Abstract
A previous study identified the peroxisome proliferator-activated receptor α (PPARα) activation biomarkers 21-steroid carboxylic acids 11β-hydroxy-3,20-dioxopregn-4-en-21-oic acid (HDOPA) and 11β,20-dihydroxy-3-oxo-pregn-4-en-21-oic acid (DHOPA). In the present study, the molecular mechanism and the metabolic pathway of their production were determined. The PPARα-specific time-dependent increases in HDOPA and 20α-DHOPA paralleled the development of adrenal cortex hyperplasia, hypercortisolism, and spleen atrophy, which was attenuated in adrenalectomized mice. Wy-14,643 activation of PPARα induced hepatic FGF21, which caused increased neuropeptide Y and agouti-related protein mRNAs in the hypothalamus, stimulation of the agouti-related protein/neuropeptide Y neurons, and activation of the hypothalamic-pituitary-adrenal (HPA) axis, resulting in increased adrenal cortex hyperplasia and corticosterone production, revealing a link between PPARα and the HPA axis in controlling energy homeostasis and immune regulation. Corticosterone was demonstrated as the precursor of 21-carboxylic acids both in vivo and in vitro. Under PPARα activation, the classic reductive metabolic pathway of corticosterone was suppressed, whereas an alternative oxidative pathway was uncovered that leads to the sequential oxidation on carbon 21 resulting in HDOPA. The latter was then reduced to the end product 20α-DHOPA. Hepatic cytochromes P450, aldehyde dehydrogenase (ALDH3A2), and 21-hydroxysteroid dehydrogenase (AKR1C18) were found to be involved in this pathway. Activation of PPARα resulted in the induction of Aldh3a2 and Akr1c18, both of which were confirmed as target genes through introduction of promoter luciferase reporter constructs into mouse livers in vivo. This study underscores the power of mass spectrometry-based metabolomics combined with genomic and physiologic analyses in identifying downstream metabolic biomarkers and the corresponding upstream molecular mechanisms.
Keywords: Cytochrome P450, Hormones, Mass Spectrometry (MS), Metabolism, Metabolomics, Nuclear Receptors, HPA, PPARα, Adrenal, Corticosterone
Introduction
Peroxisome proliferator-activated receptor α (PPARα)7 is a nuclear hormone receptor regulating a number of physiologic processes including lipid metabolism and energy homeostasis via the transactivation of various target genes (1, 2). In a previous study (3), 11β-hydroxy-3,20-dioxopregn-4-en-21-oic acid (HDOPA) and 11β,20-dihydroxy-3-oxo-pregn-4-en-21-oic acid (DHOPA), two specific biomarkers of PPARα activation, were identified through urinary ultra-performance liquid chromatography-coupled time-of-flight mass spectrometry (UPLC-TOFMS)-based metabolomics analysis of wild-type and Ppara-null mice fed either a regular diet or a diet containing the PPARα ligand Wy-14,643 (WY). However, the issue remained of defining the metabolic pathway and identifying the target genes involved in the synthesis of these two biomarkers. Although metabolomics has been actively applied to investigate physiological/pathological biomarkers via the robust analytical ability to discriminate small molecule metabolomes in biofluids, cells, tissues, and organisms (4, 5), its power for translational studies in identifying novel molecular mechanisms has not been fully appreciated.
To understand the mechanism by which PPARα increases the production of HDOPA and DHOPA, in vivo and in vitro, metabolomics combined with genomic and physiologic analyses were undertaken using several PPARα transgenic modified mouse models, including wild-type, Ppara-null, and the PPARα-humanized mice. Germ-free wild-type, and conventionally raised mice were also investigated to exclude the role of gut flora in the production of the biomarkers. The precursor (corticosterone, 21-ol), intermediate (corticosterone aldehyde hydrate, 21-gem-diol), and sequential metabolic events in the pathway to forming HDOPA and DHOPA were identified, and the involvement of PPARα-induced oxidases including cytochrome P450s (CYPs), aldehyde dehydrogenases (ALDH), and 21-hydroxysteroid dehydrogenases (21-HSD) was demonstrated. Aldh3a2 and Akr1c18, genes encoding the responsible metabolic enzymes, were identified as novel PPARα target genes. Furthermore, activation of the hypothalamic-pituitary-adrenal (HPA) axis by PPARα ligands was found, which in turn contributed to the PPARα-stimulated phenotype, e.g. adrenal cortex hyperplasia, hypercortisolism, production of 21-carboxylic acids, as well as spleen atrophy. HPA activation was linked with PPARα via the FGF21-NPY pathway, suggesting their reciprocal roles in maintaining energy homeostasis and regulating immune responses.
EXPERIMENTAL PROCEDURES
Reagents
Catalase, corticosterone, 1-aminobenzotriazole, cyanamide, and 3,5-dichlorolicylic acid were purchased from Sigma. Corticosterone aldehyde hydrate and 11-dehydrocorticosterone were purchased from Steraloids (Newport, RI). The synthesis schemes for authentic HDOPA and DHOPA and the NMR analysis identifying DHOPA diastereomers are shown in supplemental Fig. S1.
Animals
6–8-week-old male wild-type and Ppara-null mice on C57BL/6Nr background were used. The conventionally raised mice, used for controls to the germ-free mice, were on a C57BL/6J background, and the wild-type mice corresponding to the PPARα-humanized mice were on the Sv129 background. The mice used for hydrodynamic injection were 5-week-old wild-type mice on the FVB/NJ background. This strain was used because of its white coat color, which does not quench the bioluminescence output in the Xenogen IVIS Imaging 200 Series instrument. For further demonstration of the PPARα-dependent luciferase transactivation, Ppara-null and the control wild-type mice on the Sv129 background were used. Rodent grain-based diet (Bioserv, Frenchtown, NJ) was used as the control diet, and a modified diet containing either 0.1% WY-14,643 or 0.5% fenofibrate (Bioserv) was used as PPARα ligands. Germ-free mice were kept in a sterile hood before euthanizing and were confirmed sterile by a negative report on the pathogen test by cecum swab. Adrenalectomy (ADX) and the control sham surgery (SHAM) were performed on C57BL/6Nr wild-type mice, which were placed on control or WY diet 1 week post-surgery. For the fasting experiment, the fed group was fed control diet, and the fasted group was fasted for 24 h. The mice were placed in glass metabolic cages, and urine samples were collected for 24 h. All of the animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the National Cancer Institute Animal Care and Use Committee.
UPLC-TOFMS Analysis
Urine and tissue samples were extracted with acetonitrile and centrifuged to remove particulates and proteins. The samples (5 μl/injection) were subjected to chromatography on a 50 × 2.1 mm ACQUITY 1.7 μm BEH C18 column (Waters Corp., Milford, MA) using an ACQUITY UPLC system (Waters Corporation) with a gradient mobile phase comprising 0.1% formic acid and acetonitrile containing 0.1% formic acid. A 0.5 ml/min flow rate was maintained in a 10-min run as previously described (3). The eluent was introduced directly into the mass spectrometer by electrospray. Mass spectrometry was performed on a Waters Q-TOF Premier operating in positive ion mode. For mass spectrometry scanning, the data were acquired in the centroid mode from 100 to 850 m/z. For MS/MS fragmentation of target ions, collision energies ranging from 15 to 35 V were applied. MetaboLynx and QuantLynx software were applied to identify and quantify the metabolites.
cDNA Microarray Analysis
Dye-coupled cDNAs were purified with a MiniElute PCR purification kit (Qiagen) and hybridized to an Agilent 44 K mouse 60-mer oligonucleotide microarray (Agilent Technologies, Santa Clara, CA). The procedures were repeated for replicate experiments with independent hybridization and processing. The data were processed and analyzed by a Genespring GX software package (Agilent Technologies).
RNA Analysis
Quantitative real time PCR (qPCR) was performed using SYBR green PCR master mix (Superarray) in an ABI Prism 7900HT sequence detection system (Applied Biosystems). Primer sequences are listed in supplemental Table S1. The expression levels of target genes were normalized to β-actin.
In Vivo Luciferase Assays
The mouse Aldh3a2 and Akr1c18 luciferase reporter plasmids were constructed by cloning the 2.0 kb of upstream regions using primers listed in supplemental Table S2. The PCR fragments were cloned into the NheI and KpnI restriction sites in the pGL3-basic vector (Promega, Madison WI). Forty μg of Ppre-luc, pGL3-Aldh3a2-luc, pGL3-Akr1c18-luc, and pGL3-luc plasmid vectors diluted with normal saline in 2 ml were administered to 5-week-old mice by tail vein in 7 s. The diet was changed to either control or WY diet 24 h after the hydrodynamic injection. Twenty-four hours later, the mice were dosed intraperitoneally with 200 μl of d-luciferin (15 mg/ml in phosphate-buffered saline), and at 4 min following the intraperitoneal dose of d-luciferin, the mice were imaged for bioluminescence on an IVIS Imaging 200 Series (Xenogen, Hopkinton, MA). The images were acquired at a “small” binning level and the acquisition time of 5 s.
Serum Corticosterone Levels
Blood samples were taken between 0900 and 1100. The animals were anesthetized individually in a glass jar containing saturated ether vapor, and retro orbital blood was collected within 30 s from initial handling within the cage. Serum corticosterone levels were measured using the corticosterone EIA kit (Cayman, Ann Arbor, MI).
Data Analysis
The results are expressed as the means ± S.D. Statistical analysis was performed by two-tailed Student's t test or one-way ANOVA combined with Bonferroni's multiple comparisons test. p < 0.05 was considered significant.
RESULTS
Identification of HDOPA and 20α-DHOPA as Time-dependent Biomarkers of PPARα Activation by Metabolomic Analysis
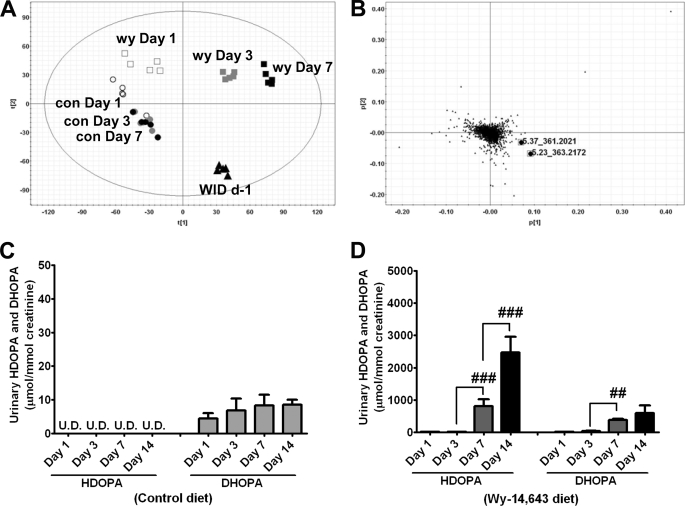
In a previous study, induction of urinary HDOPA and DHOPA by the PPARα ligand WY was only observed in wild-type mice and not in Pparα-null mice, thus establishing the role for PPARα in their production. A time course of WY administration revealed that these two biomarkers contributed to the time-dependent separation. The scores plot of principal components analysis (PCA) showed that control diet-fed mouse urine samples clustered together at all of the time points assessed. However, the 0.1% WY diet-fed urine samples separated from the control samples in a time-dependent manner following WY administration. Furthermore, urine samples collected from mice fed WY for 1 week and then placed on a control diet for 1 day began to cluster in the direction of urines from control mice (Fig. 1A). The PCA loadings plots identified HDOPA (m/z = 361.202+ at 5.37 min) and DHOPA (m/z = 363.217+ at 5.23 min) as the ions contributing to this separation (Fig. 1B). To establish identity, HDOPA and DHOPA were synthesized for tandem mass spectrometry (MS/MS) confirmation and quantification. The 20α-DHOPA was demonstrated as the predominant isomer in urine samples from WY-fed mice (supplemental Fig. S2). Although 20α-DHOPA could be detected in control urine samples, the level was much lower as compared with the urine samples from WY-fed mice, with no difference observed between individual groups collected at distinct time points (Fig. 1C). HDOPA was undetectable in the control samples (Fig. 1C), whereas levels of HDOPA and 20α-DHOPA were dramatically elevated in WY-fed mouse urine samples in a time-dependent manner (Fig. 1D). Furthermore, withdrawal of WY for 1 day significantly diminished the production of HDOPA, whereas no significant difference was found in 20α-DHOPA levels (supplemental Fig. S3A). To exclude the possible involvement of gut flora in the production of HDOPA and 20α-DHOPA, the urine metabolomes of germ-free and conventionally raised mice were assessed. PCA showed that the separation between control and WY urine samples was observed in both germ-free (supplemental Fig. S3C) and conventionally raised mice (supplemental Fig. S3E). HDOPA and 20α-DHOPA were the major ions contributing to the separation in both strains (supplemental Fig. S3, D and F). Urinary HDOPA and 20α-DHOPA were significantly increased to comparable levels in both strains after 1 week administration of the WY diet (supplemental Fig. S3B).
FIGURE 1.
Metabolomic analysis of mouse urine identifies HDOPA and 20α-DHOPA as time-dependent biomarkers of PPARα activation. A, PCA scores plot of component 1 versus component 2 for seven groups of urine samples from wild-type mice (C57BL/6Nr background), fed either control diet (con, ○) or Wy-14,643 diet (wy, □) at day 1 (white symbols), day 3 (gray symbols), day 7 (black symbols), and 1-day withdrawal from WY after 1 week of WY diet administration (WID d-1, ▴) (n = 5). B, PCA loading plot showing that HDOPA (m/z = 361.202+ at 5.37 min) and 20α-DHOPA (m/z = 363.217+ at 5.23 min) contributed to the separation between control and WY-fed groups (n = 5). Time course urinary levels of HDOPA and 20α-DHOPA in mice fed a control (C) or a WY (D) diet (n = 5). Each data bar in this figure represents the mean ± S.D. DHOPA, 20α-DHOPA; U.D., undetectable. The group differences are shown with significance values of p < 0.01 (##) and p < 0.001 (###) for one-way ANOVA.
Activation of the HPA Axis by PPARα Ligands and the Attenuation of HDOPA and 20α-DHOPA Production by Adrenalectomy
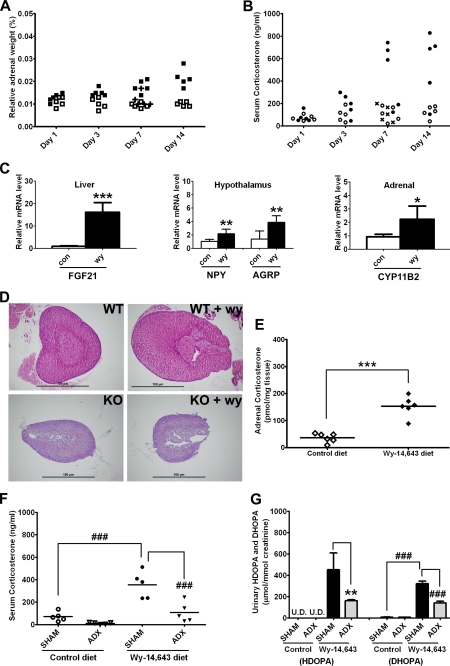
During WY administration, the increases in urinary HDOPA and 20α-DHOPA were associated with elevated adrenal weights (Fig. 2A) and circulating serum corticosterone levels (Fig. 2B) compared with the control-fed mice. Withdrawal from WY for 2 days following 1-week administration of WY significantly reversed the increases in adrenal weights and serum corticosterone levels (Fig. 2, A and B). Corticosterone is the major rodent glucocorticoid synthesized by the adrenal cortex, and the neuroendocrine system, the hypothalamic-pituitary-adrenal (HPA) axis, regulates its synthesis and secretion. The hypercortisolism found in WY-fed wild-type mice indicated an overactivation of the HPA axis; thus the markers of HPA axis were assessed. Levels of NPY and agouti-related protein (AGRP) mRNAs were significantly induced by WY administration in the mouse hypothalamus (Fig. 2C) where the two neuropeptides are produced by the AGRP/NPY neurons upon stimulating the HPA axis (6). A significant increase in the expression of the NPY stimulator, hepatic fibroblast growth factor 21 (FGF21) mRNA, was also observed (Fig. 2C). As an expected result of HPA axis activation, adrenal cortex hyperplasia (site where corticosterone is synthesized; Fig. 2D), induced adrenal CYP11B2 mRNA (encoding the enzyme responsible for corticosterone synthesis; Fig. 2C), and increased adrenal corticosterone (Fig. 2E) were demonstrated. In contrast, no significant changes in adrenal weight, adrenal cortex, serum corticosterone, and urinary HDOPA and 20α-DHOPA were found in Ppara-null mice, suggesting that PPARα-dependent adrenal cortex hyperplasia and the associated stimulated adrenal synthesis of corticosterone played a critical role in the formation of HDOPA and 20α-DHOPA. To further investigate the role of the HPA axis in the production of HDOPA and 20α-DHOPA, ADX and SHAM surgeries were performed on wild-type mice on control or WY diet. The removal of adrenal glands significantly attenuated the WY-induced production of both serum corticosterone (Fig. 2F) and urinary HDOPA and 20α-DHOPA (Fig. 2G).
FIGURE 2.
Activation of HPA axis by PPARα ligands and the attenuation of HDOPA and 20α-DHOPA production by adrenalectomy. A, time course relative adrenal weight (percentage of body weight) of wild-type mice (C57BL/6Nr background) fed either control diet (□) or WY diet (■), or withdrawal from WY for 2 days (WID) after 1 week of WY diet administration (+), 0.013% (wy) versus 0.011% (con), p > 0.5 at day 1; 0.015% (wy) versus 0.010% (con), p < 0.01 at day 3; 0.018% (wy) versus 0.009% (con), p < 0.001 at 1-week; and 0.022% (wy) versus 0.010% (con), p < 0.001 at 2-weeks of WY treatment, 0.012% (WID) versus 0.018% (wy) at 1-week, p < 0.05 (n = 5). B, time course serum corticosterone level (ng/ml) of wild-type mice fed either control diet (○) or WY diet (●) or withdrawal from WY for 2 days after 1 week of WY diet administration (×), determined by ELISA kit, 79.8 (wy) versus 71.9 (con) ng/ml, p > 0.5 at day 1; 190.0 (wy) versus99.5 (con) ng/ml, p > 0.5 at day 3; 463.9 (wy) versus 108.0 (con) ng/ml, p < 0.05 at 1-week; and 543.8 (wy) versus 126.2 (con) ng/ml, p < 0.05 at 2-weeks of WY treatment, 111.0 ng/ml (WID) versus 463.9 (wy) ng/ml at 1-week, p < 0.05 (n = 5). C, relative mRNA levels of FGF21 in livers, NPY and AGRP in hypothalamus, and CYP11B2 in adrenal glands of wild-type mice fed control (white bars) or WY (black bars) diet for 2 weeks, determined by qPCR (n = 6). D, representative histology sections of adrenal glands from wild-type (WT) and Ppara-null (KO) mice (both C57BL/6Nr background) fed control or WY diet for 2 weeks (magnification, 40×). E, adrenal corticosterone levels (pmol/mg tissue) of wild-type mice fed control diet (◇) or WY diet (♦) for 2 weeks, determined by UPLC-TOFMS (n = 6). Adrenalectomy and the control sham surgery were performed on wild-type mice, and either control or WY diet was given to both groups 1 week after the surgery. Serum and urine samples were collected at day 7 of diet administration. F, serum corticosterone level (ng/ml) of SHAM and ADX mice fed control or WY diet, determined by ELISA kit (n = 5). G, urinary HDOPA and 20α-DHOPA levels (μmol/mmol creatinine) of SHAM and ADX mice fed control or WY diet, determined by UPLC-TOFMS (n = 5). Each data bar in this figure represents the mean ± S.D., and the data points are given as individuals or individuals with mean. DHOPA, 20α-DHOPA. The group differences are shown with significance values of p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) for Student's t test and p < 0.001 (###) for one-way ANOVA.
Confirmation of Increases in Urinary 20α-DHOPA during Fasting
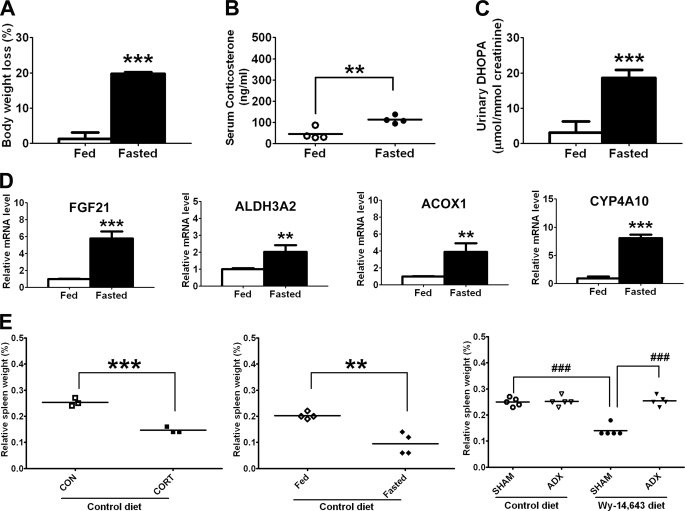
In the supposed link between PPARα and the HPA axis, one would thus expect an increase of urinary HDOPA and 20α-DHOPA during fasting, a stimulus that activates both PPARα and the HPA axis. Indeed, as expected, body weight loss (Fig. 3A) and hypercortisolism (Fig. 3B) were observed in fasted mice, accompanied by a significant increase of urinary 20α-DHOPA (Fig. 3C). However, urinary HDOPA was undetectable in both fed and fasted group, which might be due to the short term fasting being insufficient for the upstream metabolite to emerge. A 24-h fast led to an induction of several well known PPARα target genes in liver, such as Fgf21, Acox1 (acyl-coenzyme A oxidase 1), and Cyp4a10, in concert with the increase in the oxidase gene Aldh3a2 (Fig. 3D), whose transcription is controlled by PPARα as demonstrated in this study (see below). Interestingly, a similar phenotype of spleen atrophy was observed in corticosterone-treated mice, fasted mice, and WY diet-fed mice. Additionally, the effect of WY diet on spleen atrophy was abolished after removal of adrenal glands (Fig. 3E).
FIGURE 3.
Confirmation of increases in urinary 20α-DHOPA during fasting. A–C, body weight loss (A), serum corticosterone level (B), and urinary 20α-DHOPA level (C) in wild-type mice fed control diet or fasted for 24 h (n = 4). D, hepatic mRNA levels of FGF21, ALDH3A2, ACOX1, and CYP4A10 in fed or fasted mice (n = 4). E, spleen weight (percentage of body weight) of vehicle corn oil (CON) or corticosterone (CORT, 10 mg/kg) treated mice (left panel, n = 3), fed or fasted mice (middle panel, n = 4), or SHAM or adrenalectomized (ADX) mice (right panel, n = 5). Each data bar in this figure represents the mean ± S.D., and the data points are given as individuals with mean. DHOPA, 20α-DHOPA. The group differences are shown with significance values p < 0.01 (**) and p < 0.001 (***) for Student's t test and p < 0.001 (###) for one-way ANOVA.
Demonstration of Precursor and Intermediate in the Metabolic Pathway Forming HDOPA and 20α-DHOPA
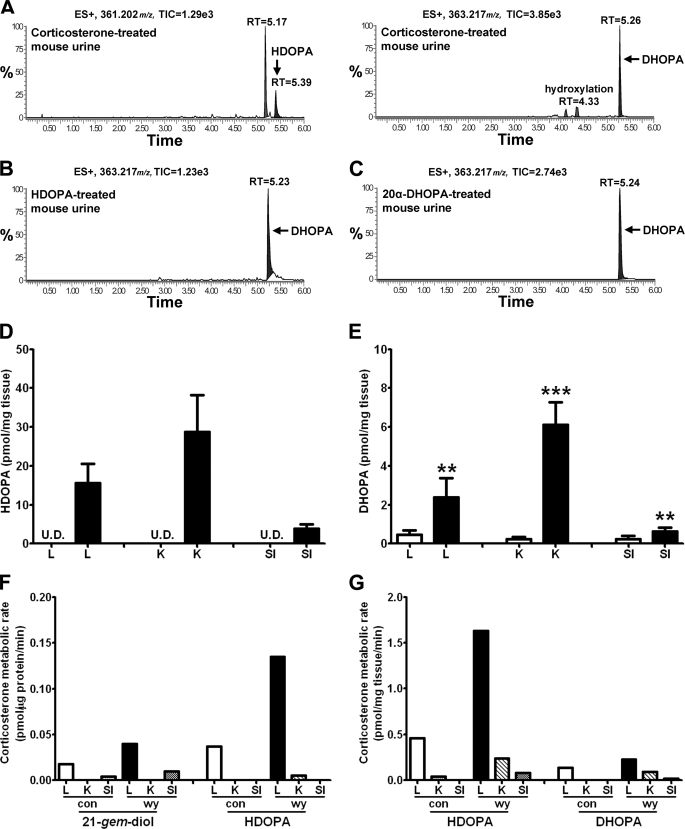
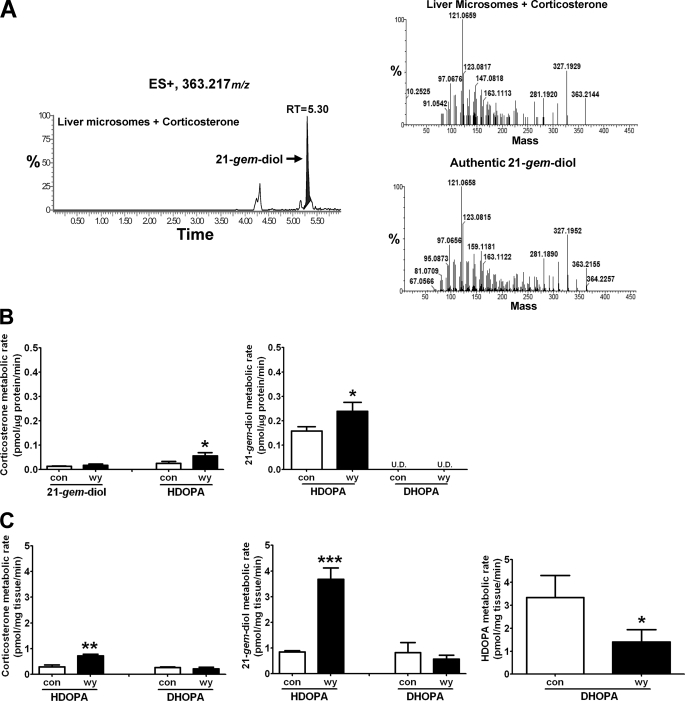
Chemical analysis of HDOPA and 20α-DHOPA established that both compounds were the carboxylic acid metabolites of a 21-steroid, corticosterone. When mice were given exogenous corticosterone along with control diet, the urine contained copious amounts of 20α-DHOPA and relatively lower amounts of HDOPA (Fig. 4A), which were identified by MetaboLynx software and further confirmed using the authentic standards and urine samples from WY-fed mice (supplemental Fig. S4, A for HDOPA and B for 20α-DHOPA), demonstrating corticosterone as the precursor of HDOPA and 20α-DHOPA. In addition, the mice were treated with HDOPA or 20α-DHOPA along with the control diet. MetaboLynx identified 20α-DHOPA as the major metabolite of HDOPA in vivo (Fig. 4B), whereas the parent 20α-DHOPA was the predominant form eliminated from the body (Fig. 4C). Therefore, the results confirmed that 20α-DHOPA was the end product formed from corticosterone. To clarify the sequential metabolic events, in vivo tissue distribution profiles of 21-carboxylic acids and in vitro metabolite profiles of corticosterone incubated with either tissue homogenates or tissue microsomes were determined by UPLC-TOFMS. Microsome preparations from different tissues were examined for their ability to convert corticosterone to 11-dehydrocorticosterone (data not shown). HDOPA was only detected in WY-fed liver, kidney, and small intestine homogenates (Fig. 4D), and 20α-DHOPA levels were significantly higher in the WY-fed tissues (Fig. 4E), whereas both were undetectable in the adrenal gland, confirming the exclusive presence of HDOPA and higher production of 20α-DHOPA in WY-fed organs, consistent with the data of the urinary metabolomes (Fig. 1, C and D). When incubating tissue microsomes (Fig. 4F) or tissue homogenates (Fig. 4G) with corticosterone, only the WY-fed livers had higher activity in the metabolism of corticosterone compared with the kidney and small intestine, suggesting that the liver is the critical tissue in metabolizing corticosterone to 21-carboxylic acids. In microsome incubation samples, a metabolite identified by MetaboLynx presented the same mass as 20α-DHOPA (m/z = 363.217+), whereas both the retention time (5.30 min) and the MS/MS spectrum were different. The ion was further confirmed as corticosterone aldehyde hydrate (21-gem-diol) when comparing the chromatograms and MS/MS spectra with the authentic standard (Fig. 5A). Liver metabolism was further investigated using homogenates and microsomes as enzyme sources, with corticosterone, 21-gem-diol, HDOPA, and 20α-DHOPA as substrates. The activities of WY-fed liver homogenates in the metabolism of corticosterone to HDOPA, and 21-gem-diol to HDOPA were both significantly higher than the control homogenates, and the rate of HDOPA production from 21-gem-diol was much higher (around 5-fold) than that from corticosterone (Fig. 5C). However, unexpectedly, the activity of WY-fed liver homogenates in the metabolism of HDOPA to 20α-DHOPA was significantly lower compared with the control homogenates, although the rates of 20α-DHOPA production were relatively lower but with no significance while using corticosterone and 21-gem-diol as the substrates (Fig. 5C). Similarly, the WY-induced higher rates of HDOPA production were found in liver microsome samples (Fig. 5B). The differences between microsome incubations and homogenate incubations, in addition to detection of the intermediate 21-gem-diol in microsome incubations, were that microsomes could not metabolize HDOPA. No 20α-DHOPA metabolite was detected in either homogenate or microsome incubations. Together these data demonstrate that corticosterone is converted to 21-carboxylic acids in the liver, via a metabolic pathway where corticosterone (21-ol) is the precursor, corticosterone aldehyde hydrate (21-gem-diol) is the intermediate, HDOPA (20-oxo-21-oic acid) as a metabolite specifically attributed to the activation of PPARα and 20α-DHOPA (20α-hydroxy-21-oic acid) as the end product. The 21-carboxylic acids produced in liver were transported and accumulated in kidney for elimination.
FIGURE 4.
Identification of corticosterone as the precursor and liver as the major organ in the metabolic pathway forming HDOPA and 20α-DHOPA. Wild-type mice (C57BL/6Nr background) were treated with either vehicle, or corticosterone, or HDOPA, or 20α-DHOPA at the dose of 10 mg/kg via gavage. Urine samples were collected during the first 24 h after the treatment. The samples were extracted and analyzed by UPLC-TOFMS. The mass spectra from treated urine samples were compared with those with vehicle-treated urine samples using MetaboLynx software to find potential metabolites. Each metabolite was represented as extracted ion chromatograms (window of 50 mDa), and mass, total ion currents (TIC), retention time (RT), putative metabolic reaction name, mass difference were discerned automatically by MetaboLynx. The accurate MS/MS spectra and retention time were compared with the authentic standards to confirm the structure. A, in vivo metabolites of corticosterone identified as HDOPA (m/z = 361.202+ at 5.39 min) and 20α-DHOPA (m/z = 363.217+ at 5.26 min). B, in vivo metabolite of HDOPA identified as the reductive metabolite 20α-DHOPA (m/z = 363.217+ at 5.23 min). C, 20α-DHOPA was eliminated from the body as the parent form (m/z = 363.217+ at 5.24 min). D and E, distributions (pmol/mg tissue) of HDOPA (D) and 20α-DHOPA (E) in livers (L), kidneys (K), and small intestines (SI) of wild-type mice fed control (white bar) or WY (black bar) diet for 1 week, determined by UPLC-TOFMS (n = 6). F and G, activities of tissue microsomes (F, pmol/μg protein/min) and tissue homogenates (G, pmol/mg tissue/min) in the metabolism of corticosterone, determined by UPLC-TOFMS (n = 1). Each data bar in this figure represents the individual value or the mean ± S.D. DHOPA, 20α-DHOPA; U.D., undetectable. The group differences are shown with significance values of p < 0.01 and p < 0.001 (***).
FIGURE 5.
Identification of 21-gem-diol as the intermediate in the metabolic pathway forming HDOPA and 20α-DHOPA. Wild-type mice were fed control or WY diet for 1 week before collecting livers. Liver microsomes and homogenates were prepared and analyzed by UPLC-TOFMS. In metabolic studies, 50 μm of corticosterone, 21-gem-diol, HDOPA, and 20α-DHOPA were incubated separately with liver microsomes or homogenates for 30 min. The mass spectra from incubation samples including both tissue and substrate were compared with the incubation solution with only the substrate using MetaboLynx to find potential metabolites. The concentration of each metabolite was obtained from QuanLynx using debrisoquine as internal standard. A, chromatogram of a corticosterone metabolite produced from liver microsomes (left panel), whose MS/MS spectrum (right top panel) and retention time (m/z = 363.217+ at 5.30 min) matched that of authentic 21-gem-diol standard (right bottom panel). B, the activities of liver microsomes in the metabolism of corticosterone (left) and 21-gem-diol (right panel) (n = 3). C, the activities of liver homogenates in the metabolism of corticosterone (left panel), 21-gem-diol (middle panel), and HDOPA (right panel) (n = 3). Each data bar in this figure represents the mean ± S.D. DHOPA, 20α-DHOPA; U.D., undetectable. The group differences are shown with significance values of p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
Identification of Aldh3a2 and Akr1c18 as PPARα Target Genes
To identify genes involved in the production of HDOPA and 20α-DHOPA, microarray and qPCR analyses were performed in liver samples from wild-type and Ppara-null mice. Messenger RNAs encoding enzymes involved in the classic reduction pathway of corticosterone metabolism, 5α-reductase encoding gene Srd5a1, 5β-reductase encoding gene Akr1d1, and 3α-HSD encoding gene Akr1c14 were significantly suppressed (supplemental Fig. S5). To determine whether these genes are direct transcriptional targets of PPARα, the mRNAs were measured after a short 4-h WY treatment. No significant change was noted in expression of these mRNAs, suggesting an indirect mechanism by which PPARα regulates these enzymes. Conversely, two genes encoding putative oxidases, aldehyde dehydrogenase (ALDH), and HSD were found by microarray analysis to be dramatically induced following a 2-week WY administration. Messenger RNAs encoding aldehyde dehydrogenase family 3, subfamily A2 (Aldh3a2), and aldo-keto reductase family 1, member C18 (Akr1c18, also known as mouse 20α-HSD), were significantly induced by WY only in the livers of wild-type mice (Fig. 6, A and B). Both genes were induced as early as 4 h after WY treatment, then increased gradually with the time, and were significantly reversed to basal levels following withdrawal of WY for 2 days. Similar inductions were observed in PPARα-humanized mice and its corresponding wild-type strain (Sv129 background) fed WY and the PPARα ligand fenofibrate. Neither ALDH3A2 nor AKR1C18 mRNAs were induced by WY in Ppara-null mice. CYPs were not significantly changed by WY treatment except for an increase in CYP4A subfamily, a well known PPARα target gene. These data demonstrate that Aldh3a2 and Akr1c18 genes are induced via a PPARα-dependent mechanism. Promoter analysis using the NUBIScan nuclear receptor binding data base revealed the presence of putative PPAR response elements (PPREs) within the promoters of the Aldh3a2 and Akr1c18 genes. Therefore, the promoter regions of the mouse Aldh3a2 and Akr1c18 encompassing their PPREs were analyzed by transient transfection of luciferase reporter constructs. When the Aldh3a2 and Akr1c18 promoter luciferase constructs were transfected into HepG2 cells along with PPARα and RXR expression plasmids, it failed to induce luciferase expression after WY treatment. To determine promoter activity in a more natural setting, the constructs were introduced to mouse liver in vivo by hydrodynamic injection through the tail vein and luciferase expression determined by imaging (7, 8). Luciferase reporter constructs under the control of the promoter region (2 kb upstream of the transcription start site) of the mouse Aldh3a2 (pGL3-Aldh3a2-luc) and Akr1c18 (pGL3-Akr1c18-luc) genes, the positive control PPRE-luciferase construct (Ppre-luc), and the negative control pGL3 vector were hydrodynamically injected into the wild-type mice (FVB/NJ background), Ppara-null mice, and its corresponding wild-type counterpart (both on the Sv129 background). In vivo imaging showed that wild-type mice administered the WY diet following injection of Ppre-luc, pGL3-Aldh3a2-luc, or pGL3-Akr1c18-luc displayed a dramatic increase in luciferase activity compared with the control diet-fed mice (Fig. 6, C and E). Furthermore, the induction of in vivo luciferase activity by WY was not observed in wild-type mice injected with the empty pGL3 vector (Fig. 6C) or in Ppara-null mice injected with Ppre-luc, pGL3-Aldh3a2-luc, or pGL3-Akr1c18-luc plasmids (Fig. 6D), demonstrating a PPARα-dependent luciferase transactivation. Successful transfection of exogenous plasmid DNA into the mouse liver was confirmed by PCR amplification of the luciferase vectors from liver DNA; amplification of the Gapdh gene was used as a control (Fig. 6, C–E). Together these data demonstrate that the PPARα can regulate the promoters of the mouse Aldh3a2 and Akr1c18 genes in vivo, thus identifying these two genes as novel PPARα targets.
FIGURE 6.
Identification of oxidase encoding genes Aldh3a2 and Akr1c18 as PPARα targets. Relative mRNA levels of ALDH3A2 (A) and AKR1C18 (B) in livers of wild-type (WT, n = 5), Ppara-null (KO, n = 4), and PPARα-humanized (PAC, n = 4) mice, determined by qPCR. In vivo luciferase activity by bioluminescent imaging were measured in wild-type mice (FVB/NJ background) (C), Ppara-null mice (Sv129 background) (D), and Sv129 wild-type mice (E) 48 h after hydrodynamic tail vein injection of 40 μg of Ppre-luc (Ppre), pGL3-Aldh3a2-luc (Aldh3a2), pGL3-Akr1c18-luc (Akr1c18), and pGL3-luc (pGL3) plasmid vectors, and 24 h following either control (−) or WY (+) diet administration. C–E, hepatic DNA copies of hydrodynamic injected vectors and the housekeeping gene Gapdh in individual mouse, demonstrated through PCR using RV3-GL2 primer and Gapdh primer respectively. Each data bar in this figure represents the mean ± S.D. The group differences are shown with significance values of p < 0.01 (**) and p < 0.001 (***) for Student's t test and p < 0.05 (#), p < 0.01 (##), and p < 0.001 (###) for one-way ANOVA. N.S. represents no statistical difference.
Involvement of CYPs, ALDH, and 21-HSD in the Metabolism of Corticosterone
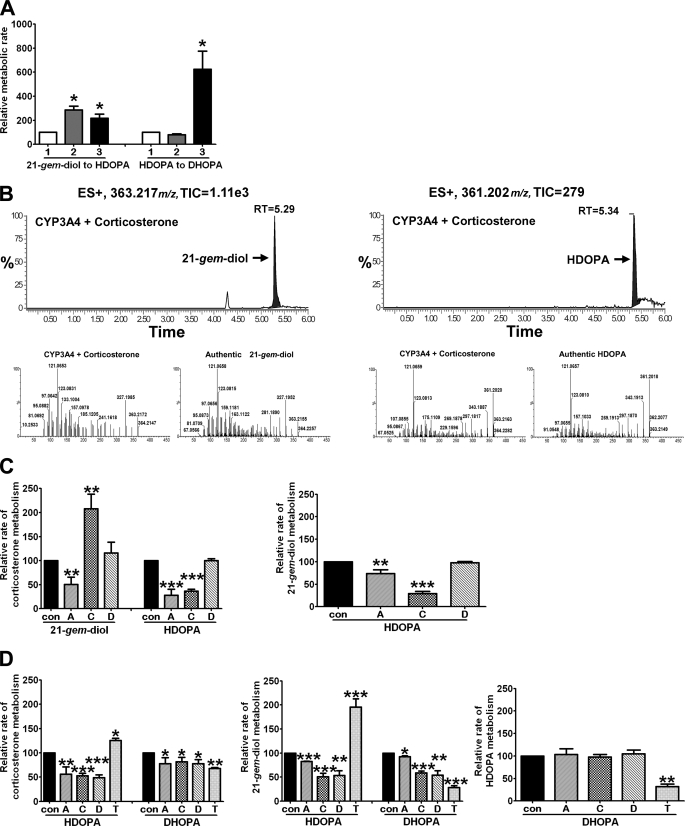
To determine whether the target genes identified by microarray, qPCR, and promoter analyses were critical in HDOPA and 20α-DHOPA formation, in vitro incubations using several cofactors and specific inhibitors were investigated. Cofactor requirements for the pathway are shown in Fig. 7A. Only NADPH and not NAD+ significantly stimulated the conversion of HDOPA to 20α-DHOPA, confirming the reaction as a NADPH-dependent reduction. In contrast, exogenous NAD+ and NADPH both significantly increased the activities of liver homogenates in the metabolism of 21-gem-diol to HDOPA compared with the control incubations containing no additional cofactors. This indicates that different enzymes requiring NAD+ and NADPH catalyze the oxidation. Considering the conventional roles of CYPs, ALDH, and HSD in steroid oxidations, and their (CYP4A subfamily, ALDH3A2, and AKR1C18) inductions in a PPARα-dependent manner, the NADPH-dependent enzymes were considered to be CYPs, and the NAD+-dependent enzymes were thought to be ALDH and HSD. A panel of recombinant CYPs was assessed, revealing that most CYPs including members of CYP2C, CYP2D, CYP3A, and CYP4A subfamilies, especially CYP3A4 and CYP3A5, could metabolize corticosterone to 21-gem-diol and HDOPA (Fig. 7B). In liver microsome samples (Fig. 7C), 1-aminobenzotriazole, a suicide inhibitor for CYPs, was the only inhibitor significantly suppressing metabolism of corticosterone to the 21-gem-diol; it also dramatically reduced metabolism of corticosterone to HDOPA and, with relatively less inhibition, metabolism of 21-gem-diol to HDOPA, suggesting that CYPs mainly serve as enzymes metabolizing corticosterone to 21-gem-diol and partly participate in the conversion of 21-gem-diol to HDOPA. Cyanamide, an ALDH specific inhibitor (9), significantly reduced the metabolism of corticosterone to HDOPA accompanied by an increased accumulation of 21-gem-diol; it also significantly suppressed the metabolism of 21-gem-diol to HDOPA, indicating that ALDH is only involved in the oxidation of 21-gem-diol to HDOPA with no effect on the upstream conversion of corticosterone to the 21-gem-diol. 3,5-Dichlorosalicylic acid, a 20α-HSD-specific inhibitor (10), had no effect on microsomal metabolism, which is in agreement with the localization of 20α-HSD as a cytosolic enzyme, whereas CYPs and ALDH are microsomal enzymes. In liver homogenate samples (Fig. 7D), all three inhibitors significantly suppressed the metabolism of corticosterone to HDOPA and 20α-DHOPA, as well as the conversion of 21-gem-diol to HDOPA and 20α-DHOPA, confirming that the metabolic roles of CYPs and ALDH coincided with the microsomal results and identifying the involvement of cytosolic 20α-HSD in metabolizing either the 21-gem-diol or both corticosterone and 21-gem-diol to HDOPA. When catalase was added, independent of what substrate was used, there was a significant increase in the formation of HDOPA and decreased production of 20α-DHOPA, suggesting that catalase favors the former oxidations performed by CYPs through providing oxygen, whereas it interferes with the latter end reduction carried out by reductases. However, no effect on the formation of 20α-DHOPA was detected when incubating with HDOPA supplemented with inhibitors, thus demonstrating that none of the three oxidases is associated with the end reduction.
FIGURE 7.
Involvement of CYPs, ALDH, and 21-HSD in the oxidation pathway of corticosterone metabolism. Wild-type mice (C57BL/6Nr background) were fed a WY diet for 1 week before collecting livers. The samples were prepared and analyzed by UPLC-TOFMS. A, effects of cofactor NAD+ (gray bar, 2), or NADPH (black bar, 3) on the activities of liver homogenates in the metabolism of 21-gem-diol and HDOPA compared with vehicle control (white bar, 1) (n = 3). B, corticosterone metabolites from recombinant human CYP3A4 identified as 21-gem-diol (left panels) and HDOPA (right panels) by chromatograms (top panels) and MS/MS spectra (bottom panels). C, inhibition on the activities of liver microsomes in the metabolism of corticosterone (left panel) and 21-gem-diol (right panel) (n = 3). D, inhibition on the activities of liver homogenates in the metabolism of corticosterone (left panel), 21-gem-diol (middle panel), and HDOPA (right panel) (n = 3). Vehicle or different inhibitors, CYPs suicide inhibitor 1-aminobenzotriazole (100 μm, bar A), ALDH inhibitor cyanamide (200 μm, bar C), AKR1C1 inhibitor 3, 5-dichlorosalicylic acid (5 μm, bar D), and catalase (750 units/ml, bar T) were added into the incubation system 10 min before the addition of substrates. Each data bar in this figure represents the mean ± S.D. TIC, total ion current; RT, retention time; DHOPA, 20α-DHOPA. The group differences between vehicle control and treatments are shown with significance values of p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
DISCUSSION
In the present study, the mechanisms of biomarker (HDOPA and 20α-DHOPA) synthesis under PPARα activation were identified as first the PPARα-stimulated adrenal cortex hyperplasia and associated hypercortisolism that provide high levels of precursor corticosterone, and second, the PPARα-induced alternative oxidative pathway that metabolizes corticosterone to 21-carboxylic acids. Corticosterone is the major adrenocortical hormone in rodents and under normal conditions with physiological levels of corticosteroids; most of the circulating form is protein-bound and predominately reduced to the tetrahydro end products through the reduction pathway (11), which is inhibited because of the remarkable down-regulation of the reductases (5α-reductase, 5β-reductase, and 3α-HSD) and the overload of reactive oxygen species that occurs under PPARα activation. In contrast, with increased or pharmacological levels of corticosteroids, for example, the hypercortisolism status under WY administration as observed in this study, a larger fraction of nonprotein-bound steroid is available for diffusion into the extravascular spaces and in coordination with the oxidases induced by PPARα, corticosterone is converted to the carboxylic acids as a major metabolic pathway.
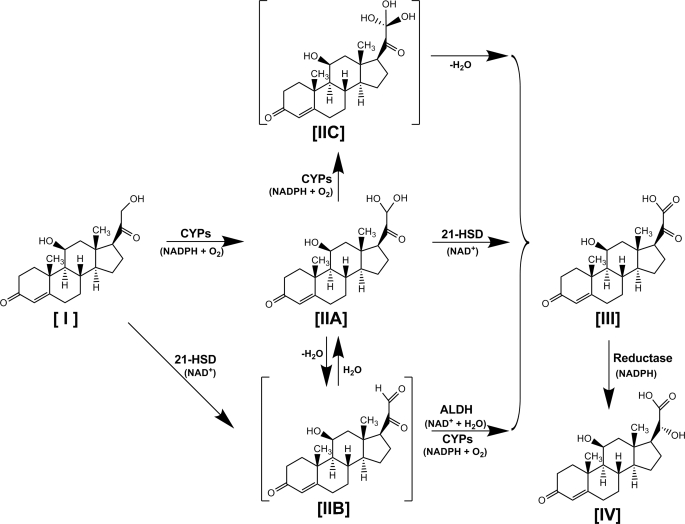
The carboxylic acid metabolites of corticosteroids (12–14), with a similar structure of 20-hydroxy-21-oic acid, were reported as primary hepatic metabolites both in vivo and in vitro. Although it is believed that transformation of the corticosteroid side chain consists of multiple consecutive oxidation steps with several putative intermediates, direct demonstration of the sequential events has not been elucidated. From the available evidence provided in the present study, the possible metabolic pathways are outlined in the scheme shown in Fig. 8. A hydroxyl group is added to the 21-ol [I] to form 21-gem-diol [IIA] by CYPs, which is then either converted to a putative intermediate 21-aldehyde [IIB] after enzymatic or nonenzymatic dehydration or further hydroxylated to a putative intermediate 21-gem-triol [IIC] by CYPs. Next, the oxidation of the 21-al, and its gem-diol, gem-triol to 20-oxo-21-oic acid [III], is mediated in turn by either dehydrogenation by ALDH3A2 or oxygenation by CYPs (I-IIA-IIB-III); dehydrogenation of 21-HSD (I-IIA-III), which was demonstrated to be AKR1C18; and dehydration (I-IIA-IIC-III). Another pathway is the dehydrogenation of 21-ol to 21-al directly by 21-HSD and then oxidation to 21-oic acid by ALDH3A2 and CYPs (I-IIB-III). The enzymes involved in the complex sequential 21-hydroxyl oxidation of corticosterone [I] to HDOPA [III] are specifically induced by PPARα activation. In contrast, the subsequent conversion of HDOPA to 20α-DHOPA [IV], a simple C20-ketone reduction, is suppressed in response to the accumulating oxidative stress triggered by PPARα activation, which explains the relatively lower specificity of 20α-DHOPA to PPARα activation compared with the oxidative product HDOPA, including levels in urine and tissue, patterns of time-dependent induction, responses to WY withdrawal, and profiles of metabolite production. None of the three intermediates proposed in this model, 21-gem-diol, 21-al, and 21-gem-triol, was detected in vivo, except for the determination of 21-gem-diol in tissue microsome samples incubated with corticosterone, which might be interpreted by the exchange of intermediates with the medium that is not uncommon in multiple step reactions. In addition, the significantly higher production of HDOPA from 21-gem-diol compared with corticosterone offered the rationale in defining it as the intermediate because of its increased affinity relative to the first substrate corticosterone.
FIGURE 8.
Proposed metabolic model for the formation of two carboxylic acids HDOPA [III] and 20α-DHOPA [IV] from corticosterone [I] via its aldehyde hydrate [IIA] and putative intermediates corticosterone aldehyde [IIB], 21-gem-triol [IIC]. CYPs (a panel of CYPs, especially the major liver enzyme CYP3A subfamily, and the PPARα-induced CYP4A subfamily), ALDH (PPARα specific induced ALDH3A2, possibly coincided with other PPARα-induced ALDH family members), and 21-HSD (PPARα-induced AKR1C18, possibly coincided with PPARα-induced HSD17B) are involved in the multi-step oxidation.
Because the oxidative stress shifts the preference of corticosterone metabolism from reduction to oxidation, PPARα-induced CYPs, ALDH3A2, and AKR1C18 participate in the 21-hydroxyl oxidation. CYPs are a large family of enzymes involved in the oxidation of a range of substrates through hydroxylation of C–H bonds. Several CYPs catalyze multistep oxidations that convert alcohol to aldehyde and, ultimately, to carboxylic acid (15–17), which offers the basis for the present observation that CYPs can metabolize corticosterone to corticosterone aldehyde hydrate and subsequently to 20-oxo-21-oic acid. ALDHs are responsible for catalyzing the irreversible oxidation of a wide spectrum of aldehydes to their corresponding carboxylic acids. As a murine 20α-HSD, although no identification of 21-HSD activity has been reported for AKR1C18, dual activity at the active site of a single enzyme is a common feature of steroidogenic oxidoreductases (18, 19). For example, in addition to the 20α-OH activity, AKR1C18 is also known for catalyzing oxidation of 3α- and 17β-hydroxysteroids (20). This is the first report of activity on a 21-OH for AKR1C18. The possibility that other ALDH and 21-HSD participate in the oxidation of corticosterone must also be considered. Indeed, other ALDH members are inducible by fibrate drugs to a certain degree (21), and 17β-HSD is a bona fide PPARα target gene (22, 23). 17β-HSD activity is favored in the oxidation direction (24), and the 21-OH dehydrogenase activity identified for 17β-HSD10 (25) suggests that the latter enzyme is another 21-HSD involved in oxidation on carbon atom 21, resulting in formation of HDOPA.
Human ALDH3A2 and AKR1C1 genes were both reported to be potently induced by reactive oxygen species. The induction of ALDH3A2 was even demonstrated to be only by PPARα ligands (21) and only in wild-type mice (26, 27). However, the presence of PPREs within the promoters of ALDH3A2 and AKR1C1 has not been determined. In the present study, mouse hepatic ALDH3A2 and AKR1C18 mRNAs were induced by PPARα ligands within 4 h after administration of the agonist and in a PPARα-dependent manner. Transfection of luciferase reporter constructs of 5′-flanking sequences encompassing the putative PPRE into mouse through hydrodynamic sheer injection demonstrated the presence of functional PPREs in the promoters of both genes. Although hydrodynamic injection was originally and predominately used as a powerful means for gene therapy (28–30), an additional important application developed more recently is in vivo transfection for biological studies (31, 32). Interestingly, HepG2 cells were unable to generate an induction of these promoters, suggesting the possible involvement of coactivators or other factors required for induction not present in the cultured cells but present in liver. Delineation of the exact PPRE responsible for induction of the Aldh3a2 and Akr1c18 promoter by deletion or mutation strategy requires further study.
Understanding the pathways by which PPARα increases the synthesis of HDOPA and 20α-DHOPA using a metabolomic approach has uncovered a novel role for PPARα in activating the HPA axis. The question remains as to why the HPA axis is activated by PPARα ligands. Activation of HPA axis is triggered by metabolic signals, for example, fasting-induced glycopenia (33) via neuropeptides involved in appetite stimulation and energy expenditure inhibition, such as NPY and AGRP (34, 35). This is a fundamental adaptive process in mammals where stimulating secretion of glucocorticoids, the end products of the HPA axis, leads to multiple metabolic effects on gluconeogenesis and fatty acid release. PPARα has also been implicated in a variety of metabolic processes including lipid metabolism and energy homeostasis (1, 2), affecting many of the pathways in response to fasting, such as gluconeogenesis, ketogenesis, and fatty acid metabolism. Recent studies showed that FGF21, the endocrine hormone synthesized from a liver PPARα target gene (36), is able to enter into the brain through the blood-brain barrier and up-regulate hypothalamic NPY (37, 38). In the present study, fasting and PPARα agonist administration both induced liver FGF21, which confirms previous observations (2, 39). Additionally, a critical role of FGF21 in the adaptation of the body to fasting was demonstrated (2, 39). However, the mechanism by which FGF21 stimulates lipolysis and ketogenesis has not been determined. In this study, several lines of evidence indicate a link between PPARα and the HPA axis. First, PPARα agonists and fasting activated both PPARα and HPA axis, via the FGF21-NPY pathway, thereby resulting in hypercortisolism and excretion of urinary 21-carboxylic acid metabolites, HDOPA and 20α-DHOPA, through an induced oxidative metabolism involving several PPARα target oxidases such as CYP4A, ALDH3A2, AKR1C18, and HSD17B. Second, adrenal cortex hyperplasia, hypercortisolism, and urinary 21-carboxylic acid metabolites were not observed in Ppara-null mice fed PPARα agonists. Third, these PPARα-activated features were alleviated in adrenalectomized wild-type mice. Lastly, a similar phenotype of spleen atrophy was observed in corticosterone-treated mice, fasted mice, and PPARα agonist-fed mice, and the latter could be abolished by ADX. The similar effects of PPARα ligands (both endogenous ligands under fasting and exogenous agonists) and corticosterone on these parameters, together with the reverse by adrenalectomy, suggest that corticosterone contributes to the pleiotropic actions of PPARα.
It is noteworthy to mention that PPARα and the HPA axis end product glucocorticoids both can modulate the immune system. Administration of PPARα ligands to mice results in a significant reduction of body weight as well as of thymus and spleen weights, which occurs in a PPARα-dependent manner (40). Glucocorticoids suppress the immune system leading to thymic and splenic atrophy. Although the data suggest that they share a similar immunosuppression mechanism involving lymphocyte and thymocyte apoptosis, as well as cell cycle arrest in G0/G1 phase (41–43), no direct proof has been reported for the involvement of glucocorticoids in immunosuppression by PPARα ligands. Using the adrenalectomized model, spleen atrophy was only observed in the WY-fed sham group, whereas the removal of adrenal glands totally depleted the WY-induced spleen atrophy, implicating a critical role for corticosterone on the immunosuppression of PPARα ligands.
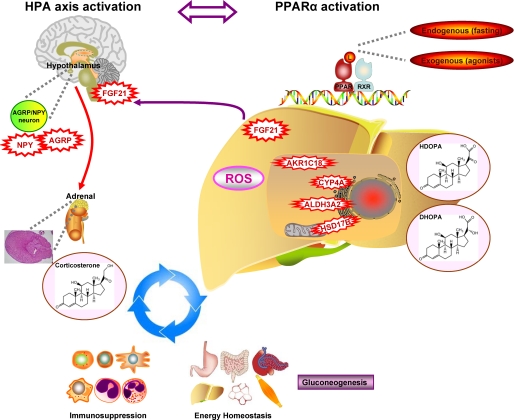
Based on all of the data, the following model is proposed for how PPARα cooperates with HPA axis in regulating energy homeostasis and immune response (Fig. 9). During fasting or the fasting-like state, activation of PPARα by endogenous ligands or exogenous agonists result in increased expression of FGF21 that is secreted from liver, enters the brain, and stimulates the HPA axis leading to the adrenal cortex hyperplasia and hypercortisolism, which in turn contributes to the role of PPARα in gluconeogenesis. PPARα also directly stimulates transcription of genes involved in fatty acid oxidation and ketogenesis. The synergic result of this bipartite mechanism is the mobilization of liver glycogen, adipose tissue triglycerides, and muscle protein, thereby providing glycerol, fatty acids, ketones, and alanine as sources for gluconeogenesis to maintain the energy balance. This finding reveals a link between PPARα and HPA axis, which acts in an autocrine-neuro-endocrine (FGF21-NPY-corticosterone) fashion to regulate diverse components of the adaptive metabolic response. The data presented herein demonstrate the power of mass spectrometry-based metabolomic analysis in combination with genomic and physiologic studies using transgenic mouse models to identify downstream metabolic biomarkers and uncover novel upstream molecular mechanisms.
FIGURE 9.
Reciprocal roles between PPARα activation and HPA axis activation in stimulation of gluconeogenesis, suppression of immune responses, and the production of 21-carboxylic acids. Red represents stress (reactive oxygen species (ROS)), enzymes (CYP4A, ALDH3A2, AKR1C18, and HSD17B), growth factor (FGF21), and neuropeptides (NPY and AGRP) induced by PPARα ligands (L). Purple indicates metabolic process (gluconeogenesis) stimulated under the crosstalk between PPARα activation and HPA axis activation.
Supplementary Material
Acknowledgments
We thank Susan L. Tonkonogy for providing the germ-free mice. We also appreciate the expert assistance of John Buckley, Linda Byrd, Jessica Bonzo, Aijuan Qu, Jiancun Larry Zhang, and Dong Wook Kang.
This work was supported, in whole or in part, by National Institutes of Health NCI Intramural Research Program. This work was also supported by Center for Gastrointestinal Biology and Disease Grant P30 DK34987.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- PPAR
- peroxisome proliferator-activated receptor
- ADX
- adrenalectomy
- AGRP
- agouti-related protein
- AKR1C18
- aldo-keto reductase family 1, member C18
- ALDH
- aldehyde dehydrogenases
- ALDH3A2
- aldehyde dehydrogenase family 3, subfamily A2
- CYP
- cytochrome P450
- DHOPA
- 11β,20-dihydroxy-3-oxo-pregn-4-en-21-oic acid, (20-hydroxy-21-oic acid)
- FGF21
- fibroblast growth factor 21
- HDOPA
- 11β-hydroxy-3,20-dioxopregn-4-en-21-oic acid, (20-oxo-21-oic acid)
- HPA
- hypothalamic-pituitary-adrenal axis
- HSD
- hydroxysteroid dehydrogenase
- MS/MS
- tandem mass spectrometry
- NPY
- neuropeptide Y
- PCA
- principal components analysis
- PPRE
- peroxisome proliferator response element
- UPLC-TOFMS
- ultra-performance liquid chromatography-coupled time-of-flight mass spectrometry
- WY
- Wy-14,643
- SHAM
- control sham surgery
- qPCR
- quantitative real time PCR
- ANOVA
- analysis of variance
- WID
- withdrawal from diet containing WY.
REFERENCES
- 1.Desvergne B., Wahli W. (1999) Endocr. Rev. 20, 649–688 [DOI] [PubMed] [Google Scholar]
- 2.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V., Elmquist J. K., Gerard R. D., Burgess S. C., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. (2007) Cell Metab. 5, 415–425 [DOI] [PubMed] [Google Scholar]
- 3.Zhen Y., Krausz K. W., Chen C., Idle J. R., Gonzalez F. J. (2007) Mol. Endocrinol. 21, 2136–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson J. K., Lindon J. C., Holmes E. (1999) Xenobiotica 29, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 5.Griffin J. L. (2006) Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen Y. S., Adrian T. E., Allen J. M., Tatemoto K., Crow T. J., Bloom S. R., Polak J. M. (1983) Science 221, 877–879 [DOI] [PubMed] [Google Scholar]
- 7.Liu F., Song Y., Liu D. (1999) Gene Ther. 6, 1258–1266 [DOI] [PubMed] [Google Scholar]
- 8.Zhang G., Budker V., Wolff J. A. (1999) Human Gene Ther. 10, 1735–1737 [DOI] [PubMed] [Google Scholar]
- 9.Loomis C. W., Brien J. F. (1983) Can. J. Phys. Pharmacol. 61, 431–435 [DOI] [PubMed] [Google Scholar]
- 10.Dhagat U., Endo S., Sumii R., Hara A., El-Kabbani O. (2008) J. Med. Chem. 51, 4844–4848 [DOI] [PubMed] [Google Scholar]
- 11.Peterson R. E., Pierce C. E. (1960) J. Clin. Invest. 39, 741–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradlow H. L., Zumoff B., Monder C., Lee H. J., Hellman L. (1973) J. Clin. Endocrinol. Metab. 37, 811–818 [DOI] [PubMed] [Google Scholar]
- 13.Monder C., Purkaystha A. R., Farhi R. L., Han C. A. (1982) J. Steroid Biochem. 16, 613–616 [DOI] [PubMed] [Google Scholar]
- 14.Han A., Marandici A., Monder C. (1983) J. Biol. Chem. 258, 13703–13707 [PubMed] [Google Scholar]
- 15.Yamazaki T., Ohno T., Sakaki T., Akiyoshi-Shibata M., Yabusaki Y., Imai T., Kominami S. (1998) Biochemistry 37, 2800–2806 [DOI] [PubMed] [Google Scholar]
- 16.Imai T., Yamazaki T., Kominami S. (1998) Biochemistry 37, 8097–8104 [DOI] [PubMed] [Google Scholar]
- 17.Cali J. J., Russell D. W. (1991) J. Biol. Chem. 266, 7774–7778 [PubMed] [Google Scholar]
- 18.Strickler R. C., Tobias B., Covey D. F. (1981) J. Biol. Chem. 256, 316–321 [PubMed] [Google Scholar]
- 19.de Launoit Y., Simard J., Durocher F., Labrie F. (1992) Endocrinology 130, 553–555 [DOI] [PubMed] [Google Scholar]
- 20.Penning T. M., Burczynski M. E., Jez J. M., Hung C. F., Lin H. K., Ma H., Moore M., Palackal N., Ratnam K. (2000) Biochem. J. 351, 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alnouti Y., Klaassen C. D. (2008) Toxicol. Sci. 101, 51–64 [DOI] [PubMed] [Google Scholar]
- 22.Fan L. Q., Cattley R. C., Corton J. C. (1998) J. Endocrinol. 158, 237–246 [DOI] [PubMed] [Google Scholar]
- 23.Yokoi Y., Horiguchi Y., Araki M., Motojima K. (2007) FEBS J. 274, 4837–4847 [DOI] [PubMed] [Google Scholar]
- 24.Brereton P., Suzuki T., Sasano H., Li K., Duarte C., Obeyesekere V., Haeseleer F., Palczewski K., Smith I., Komesaroff P., Krozowski Z. (2001) Mol. Cell Endocrinol. 171, 111–117 [DOI] [PubMed] [Google Scholar]
- 25.Shafqat N., Marschall H. U., Filling C., Nordling E., Wu X. Q., Björk L., Thyberg J., Mårtensson E., Salim S., Jörnvall H., Oppermann U. (2003) Biochem. J. 376, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashibe B., Hirai T., Higashi K., Sekimizu K., Motojima K. (2007) J. Biol. Chem. 282, 20763–20773 [DOI] [PubMed] [Google Scholar]
- 27.Burczynski M. E., Lin H. K., Penning T. M. (1999) Cancer Res. 59, 607–614 [PubMed] [Google Scholar]
- 28.Yang J., Chen S., Huang L., Michalopoulos G. K., Liu Y. (2001) Hepatology 33, 848–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai C., Li Y., Yang J., Liu Y. (2003) J. Biol. Chem. 278, 27080–27087 [DOI] [PubMed] [Google Scholar]
- 30.Chen L., Woo S. L. (2005) Proc. Nat. Acad. Sci. U.S.A. 102, 15581–15586 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Dentin R., Liu Y., Koo S. H., Hedrick S., Vargas T., Heredia J., Yates J., 3rd, Montminy M. (2007) Nature 449, 366–369 [DOI] [PubMed] [Google Scholar]
- 32.Dentin R., Hedrick S., Xie J., Yates J., 3rd, Montminy M. (2008) Science 319, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 33.Goldstein D. S., Garty M., Bagdy G., Szemeredi K., Sternberg E. M., Listwak S., Pacak K., Deka-Starosta A., Hoffman A., Chang P. C., et al. (1993) J. Neuroendocrinol. 5, 475–486 [DOI] [PubMed] [Google Scholar]
- 34.Hanson E. S., Dallman M. F. (1995) J. Neuroendocrinol. 7, 273–279 [DOI] [PubMed] [Google Scholar]
- 35.Wahlestedt C., Skagerberg G., Ekman R., Heilig M., Sundler F., Håkanson R. (1987) Brain Res. 417, 33–38 [DOI] [PubMed] [Google Scholar]
- 36.Lundåsen T., Hunt M. C., Nilsson L. M., Sanyal S., Angelin B., Alexson S. E., Rudling M. (2007) Biochem. Biophys. Res. Commun. 360, 437–440 [DOI] [PubMed] [Google Scholar]
- 37.Chikahisa S., Tominaga K., Kawai T., Kitaoka K., Oishi K., Ishida N., Rokutan K., Séi H. (2008) Endocrinology 149, 5262–5271 [DOI] [PubMed] [Google Scholar]
- 38.Ishida N. (2009) PPAR Res. 2009, 412949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badman M. K., Pissios P., Kennedy A. R., Koukos G., Flier J. S., Maratos-Flier E. (2007) Cell Metab. 5, 426–437 [DOI] [PubMed] [Google Scholar]
- 40.Yang Q., Xie Y., Alexson S. E., Nelson B. D., DePierre J. W. (2002) Biochem. Pharmacol. 63, 1893–1900 [DOI] [PubMed] [Google Scholar]
- 41.Cunard R., DiCampli D., Archer D. C., Stevenson J. L., Ricote M., Glass C. K., Kelly C. J. (2002) J. Immunol. 169, 6806–6812 [DOI] [PubMed] [Google Scholar]
- 42.Liberman A. C., Druker J., Perone M. J., Arzt E. (2007) Cytokine Growth Factor Rev. 18, 45–56 [DOI] [PubMed] [Google Scholar]
- 43.Reisman D., Thompson E. A. (1995) Mol. Endocrinol. 9, 1500–1509 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.