLs were present on the membrane surface at these time points. As the incubation time was increased, the bADDL signal became acid-resistant and remained in the dendrites and the soma after stripping (Fig. 3A, panel 4), suggesting transport of bADDLs to the interior of dendrites. Quantification of >50 neurons (Fig. 3A, panel 5) showed significant differences in bADDL binding between stripped and non-stripped neurons at 5 min post-binding (p < 0.05).
FIGURE 3.
Synaptic uptake of bADDLs. After being treated with 500 nm bADDLs for various lengths of time, neurons were subjected to high salt acid stripping to remove the membrane surface bADDLs. Cells were then fixed and permeabilized before stained with Alexa Fluor 555-labeled-streptavidin. A, panels 1–4, bADDL staining from pre- and post-stripping ADDL binding; panel 5, summary of bADDL binding quantification. *, p < 0.05. Bar scale: 10 μm. B, bADDL trafficking revealed by double staining with CTb-Alexa 488, a lipid raft marker, and streptavidin-Alexa 555. Representative dendritic segments show that at 1 min following bADDL treatment, CTb (green) binding on dendritic spines is highly colocalizated with bADDL (red) staining, displayed as yellow-colored (panel 1). After stripping off of the surface bADDLs, the internalized bADDLs (red) is seen in the internal area of the dendrites (panel 2). At 30 min following bADDL treatment (panels 3 and 4), bADDL stays colocalized with CTb after stripping (panel 4), indicating that the majority of bADDLs has been transported to intraspine compartments. Bar scale: 3 μm. C, bound and internalized bADDLs were highly colocalized with cpg2. D, FK506-blocked bADDL internalization correlated with the length of bADDL treatment time. Scale bar: 10 μm. **, p < 0.001.
To further visualize bADDL trafficking, we used cholera toxin B (CTb) as a dendritic membrane marker, because it binds specifically and with high affinity to sphingolipids, a major component of lipid rafts that are enriched in postsynaptic densities (54, 55). Within 1 min of incubation, bADDL staining was observed in close colocalization with CTb, generating a gold color after the images were merged (Fig. 3B). The internalization of bADDLs took place rapidly, as streptavidin staining was visible in the inner layer of dendrites after acid stripping even at 1 min post-binding (Fig. 3B, panels 1 and 2). By 30 min post-binding, a significant amount of bADDLs remained along the dendrites after stripping and was entirely colocalized to CTb (Fig. 3B, panels 3 and 4). To confirm this apparent internalization of bADDLs, we performed immunofluorescent colocalization experiments with antibodies to cpg2 (candidate plasticity gene 2), which is localized to the endocytic zone of excitatory synapses (56). If bADDLs are internalized into endosomes, then bADDL immunoreactivity would colocalize with this marker in a time-dependent manner. We observed that bADDLs colocalized with cpg2 immunoreactivity, with the extent of colocalization increasing over time (Fig. 3C). The internalization was blocked by FK506, a selective inhibitor of calcineurin, at a dose range of 2 nm to 4 μm (Fig. 3D). This finding is in agreement with the siRNA data suggesting that bADDLs are internalized via a calcineurin-dependent mechanism.
Aβ Oligomers Cause Removal of Surface AMPAR via Calcineurin-dependent Endocytosis
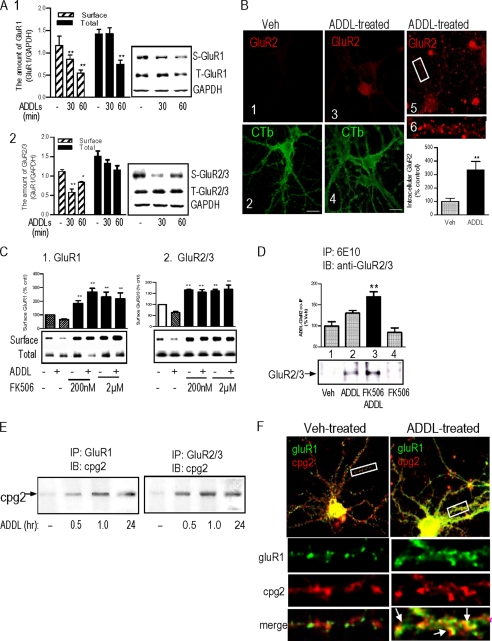
Because the synaptic localization of AMPARs is a highly regulated dynamic process, we next tested the effects of ADDLs on AMPAR surface expression. Primary neurons were treated with bADDLs for the indicated periods of time, and surface proteins were labeled by treatment with a cell-impermeable biotin-conjugated cross-linking reagent. ADDL treatment caused a decrease in biotinylated surface GluR1 (S-GluR1) first seen at 30 min post-treatment, whereas the total GluR1 (T-GluR1) remained unchanged (Fig. 4A). By 60 min, biotinylated S-GluR1 was reduced to 50% of untreated controls (p < 0.01), accompanied by a significant loss (p < 0.01) of T-GluR1 protein (Fig. 4A, panel 1). The ADDL-induced surface GluR2/3 loss (S-GluR2/3) (Fig. 4A, panel 2) was most obvious at 30 min after treatment (p < 0.01) and remained significant at 60 min post-treatment (p < 0.05). In contrast, the decrease in surface GluR4 was apparent at 30 min, and the amount of the surface GluR4 returned to base-line levels at 60 min (data not shown).
FIGURE 4.
ADDL-induced AMPAR loss. A, panel 1, rat hippocampal neurons were treated with 500 nm ADDLs for 30 and 60 min, respectively. Changes in the surface and the total amount of GluR1 and GluR2/3 were examined by surface biotinylation. The results indicate ADDL-induced reductions of the surface and total amounts of GluR1 (panel 1) and reduction of surface GluR2/3 (panel 2). S-GuR1, surface GluR1; T-GluR1, total GluR1; S-GluR2/3, surface GluR2/3; T-GluR2/3, total GluR2/3; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, immunocytochemistry detecting ADDL-induced GluR2 endocytosis (see “Experimental Procedures” for details). The internalized GluR2 was detected with an anti-mouse IgG-Alexa Fluor 555 (1:500). CTb-Alexa Fluor 488 (1:10,000) was used to label the neuronal membrane. Thirty neurons from 14 vehicle (Veh)-treated image planes and 35 neurons from 16 ADDL-treated image planes were quantified. C, effects of FK506 on AMPAR internalization. Hippocampal neurons were pretreated with 200 nm and 2 μm FK506 followed by treatment with 500 nm ADDLs for 1 h. The amounts of surface AMPAR subunits were assessed by surface biotinylation. The results showed that FK506 increased the surface GluR and prevented ADDL-induced surface GluR loss. D, detection of ADDL-GluR2/3 complex by co-IP. The ADDL-treated hippocampal neuronal lysates in the presence and absence of FK506 were precipitated by 6E10 followed by detection of AMPAR on Western blots (IB) by anti-GluR antibodies. The results showed that GluR2/3 was co-precipitated with ADDLs, the amount of which was increased under inhibition of endocytosis by FK506. A–D, **, p < 0.01. Scale bar, 8 μm. E, ADDL-induced association of GluRs with cpg2. Hippocampal neurons were treated with 500 nm ADDLs for various lengths of time. The treated neuronal lysates were precipitated by anti-GluR1 and anti-GluR2/3 antibodies, respectively. The complexes were then resolved on SDS-PAGE, and co-IP of cpg2 was detected on Western blots with an anti-cpg2 antibody. The results showed that the 500 nm ADDL treatment induced associations of cpg2 with GluR1 and Glur2/3 that lasted as long as 24 h after treatment. F, ADDL-induced colocalization of GluR1 with cpg2 detected with immunocytochemistry. The results showed that cpg2 and GluR1 immunostaining signals were separated localized in untreated hippocampal neurons, but they became partially colocalized following ADDL treatment.
To confirm these cell surface biotinylation data, we tested ADDL-induced AMPAR internalization using the surface stripping method (43). In these experiments, antibodies to GluR2 and GluR3 subunits were added to the medium and allowed to bind to their target receptors. If the antibody was internalized it would become acid-resistant; surface localized antibodies, in contrast, would be stripped from the cell by acid solution. Under control conditions there was relatively low GluR2 internalization as evidenced by antibody sensitivity to acid solution (Fig. 4B, panel 1). Significantly increased internalization (p < 0.01) was detected after ADDL treatment (Fig. 4B, panels 3, 5, and 6). ADDL-induced internalization was also observed with GluR3, with the internalized receptors distributed mostly along the dendrites (supplemental Fig. 2). FK506 (200 nm) pretreatment increased the surface AMPARs and completely blocked the ADDL-induced surface GluR losses (Fig. 4C).
We next tested whether inhibition of AMPAR internalization by FK506 would lead to ADDLs forming stable complexes with GluR subunits (or closely associated proteins) using co-immunoprecipitation (co-IP). After the application of ADDLs to neurons, protein complexes were solubilized, and those containing Aβ or APP were collected using the antibody 6E10. Co-precipitated AMPARs were then detected by Western blot. No co-IP of GluRs was seen in the control neurons (Fig. 4D, lane 1). Unlike GluR1 and GluR4, GluR2/3 was pulled down by 6E10 from ADDL-treated hippocampal primary neuron extracts (Fig. 4D, lane 2). In the presence of FK506 and ADDLs, the amount of immunoprecipitated GluR2/3 was significantly increased (Fig. 4D, lane 3), whereas samples treated with FK506 alone were negative (Fig. 4D, lane 4). Reverse IP experiments were also performed to verify the results. Neurons were treated with bADDLs in the presence or absence of FK506 and immunoprecipitated with an anti-GluR1 or an anti-gluR2/3 antibody. This IP was followed by detection of Aβ oligomers on Western blots with streptavidin conjugated with horseradish peroxidase. Although the anti-GluR1 antibody did not pull down detectable bADDL signals (data not shown), bADDL species were detected by streptavidin in the complex pulled down by the GluR2/3 antibody, which was most obvious in the FK506-treated samples (supplemental Fig. 3). Additionally, these samples were assessed by dot blotting using the high affinity anti-ADDL antibody ACU-954 (15), which gave consistent results (supplemental Fig. 3B). These results suggest that ADDLs form complexes with GluR2/3 on the neuronal surface, the detection of which is increased by the inhibition of ADDL and AMPAR internalization by FK506.
We then asked whether ADDL treatment would increase the amount of AMPAR associated with the postsynaptic endosomal marker, cpg2. Hippocampal neurons were treated with 500 nm ADDLs for different period of times; then the cells were lysed and immunoprecipitated with GluR subunit antibodies, and cpg2 was detected by Western blot. The results (Fig. 4E) showed that although little or no GluR1 or GluR2/3 was associated with cpg2 in normal cultured hippocampal neurons, ADDL treatment induced cpg2 co-IP with GluR1 or GluR2/3 antibodies. Likewise, immunostaining showed limited colocalization of GluR1 with cpg2 in dendritic spines in normal neurons (Fig. 4F, Veh-treated), whereas colocalization between GluR1 and cpg2 was increased following ADDL treatment (Fig. 4F, arrows). These results further indicate the translocation of AMPAR from the membrane surface to neuronal endosomal loci in response to ADDL treatment.
Removal of Surface AMPARs by Cell-derived Aβ Oligomers
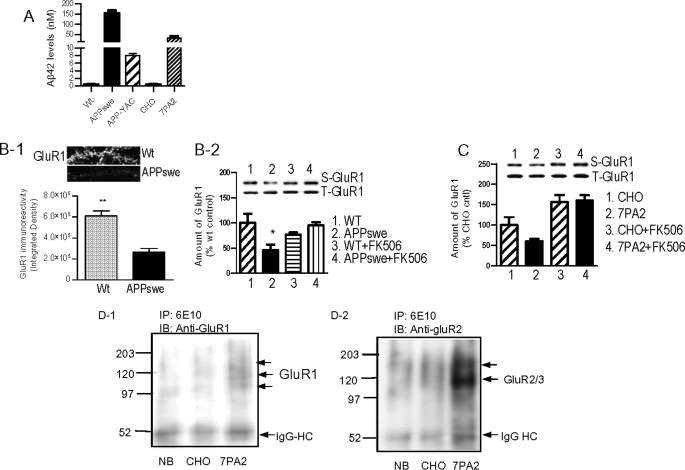
Because ADDLs are prepared from synthetic Aβ peptides, it was important to determine whether the observed effects were also generated by endogenously produced Aβ oligomers. We thus collected media from primary neuronal cultures from transgenic mice expressing wild type human APP (APP-YAC) (57) and the APPswe mutant (58, 59). Conditioned medium (CM) was also collected from the 7PA2 cell line, which contains secreted soluble Aβ oligomers (60, 61). ELISA measurements detected Aβ42 in the CM of all APP-expressing cells, with Aβ42 levels being APPswe > 7PA2 > APP-YAC (Fig. 5A). Treating the rat hippocampal neurons with APPswe CM, but not with CM from wild type neurons, for 6 h caused a marked loss of the GluR1 in dendritic spines (Fig. 5B, panel 1). A reduction in the surface GluR1 was observed at 1 h following the APPswe CM treatment, which was blocked by FK506 (Fig. 5B, panel 2). Similarly, incubation of rat hippocampal neurons with CM from 7PA2 cells also reduced the surface GluR1 in an FK506-sensitive manner (Fig. 5C). Thus the removal of AMPARs via calcineurin is a common feature shared by synthetic and cell-derived Aβ oligomers.
FIGURE 5.
Surface AMPAR removal by cell-derived Aβ oligomers. A, ELISA show high levels of Aβ42 from APPswe neurons and 7PA2 cells. Wt, wild type. B, panel 1, immunocytochemical results showing APPswe medium cause a striking loss of dendritic GluR1 in rat hippocampal neurons after 6-h incubation. Twelve dendrites from five neurons in total were quantified and compared with a t test. **, p < 0.001. Panel 2, measured by surface biotinylation, APPswe medium caused significant reduction in the surface GluR1 in rat hippocampal neurons after 1 h of incubation. The APPswe-meduated surface GluR1 loss was prevented by FK506. *, p < 0.05. C, reduction of surface GluR1 caused by treatment with condition medium from 7PA2 cells for 6 h. FK506 prevented the effect and increased surface GluR1. D, photoreactive cross-link experiments show that cell-derived Aβ and/or Aβ oligomers preferentially interact with GluR2. IB, immunoblotting; IgG-HC, IgG heavy chain; NB, neurobasal culture medium.
We next used photoreactive amino acid cross-linking to further test whether the endogenous Aβ oligomers interact physically with GluR-containing protein complexes. 7PA2 and parent wild type CHO cells were cultured with a medium containing photoreactive derivatives l-photo-methionine and l-photo-leucine. These amino acids were incorporated into all proteins during protein synthesis including APP. The media collected from 7PA2 and CHO cells were applied to primary rat hippocampal neurons followed by UV activation, which caused a covalent cross-link of the l-photo-methionine- and l-photo-leucine-labeled Aβ/Aβ oligomers (as well as other labeled proteins) to neuronal proteins in close proximity (62). Solubilized protein complexes from treated hippocampal neurons were then immunoprecipitated by 6E10 and analyzed by Western blotting with anti-GluR antibodies. As shown in Fig. 5D, panels 1 and 2, GluR1 was co-precipitated by 6E10 from 7PA2 medium-treated neurons, but not from the neurobasal or parent CHO medium-treated neurons, after photoreactive cross-linking. The GluR1 antibody detected multiple protein bands that migrated at an apparent molecular mass of ∼105–130 kDa (Fig. 5D, panel 1). This banding pattern potentially resulted from GluR1 cross-linking with different unrelated proteins or perhaps distinct species of Aβ oligomers. A more intense protein band was detected by GluR2/3 antibody (Fig. 5D, panel 2). Although the major band migrated at ∼110 kDa, an additional weaker bank appeared at a higher molecular mass (∼150 kDa) position. Such results were not observed with GluR4 (data not shown). Conversely, the cross-linked neuronal lysates were precipitated using an anti-GluR2/3 antibody followed by detection of Aβ species on Western blots or dot blots with a mixture of the 6E10 and ACU-954 antibodies. Positive Aβ oligomer signals were precipitated by the anti-GluR2/3 antibody from neuronal samples cross-linked with 7PA2 medium containing l-p-Met- and l-p-Leu-labeled Aβ species (supplemental Fig. 4). Cross-linking of endogenous proteins with l-photo-methionine and l-photo-leucine occurs at 0 distance between two interacted proteins; these results suggest that the cellular Aβ interacts with GluR2/3 or with proteins complexed to these receptors.
Effects of AMPAR Antagonists on bADDL Binding
We next asked whether AMPAR inhibitors would alter bADDL binding. Hippocampal neurons were pretreated with several classes of antagonists including the competitive inhibitor CNQX, the calcium-permeable AMPAR inhibitor IEM1064, and an allosteric negative modulator GYKI52466 prior to bADDL binding. Rapid bADDL binding was observed on dendrites as seen previously (Fig. 6A, panel 1). GYKI52466 inhibited bADDL binding such that it induced a marked reduction in bADDL binding at 10 min post-bADDL application (Fig. 6A) in a dose-dependent manner (A and B). The calcium-permeable AMPAR inhibitor IEM1460 also showed a transient but much milder effect, whereas CNQX caused a more lasting although moderate reduction in ADDL binding (Fig. 6B, panel 1). Two-way analysis of variance revealed significant compound treatment (F4, 159 = 8.764, p < 0.0001) and time (F1, 159 = 16.75, p < 0.001) effects, as well as a significant interactions (F4, 159 = 6.796, p < 0.001). In summary, these results indicate that AMPAR inhibitors modulate the association of bADDLs with hippocampal neurons.
FIGURE 6.
Effect of AMPA receptor antagonists on bADDL synaptic binding. Rat hippocampal neurons (21 days in vitro) were incubated with 500 nm bADDLs for 10 and 60 min in the presence of GYKI52466 (50 μm or 100 μm), IEM1460 (100 μm), or CNQX (50 μm). A, representative neuronal images showing changes in bADDL binding in the absence (panel 1) or presence (panel 2) of GYKI51466. B-1, bar graph summarizing the quantification of bADDL binding intensities from each treatment (n ≥ 30 neurons). B-2, dose-dependent changes in bADDL binding by GYKI52466 (B-1 and B-2, *, p < 0.05; **, p < 0.001). Veh, vehicle.
GYKI52466 and CNQX Prevented ADDL-induced Surface AMPA Receptor Loss
We next tested whether AMPAR antagonists impact ADDL-induced surface receptor loss using biotinylation. As shown in Fig. 7A, GYKI52466 and CNQX prevented the loss of most surface GluRs. In contrast, the Ca2+-permeable AMPA receptor inhibitor IEM1460 showed no effect. GYKI52466 alone did not have any effect on the surface AMPA receptor expression (Fig. 7B), suggesting that its bADDL binding inhibitory effect is not attributable to receptor internalization, unlike the reduced bADDL binding under the actions of AMPA and glutamate in Fig. 2C. Furthermore, immunocytochemical experiments showed that ADDL-induced loss of AMPAR-containing dendritic spines was overcome by GYKI52466 (Fig. 7C). Thus this AMPAR inhibitor protected neurons against ADDL-induced spine loss and AMPAR internalization.
FIGURE 7.
AMPA receptor antagonists prevent ADDL-induced surface AMPA receptor and spine loss. A, rat primary hippocampal cultures were treated with ADDLs (500 nm) for 1 h in the presence or absence of GYKI52466 (100 μm), IEM1460 (100 μm), and CNQX (100 μm). Changes in surface AMPA receptors were examined by surface biotinylation. Both GYKI52466 and CNQX were shown to reduce ADDL-induced GluR1 and GluR2/3 loss. B, GYKI52466 alone has no effect on surface AMPA receptor expression. C, GYKI52466 prevented ADDL-induced spine loss. Veh, vehicle. A–C, *, p < 0.05; **, p < 0.01.
DISCUSSION
The current study demonstrates that in cell cultures soluble Aβ42 oligomers selectively bind to neurons with excitatory synapses, where they cause the loss of surface AMPARs. High throughput siRNA screening followed by biochemical and pharmacological verification with primary hippocampal cultures identified two essential components in this detrimental interaction: 1) AMPARs (particularly those containing GluR2) at the dendritic surface and 2) an endocytic process responsible for the internalization of ADDLs and AMPARs that is driven by calcineurin activity. Inhibition of either component is sufficient to prevent ADDL-induced surface AMPAR loss. bADDLs interact predominantly with dendritic spines harboring excitatory synaptic proteins. Colocalization of ADDLs with cpg2 further indicates the selectivity of Aβ oligomers to glutamatergic synapses. This observation implicates the excitatory glutamatergic synapses as the primary site of AD pathogenesis. Supporting this notion are findings from Palop et al. (63) showing that high levels of Aβ in the brain of transgenic mice expressing human APP cause aberrant excitatory neuronal activity, which can be mimicked by excitotoxic treatments and prevented by blocking overexcitation. In a separate study Cirrito et al. (64), using a different human APP-expressing transgenic mouse (tg2576) model, report that interstitial fluid Aβ level is elevated by excitatory (glutamatergic) synaptic activity. Given the data presented here and elsewhere, increased Aβ could negatively feed back to inhibit the excitatory transmission at certain synapses.
The current data suggest a role for the surface AMPARs in bADDLs binding to spines because 1) pharmacological removal of surface AMPARs reduces bADDL binding, 2) GluR2/3 can be recovered with antibodies to Aβ after oligomer treatment, and 3) AMPAR inhibitors interfere with ADDL binding to neurons. However, previous studies have not found AMPAR to be a direct binding target for Aβ oligomers (10, 17, 21). However, synaptic AMPAR complexes exhibit highly dynamic mobility and undergo rapid internalization in response to glutamate transmission and ADDL binding. By including 4% sucrose to stabilize the membrane surface proteins, we observed higher level colocalization of bADDL binding with GluR2 than with other GluR subunits. Inhibition of receptor internalization by FK506 allows greater availability of AMPARs on the membrane surface and significantly increases the amount of GluR2/3, but not GluR1 and GluR4, co-precipitated with ADDLs. Further, photoreactive amino acid cross-linking revealed a close association between cell-derived Aβ and GluR2/3. AMPARs play a critical role in the dynamics of dendritic spines. Overexpression of GluR2 (65) or insertion of the GluR1 C-terminal fragment increases and stabilizes spines size (66), whereas removal of the surface AMPAR leads to spine loss (18). Thus, although our current evidence has yet to show conclusively that GluR2 is a direct binding partner for Aβ oligomers, it is in line with the notion of GluR2 influencing bADDL binding via its role in regulating spine expression. Removal of GluR2-containing surface AMPARs via pharmacological endocytosis not only could reduce the number of potential GluR2 direct or indirect GluR2 binding sites but also could cause a decrease in spine numbers together with surface expression of other synaptic proteins such as α7-nAchR, NMDAR (10, 17, 22), insulin receptor (23, 24), and the cellular prion protein (21), all of which have been show to bind Aβ oligomers.
We have also shown that AMPAR antagonists GYKI52466 and CNQX reduce bADDL binding and prevent ADDL-induced surface receptor loss. It is unlikely that both compounds reduce bADDL binding by directly competing with bADDLs for the binding site, because GYKI5246 and CNQX bind distinct sites on the AMPAR. Thus the reduction in bADDL binding is more likely related to either a functional or morphological state of AMPAR influenced by the antagonists. Changes in these receptor states could have an impact either directly on the binding of the Aβ oligomer to GluR2/3 or indirectly via associations with an intermediate receptor-binding protein. It is interesting that GYKI52466 produced the most profound inhibitory effect on bADDL binding. CNQX, although a more potent and competitive inhibitor, showed only a modest binding inhibition. The Ca2+-permeable AMPAR inhibitor IEM1460 did not protect the surface receptor, suggesting that the primary target might be the non-Ca2+-permeable GluR2 subunit; this is supported by various data in the present study. GYKI52466 is a 2,3-benzodiazepine that selectively inhibits AMPA receptors by binding to an allosteric region distinct from the glutamate recognition site (67–70). It remains to be determined why interaction with this binding site has the stronger effect on ADDL binding to neurons. The GYKI class AMPAR antagonists are neuroprotective in a variety of animal disease models (70, 71, 73, 74). As increased epileptiform activities develop in transgenic mice, presumably because of accumulated Aβ levels in the brain (75), it will be important to determine whether GYKI25466 can protect AD transgenic mice through its inhibitory effect of ADDL binding-AMPAR interaction, which if true would offer a potential approach for treating the neuronal dysfunction and cognitive deficits in AD.
To date various neuronal surface receptors and signaling proteins have been reported to bind Aβ oligomers (10, 12, 21, 23, 24), and multiple Aβ oligomer species produced both synthetically and in vivo have been shown to be toxic to synapses (6, 13, 14, 76). However, because of the instability of Aβ oligomer assemblies (77, 78), it has remained challenging to unequivocally determine whether the synaptotoxicity is due to the action of a specific species or to a combination of multiple species. Furthermore, it remains to be addressed why Aβ oligomers bind to multiple targets and whether selectivity exists between different oligomer species and different neuronal receptors/proteins.
Despite the current lack of answers to the above questions, different studies including our current one show Aβ oligomers inducing surface receptor loss via endocytosis driven by calcineurin (17, 18). In an AD animal model, inhibition of calcineurin by FK506 improved cognitive function (79). Upon binding, ADDLs rapidly gain entry to the endocytic zone within dendritic spines again via calcineurin-regulated endocytosis. Aβ oligomers are found both extracellularly and intracellularly (80, 81). It is possible in vivo that the Aβ released from presynaptic terminals binds to glutamatergic synapses and triggers the activation of calcineurin leading to Aβ internalization, where perhaps its toxic effects are manifest. Thus, the benefit of FK506 in the AD brain may include reducing intracellular Aβ oligomers via blocking their internalization.
Internalization of AMPA receptor is a critical mechanism underlying inhibition of synaptic efficacy such as long-term depression (LTD) mediated by NMDAR and/or mGluRs (82, 83). While GluR1 is associated with NMDA receptor-mediated LTD, GluR2 has been shown to be linked to mGluR-dependent LTD (84). It is possible that this pathway is shared by those underlying Aβ oligomer actions, as all of the involved components have parallel changes induced by Aβ oligomers, such as the involvement of calcineurin activation and AMPA receptor internalization. Indeed, Aβ oligomers enhance both NMDAR- and mGluR-mediated LTD (18, 85), and Aβ partially occludes mGluR-dependent LTD (18). Loss of AMPAR has been repeatedly observed in AD transgenic mice (18, 36–38) and in the human AD brain. Downscaling of AMPAR precedes neurofibrillary tangles (34), and loss of GluR1 and GluR2/3 occurs selectively in the entorhinal cortex and CA1 of the hippocampus, the regions that are most vulnerable to AD pathogenesis (29–33). It is therefore possible that persistent decreases in synaptic efficacy occur in those brain regions. The presence of GluR2 in the heteromeric AMPAR complex determines the Ca2+ permeability of the receptor channel. Neurons from GluR2−/− and GluR2/3−/− mice brain show enhanced dendritic AMPAR internalization in response to AMPA, which is attributed to increased voltage-dependent Ca2+ influx (43, 72). Thus, by targeting GluR2, Aβ oligomers could potentially perturb the normal Ca2+ impermeability of the AMPAR channel leading to increased Ca2+ leakage into dendrites.
In conclusion, the current study has suggested a pathway by which Aβ oligomers specifically target excitatory GluR2-containing synapses, where they cause the removal of surface AMPARs leading to synaptic loss. This pathway might be shared by those underlying NMDA- and mGluR-mediated LTDs (Fig. 8). On the other hand, perturbation of the GluR2-containing AMPAR channel could cause increased Ca2+ permeability, leading to an aberrant Ca2+ signal, which further enhances the removal of the dendritic surface AMPARs (Fig. 8). Although the calcineurin-induced AMPAR endocytic cascades could be involved in normal physiological feedback processes, aberrant excitatory transmission and accumulation of high levels of Aβ oligomers could cause abnormal augmentation and/or dysregulation of these processes leading to AD-related synaptic pathogenesis. When this happens, FK506, or a negative modulator of AMPAR, might be beneficial in preventing massive synaptic loss by oligomers and the subsequent cognitive deficits associated with AD.
FIGURE 8.
A schematic diagram showing hypothetic cascades through which Aβ oligomers induce surface AMPAR loss leading to inhibition of synaptic efficacy. Binding of Aβ oligomers to receptors on dendritic spines triggers activation of calcineurin (PP2B), probably via Ca2+/calmodulin-dependent kinase II (CaMKII) activity. The calcineurin activity activates the clathrin-dependent endocytosis of AMPARs and/or NMDARs via the involvement of dynamin. Activation of the group I metabotropic glutamate receptors (mGluR1/5) is involved in this cascade, which in turn facilitates the internalization of AMPARs. On the other hand, the interaction of Aβ oligomers with GluR2 may potentially interfere with the Ca2+ impermeability of the AMPAR channel leading to increased voltage-dependent Ca2+ influx, thus generating aberrant Ca2+ toxicity. The abnormal Ca2+ signals could enhance the internalization of dendritic AMPAR. Removal of the surface AMPAR and NMDA lead to LTD and reduced spine numbers. If LTD and loss of spine structure persist, the resulting synaptic transmission failure would lead to memory impairment.
Supplementary Material
Acknowledgments
We thank Renee Gaspar for technical assistance with confocal microscopy and Robert Hepler and Joseph Joyce for assistance with ADDL preparation and characterization for the siRNA screen.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- AD
- Alzheimer disease
- Aβ
- amyloid β
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
- AMPA receptor
- APP
- amyloid precursor protein
- NMDA
- N-methyl-d-aspartic acid
- NMDAR
- NMDA receptor
- SSMD
- strictly standardized mean difference
- CHO
- Chinese hamster ovary
- siRNA
- small interfering RNA
- bADDL
- biotin-labeled amyloid β-derived diffusible ligand
- CTb
- cholera toxin B
- IP
- immunoprecipitation
- CM
- conditioned medium
- LTD
- long-term depression
- PBS
- phosphate-buffered saline
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Selkoe D. J. (2002) Science 298, 789–791 [DOI] [PubMed] [Google Scholar]
- 2.Coleman P. D., Yao P. J. (2003) Neurobiol. Aging 24, 1023–1027 [DOI] [PubMed] [Google Scholar]
- 3.Lassmann H., Fischer P., Jellinger K. (1993) Ann. N.Y. Acad. Sci. 695, 59–64 [DOI] [PubMed] [Google Scholar]
- 4.Mucke L., Masliah E., Yu G. Q., Mallory M., Rockenstein E. M., Tatsuno G., Hu K., Kholodenko D., Johnson-Wood K., McConlogue L. (2000) J. Neurosci. 20, 4050–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terry R. D., Masliah E., Salmon D. P., Butters N., DeTeresa R., Hill R., Hansen L. A., Katzman R. (1991) Ann. Neurol. 30, 572–580 [DOI] [PubMed] [Google Scholar]
- 6.Gong Y., Chang L., Viola K. L., Lacor P. N., Lambert M. P., Finch C. E., Krafft G. A., Klein W. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10417–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsend M., Shankar G. M., Mehta T., Walsh D. M., Selkoe D. J. (2006) J. Physiol. 572, 477–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 9.Lacor P. N., Buniel M. C., Chang L., Fernandez S. J., Gong Y., Viola K. L., Lambert M. P., Velasco P. T., Bigio E. H., Finch C. E., Krafft G. A., Klein W. L. (2004) J. Neurosci. 24, 10191–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacor P. N., Buniel M. C., Furlow P. W., Clemente A. S., Velasco P. T., Wood M., Viola K. L., Klein W. L. (2007) J. Neurosci. 27, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert M. P., Barlow A. K., Chromy B. A., Edwards C., Freed R., Liosatos M., Morgan T. E., Rozovsky I., Trommer B., Viola K. L., Wals P., Zhang C., Finch C. E., Krafft G. A., Klein W. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6448–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shankar G. M., Bloodgood B. L., Townsend M., Walsh D. M., Selkoe D. J., Sabatini B. L. (2007) J. Neurosci. 27, 2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 15.Shughrue P. J., Acton P. J., Breese R. S., Zhao W. Q., Chen-Dodson E., Hepler R. W., Wolfe A. L., Matthews M., Heidecker G. J., Joyce J. G., Villarreal S. A., Kinney G. G. (2010) Neurobiol. Aging 31, 189–202 [DOI] [PubMed] [Google Scholar]
- 16.Chin J., Massaro C. M., Palop J. J., Thwin M. T., Yu G. Q., Bien-Ly N., Bender A., Mucke L. (2007) J. Neurosci. 27, 2727–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyder E. M., Nong Y., Almeida C. G., Paul S., Moran T., Choi E. Y., Nairn A. C., Salter M. W., Lombroso P. J., Gouras G. K., Greengard P. (2005) Nat. Neurosci. 8, 1051–1058 [DOI] [PubMed] [Google Scholar]
- 18.Hsieh H., Boehm J., Sato C., Iwatsubo T., Tomita T., Sisodia S., Malinow R. (2006) Neuron 52, 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H. W., Pasternak J. F., Kuo H., Ristic H., Lambert M. P., Chromy B., Viola K. L., Klein W. L., Stine W. B., Krafft G. A., Trommer B. L. (2002) Brain Res. 924, 133–140 [DOI] [PubMed] [Google Scholar]
- 20.Cleary J. P., Walsh D. M., Hofmeister J. J., Shankar G. M., Kuskowski M. A., Selkoe D. J., Ashe K. H. (2005) Nat. Neurosci. 8, 79–84 [DOI] [PubMed] [Google Scholar]
- 21.Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009) Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshpande A., Kawai H., Metherate R., Glabe C. G., Busciglio J. (2009) J. Neurosci. 29, 4004–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W. Q., De Felice F. G., Fernandez S., Chen H., Lambert M. P., Quon M. J., Krafft G. A., Klein W. L. (2008) FASEB J. 22, 246–260 [DOI] [PubMed] [Google Scholar]
- 24.Townsend M., Mehta T., Selkoe D. J. (2007) J. Biol. Chem. 282, 33305–33312 [DOI] [PubMed] [Google Scholar]
- 25.Bredt D. S., Nicoll R. A. (2003) Neuron 40, 361–379 [DOI] [PubMed] [Google Scholar]
- 26.Malinow R., Malenka R. C. (2002) Annu. Rev. Neurosci. 25, 103–126 [DOI] [PubMed] [Google Scholar]
- 27.Morris R. G. (2006) Eur. J. Neurosci. 23, 2829–2846 [DOI] [PubMed] [Google Scholar]
- 28.Reisel D., Bannerman D. M., Schmitt W. B., Deacon R. M., Flint J., Borchardt T., Seeburg P. H., Rawlins J. N. (2002) Nat. Neurosci. 5, 868–873 [DOI] [PubMed] [Google Scholar]
- 29.Geddes J. W., Brunner L., Cotman C. W., Buzsáki G. (1992) Exp. Neurol. 115, 271–281 [DOI] [PubMed] [Google Scholar]
- 30.Armstrong D. M., Ikonomovic M. D., Sheffield R., Wenthold R. J. (1994) Brain Res. 639, 207–216 [DOI] [PubMed] [Google Scholar]
- 31.Carter T. L., Rissman R. A., Mishizen-Eberz A. J., Wolfe B. B., Hamilton R. L., Gandy S., Armstrong D. M. (2004) Exp. Neurol. 187, 299–309 [DOI] [PubMed] [Google Scholar]
- 32.Yasuda R. P., Ikonomovic M. D., Sheffield R., Rubin R. T., Wolfe B. B., Armstrong D. M. (1995) Brain Res. 678, 161–167 [DOI] [PubMed] [Google Scholar]
- 33.Ikonomovic M. D., Sheffield R., Armstrong D. M. (1995) Hippocampus 5, 469–486 [DOI] [PubMed] [Google Scholar]
- 34.Ikonomovic M. D., Mizukami K., Davies P., Hamilton R., Sheffield R., Armstrong D. M. (1997) J. Neuropathol. Exp. Neurol. 56, 1018–1027 [DOI] [PubMed] [Google Scholar]
- 35.Ting J. T., Kelley B. G., Lambert T. J., Cook D. G., Sullivan J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cha J. H., Farrell L. A., Ahmed S. F., Frey A., Hsiao-Ashe K. K., Young A. B., Penney J. B., Locascio J. J., Hyman B. T., Irizarry M. C. (2001) Neurobiol. Dis. 8, 90–102 [DOI] [PubMed] [Google Scholar]
- 37.Almeida C. G., Tampellini D., Takahashi R. H., Greengard P., Lin M. T., Snyder E. M., Gouras G. K. (2005) Neurobiol. Dis. 20, 187–198 [DOI] [PubMed] [Google Scholar]
- 38.Chang E. H., Savage M. J., Flood D. G., Thomas J. M., Levy R. B., Mahadomrongkul V., Shirao T., Aoki C., Huerta P. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takumi Y., Ramírez-León V., Laake P., Rinvik E., Ottersen O. P. (1999) Nat. Neurosci. 2, 618–624 [DOI] [PubMed] [Google Scholar]
- 40.Zhang X. D. (2008) J. Biomol. Screen. 13, 363–377 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X. D., Kuan P. F., Ferrer M., Shu X., Liu Y. C., Gates A. T., Kunapuli P., Stec E. M., Xu M., Marine S. D., Holder D. J., Strulovici B., Heyse J. F., Espeseth A. S. (2008) Nucleic Acids Res. 36, 4667–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X. D. (2007) J. Biomol. Screen. 12, 645–655 [DOI] [PubMed] [Google Scholar]
- 43.Beattie E. C., Carroll R. C., Yu X., Morishita W., Yasuda H., von Zastrow M., Malenka R. C. (2000) Nat. Neurosci. 3, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 44.Carroll R. C., Beattie E. C., von Zastrow M., Malenka R. C. (2001) Nat. Rev. Neurosci. 2, 315–324 [DOI] [PubMed] [Google Scholar]
- 45.Haigler H. T., Maxfield F. R., Willingham M. C., Pastan I. (1980) J. Biol. Chem. 255, 1239–1241 [PubMed] [Google Scholar]
- 46.Wilde A., Beattie E. C., Lem L., Riethof D. A., Liu S. H., Mobley W. C., Soriano P., Brodsky F. M. (1999) Cell 96, 677–687 [DOI] [PubMed] [Google Scholar]
- 47.Hepler R. W., Grimm K. M., Nahas D. D., Breese R., Dodson E. C., Acton P., Keller P. M., Yeager M., Wang H., Shughrue P., Kinney G., Joyce J. G. (2006) Biochemistry 45, 15157–15167 [DOI] [PubMed] [Google Scholar]
- 48.Lambert M. P., Velasco P. T., Chang L., Viola K. L., Fernandez S., Lacor P. N., Khuon D., Gong Y., Bigio E. H., Shaw P., De Felice F. G., Krafft G. A., Klein W. L. (2007) J. Neurochem. 100, 23–35 [DOI] [PubMed] [Google Scholar]
- 49.Allen P. B., Ouimet C. C., Greengard P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9956–9961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S. H., Simonetta A., Sheng M. (2004) Neuron 43, 221–236 [DOI] [PubMed] [Google Scholar]
- 51.Lissin D. V., Carroll R. C., Nicoll R. A., Malenka R. C., von Zastrow M. (1999) J. Neurosci. 19, 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin J. W., Ju W., Foster K., Lee S. H., Ahmadian G., Wyszynski M., Wang Y. T., Sheng M. (2000) Nat. Neurosci. 3, 1282–1290 [DOI] [PubMed] [Google Scholar]
- 53.Man H. Y., Lin J. W., Ju W. H., Ahmadian G., Liu L., Becker L. E., Sheng M., Wang Y. T. (2000) Neuron 25, 649–662 [DOI] [PubMed] [Google Scholar]
- 54.Hering H., Lin C. C., Sheng M. (2003) J. Neurosci. 23, 3262–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao W. Q., Waisman D. M., Grimaldi M. (2004) J. Neurochem. 90, 609–620 [DOI] [PubMed] [Google Scholar]
- 56.Cottrell J. R., Borok E., Horvath T. L., Nedivi E. (2004) Neuron 44, 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamb B. T., Sisodia S. S., Lawler A. M., Slunt H. H., Kitt C. A., Kearns W. G., Pearson P. L., Price D. L., Gearhart J. D. (1993) Nat. Genet. 5, 22–30 [DOI] [PubMed] [Google Scholar]
- 58.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Science 274, 99–102 [DOI] [PubMed] [Google Scholar]
- 59.Mullan M., Crawford F., Axelman K., Houlden H., Lilius L., Winblad B., Lannfelt L. (1992) Nat. Genet. 1, 345–347 [DOI] [PubMed] [Google Scholar]
- 60.Podlisny M. B., Ostaszewski B. L., Squazzo S. L., Koo E. H., Rydell R. E., Teplow D. B., Selkoe D. J. (1995) J. Biol. Chem. 270, 9564–9570 [DOI] [PubMed] [Google Scholar]
- 61.Walsh D. M., Klyubin I., Fadeeva J. V., Rowan M. J., Selkoe D. J. (2002) Biochem. Soc. Trans. 30, 552–557 [DOI] [PubMed] [Google Scholar]
- 62.Suchanek M., Radzikowska A., Thiele C. (2005) Nat. Methods 2, 261–267 [DOI] [PubMed] [Google Scholar]
- 63.Palop J. J., Chin J., Roberson E. D., Wang J., Thwin M. T., Bien-Ly N., Yoo J., Ho K. O., Yu G. Q., Kreitzer A., Finkbeiner S., Noebels J. L., Mucke L. (2007) Neuron 55, 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cirrito J. R., Yamada K. A., Finn M. B., Sloviter R. S., Bales K. R., May P. C., Schoepp D. D., Paul S. M., Mennerick S., Holtzman D. M. (2005) Neuron 48, 913–922 [DOI] [PubMed] [Google Scholar]
- 65.Passafaro M., Nakagawa T., Sala C., Sheng M. (2003) Nature 424, 677–681 [DOI] [PubMed] [Google Scholar]
- 66.Kopec C. D., Real E., Kessels H. W., Malinow R. (2007) J. Neurosci. 27, 13706–13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donevan S. D., Rogawski M. A. (1993) Neuron 10, 51–59 [DOI] [PubMed] [Google Scholar]
- 68.Donevan S. D., Yamaguchi S., Rogawski M. A. (1994) J. Pharmacol. Exp. Ther. 271, 25–29 [PubMed] [Google Scholar]
- 69.Zorumski C. F., Yamada K. A., Price M. T., Olney J. W. (1993) Neuron 10, 61–67 [DOI] [PubMed] [Google Scholar]
- 70.Balannik V., Menniti F. S., Paternain A. V., Lerma J., Stern-Bach Y. (2005) Neuron 48, 279–288 [DOI] [PubMed] [Google Scholar]
- 71.Abrahám G., Sólyom S., Csuzdi E., Berzsenyi P., Ling I., Tarnawa I., Hámori T., Pallagi I., Horváth K., Andrási F., Kapus G., Hársing L. G., Jr., Király I., Patthy M., Horváth G. (2000) Bioorg. Med. Chem. 8, 2127–2143 [DOI] [PubMed] [Google Scholar]
- 72.Biou V., Bhattacharyya S., Malenka R. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Belayev L., Alonso O. F., Liu Y., Chappell A. S., Zhao W., Ginsberg M. D., Busto R. (2001) J. Neurotrauma 18, 1031–1038 [DOI] [PubMed] [Google Scholar]
- 74.Catarzi D., Colotta V., Varano F. (2007) Med. Res. Rev. 27, 239–278 [DOI] [PubMed] [Google Scholar]
- 75.Palop J. J., Mucke L. (2009) Arch. Neurol. 66, 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walsh D. M., Selkoe D. J. (2007) J. Neurochem. 101, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 77.Ono K., Condron M. M., Teplow D. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14745–14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roychaudhuri R., Yang M., Hoshi M. M., Teplow D. B. (2009) J. Biol. Chem. 284, 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dineley K. T., Hogan D., Zhang W. R., Taglialatela G. (2007) Neurobiol. Learn. Mem. 88, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cirrito J. R., May P. C., O'Dell M. A., Taylor J. W., Parsadanian M., Cramer J. W., Audia J. E., Nissen J. S., Bales K. R., Paul S. M., DeMattos R. B., Holtzman D. M. (2003) J. Neurosci. 23, 8844–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gouras G. K., Almeida C. G., Takahashi R. H. (2005) Neurobiol. Aging 26, 1235–1244 [DOI] [PubMed] [Google Scholar]
- 82.Carroll R. C., Lissin D. V., von Zastrow M., Nicoll R. A., Malenka R. C. (1999) Nat. Neurosci. 2, 454–460 [DOI] [PubMed] [Google Scholar]
- 83.Xiao M. Y., Zhou Q., Nicoll R. A. (2001) Neuropharmacology 41, 664–671 [DOI] [PubMed] [Google Scholar]
- 84.Gladding C. M., Collett V. J., Jia Z., Bashir Z. I., Collingridge G. L., Molnár E. (2009) Mol. Cell. Neurosci. 40, 267–279 [DOI] [PubMed] [Google Scholar]
- 85.Li S., Hong S., Shepardson N. E., Walsh D. M., Shankar G. M., Selkoe D. (2009) Neuron 62, 788–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.