Abstract
RNA interference screen previously revealed that a HECT-domain E3 ubiquitin ligase, neuronal precursor cell expressed, developmentally down-regulated 4-2 (Nedd4-2), is necessary for ubiquitination and endocytosis of the dopamine transporter (DAT) induced by the activation of protein kinase C (PKC). To further confirm the role of Nedd4-2 in DAT ubiquitination and endocytosis, we demonstrated that the depletion of Nedd4-2 by two different small interfering RNA (siRNA) duplexes suppressed PKC-dependent ubiquitination and endocytosis of DAT in human and porcine cells, whereas knock-down of a highly homologous E3 ligase, Nedd4-1, had no effect on DAT. The abolished DAT ubiquitination in Nedd4-2-depleted cells was rescued by expression of recombinant Nedd4-2. Moreover, overexpression of Nedd4-2 resulted in increased PKC-dependent ubiquitination of DAT. Mutational inactivation of the HECT domain of Nedd4-2 inhibited DAT ubiquitination and endocytosis. Structure-function analysis of Nedd4-2-mediated DAT ubiquitination revealed that the intact WW4 domain and to a lesser extent WW3 domain are necessary for PKC-dependent DAT ubiquitination. Moreover, a fragment of the Nedd4-2 molecule containing WW3, WW4, and HECT domains was sufficient for fully potentiating PKC-dependent ubiquitination of DAT. Analysis of DAT ubiquitination using polyubiquitin chain-specific antibodies showed that DAT is mainly conjugated with Lys63-linked ubiquitin chains. siRNA analysis demonstrated that this polyubiquitination is mediated by Nedd4-2 cooperation with UBE2D and UBE2L3 E2 ubiquitin-conjugating enzymes. The model is proposed whereby each ubiquitinated DAT molecule is modified by a single four-ubiquitin Lys63-linked chain that can be conjugated to various lysine residues of DAT.
Keywords: Neurotransmitters, Protein/Intracellular Trafficking, Protein/Post-translational Modification, Protein/Sorting, Transport/Neurotransmitter, Nedd4-2, Ubiquitination
Introduction
Post-translational modification of proteins by ubiquitination has recently emerged as an important mechanism governing membrane trafficking processes. Ubiquitin moieties attached to integral membrane proteins serve to target these proteins for rapid internalization at the cell surface and, after internalization, to lysosomes for degradation (1–3). In addition, ubiquitination of the components of internalization and endosomal sorting machineries regulate interactions and turnover of these proteins (1, 4–5).
Ubiquitination is accomplished through sequential actions of the E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase, the latter typically determining the substrate specificity of the reaction (6–7). Proteins can be mono- and polyubiquitinated. Polyubiquitin chains can be formed by conjugation of ubiquitin to any of seven lysines in the ubiquitin molecule, although chains linked through Lys48 and Lys63 are most commonly found. Monoubiquitination and Lys63 polyubiqutination of transmembrane proteins are thought to represent the major sorting signals controlling their trafficking (2, 8–12).
We have recently demonstrated that ubiquitination mediates protein kinase C (PKC)2-dependent endocytosis of the human dopamine transporter (DAT) (13–15). Upon PKC activation by phorbol esters, heterologously expressed DAT is internalized via a clathrin-dependent endocytic pathway and degraded in lysosomes (16, 17). Ubiquitination of three lysines in the amino-terminal domain of DAT is critical for PKC-dependent DAT endocytosis. RNA interference (RNAi) screening analysis revealed the importance of neuronal precursor cell expressed, developmentally down-regulated 4-2 (Nedd4-2) for PKC-dependent ubiquitination and endocytosis of DAT (15). However, the mechanisms by which Nedd4-2 mediates PKC-dependent ubiquitination of DAT are not understood.
Nedd4-2 belongs to the NEDD4 family of E3 ligases that contain catalytic HECT (homolog to e6-AP C-term) domain in their carboxyl terminus (18). The NEDD4 family of E3 ligases has been implicated in ubiquitination and regulation of endocytosis of many channels, receptors, and transporters. Interestingly, despite high homology to other family members, Nedd4-2 has been proposed to be a specific and essential regulator of several mammalian channel and transport proteins (14, 18). The regulation of ubiquitination and function of amiloride-sensitive epithelial Na+ channel (ENaC) by Nedd4-2 is most well studied (19–26). Yeast has a single member of the NEDD4 family, Rsp5. This E3 ligase is involved in ubiquitination and sorting of a variety of transporters, receptors, and other endocytic cargo as well as the components of endocytic machinery (27, 28).
The NEDD4 family of E3 ligases typically has the C2 (Ca2+/lipid binding) domain at the amino terminus, 2–4 WW domains, and the HECT domain that can be conjugated to ubiquitin and transfer ubiquitin to the substrate. NEDD4 ligases bind to their substrates via WW domains that recognize the (L/P)PXY sequence motifs. However, a number of NEDD4 family substrates do not contain this sequence and interact with these E3s either through other mechanisms or indirectly via an adaptor protein containing (L/P)PXY motifs (29–32).
DAT represents an example of a substrate that does not have conventional WW binding sequences, and the role of WW domains of Nedd4-2 as well as a general mechanism of DAT ubiquitination by Nedd4-2 are unclear. Hence, we exploited the model system of the PKC-induced DAT ubiquitination and endocytosis to further elucidate the mechanisms by which Nedd4-2 mediates ubiquitination and endocytosis of a cargo such as DAT. Using a combination of RNAi and Nedd4-2 mutant overexpression approaches, the role of WW3 and WW4 domains of Nedd4-2 in DAT ubiquitination was demonstrated. Furthermore, we propose that Nedd4-2 utilizes UBE2D and UBE2L3 E2 ubiquitin-conjugating enzymes to transfer a single tetra-ubiquitin Lys63-linked chain onto the DAT molecule.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies were purchased from the following sources: monoclonal rat antibody against the amino terminus of DAT (Nt-DAT) and polyclonal rabbit antibody against the C-terminal DAT (Ct-DAT) from Chemicon (Temecula, CA); monoclonal mouse antibody P4D1 to ubiquitin, polyclonal goat to human Nedd4-1 (A-16), and goat polyclonal to UBE2D1–3 (C-15) were from Santa Cruz Biotechnology (Santa Cruz, CA); monoclonal mouse to green fluorescent protein (GFP) from Zymed Laboratories Inc., Inc. (San Francisco, CA); mouse monoclonal antibody to hemagglutinin epitope HA11 (16B12) from Covance (Berkley, CA); donkey anti-mouse antibodies conjugated with Cy5 or Cy3 from Jackson ImmunoResearch (West Grove, PA); rabbit polyclonal antibody to NEDD4-2 was a kind gift from Dr. Oliver Staub (University of Lausanne, Lausanne, Switzerland). Phorbol 12-myristate 13-acetate (PMA) and rabbit polyclonal anti-ubiquitin (U5379) were purchased from Sigma. Monoclonal humanized antibodies specific to Lys63-linked (Apu3.A8) or Lys48-linked polyubiquitin chains (Apu2.07) were a kind gift from Genentech, Inc. Monoclonal antibody to clathrin heavy chain TD.1 were from American Type Culture Collections, Inc. (ATCC). Recombinant four-ubiquitin chain standards were a gift from Dr. Changwei Liu (University of Colorado, Denver, CO). Affinity purified rabbit polyclonal antibody to UBE2L3 was from Boston Biochem (Cambridge, MA). Rabbit polyclonal antibody to UBE2E3 was a kind gift from Dr. Scott Plafker.
Plasmids
Cyan fluorescent protein (CFP) and HA-tagged DAT was generated by site-point mutagenesis using a CFP-DAT plasmid as described previously (15). The human NEDD4-2-GFP construct was kindly provided by Dr. Christie P. Thomas (University of Iowa, Iowa City, IA). The YFP-Nedd4-2 construct was made by cutting NEDD4-2-GFP with KpnI and HindIII and inserting it into pEYFP-C1 (Clontech). YFP-FLAG-His-Nedd4-2 (YFP-FH-Nedd42) was generated by replacing the DAT sequence in the FLAG-(His)10-DAT construct (13) with the full-length Nedd4-2 sequence, and cloning the entire FLAG-His-Nedd4-2 into BglII and HindIII of pEYFP-C1. Single and multiple amino acid substitutions were made using the YFP-FH-Nedd4-2 as a template and a QuikChange site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene Cloning Systems, La Jolla, CA). The mutations were verified by automatic dideoxynucleotide sequencing. Fragments of the hNedd4-2 protein tagged with YFP were generated by PCR amplification and inserting into specific digestion sites of pEYFP-C1. YFP-WW1-2, YFP-WW3-4, and YFP-WW3-4-HECT correspond to residues 195–397, 475–560, and 475–955, respectively. To generate the YFP-WW3-1-HECT construct, in which the WW4 domain was converted into WW1 (LPPGWEEKVDNLGRTYYVNHNNRTTQWHRPSL), the WW4 sequence, 5′-GAATTCACTTGGATGGCCGAACGTTTTATATTGATCATAATAGCAAAATTACTCAGTGGGAAGAC-3′, was changed to 5′-AAGTGGACAATTTAGGCCGAACGTACTATGTCAACCACAACAACCGGACCACTCAGTGGCACAGA-3′ by site-directed mutagenesis. To generate the YFP-FH-Nedd42 DNA construct that is not sensitive to siRNA duplex 4 (YFP-Nedd4-2-rf), several site-point mutations were made that did not change the amino acid sequence of Nedd4-2. The same mutations were made to generate siRNA-resistant WW mutants of YFP-FH-Nedd42.
Cell Culture and Transfections
Human HEK293 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (HyClone, Logan, UT) and antibiotics. Porcine aortic endothelial (PAE) cells were grown in Ham's F-12 medium containing 10% fetal bovine serum and antibiotics. Single cell clones of HEK293 and PAE cells stably expressing CFP-HA-DAT or FLAG-His-DAT were generated by selection in the presence of G418 (400 μg/ml).
An Effectene method (Qiagen, Hilden, Germany) was used for DNA transfections of HEK293 and PAE cells. The cells were used in transient-expression experiments for experiments on the second or third day.
siRNA transfections were performed with DharmaFECT® Transfection Reagent 1 from Dharmacon RNA Technologies (Lafayette, CO) following the manufacturer's protocol. All siRNAs were purchased from Dharmacon. For depleting Nedd4-2 siGENOME SMARTpool (SP) and duplexes 1, 3, 4, and 5 were used. siGENOME SMARTpool catalog number M-007178-01 was used to deplete Nedd4-1. To deplete UBE2D2/3 and UBE2D1 ON-TARGET siRNAs HUAFB-000047 and HUAFB-000049, respectively, were used. For UBE2L3 depletion, siGENOME duplex 1 (D-010384). siGENOME D-008845-03 was used to deplete UBE2E3. Clarthin heavy chain was depleted as previously described (17). siGENOME NonTargeting duplex 3 (NT) was used as a control.
For siRNA knock-down/rescue experiments, the cells were plated in 35-mm dishes, transfected with siRNA the next day at 30% confluence, transfected with the appropriate DNA plasmid on day 2, and then transfected again with siRNA on day 3. The cells were lysed 2 days after the last transfection.
Antibody Feeding Endocytosis Assay
The endocytosis assay using HA11 antibody was performed similarly as described in Ref. 15. Briefly, the cells grown on glass coverslips were incubated with 1 μg/ml of HA11 in binding medium (Dulbecco's modified Eagle's medium or F-12, 0.1% bovine serum albumin) for 15 min and then with Me2SO (vehicle) or PMA (1 μm), all at 37 °C, for the indicated times. The cells were then washed with ice-cold Ca2+,Mg2+-free PBS (CMF-PBS) and fixed with freshly prepared 4% paraformaldehyde for 15 min at room temperature. The cells were stained with secondary anti-mouse antibody conjugated with Cy5 (5 μg/ml) in CMF-PBS containing 0.5% bovine serum albumin at room temperature for 1 h to occupy surface HA11. After washing, the cells were permeabilized by a 5-min incubation in CMF-PBS containing 0.1% saponin, 0.5% bovine serum albumin at room temperature, and then incubated with the same secondary conjugated with Cy3 (1 μg/ml) in CMF-PBS containing 0.1% saponin, 0.5% bovine serum albumin for 45 min to stain internalized HA11. Both primary and secondary antibody solutions were precleared by centrifugation at 100,000 × g for 20 min. After staining, the coverslips were mounted in Mowiol (Calbiochem, La Jolla, CA).
Fluorescence Microscopy
To obtain high resolution three-dimensional images of the cells, a Z-stack of images was acquired through Cy5, Cy3, and CFP filter channels using a Marianas Imaging work station and SlideBook 4.2 software (Intelligent Imaging Innovation, Denver, CO) as described previously (15). Typically, 15–20 serial two-dimensional images were recorded at 300-nm intervals. All image acquisition settings were identical in each experiment. The Z-stack of images obtained was deconvoluted using a nearest neighbor algorithm of SlideBook4.2. Quantification of the relative amount of Cy5 and Cy3 fluorescence in the cell was performed using the statistics module of the SlideBook4.2. The background-subtracted three-dimensional images were segmented using a minimal intensity of CFP as a low threshold. New images were generated using this mask so that all voxels that did not contain CFP were assigned a value of zero. The integrated voxel intensity of Cy5 and Cy3 in each cell in the new image was then quantitated.
Some Cy3 images are presented in a pseudocolor mode. Cy3 intensity is displayed stretched between the low and high renormalization values, according to a temperature-based lookup table with blue (cold) indicating low values and red (hot) indicating high values. To eliminate distracting data from regions outside of cells, the CFP channel (total CFP-HA-DAT) was used as a saturation channel, and the Cy3 images were displayed as CFP intensity-modulated images. In these images, data with CFP values greater than the high threshold of the saturation (CFP) channel are displayed at full saturation, whereas data values below the low threshold are displayed with no saturation (i.e. black).
Immunoprecipitation
The cells grown in 35-mm dishes were placed on ice and washed with CMF-PBS, and the proteins were solubilized in Triton X-100/glycerol/HEPES lysis buffer supplemented with 10 mm N-ethylmaleimide and protease inhibitors for 20 min at 4 °C (13). The lysate was then centrifuged at 100,000 × g for 20 min to remove insoluble material. Lysates were incubated with appropriate antibodies overnight and antibodies were precipitated with protein A- or protein G-Sepharose. Immunoprecipitates and aliquots of cell lysates were denatured in sample buffer at 95 °C, resolved by electrophoresis, and probed with various antibodies followed by chemiluminescence detection. Several x-ray films exposed for different times were analyzed to determine the linear range of the chemiluminescence signals, and the quantifications were performed using densitometry and ImageJ software analysis.
Statistical Analysis
The statistical significance of the data were analyzed by unpaired or paired t tests. Significant differences were defined as those with p < 0.05.
RESULTS
Nedd4-2 Is Essential for PKC-dependent DAT Ubiquitination and Endocytosis in HEK293 and PAE Cells
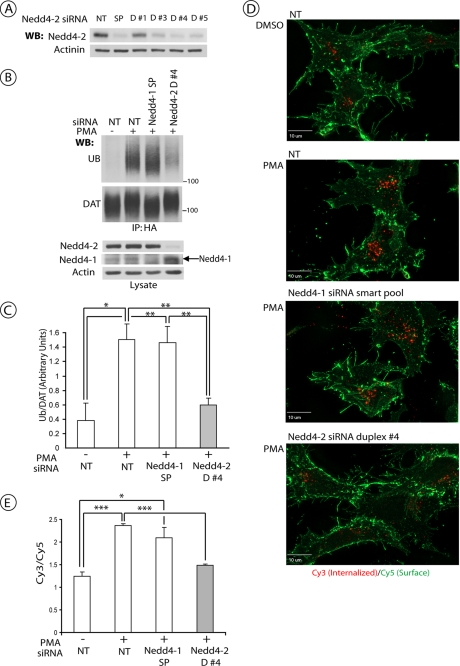
Our previous studies demonstrated that transfection of human HeLa cells with the pool of siRNA duplexes that target human Nedd4-2 inhibits ubiquitination and endocytosis of ectopically expressed human DAT (15). Recent years exposed major weaknesses of the RNAi methodology, mainly associated with a high probability of off-target effects of siRNAs, and established the criteria for performing and interpreting RNAi experiments, such as a similar functional effect of multiple siRNAs and a rescue of siRNA effects by protein replacement. Therefore, Nedd4-2 was depleted with several individual siRNA duplexes in two other cell lines. In human HEK293 cells stably expressing CFP-HA-DAT, several duplexes efficiently depleted Nedd4-2 (Fig. 1A). siRNA duplex 4 was the most effective and, therefore, this siRNA was chosen for subsequent experiments in HEK293 cells. As shown in Fig. 1, B and C, siRNA duplex 4 strongly inhibited PMA-stimulated, PKC-dependent DAT ubiquitination. The effect of this siRNA duplex on the PMA-induced endocytosis of CPF-HA-DAT was examined using an HA11 antibody uptake assay (15). In this assay, HA11·CFP-HA-DAT complexes located at the plasma membrane or in endosomes are detected using, respectively, Cy5- or Cy3-conjugated secondary antibodies. PMA treatment of control cells led to accumulation of Cy3-marked CFP-HA-DAT in endosomes and an increase in the Cy3/Cy5 fluorescence ratio (Fig. 1, D and E). In contrast, siRNA duplex 4 reduced the accumulation of CFP-HA-DAT in endosomes and the Cy3/Cy5 ratio in PMA-treated cells to the levels observed in cells not treated with PMA.
FIGURE 1.
Nedd4-2 siRNA inhibits PKC-dependent DAT ubiquitination and endocytosis in HEK293 cells. A, HEK293 cells stably expressing CFP-HA-DAT (HEK/CFP-HA-DAT) were transfected with non-targeting siRNA (NT), SmartPool™ (SP), or four individual siRNA duplexes targeting human Nedd4-2. Equal amounts of cell lysates were electrophoresed, and the amounts of Nedd4-2 and α-actinin (loading control) were determined by Western blotting (WB). B, HEK/CFP-HA-DAT cells were transfected with NT siRNA, SP targeting Nedd4-1, or duplex 4 (D#4) siRNA targeting Nedd4-2. Cells were treated with 1 μm PMA or Me2SO for 15 min, lysed, and DAT was precipitated with the HA11 antibody. Following electrophoresis and transfer, DAT immunoprecipitates were probed with monoclonal antibodies against ubiquitin (P4D1) and, consecutively, with the monoclonal GFP antibody. Total cell lysates were blotted with antibodies to Nedd4-2, Nedd4-1, and actin (loading control). C, quantification of three experiments performed identically to the experiment presented in B. Bars represent the mean amounts (±S.E.) of ubiquitinated DAT normalized to the amount of total DAT (arbitrary units). *, p < 0.05; **, p < 0.01. D, representative images of HEK/CFP-HA-DAT cells treated with NT, SP of siRNAs targeting Nedd4-1, or duplex 4 siRNA targeting Nedd4-2. HA11 endocytosis assay was performed as described under “Experimental Procedures.” Merged images of Cy5 immunofluorescence (green, surface CFP-HA-DAT) and Cy3 immunofluorescence (red, internalized CFP-HA-DAT) are presented. Identical intensity scales are used. Scale bars, 10 μm. E, quantification of the Cy3/Cy5 ratio in cells from the experiments represented in D. Bars represent mean values (±S.E.) of a minimum of 6 images per condition. *, p < 0.05; **, p < 0.001.
In porcine (PAE) cells stably expressing FH-DAT, siRNA duplex 3 efficiently depleted Nedd4-2 and did not cause significant toxicity (see supplemental Fig. S1A). This siRNA duplex 3 strongly inhibited PKC-dependent ubiquitination of FH-DAT stably expressed in these cells (supplemental Fig. S1, B and C). siRNA duplex 3 also inhibited PKC-induced endocytosis of FH-DAT as revealed by immunofluorescence microscopy (supplemental Fig. S1D). Whereas PMA caused accumulation of the FH-DAT immunoreactivity in the vesicular compartments in the perinuclear area of the cell and dramatic decrease in the amount of DAT localized at cell edges (plasma membrane), depletion of Nedd4-2 prevented this PMA-dependent re-distribution of FH-DAT. Thus, the experiments in Figs. 1 and supplemental S1 demonstrated that at least two siRNAs targeting human Nedd4-2 had similar effects on DAT ubiquitination and endocytosis in human and porcine cells.
To further control for the specificity of Nedd4-2 depletion effects, HEK293/CFP-HA-DAT cells were depleted of Nedd4-1 protein, the closest homolog to Nedd4-2 among E3 ligases of this family (33, 34). Fig. 1, B–D, shows that knock-down of Nedd4-1 did not affect CFP-HA-DAT ubiquitination and endocytosis, thus indicating that specific properties of Nedd4-2 are responsible for the E3 ligase function of this protein during DAT ubiquitination.
Overexpression of Catalytically Active Nedd4-2 Promotes PKC-dependent DAT Ubiquitination
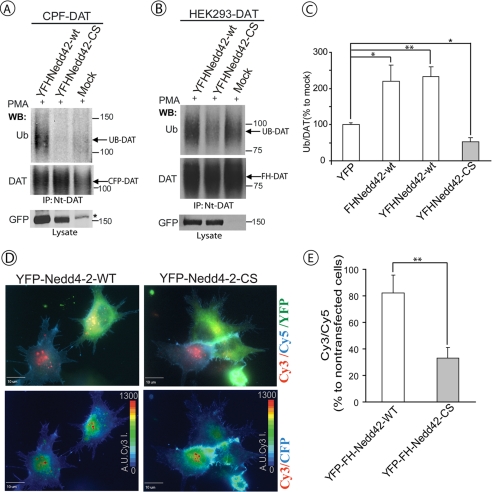
Because knock-down of Nedd4-2 inhibits DAT ubiquitination, we examined whether overexpression of Nedd4-2 would result in the opposite effect, increased DAT ubiquitination. To this end, full-length Nedd4-2 fused to YFP and FLAG-His epitope tags (YFP-FH-Nedd4-2) was transiently co-expressed with CFP-DAT in HEK293 cells. Fig. 2A shows that overexpression of the YFP fusion Nedd4-2 protein increased PMA-induced ubiquitination of CFP-DAT.
FIGURE 2.
HECT domain activity of Nedd4-2 is necessary for PKC-dependent DAT ubiquitination and endocytosis. A, HEK293T cells were co-transfected with CFP-DAT and wild-type YFP-FH-Nedd4-2 (YFHNedd42-wt), HECT domain mutant of Nedd4-2 (YFHNedd42-CS), or empty vector (mock-transfected). After 2 days, the cells were treated with 1 μm PMA for 15 min, lysed, and CFP-DAT was immunoprecipitated with Nt-DAT antibody. The immunoprecipitates were probed by Western blotting (WB) with ubiquitin (P4D1) and, subsequently, Nt-DAT antibodies. YFP-tagged Nedd4-2 constructs were detected in cell lysates using GFP antibodies. Star indicates the nonspecific band. B, HEK293T cells stably expressing human DAT (HEK293/DAT) were transfected with YFHNedd42-wt, YFHNedd42-CS, or empty vector (mock). After 2 days the cells were treated with 1 μm PMA for 15 min and lysed. DAT was immunoprecipitated using Nt-DAT antibody, and the immunoprecipitates were probed with ubiquitin (P4D1) and, subsequently, Nt-DAT antibodies. YFP-tagged Nedd4-2 constructs were detected in cell lysates using GFP antibody. C, quantification of several experiments performed identically to the experiment presented in B. Bars represent the mean amounts (±S.E.) of ubiquitinated DAT normalized to the amount of total DAT in arbitrary units (Ub/DAT). All Ub/DAT values are calculated as percent of the Ub/DAT value in mock-transfected cells. *, p < 0.05; **, p < 0.01 (n = 3). D, HEK293 cells stably expressing CFP-HA-DAT (HEK/CFP-HA-DAT) were transfected with YFHNedd42-wt or the YFHNedd42-CS mutant. HA11 endocytosis assay was performed as described under “Experimental Procedures.” HA11 preincubated cells were exposed to 1 μm PMA for 30 min. Merged images (top row) of Cy5 immunofluorescence (blue, cell surface CFP-HA-DAT), Cy3 immunofluorescence (red, internalized CFP-HA-DAT), and YFP (green, YFP-FH-Nedd4-2) are presented. Pseudocolor images (Cy3/CFP) were generated as described under “Experimental Procedures” are shown in the bottom row. Identical intensity scales are used. Scale bars, 10 μm. E, quantification of the Cy3/Cy5 ratio in cells expressing and not expressing YFP-FH-Nedd4-2 (wild-type or CS mutant) from the experiments performed similarly to the experiment presented in D. Bars represent mean values (± S.E.) of a minimum of 6 cells from three independent experiments. **, p < 0.01 (n = 18).
To test for the involvement of the E3 ligase activity of Nedd4-2 in its effects on DAT, the catalytic cysteine residue in the HECT domain was mutated to serine (CS mutation). As shown in Fig. 2A, the YFP-FH-Nedd4-2 CS mutant did not increase PMA-induced ubiquitination of CFP-DAT and even slightly decreased this ubiquitination as compared with mock-transfected cells. Essentially similar results were obtained in experiments where wild-type YFP-FH-Nedd4-2 protein or the CS mutant were transiently expressed in HEK293 cells that constitutively express DAT (Fig. 2, B and C). In these experiments the expression of the CS mutant resulted in a statistically significant decrease in the extent of PKC-dependent DAT ubiquitination, suggesting that this mutant interferes with the function of the endogenous Nedd4-2. Expression of FLAG/His-tagged Nedd4-2 increased DAT ubiquitination to a similar extent as did YFP-FH-Nedd4-2 (Fig. 2C), suggesting that the amino-terminal attached YFP did not affect Nedd4-2 activity. The data presented in Fig. 2, A–C, suggested that the DAT ubiquitination reaction is controlled by intracellular levels of the E3 ligase, Nedd4-2, and that HECT domain activity is necessary for the Nedd4-2-mediated ubiquitination of DAT.
Despite increasing DAT ubiquitination, overexpression of the wild-type Nedd4-2 fusion did not accelerate PKC-dependent endocytosis of CFP-HA-DAT in HEK cells, suggesting that endogenous levels of Nedd4-2-mediated DAT ubiquitination are sufficient for the maximum high rate of DAT endocytosis (Fig. 2, D and E). The CS mutant of Nedd4-2 significantly decreased CFP-HA-DAT endocytosis, thus indicating that the catalytically inactive mutant has a dominant-negative effect on PKC-dependent DAT endocytosis (Fig. 2, D and E).
Nedd4-2 WW3 and WW4 Domains Are Essential for PKC-dependent DAT Ubiquitination
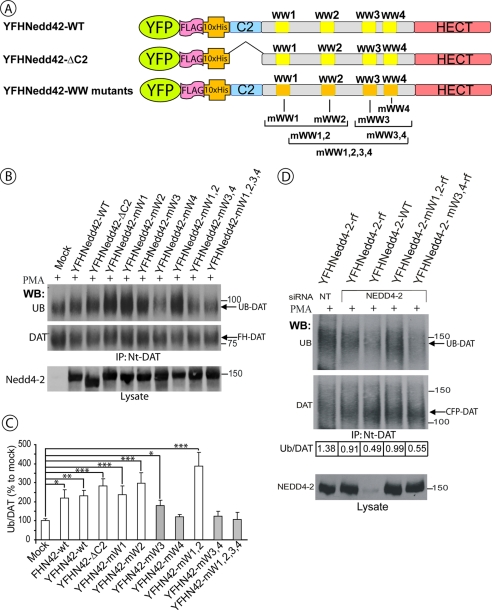
To define the role of different domains of Nedd4-2 in its regulation of DAT ubiquitination, several site-point and deletion mutants of YFP-FH-Nedd4-2 were generated (Fig. 3A). Each individual WW domain or simultaneously several WW domains were inactivated in the full-length Nedd4-2 by substituting two tryptophan and one proline residues, which are known to be critical for interaction with PPXY motifs, by two alanines and a phenylalanine, respectively (35, 36). To test for the importance of the C2 domain, this domain was deleted. Although overexpression of the wild-type Nedd4-2 fusion protein or a Nedd4-2 with deleted C2 domain in HEK293/CFP-HA-DAT cells promoted PKC-dependent ubiquitination of CFP-HA-DAT, overexpression of the Nedd4-2 mutant with all WW domains mutated did not increase DAT ubiquitination (Fig. 3, B and C). Interestingly, Nedd4-2 mutants lacking functional WW4 alone or together with WW3 did not increase DAT ubiquitination (Fig. 3B). Mutation of WW3 alone partially inhibited Nedd4-2-mediated DAT ubiquitination. In contrast, inactivation of WW1 and WW2 domains did not affect the ability of Nedd4-2 to elevate DAT ubiquitination, although a tendency of increased DAT ubiquitination, as compared with the wild-type Nedd4-2, was observed (Fig. 3, B and C).
FIGURE 3.
WW3 and WW4 domains of Nedd4-2 are necessary for DAT ubiquitination. A, schematic representation of wild-type and mutant YFP-FH-Nedd4-2 constructs. B, HEK293T cells stably expressing FH-DAT (HEK293/FH-DAT) were transfected with empty vector (mock), wild-type, or mutant YFP-FH-Nedd4-2. After 2 days the cells were treated with 1 μm PMA for 15 min and lysed. DAT was immunoprecipitated using Nt-DAT antibody, and the immunoprecipitates were probed by Western blotting (WB) with ubiquitin (P4D1) and, subsequently, Nt-DAT antibodies. YFP-tagged Nedd4-2 constructs were detected in cell lysates using the Nedd4-2 antibody. C, quantification of several experiments performed identically to the experiment presented in B. Bars represent the mean values (±S.E.) of ubiquitinated DAT normalized to the amount of total DAT in arbitrary units (Ub/DAT). All Ub/DAT values are calculated as percent of the Ub/DAT value in mock-transfected cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (n = 3). D, HEK/CFP-DAT cells were co-transfected with either non-targeting (NT) or duplex 4 siRNA (targeting Nedd4-2) and various DNA plasmids: wild-type YFP-FH-Nedd4-2, YFP-FH-Nedd4-2 resistant to siRNA duplex 4 (resistant form, rf), and YFP-FH-Nedd4-2-mWW1,2-rf or YFP-FH-Nedd4-2-mWW3,4-rf. After 2 days the cells were treated with 1 μm PMA for 15 min and lysed. DAT was immunoprecipitated using the Nt-DAT antibody, and the immunoprecipitates were probed with ubiquitin (P4D1) and, subsequently, Nt-DAT antibodies. YFP-tagged Nedd4-2 was detected in cell lysates using Nedd4-2 antibody. The amount of ubiquitinated DAT normalized to the amount of total DAT (Ub/DAT) is indicated for each experimental variant. The experiment is representative of two independent experiments with similar results.
To further support the model of the essential role of WW3 and WW4 domains but not WW1 and WW2 domains in DAT ubiquitination, siRNA knock-down and rescue experiments were performed. Transfection of HEK293/CFP-HA-DAT cells with Nedd4-2 siRNA duplex 4 alone (Fig. 1) or together with the YFP-FH-Nedd4-2 construct significantly inhibited PKC-induced DAT ubiquitination (Fig. 3D). Expression of the Nedd4-2 mutant, which contained mismatching mutations that make Nedd4-2 mRNA resistant to siRNA duplex 4 (Nedd4-2-rf), resulted in the recovery of DAT ubiquitination in cells depleted of Nedd4-2. The rescue effect was partial because only a population of cells (∼50%) expressed a sufficient amount of Nedd4-2-rf to compensate for the loss of endogenous Nedd4-2. Nedd4-2-rf bearing WW1 and WW2 mutations rescued DAT ubiquitination to the extent similar to that observed in cells transfected with the “wild-type” version of Nedd4-2-rf (Fig. 3D). In contrast, a mutant of Nedd4-2-rf, in which both WW3 and WW4 domains have been inactivated, did not rescue DAT ubiquitination (Fig. 3D). Altogether, the data presented in Fig. 3 strongly indicate that the WW4 domain and to a lesser extent WW3 domain of Nedd4-2 are necessary for PKC-dependent ubiquitination of DAT whereas WW1, WW2, and C2 domains are not required.
HECT, WW3, and WW4 Domains of Nedd4-2 Are Sufficient for DAT Ubiquitination
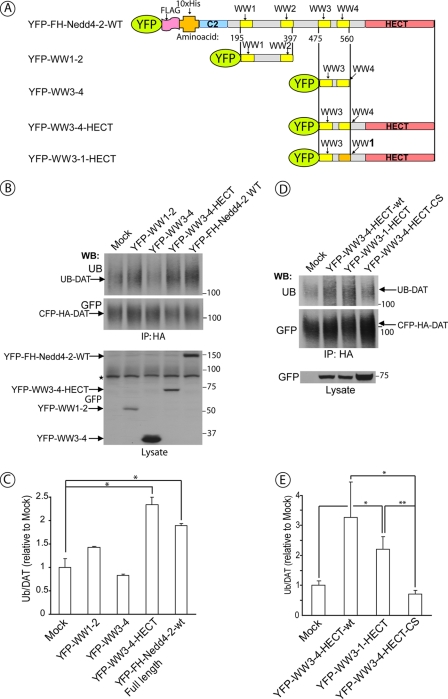
The data presented in Fig. 3 demonstrate that deletion of the C2 domain and inactivation of WW1 and WW2 domains of Nedd4-2 do not affect the ability of Nedd4-2 to ubiquitinate DAT. Therefore, we tested whether the entire amino-terminal section of Nedd4-2, encompassing the C2, WW1, and WW2 domains, were not required for DAT ubiquitination. Therefore, several fusion proteins containing a combination of WW and HECT domains of Nedd4-2 were transiently expressed in HEK293/CFP-HA-DAT cells (Fig. 4A). In agreement with the mutagenesis data (Fig. 3), a COOH-terminal fragment of the Nedd4-2 protein consisting of WW3, WW4, and HECT domains (WW3–4-HECT) was capable of mimicking the effect of the full-length Nedd4-2 in elevating DAT ubiquitination (Fig. 4, B and C). Fusion proteins that lacked the HECT domain and consisted of the region of the Nedd4-2 molecule containing either WW3 and WW4 domains or WW1 and WW2 domains did not significantly affect DAT ubiquitination. In summary, the data presented in Figs. 2, 3, and 4, A–C, suggest that the catalytic activity of the HECT domain together with functional WW3 and WW4 domains are necessary and sufficient for PKC-dependent ubiquitination of DAT.
FIGURE 4.
WW3, WW4 and HECT domains of Nedd4-2 are sufficient for DAT ubiquitination. A, schematic representation of the deletion mutants of YFP-Nedd4-2 and a fragment of Nedd4-2 in which the WW4 domain was converted to the WW1 domain by multisite point mutagenesis. B, HEK293T/CFP-HA-DAT cells were transfected with empty vector (mock), wild-type YFP-FH-Nedd4-2 or deletion mutants of YFP-Nedd4-2 depicted in A. After 2 days the cells were treated with 1 μm PMA for 15 min and lysed. CFP-HA-DAT was immunoprecipitated using HA11 antibody, and the immunoprecipitates were probed with ubiquitin (P4D1) and, subsequently, GFP antibodies. YFP-tagged Nedd4-2 was detected in cell lysates using the GFP antibody. Star indicates the nonspecific band. C, quantification of several experiments performed identically to the experiment presented in B. Bars represent the mean values (±S.E., n = 3) of ubiquitinated DAT normalized to the amount of total DAT (arbitrary units). *, p < 0.05. D, HEK293T cells were co-transfected with CFP-HA-DAT and YFP-WW3-4-HECT, YFP-WW3-1-HECT, YFP-WW3-4-HECT-CS, or empty vector (mock-transfected). After 2 days the cells were treated with 1 μm PMA for 15 min and lysed. CFP-HA-DAT was immunoprecipitated using HA11 antibody, and the immunoprecipitates were probed with ubiquitin (P4D1) and, subsequently, GFP antibodies. YFP-tagged Nedd4-2 mutants were detected in cell lysates using the GFP antibody. E, quantification of several experiments performed identically to the experiment presented in A. Bars represent the mean values (±S.E., n = 5) of ubiquitinated DAT normalized to the amount of total DAT (arbitrary units). *, p < 0.05; **, p < 0.01. WB, Western blot.
Interestingly, conversion of the WW4 domain into the WW1 domain by multiple single amino acid substitutions in the YFP-WW3-4-HECT construct resulted in generation of a functional ubiquitin ligase capable of ubiquitinating DAT (YFP-WW3-WW1-HECT; Fig. 4, D and E). Therefore, it is likely that the position of the COOH-terminal WW domain proximal to the HECT domain within the Nedd4-2 molecule is critical for Nedd4-2 function.
UBE2D and UBE2L3 E2-conjugating Enzymes Are Necessary for Lys63-linked Polyubiquitination of DAT
To gain further insight in the mechanism of DAT ubiquitination, we examined which E2 ubiquitin-conjugating enzymes are involved. Previous in vitro analysis implicated UBE2D2 as the preferred E2 enzyme used by Nedd4-2 (37). UBE2D2 has two highly homologous E2s (UBE2D1 and -3), which were also capable of supporting Nedd4-2-mediated ubiquitination in the latter study. To assess the importance of UBE2D enzymes in intact cells, siRNA duplex targeting human UBE2D2/3 was transfected into HEK293/CFP-HA-DAT cells. Depletion of UBE2D2/3 caused significant inhibition of PKC-dependent ubiquitination of DAT (Fig. 5). siRNA knockdown of UBE2D1 did not significantly affect DAT ubiquitination (data not shown). Depletion of UBE2D1 by specific siRNA was not apparent from immunoblotting detection by the UBE2D1–3 antibody (data not shown), suggesting that the UBE2D1 isozyme is present in low abundance in HEK293 cells. The inhibitory effect of the depletion of UBE2D2/3 on PMA-induced DAT ubiquitination (Fig. 5B) and endocytosis (data not shown) was comparable with those effects of the Nedd4-2 knockdown.
FIGURE 5.
UBE2D1-3 and UBE2L3 ubiquitin-conjugating enzymes are involved in DAT ubiquitination. A, HEK/CFP-HA-DAT cells were transfected with NT siRNA, Nedd4-2 siRNA, siRNA targeting UBE2D2-3, UBE2E3, UBE2L3, or a mixture of siRNAs targeting UBE2D2-3 and UBE2L3. Cells were treated with 1 μm PMA for 15 min, lysed, and DAT was precipitated with the HA11 antibody. Following electrophoresis and transfer, DAT immunoprecipitates (IP) were probed with antibodies against ubiquitin (P4D1) and, consecutively, with the monoclonal GFP antibody. Total cell lysates were blotted with antibodies to Nedd4-2, E2 enzymes, and actin (loading control). B, quantification of several siRNA experiments exemplified in A. Bars represent the mean values (±S.E., n = 3) of ubiquitinated DAT normalized to the amount of total DAT in arbitrary units (Ub/DAT). *, p < 0.05; **, p < 0.01. WB, Western blot.
Two other E2 enzymes, UBE2E3 and UBE2L3, have also been implicated in Nedd4-2-mediated ubiquitination in in vitro experiments (37–38). Knockdown of UBE2E3 did not inhibit DAT ubiquitination (Fig. 5). In contrast, UBE2L3 siRNA reduced DAT ubiquitination alone and in combination with UBE2D2/3 siRNAs (Fig. 5). Together, these data suggested that whereas UBE2D2/3 are the major ubiquitin-conjugating enzymes coupled to Nedd4-2, there is a redundancy in this reaction and another E2, UBE2L3, is additionally involved.
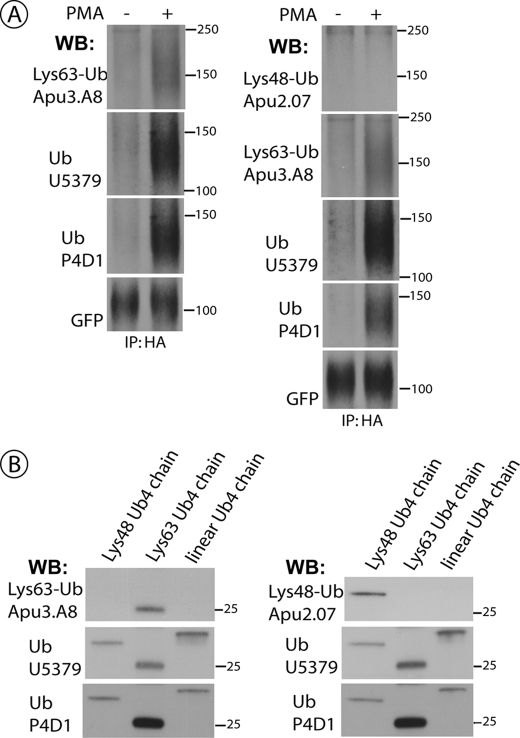
Interestingly, both UBE2D2/3 and UBE2L3 have been implicated in ubiquitination of the epidermal growth factor receptor that is mediated by the RING-domain E3 ligase, Cbl (39, 40). Epidermal growth factor receptor is modified by Lys63-linked polyubiquitin chains (41). In our previous mass spectrometry analysis of DAT modifications, mostly Lys63-linked chains and traces of Lys48 chains were detected (13). Therefore, we examined DAT ubiquitination using recently developed antibodies that recognize two major types of ubiquitin chains, Lys48- or Lys63-linked chains, with high selectivity (42). As shown in Fig. 6A, no signal corresponding to Lys48 chains was detected in DAT immunoprecipitates whereas a strong Lys63 chain ubiquitination was associated with DAT that was recovered from PMA-treated cells. In control experiments, both chain-specific (Lys63- or Lys48-linked) antibodies demonstrated selective recognition of corresponding ubiquitin chain standards (Fig. 6B). The efficiency of recognition of the recombinant Lys63-linked chains by the pan-ubiquitin antibody P4D1 was substantially higher than that of the Lys63 chain-specific antibody Apu3.A8. However, the ratio of the intensity of P4D1 and Apu3.A8 Western blot signals was similar (∼20-fold) for DAT immunoprecipitates from PMA-treated cells and recombinant Lys63 chains when blotting with these antibodies (best signal-to-noise ratio for each antibody) was performed in parallel and under identical experimental conditions (supplemental Fig. S2). Altogether, the data in Figs. 6 and supplemental S2 suggest that DAT is predominantly ubiquitinated by Lys63-linked chains.
FIGURE 6.
DAT is ubiquitinated by Lys63-linked polyubiquitin chains. A, HEK/CFP-HA-DAT cells were treated with 1 μm PMA or Me2SO for 15 min, lysed, and DAT was precipitated with the HA11 antibody. Following electrophoresis and transfer, DAT immunoprecipitates (IP) were probed by Western blotting (WB) with antibodies against Lys63-Ub chains or Lys48-Ub chains, followed by goat polyclonal anti-ubiquitin (U5379), mouse monoclonal anti-ubiquitin (P4D1), and finally with the monoclonal GFP antibody. B, the recombinant four-ubiquitin Lys63-linked, Lys48-linked, and linear chains were electrophoresed and blotted with the same antibodies that were used to probe DAT immunoprecipitates in A.
Analysis of numerous Western blots revealed that ubiquitinated DAT runs on SDS-PAGE as a single smeared band that is migrated slower than a non-ubiquitinated DAT form by a strikingly consistent distance. Measurement of this shift yielded 26 ± 1 kDa (S.E.). Based on electrophoretic analysis of recombinant polyubiquitin chains, this difference in molecular mass corresponds to the four-ubiquitin Lys63-linked chain but does not match the apparent size of three- or four-ubiquitin Lys48-linked chains (Fig. 6) (42). Thus, we propose that each ubiquitinated DAT molecule is modified by four ubiquitins, at least some of which are Lys63-linked oligomers.
DISCUSSION
In this study we investigated the mechanisms of Nedd4-2-mediated ubiquitination of transmembrane proteins using DAT as an experimental model. Nedd4-2 depletion by different siRNA duplexes inhibited PKC-induced DAT ubiquitination in various cellular backgrounds (see Figs. 1 and supplemental S1). DAT ubiquitination was rescued by reconstitution of Nedd4-2 in siRNA-depleted cells (see Fig. 3). Moreover, the increased ubiquitination of DAT by overexpressed Nedd4-2 showed that Nedd4-2 is capable of catalyzing DAT ubiquitination, which supports the role of Nedd4-2 in DAT ubiquitination originally revealed in RNAi screening experiments (15).
Among several members of the mammalian HECT domain E3 ligase family, Nedd4-2 is most frequently implicated in ubiquitination and regulation of endocytic trafficking of transmembrane proteins (18). Thus, we utilized a model system of ubiquitination and endocytosis of DAT to gain insights into the mechanisms of this Nedd4-2 function. Although the requirement of the HECT domain activity for DAT ubiquitination and endocytosis is not surprising, the lack of a role for the C2 domain observed for the DAT ubiquitination reaction suggests that this Nedd4-2 function does not require Ca2+-dependent association of Nedd4-2 with the membrane. Likewise, naturally occurring splicing variants of human Nedd4-2 lacking the C2 domain ubiquitinate and down-regulate ENaC with the same or even higher efficiency than the wild-type Nedd4-2 (43, 44).
Despite that all four WW domains in Nedd4-2 are highly homologous to each other, these domains show selectivity in their binding properties in vitro, as evidenced, for example, by different binding affinities of these WW domains to individual subunits of ENaC (35). Our analysis uncovered a considerable level of specificity and selectivity in the function of these domains during DAT ubiquitination in intact cells. WW4 domain and to a lesser extent the WW3 domain were found to be necessary and sufficient for DAT ubiquitination (Figs. 3 and 4). Similarly, WW3 and WW4 domains were required for ubiquitination of ENaC; however, in this case WW3 was equally or more important than WW4 (35, 45–47). Our observation of interchangeability of WW4 and WW1 domains (Fig. 4) in mediating DAT ubiquitination suggests that the position and/or orientation of WW domains within the Nedd4-2-substrate complex rather than relative affinities of WW domains are critical for the efficient transfer of the ubiquitin to the substrate. This observation supports the view that the substrate specificity of the Nedd4 family E3 ligases is mainly determined by the linker sequences between WW domains rather than by the WW domains themselves.
It should be noted that DAT does not have conventional WW domain binding (L/P)PXY motifs, and the binding site for Nedd4-2 in DAT has not been mapped. Weak co-immunoprecipitation of Nedd4-2 with DAT is indicative of the transient interaction through non-canonical sequences or an indirect interaction (15). The mechanism of the PKC effect on Nedd4-2-mediated ubiquitination also remains unclear. We have been unable to detect PKC-induced phosphorylation of Nedd4-2 using the same antibody that was utilized for the detection of TrkA-dependent phosphorylation of Nedd4-2 (48). It is possible that PKC facilitates the accessibility of DAT to Nedd4-2 rather than directly modifies the Nedd4-2 activity. Despite all uncertainties in the interactions between PKC, Nedd4-2, and DAT, the observation that WW3 and WW4 domains are important for ubiquitination of both DAT and ENaC indicates the substantial similarity of the molecular mechanisms of ubiquitination of these two substrates by Nedd4-2.
Whereas overexpression studies revealed a role for individual WW domains in DAT ubiquitination, mutant Nedd4-2 proteins with inactivated WW3 and WW4 (Fig. 3) or containing both domains but lacking the HECT domain (Fig. 4) did not inhibit PKC-dependent DAT ubiquitination and endocytosis. Because WW domains are necessary for binding to the substrate, it is expected that expression of the Nedd4-2 protein with mutations in these domains would not interfere with endogenous Nedd4-2 interactions with the substrate. On the other hand, a fragment containing WW3 and WW4 domains (Fig. 4) was expected to compete with endogenous Nedd4-2 for binding to the substrate. However, it is possible that this fragment did not efficiently bind to DAT or an intermediate adaptor and, therefore, did not interfere with the functional interactions of endogenous Nedd4-2. On the other hand, the YFP-WW3-4-HECT-CS mutant had a dominant-negative effect on DAT ubiquitination (Fig. 4), suggesting the role of the HECT domain, and possibly linker sequences, in the ability of WW3 and WW4 domains to bind DAT. This notion is consistent with the recent demonstration of the direct interaction of the HECT domain of Rsp5 with the substrate (49).
Surprisingly, increased DAT ubiquitination in cells overexpressing Nedd4-2 did not translate into increased endocytosis. Presumably, the endogenous DAT ubiquitination level is sufficient for the maximal rate of PKC-induced endocytosis of DAT and further increase in the endocytic rate is prevented due to limitation at the stages of endocytosis other than DAT ubiquitination. In general, limited capacity of the endocytic pathway is a characteristic feature of the stimulus-induced endocytosis of an ubiquitinated cargo such as, for instance, epidermal growth factor receptor (8, 41). In contrast, constitutive endocytosis of various channels and transporters is typically accelerated by Nedd4-2 overexpression (50–52).
The mechanisms of ubiquitination that determine the type of polyubiquitin chains are not well understood. It has been proposed that the structure of the HECT domain defines whether Lys63- or Lys48-linked chains are attached to the substrate (53), although a role for specific E2s in this decision is proposed in the case of RING-domain E3 ligases (54). Our demonstration of the involvement of UBE2D2/3 in Nedd4-2-mediated Lys63-polyubiquitination of DAT (Figs. 5 and 6) support the model that (i) E3 ligases of the Nedd4 family preferentially mediate attachment of Lys63-linked ubiquitin chains; and (ii) UBE2D enzymes can be coupled to both HECT and RING domain (Cbl) E3 ligases in promoting Lys63 polyubiquitination (12, 39, 41, 55–57).
Based on the shift in the apparent molecular mass of ubiquitinated DAT species (26 kDa) and predominant Lys63-linked polyubiquitination of DAT (Fig. 6) we propose that 4 ubiquitins are conjugated to one DAT molecule. Analysis of PKC-dependent ubiquitination of several DAT lysine mutants showed that regardless of whether one, two, or all three major amino-terminal conjugation sites are mutated, or additional 4 carboxyl-terminal lysines are mutated (presumably the cryptic conjugation sites are then ubiquitinated), the shift between the non-ubiquitinated and ubiquitinated DAT forms is the same (14). Moreover, the same molecular weight shift is observed in all experiments with various Nedd4-2 mutants (Figs. 3 and 4). Thus, given that a single form of ubiquitinated DAT is present under all these conditions, a constant number of lysines must be conjugated to ubiquitin in each ubiquitinated DAT molecule. The most likely scenario is that only a single lysine (or an amino-terminal amino group) is conjugated with a four-ubiquitin Lys63-linked chain per one molecule of wild-type or mutant DAT. The size of the chain can be limited by non-covalent interaction of ubiquitins with the N-lobe of the HECT domain (58). However, why after attachment of the ubiquitin chain to one site in the DAT molecule further ubiquitin conjugation to other sites is prevented is unknown. Such inhibition of ubiquitination could be due to the loss of accessibility of ubiquitinated DAT to Nedd4-2 because of rapid endocytosis or due to sterical hindrance. The former model would assume that if the endocytosis is inhibited, multiple sites would be ubiquitinated and multiple forms of ubiquitinated DAT would exist. However, inhibition of DAT endocytosis by clathrin heavy chain siRNA did not result in the appearance of an additional species of ubiquitinated DAT although it increased the amount of ubiquitinated DAT (supplemental Fig. S3). Although future studies should fully address the peculiar mechanisms controlling the process of DAT ubiquitination by Nedd4-2, the data presented in this study support the emerging concept that Lys63-linked polyubiquitin chains are the major molecular signals mediating internalization and intracellular sorting of integral membrane proteins.
Supplementary Material
Acknowledgments
We thank Melissa Adams for help in preparation of the manuscript. We are grateful to Drs. C. Thomas, O. Staub, C. Liu, S. Plafker, and Genentech Inc. for generous gifts of reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant DA014204 from the NIDA.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PKC
- protein kinase C
- CMF-PBS
- Ca2+,Mg2+-free phosphate-buffered saline
- DAT
- dopamine transporter
- ENaC
- amiloride-sensitive epithelial Na+ channel
- GFP, CFP, and YFP
- green, cyan, and yellow fluorescent protein
- Nedd4-2
- neuronal precursor cell expressed, developmentally down-regulated 4-2
- RNAi
- RNA interference
- siRNA
- small interfering RNA
- HA
- hemagglutinin
- PAE
- porcine aortic endothelial
- PBS
- phosphate-buffered saline
- PMA
- phorbol 12-myristate 13-acetate
- HEK
- human embryonic kidney
- Nt-DAT
- amino terminus of DAT
- Ct-DAT
- carboxyl terminus of DAT.
REFERENCES
- 1.Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 2.Staub O., Rotin D. (2006) Physiol. Rev. 86, 669–707 [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 4.Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P. P., Polo S. (2006) Nat. Cell Biol. 8, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 5.Mosesson Y., Yarden Y. (2006) Isr. Med. Assoc. J. 8, 233–237 [PubMed] [Google Scholar]
- 6.Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 7.d'Azzo A., Bongiovanni A., Nastasi T. (2005) Traffic 6, 429–441 [DOI] [PubMed] [Google Scholar]
- 8.Miranda M., Sorkin A. (2007) Mol. Interv. 7, 157–167 [DOI] [PubMed] [Google Scholar]
- 9.Ikeda F., Dikic I. (2008) EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan L. M., Piper S., Dodd R. B., Saville M. K., Sanderson C. M., Luzio J. P., Lehner P. J. (2006) EMBO J. 25, 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauwers E., Jacob C., André B. (2009) J. Cell Biol. 185, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., Belgareh-Touzé N. (2008) Traffic 9, 1372–1391 [DOI] [PubMed] [Google Scholar]
- 13.Miranda M., Wu C. C., Sorkina T., Korstjens D. R., Sorkin A. (2005) J. Biol. Chem. 280, 35617–35624 [DOI] [PubMed] [Google Scholar]
- 14.Miranda M., Dionne K. R., Sorkina T., Sorkin A. (2007) Mol. Biol. Cell 18, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorkina T., Miranda M., Dionne K. R., Hoover B. R., Zahniser N. R., Sorkin A. (2006) J. Neurosci. 26, 8195–8205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels G. M., Amara S. G. (1999) J. Biol. Chem. 274, 35794–35801 [DOI] [PubMed] [Google Scholar]
- 17.Sorkina T., Hoover B. R., Zahniser N. R., Sorkin A. (2005) Traffic 6, 157–170 [DOI] [PubMed] [Google Scholar]
- 18.Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 19.Flores S. Y., Debonneville C., Staub O. (2003) Pflugers Arch. 446, 334–338 [DOI] [PubMed] [Google Scholar]
- 20.Chan W., Tian R., Lee Y. F., Sit S. T., Lim L., Manser E. (2009) J. Biol. Chem. 284, 8185–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edinger R. S., Lebowitz J., Li H., Alzamora R., Wang H., Johnson J. P., Hallows K. R. (2009) J. Biol. Chem. 284, 150–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almaca J., Kongsuphol P., Hieke B., Ousingsawat J., Viollet B., Schreiber R., Amaral M. D., Kunzelmann K. (2009) Pflugers Arch. 458, 713–721 [DOI] [PubMed] [Google Scholar]
- 23.Kabra R., Knight K. K., Zhou R., Snyder P. M. (2008) J. Biol. Chem. 283, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 24.Zhou R., Patel S. V., Snyder P. M. (2007) J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 25.Malik B., Yue Q., Yue G., Chen X. J., Price S. R., Mitch W. E., Eaton D. C. (2005) Am. J. Physiol. Renal Physiol. 289, F107–F116 [DOI] [PubMed] [Google Scholar]
- 26.Snyder P. M. (2005) Endocrinology 146, 5079–5085 [DOI] [PubMed] [Google Scholar]
- 27.Kaliszewski P., Zoładek T. (2008) Acta Biochim. Pol. 55, 649–662 [PubMed] [Google Scholar]
- 28.Belgareh-Touzé N., Léon S., Erpapazoglou Z., Stawiecka-Mirota M., Urban-Grimal D., Haguenauer-Tsapis R. (2008) Biochem. Soc. Trans. 36, 791–796 [DOI] [PubMed] [Google Scholar]
- 29.Vecchione A., Marchese A., Henry P., Rotin D., Morrione A. (2003) Mol. Cell. Biol. 23, 3363–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 31.Bhandari D., Robia S. L., Marchese A. (2009) Mol. Biol. Cell 20, 1324–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikko E., Sullivan J. A., Pelham H. R. (2008) EMBO Rep. 9, 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conkright J. J., Apsley K. S., Martin E. P., Ridsdale R., Rice W. R., Na C. L., Yang B., Weaver T. E. (2010) Am. J. Respir. Cell Mol. Biol. 42, 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingham R. J., Gish G., Pawson T. (2004) Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 35.Fotia A. B., Dinudom A., Shearwin K. E., Koch J. P., Korbmacher C., Cook D. I., Kumar S. (2003) FASEB J. 17, 70–72 [DOI] [PubMed] [Google Scholar]
- 36.Macias M. J., Hyvönen M., Baraldi E., Schultz J., Sudol M., Saraste M., Oschkinat H. (1996) Nature 382, 646–649 [DOI] [PubMed] [Google Scholar]
- 37.Fotia A. B., Cook D. I., Kumar S. (2006) Int. J. Biochem. Cell Biol. 38, 472–479 [DOI] [PubMed] [Google Scholar]
- 38.Debonneville C., Staub O. (2004) Mol. Cell. Biol. 24, 2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umebayashi K., Stenmark H., Yoshimori T. (2008) Mol. Biol. Cell 19, 3454–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 41.Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 42.Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 43.Itani O. A., Stokes J. B., Thomas C. P. (2005) Am. J. Physiol. Renal Physiol. 289, F334–F346 [DOI] [PubMed] [Google Scholar]
- 44.Raikwar N. S., Thomas C. P. (2008) Am. J. Physiol. Renal Physiol. 294, F1157–F1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itani O. A., Campbell J. R., Herrero J., Snyder P. M., Thomas C. P. (2003) Am. J. Physiol. Renal Physiol. 285, F916–F929 [DOI] [PubMed] [Google Scholar]
- 46.Asher C., Sinha I., Garty H. (2003) Biochim. Biophys. Acta 1612, 59–64 [DOI] [PubMed] [Google Scholar]
- 47.Henry P. C., Kanelis V., O'Brien M. C., Kim B., Gautschi I., Forman-Kay J., Schild L., Rotin D. (2003) J. Biol. Chem. 278, 20019–20028 [DOI] [PubMed] [Google Scholar]
- 48.Arévalo J. C., Waite J., Rajagopal R., Beyna M., Chen Z. Y., Lee F. S., Chao M. V. (2006) Neuron 50, 549–559 [DOI] [PubMed] [Google Scholar]
- 49.Lee J. R., Oestreich A. J., Payne J. A., Gunawan M. S., Norgan A. P., Katzmann D. J. (2009) J. Biol. Chem. 284, 32126–32137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatanaka T., Hatanaka Y., Setou M. (2006) J. Biol. Chem. 281, 35922–35930 [DOI] [PubMed] [Google Scholar]
- 51.Ekberg J., Schuetz F., Boase N. A., Conroy S. J., Manning J., Kumar S., Poronnik P., Adams D. J. (2007) J. Biol. Chem. 282, 12135–12142 [DOI] [PubMed] [Google Scholar]
- 52.Boehmer C., Palmada M., Rajamanickam J., Schniepp R., Amara S., Lang F. (2006) J. Neurochem. 97, 911–921 [DOI] [PubMed] [Google Scholar]
- 53.Kim H. C., Huibregtse J. M. (2009) Mol. Cell. Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.VanDemark A. P., Hofmann R. M., Tsui C., Pickart C. M., Wolberger C. (2001) Cell 105, 711–720 [DOI] [PubMed] [Google Scholar]
- 55.Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. (2009) J. Biol. Chem. 284, 19228–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galan J. M., Haguenauer-Tsapis R. (1997) EMBO J. 16, 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soetens O., De Craene J. O., Andre B. (2001) J. Biol. Chem. 276, 43949–43957 [DOI] [PubMed] [Google Scholar]
- 58.French M. E., Kretzmann B. R., Hicke L. (2009) J. Biol. Chem. 284, 12071–12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.