Abstract
The target of rapamycin (TOR) is a conserved eukaryotic Ser/Thr kinase that regulates cellular growth in response to the nutrient and energy state. TOR signaling plays an important role in the development of diseases such as cancer, obesity, and diabetes and in different redox-sensitive processes (hypoxia, apoptosis, and aging). Because TOR has been detected at different cellular membranes and in the nucleus, its localization may influence the specific signaling readout. To better understand how TOR can associate with different membranes, the lipid-binding properties of the redox-sensitive yeast TOR1 FATC domain (y1fatc) have been characterized by solution NMR spectroscopy. Binding studies with different lipids indicate that y1fatc interacts specifically with a membrane-mimetic environment but appears not to recognize a specific lipid headgroup. In both, the structures of oxidized and reduced micelle-bound y1fatc, residues Ile-2456 to Trp-2470 of the lipid-binding motif form a hydrophobic bulb that has a rim of charged residues. The diffusion constants for both micelle-bound states are consistent with the rotational correlation times from the analysis of the 15N relaxation data. Based on the Kd values, the oxidized form (Kd ∼ 0.31 mm) binds dodecyl phosphocholine micelles slightly tighter than the reduced form (Kd ∼ 1.86 mm). Binding studies with y1fatc in which one or both tryptophans (Trp-2466 and Trp-2470) were replaced by alanine suggest that these residues are important for the exact positioning in the membrane and that the other aromatic (His-2462, Tyr-2463, and Phe-2469) and aliphatic residues (Ile-2456, Leu-2459, Ile-2464, and Pro-2468) in the lipid-binding motif contribute significantly to the affinity.
Keywords: Methods/NMR, Protein/Binding/Lipid, Protein/Structure, Signal Transduction, TOR, FATC, Bicelle, Dodecyl Phosphocholine, Membrane-mimetic, Micelle
Introduction
The target of rapamycin (TOR)2 is a conserved Ser/Thr kinase that regulates cell growth and metabolism of eukaryotic organisms in response to nutrients, growth factors, the cellular energy state, and stress (1, 2). TOR has been shown to function in two distinct molecular complexes termed TORC1 and -2 that integrate signals from several different effector pathways (1). TORC1 is specifically inhibited by the inhibitor complex rapamycin-FKBP12 (FK506-binding protein of 12 kDa), whereas TORC2 is not (1) and only dissociates if exposed to rapamycin for a long time (2, 3). TOR signaling plays a key role for the development of an organism and influences its aging and lifespan (1, 4, 5). Misregulation of TOR signaling is involved in diseases like obesity, diabetes, cancer, and cardiovascular disorders (6–10).
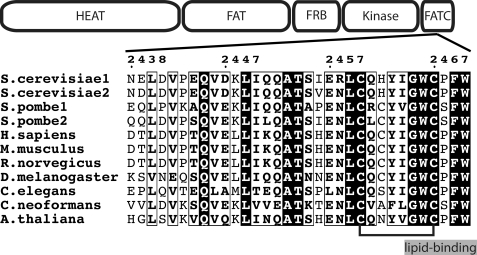
TOR proteins have a molecular mass of ∼280 kDa and share a modular structure (Fig. 1). The N-terminal HEAT repeat region and the following FAT domain were suggested to form long rod-like α-helical assemblies that mediate protein-protein interactions (11, 12). The FKBP12-rapamycin binding (FRB) domain is N-terminal of the catalytic domain. The C-terminal FATC domain occurs in all phosphoinositol kinase-related kinases in tandem with the FAT domain and has been shown to play an important role for the regulation of TOR function (12–14). The FATC domain has been highly conserved in organisms ranging from yeast to man (see Fig. 1). The solution structure of the oxidized form consists of an α-helix and a disulfide-bonded loop. Based on the additionally determined redox potential and in vivo mutagenesis data from yeast cells, it could be shown that the cellular stability of TOR is influenced by the redox state of the FATC domain (15). Subsequent studies by Sarbassov and Sabatini showed that signaling via the Raptor-mTOR complex is redox-regulated and that the oxidizing agent phenyl arsin oxide induced the dissociation of the purified Raptor-mTOR complex (16). TOR signaling has been shown to be involved in different redox-sensitive processes. Several studies suggested that the mTOR pathway positively regulates hypoxia-inducible factor 1-α and that hypoxia down-regulates mTORC1 signaling (reviewed in Ref. 2). mTOR further controls mitochondrial oxidative function and senses osmotic stress via mitochondrial dysfunction (17–19). Finally, there is evidence that TOR plays a role in apoptosis (20).
FIGURE 1.
Domain structure and sequence conservation of the TOR FATC domain. TOR proteins share several functional regions. The N-terminal region ∼1200 amino acids long is composed of HEAT repeats (huntingtin, elongation factor 3, and regulatory subunit A of PP2A) and is followed by the ∼550-residue FAT (FRAP, ATM, and TTRAP) domain. The ∼100-amino acid FKBP12-rapamycin binding (FRB) domain is N-terminal of the ∼250-amino acid Ser/Thr kinase domain. The highly conserved FATC (FAT C-terminal) domain encompasses ∼35 residues. The redox state of the two conserved cysteines Cys-2460 and Cys-2467 (connected by a black line) influences the cellular stability of TOR (15). The potential C-terminal lipid-binding motif (GWC … W) is highlighted. The numbering at the top refers to yeast TOR1 (= S. cerevisiae1). All sequence alignment representations were generated using the program ESPript (54).
It was suggested that the specific output of the TOR signaling network is influenced by the cellular localization of the two TOR complexes (21). Early studies in yeast indicated that TOR proteins are mainly localized at the plasma membrane and a second presumably vesicular fraction (22, 23). Based on a very recent study TORC1 localizes exclusively to the vacuolar membrane, whereas TORC2 localizes dynamically to a new plasma membrane domain named MCT (membrane compartment containing TORC2) (24). The results for the localization of mTOR are much more diverse. Initial studies with insulin-responsive cells showed that mTOR localized predominately to microsomal membranes with very little present in cytosol and plasma membrane fractions (25). Using different cell lines, mTOR was later detected in the endoplasmic reticulum and the Golgi fractions (21), in the nucleus (26), and in membrane raft structures (27). Several other studies showed that mTOR can associate with mitochondria (17, 18). Finally, TOR from female mosquito fat body cells has been observed to shift from the cytoplasm to the plasma membrane following a blood meal (28).
An analysis of the amino acid sequence of various phosphoinositol kinase-related kinases with the program eMotif, which was done by G. G. Tevzadze (University of Chicago),3 revealed a potential lipid-binding motif GWC … W in the C-terminal FATC domain (Fig. 1). Here, the NMR characterization of the interaction of the redox-sensitive FATC domain of yeast TOR1 (y1fatc) with different lipids; the structures, backbone dynamics, diffusion properties, and thereof derived dissociation constants of the oxidized and reduced forms bound to dodecyl phosphocholine (DPC) micelles; and the analysis of the effect of W → A mutations on the association with micelles are presented.
EXPERIMENTAL PROCEDURES
Protein Expression and NMR Sample Preparation
The FATC domain of yeast TOR1 (2438–2470, y1fatc) was expressed and purified as described previously (15). 1,2-Dioleoyl-sn-glycero-3-phosphate (DOPA), 1,2-dioctanoyl-sn-glycero-3-phosphate (Dioct-PA), 1,2-dihexanoyl-sn-glycero-3-(phosphoinositol-3,4,5-trisphosphate) (Dihex-PIP3), 1,2-diheptanoyl-sn-glycero-3-phosphocholine, and DPC were purchased from Avanti Polar Lipids. 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was bought from Genzyme Pharmaceuticals. Deuterated DPC (d38-DPC) was obtained from Cambridge Isotope Laboratories.
For the titrations of y1fatc with different lipids or lipid mixtures, the protein concentration ranged from 70–130 μm in 50 mm Tris, 100 mm NaCl, 95% H2O/5% D2O, pH 6.5. For the structure determinations the protein concentration in the different samples ranged between 0.4 and 0.5 mm. Lipid stock solutions for the titrations or NMR samples of y1fatc with a defined lipid concentration were prepared as follows. A defined amount of lipid from a concentrated stock in chloroform was placed in a glass vial and tried under a stream of argon gas. The dried lipid was then dissolved in buffer or a protein sample, and the pH was adjusted to 6.5. A sample of 15N-y1fatc with DMPC/DHepPC bicelles (q = 0.2, [DMPC] = 0.04 m, and [DHepPC] = 0.20 m) was prepared as follows. The needed amount of a DMPC stock solution in chloroform was placed in a glass vial and dried under a stream of argon gas. The dried lipid was then stepwise dissolved by adding more and more of the appropriate amount of a DHepPC stock solution in buffer, always followed by vigorous vortexing. Last, the protein solution was added, and the pH was adjusted to 6.5. The final protein concentration was 0.11 mm.
For the structure determination of oxidized y1fatc bound to DPC micelles, samples contained either 190–220 mm d38-DPC or 140 mm d38-DPC/10 mm DPC. For the structure determination of reduced y1fatc bound to DPC micelles, the protein was first reduced with 20 mm tris(2-carboxyethyl)phosphine for ∼10–30 min, before 170 mm d38-DPC were added.
For the diffusion measurements, the protein concentration was 0.063 mm for oxidized and 0.409 mm for reduced y1fatc with 170 mm DPC; 0.051 mm for both with 30 mm DPC; 0.409 mm for free oxidized y1fatc and 0.051 mm for the free reduced form. The diffusion properties of the micelle alone were determined from a sample of 170 mm d38-DPC.
NMR Spectroscopy
NMR spectra were acquired at 298 or 318 K on Bruker DRX600 and DRX800 spectrometers; the latter was equipped with a cryogenic probe. The data were processed with NMRPipe (29) and analyzed using NMRView (30). Assignments for the 13C, 15N, and 1H nuclei were based on three-dimensional HNCA, three-dimensional HCCH-TOCSY, and 15N- and 13C-edited NOESY spectra. Stereo-specific assignments for β-methylene and valine γ-methyl groups and dihedral angle information for the side chain angle χ1 were derived from three-dimensional HACAHB-COSY, three-dimensional HNHB, 13CO-{13Cγ}-SED-, and 15N-{13Cγ}-SED-15N-1H HSQC (only for oxidized y1fatc bound to DPC micelles), and 13C-{13CΟ}-SED- and 13C-{15Ν}-SED-13C-1H HSQC spectra. Distance restraints were obtained from 15N- and 13C-edited NOESY spectra. Information about the backbone dynamics was derived from the measurement of 15N relaxation experiments, including T1, T2, and {1H}-15N NOE. If not explicitly mentioned elsewhere, references for all experiments are given in Ref. 31. The diffusion properties of free and micelle-bound protein were determined using a pulsed-field gradient stimulated echo sequence (32) that contained additional water-suppression pulses.
Kd Determination
The dissociation constant for oxidized and reduced y1fatc bound to DPC micelles was determined using an approach similar to the one described in previous studies (33, 34). If the protein-micelle complex is on fast exchange on the diffusion time scale, the measured diffusion coefficient D is a weighted average given by: Dobs,p = fb,p·Db,p + (1 − fb)·Dfree,p, where fb,p is the fraction of micelle-bound protein. Dfree,p and Db,p are the diffusion constants of the free and micelle-bound protein that were determined at 0 and 170 mm DPC. Dobs,p is the diffusion constant at intermediate micelle concentrations, here 30 mm DPC. Resolving the equation for fb,p gives: fb,p = (Dobs,p − Dfree,p)/(Dbound,p − Dfree,p). The dissociation constant is described by: Kd = ([free protein]·[free micelle])/[protein-micelle complex] = ((1 − fb,p)·[free micelle])/fb,p. To calculate the Kd for the protein-micelle complex, fb,p was derived from the measured diffusion constants as described above (33, 34). The concentration of free micelles was estimated in the following way. Above the CMC, the concentration of free lipid is approximately equal to the CMC, which is 1.1 mm for DPC (35). Using an average aggregation number of ∼54 (36), the total concentration of DPC micelles ([mict]) can be estimated by: [mict] = ([DPCt] − CMC)/aggregation number. In our case Dobs,p was measured at 30 mm DPC, therefore the total micelle concentration was estimated to be (30 mm − 1.1 mm)/54 = 0.54 mm. Assuming a 1:1 stoichiometry of protein to micelle, the concentration of free DPC micelles ([micf]) can be calculated by: [micf] = [mict] − fb,p·[protein concentration].
Structure Calculation
All structure calculations were performed with XPLOR-NIH (37) using molecular dynamics in torsion angle and Cartesian coordinate space. Distance restraints were generated in NMRView (30) and classified according to NOE-cross-peak intensities. Upper bounds were 2.8 Å, 3.5 Å, 4.5 Å, and 5.5 Å. The lower bound was always 1.8 Å. For all NOE restraints r−6 sum averaging was used. Backbone dihedral angle restraints for ϕ and ψ were derived based on the determined 13Cα chemical shifts and on initial structure calculations. Based on 3JHαHβ2/3 and 3JNβ2/3 coupling constants and NOE data, side chain χ1 angles were restrained to one of the staggered conformations (60°, 180°, and −60°) ± 30°.
RESULTS
y1Fatc Binds Lipids Only above the CMC
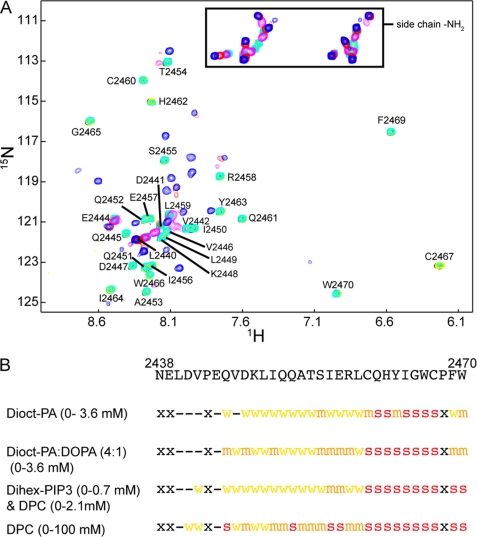
The interaction of oxidized y1fatc with different lipids was characterized by recording 1H,15N HSQC spectra as a function of the lipid concentration. Fig. 2A shows the result for DPC. Up to a DPC concentration of 0.76 mm no significant spectral changes could be observed. Only at the next titration point (1.58 mm), which is just above the CMC of DPC (1.1 mm), several resonances disappeared or were strongly weakened. The resonances that disappeared include not only the C-terminal region that harbors the potential lipid binding motif (Fig. 1) but basically the whole disulfide bonded loop from Ile-2456 to Trp-2470. At 3.27 mm DPC, only the most N-terminal resonances were still visible. At high DPC concentrations (100 mm) several new resonances appeared. At this condition the equilibrium between free and micelle-associated DPC was driven to the micelle state, which allowed detection of the micelle-bound y1fatc resonances.
FIGURE 2.
The FATC domain binds lipids above their CMC. A, 1H,15N HSQC spectrum of oxidized y1fatc in the presence of different concentrations of DPC (black, 0 mm; green, 0.16 mm; yellow, 0.40 mm; cyan, 0.76 mm; magenta, 1.58 mm; red, 3.27 mm; and blue, 117.23 mm). Assignments for the free form are indicated (15). NMR titrations of y1fatc were also performed with Dioct-PA, a 1:4 mixture of DOPA and Dioct-PA (0–4 mm), and with Dihex-PIP3/DPC. The 1H,15N HSQC spectra showing those titrations are given in supplemental Fig. 6. B, summary of the NMR titrations of y1fatc with different lipids. In all cases spectral changes were only observed above the estimated CMC. Residues that disappeared just above the CMC are marked with a red s. Residues that disappeared or shifted significantly at medium lipid concentration are labeled with an orange m, and those that disappeared or shifted at higher lipid concentrations are labeled with a yellow w. Residues that were not affected by the addition of lipids are marked with “-” and those that show no 1H,15N HSQC peak with “x.”
The interaction of y1fatc with phosphatidic acid (PA) and phosphoinositol 3,4,5-trisphosphate (PIP3) was characterized in the same manner. Both are negatively charged lipids that play important roles during signal transduction. PA has further been shown to influence TOR signaling through interactions with the FRB domain (38) and to regulate the assembly of the two TOR complexes (39). The spectral changes observed for dioctanoyl-PA (Dioct-PA) and a 4:1 mixture of Dioct-PA and DOPA were very similar (supplemental Fig. 6) to each other and to the one with DPC and are summarized in Fig. 2B. The 4:1 mixture of Dioct-PA:DOPA is expected to form micelles at a lower lipid concentration, because the CMC of DOPA is much smaller than that for Dioct-PA due the longer fatty acid chain (C18 versus C8). Accordingly, the titration with Dioct-PA showed strong spectral changes at ∼1.11 mm and that for 4:1 Dioct-PA:DOPA already at ∼0.39–0.74 mm. Values for the CMCs of the used PA molecules could not be found in the literature. However, the value for Dioct-PA should be in the same range as for dioctanoyl-PC (0.27 mm) and dioctanoylphosphoglycerol (1.21 mm) and that for DOPA alone should be similar or smaller than the values for dipalmitoyl-PC (0.46 nm) and dimyristoylphosphoglycerol (0.011 mm). All phospholipid CMC values were taken from a table provided on the website of Avanti Polar Lipids.
If y1fatc was titrated with dihexanoyl-PIP3 (Dihex-PIP3) to a concentration of ∼0.7 mm, there were no significant changes visible. Due to the short fatty acid chain length the CMC should be similarly high as for dihexanoyl-PC (15 mm). If in addition to Dihex-PIP3, DPC was added at a concentration of 0.58 mm or higher to induce micelle formation, the same resonances began to disappear as for the other tested lipids (Fig. 2 and supplemental Fig. 6).
In summary, y1fatc bound to all tested lipids above their CMC. This indicates that y1fatc does not recognize a specific lipid headgroup but binds specifically to a membrane-like environment.
The 1H-15N HSQC Spectra of Oxidized and Reduced y1fatc Bound to DPC Micelles Look very Different
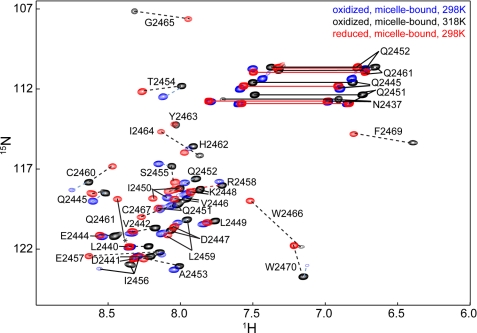
Based on the structural characterization of free oxidized y1fatc, it can form a regulatory disulfide bond between two conserved cysteines (Cys-2460, Cys-2467) (15). It was therefore analyzed if the redox state influences the interaction with DPC micelles. Fig. 3 shows a superposition of the NMR spectra of oxidized and reduced y1fatc in the presence of DPC at concentrations typically used for structural studies (∼150–250 mm) (40–42). The spectra of micelle-bound oxidized and reduced y1fatc appeared very different from each other (Fig. 3) and from the ones of the respective free forms (Fig. 2A and Ref. 15).
FIGURE 3.
Superposition of the 1H,15N HSQC spectra of DPC micelle-bound oxidized (blue, 298 K; black, 318 K) and reduced (red, 298 K) y1fatc. Black dotted lines indicate the chemical shift variation between the micelle-bound reduced form at 298 K and the micelle-bound form oxidized at 318 K. Blue dotted lines indicate the shift of peaks for micelle-bound oxidized y1fatc that is induced by raising the temperature from 298 to 318 K. Note that the resonances of residues 2463–2469 of micelle-bound oxidized y1fatc become only visible if the temperature is raised from 298 to 318 K.
For oxidized micelle-bound y1fatc, the number of peaks at 298 K was smaller than expected based on the number of exchangeable amide groups. Resonance assignment of the visible peaks revealed that those of the presumably membrane-bound stretch were very weak or not visible. Raising the temperature to 318 K made the missing resonances detectable. In contrast, reduced micelle-bound y1fatc shows already strong resonances for the membrane-binding region at 298 K. This suggests that the increased conformational flexibility in the C-terminal region of the reduced form influences the affinity and/or dynamic properties of the association with DPC micelles.
y1fatc Can also Associate with DMPC/DHepPC Bicelles
The top representation in supplemental Fig. 7 shows a superposition of the spectra of oxidized y1fatc in the free form, bound to DPC micelles and bound to DMPC/DHepPC bicelles at 298 K. The bottom representation shows a superposition of the micelle- and bicelle-bound states at 318 K. These data demonstrate that y1fatc can also bind to the rather planar lipid bilayer presented by the bicelles. The different fluidity and curvature of the two membrane-mimetic environments appear not to have a significant effect on the affinity, however, they may influence the immersion depth and/or angle, which may explain the observed spectral differences between micelle- and bicelle-bound y1fatc.
The NMR Structures of Micelle-bound Oxidized and Reduced y1fatc
Because the spectra of micelle-bound oxidized and reduced y1fatc looked very different from each other, the structures of both were determined by NMR spectroscopy. The structural statistics are given in Table 1. DPC micelles were used as a membrane-mimetic system, because they are stable over a broad temperature range and because they are well established for NMR structural studies of membrane-binding peptides and proteins (40–42).
TABLE 1.
Statistics for the 20 final structures of y1fatc bound to DPC micelles
None of the structures had distance restraints violations > 0.5 Å or dihedral angle violations > 5°. r.m.s.(d.) = root mean square (deviation); bb = backbone.
| Redox state and sample temperature | Oxidized, 318 K | Reduced, 298 K |
|---|---|---|
| Distance restraints | All (assigned + ambiguous) | All (assigned + ambiguous) |
| Total | 932 (857 + 75) | 1167 (1027 + 140) |
| 15N NOESY | 249 (217 + 32) | 311 (258 + 53) |
| Aliphatic 13C NOESY | 630 (590 + 40) | 781 (700 + 81) |
| Aromatic 13C NOESY | 53 (50 + 3) | 75 (69 + 6) |
| φ Angle restraints | 25 | 21 |
| ψ Angle restraints | 21 | 20 |
| χ1 Angle restraints | 15 | 16 |
| r.m.s.d. values from experimental restraints | ||
| Distance (Å) | 0.0424 ± 0.0012 | 0.0379 ± 0.0011 |
| Dihedral angle (°) | 0.458 ± 0.114 | 0.204 ± 0.049 |
| r.m.s.d. values from idealized geometry | ||
| Bonds | 0.0053 ± 0.0001 | 0.0050 ± 0.0001 |
| Angles | 0.610 ± 0.018 | 0.673 ± 0.011 |
| Improper | 0.430 ± 0.022 | 0.460 ± 0.018 |
| Lennard-Jones energy (kcal mol−1) | −89.1 ± 11.4 | −99.9 ± 8.9 |
| Procheck (57) statistics for residues 2442–2470, residues in | ||
| Most favored regions | 65.8 | 75.8 |
| Additional allowed regions | 33.8 | 24.2 |
| Generously allowed regions | 0.4 | 0 |
| Disallowed regions | 0 | 0 |
| Average r.m.s.d. to mean structure (Å) | ||
| Residues 2443–2470 (bb/heavy) | 1.35/1.97 | 0.70/1.05 |
| Residues 2443–2453 (bb/heavy) | 0.25/0.93 | 0.21/0.57 |
| Residues 2454–2470 (bb/heavy) | 0.73/1.52 | 0.33/0.81 |
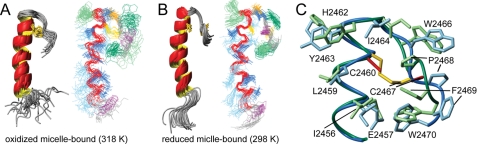
The structure of oxidized micelle-bound y1fatc (Fig. 4A) consists of the one of the free form of a long N-terminal α-helix and a disulfide-bonded loop that folds the protein chain back onto itself. The structure is overall well defined with 65.8% of the residues in the structured region (Val-2442 through Trp-2470) occupying the most favored region of the Ramachandran plot (Table 1, middle column). Compared with the free form, the α-helix extends not only from residue Glu-2444 to Cys-2460 but to His-2462. Further, it is significantly less well defined around residues Ala-2453 to Ser-2454. This is reflected in a rather high root mean square deviation for the full structured region (2443–2470, 1.35 Å) and a significantly lower one for only the helical region up to Ala-2453 (0.25 Å, Table 1, middle column). A very low propensity for α-helical secondary structure for residue Gln-2452 and rather low ones for residues Ala-2453 to Ser-2454 are evident from the secondary Cα-chemical shifts (supplemental Fig. 8) and from a lack of several α-helix typical NOEs in this region (e.g. 2455Hβ–2452Hα, 2456HN–2453Hα). Based on the titration with DPC (Fig. 2), this region may be at the micelle-solvent interface.
FIGURE 4.
The NMR structures of DPC-micelle bound oxidized (A) and reduced (B) y1fatc. Both plots show a ribbon representation of the superposition of the 20 lowest energy structures to the left and a line representation to the right. The side chains of the disulfide bonding forming cysteines (Cys-2460 and Cys-2467) are colored yellow. In the line representation the backbone of the α-helical stretch is depicted in red. Aromatic side chains are shown in light (F, H, Y) and dark (W) green, hydrophilic side chains in light blue, and those containing methyl groups in dark blue. Prolines are colored purple, and the conserved glycine (Gly-2465) that facilitates the chain reversal is in orange. C shows a superposition of the membrane-binding region of reduced (green, cysteine side chains in red) and oxidized (blue, cysteine side chains in yellow) y1fatc bound to DPC micelles. The side chains of Arg-2458 and Gln-2461 have been omitted for clarity. All structure images were made with the programs MolMol (55) and POV-Ray.
The structure of micelle-bound reduced y1fatc (Fig. 4B) is significantly better defined than that of the micelle-bound oxidized form (Fig. 4A), which is reflected in the larger number of detected NOE restraints, lower r.m.s.d. values, and better Procheck statistics (Table 1, right column). 75.8% of residues in the structured region occupy the most favored region of the Ramachandran plot. Compared with the free and micelle-bound oxidized forms the N-terminal α-helix extends even further to Tyr-2463. Similar to the oxidized micelle-bound form, but to a lower extent, the α-helix is less well defined around residue Gln-2452 consistent with the low α-helix propensity indicated by the 13Cα secondary shift (supplemental Fig. 8). In free reduced y1fatc, the region around the C-terminal loop undergoes conformational exchange, which broadens the respective resonances beyond detection (15). In the micelle-bound form the C terminus, although not restricted by a disulfide bond between Cys-2460 and Cys-2467, folds back onto the α-helix, thereby maintaining the hydrophobic interactions between Trp-2470 and Ile-2456 that were observed in free and micelle-bound oxidized y1fatc.
Comparison of the Structures of Micelle-bound Oxidized and Reduced y1fatc
The backbone dihedral angles of oxidized and reduced micelle-bound y1fatc target on average similar areas in the Ramachandran plot. However, those of the calculated ensemble of the oxidized form bound to DPC micelles occupy a larger ϕ/ψ space, which is reflected in increased conformational heterogeneity (Fig. 4, A and B). Also the side-chain conformations of Pro-2468, Phe-2469, and Trp-2470 are less well restrained by the NMR data for the oxidized form, which however could only be obtained at a 20 K higher temperature (318 K) than that for the reduced form (298 K). The influence of the temperature on the affinity and dynamics of the association reaction is discussed in the following two sections. There are further differences in the orientations of several side chains in the C-terminal loop (Fig. 4C). In the oxidized form the planar side of the aromatic ring of Trp-2470 faces the nearby side chain of Glu-2457, which is evident from the NOE data and from the low, due to ring current shifts, Hβ (1.20 ppm) and Hγ (1.83 ppm) chemical shifts of Glu-2457. In the reduced form the edges of the indole ring point toward Glu-2457 (Hβ 2.33/2.21 ppm, Hγ 2.41/2.29 ppm). Reduction of the single disulfide bond changes also the side-chain orientation of Cys-2467, which brings the sulfide group in proximity of the C-terminal carboxylate.
Backbone Dynamics of Oxidized and Reduced Micelle-associated y1fatc
Binding of y1fatc to a large object should alter the dynamic properties of the protein. The top part of supplemental Fig. 9 shows the 15N relaxation data for free oxidized (15) and micelle-bound oxidized and reduced y1fatc at 298 K. A comparison of the data for oxidized micelle-bound y1fatc at 298 and 318 K is shown in the bottom part. Average values for the dynamic parameters for the well structured region (Glu-2444 through Trp-2470) are listed in Table 2.
TABLE 2.
Average values for the for dynamic parameters derived from 15N relaxation data (supplemental Fig. 9) for the well structured region (Glu-2444 through Trp-2470) of y1fatc
| Sample | Free oxidized, 298 K | Micelle-bound reduced, 298 K | Micelle-bound oxidized, 298 K | Micelle-bound oxidized, 318 K |
|---|---|---|---|---|
| 15N T1 (ms) | 565 ± 72 | 837 ± 39 | 775 ± 37 | 610 ± 40 |
| 15N T2 (ms) | 96 ± 38 | 67 ± 8 | 57 ± 14 | 102 ± 34 |
| {1H},15N NOE | 0.20 ± 0.18 | 0.59 ± 0.10 | 0.53 ± 0.14 | 0.53 ± 0.12 |
| S2a | 0.64 ± 0.10 | 0.88 ± 0.03 | 0.87 ± 0.07 | 0.79 ± 0.05 |
| τc (ns)a | 1.65b | 10.1 | 10.5 | 6.5 |
a From the analysis of the 15N relaxation data with the program TENSOR2 (44) according to a model-free approach.
b For free oxidized y1fatc, the rotational correlation time was estimated based on the molecular weight.
Binding of reduced and oxidized y1fatc to DPC micelles results in an increase in the T1 times and {1H}-15N NOE values and in a decrease in the T2 times compared with free y1fatc (all at 298 K, Table 2). For free and membrane-bound y1fatc under all conditions, the structural and the 15N relaxation data indicate a random coil conformation for residues N-terminal of Val-2442, which is evident from the decrease of the {1H}-15N NOE and the increase of the T1 and T2 times. The range of the measured 15N relaxation parameters agrees with those reported for the small protein crambin bound to DPC micelles (41, 43). A more extended analysis of the data using the program TENSOR2 (44) and an isotropic diffusion tumbling model provide overall rotational correlation times (τc) of around 10 ns for the micelle-bound forms at 298 K (Table 2). Assuming a particle size of ≥23 kDa, 4 kDa for y1fatc and ≥19 kDa for a DPC micelle with an aggregation number of ≥54 (36), this fits well with the τc value estimated based on the molecular weight (≥9.6 ns). Thereby the 15N relaxation data confirm that both, oxidized and reduced y1fatc, associate well with membrane-mimetic DPC micelles.
Because several of the backbone 1H,15N signals of micelle-bound oxidized y1fatc are not visible in the 1H,15N HSQC spectra at 298 K (Fig. 3), a second set of 15N relaxation data was recorded at the same temperature used for the structure determination (318 K). The observed increase in T2 times (Table 2) and the reduction of the rotational correlation time are consistent with the estimated molecular weight and a temperature of 318 K (43).
The analysis of the 15N relaxation data with the program TENSOR2 (44) according to a model-free approach provided also the subnanosecond order parameter S2, the parameter Rex, which contains information about the contribution of conformational exchange to the relaxation process, and the parameter τe, which is a measure for internal mobility (supplemental Fig. 9, bottom three panels). The average S2 values (Table 2) for residues Glu-2444 to Trp-2470 for micelle-bound oxidized or reduced y1fatc indicate that both forms are overall well structured. Consistent with an increase in the temperature, the S2 values for micelle-bound oxidized y1fatc at 318 K are slightly lower. Micelle-bound oxidized y1fatc shows Rex contributions for residues Tyr-2463 to Trp-2466 in the data set recorded at 318 K. These are consistent with the lower structural precision in the disulfide-bonded loop of the oxidized micelle-bound form at 318 K (Fig. 4, A and B).
Diffusion Data for Micelle-associated Oxidized and Reduced y1fatc and Thereof Estimated Kd Values
The diffusion constants for oxidized and reduced y1fatc in the free form, in the partly and fully DPC micelle-bound form, as well as the value for the free DPC micelle are listed in Table 3. Consistent with the 15N relaxation data, the association of both oxidized and reduced y1fatc with DPC micelles lowered significantly the measured diffusion constants. The diffusion constant for reduced micelle-bound y1fatc is slightly lower than that for the oxidized micelle-bound form. This may be explained by a larger size and/or a different shape of the complex as well as by different association dynamics.
TABLE 3.
Diffusion coefficients determined from pulse-field gradient NMR measurements
| Temperature/y1fatc redox state | Free micellea | Free y1fatc | Partially micelle-bound y1fatcb | Micelle-bound y1fatca |
|---|---|---|---|---|
| m2/s | ||||
| 298 K/reduced | 1.10 × 10−10 | 1.53 × 10−10 | 1.32 × 10−10 | 0.59 × 10−10 |
| 298 K/oxidized | 1.10 × 10−10 | 1.80 × 10−10 | 1.13 × 10−10 | 0.72 × 10−10 |
| 318 K/oxidized | 1.91 × 10−10 | 3.57 × 10−10 | 3.04 × 10−10 | 1.15 × 10−10 |
a Measured in samples containing 170 mm DPC.
b Measured in samples containing 30 mm DPC.
The Kd values that were estimated from the analysis of the diffusion data are listed in Table 4. The Kd for the association of oxidized y1fatc with DPC micelles at 298 K (0.31 mm) is slightly lower than the one for the reduced form (1.86 mm). As expected for an exothermic association reaction, increasing the temperature from 298 to 318 K increased the Kd for the oxidized micelle-bound form. The slightly higher affinity of the oxidized compared with the reduced form for DPC micelles may be explained by a lower entropic cost for the accommodation of the well structured disulfide bonded loop in the micellar environment.
TABLE 4.
Thermodynamic parameters derived from the NMR-derived diffusion data (Table 3)
| Temperature/y1fatc redox state | Fraction of micelle-bound y1fatc (fb,p) at 30 mm DPCa | Total micelle concentration at 30 mm DPC ([mict])a,b | Fraction of free micelles at 30 mm DPC ([micf])a | Estimated dissociation constant (Kd)a |
|---|---|---|---|---|
| mm | mm | mm | ||
| 298 K/reduced | 0.22 | 0.54 | 0.524 | 1.86 |
| 298 K/oxidized | 0.62 | 0.54 | 0.504 | 0.31 |
| 318 K/oxidized | 0.22 | 0.54 | 0.524 | 1.86 |
a The used equations are given under “Experimental Procedures.”
b Assuming an aggregation number of 54 for the DPC micelles (36).
Derivation of a Model of the Micelle Association of Oxidized and Reduced y1fatc
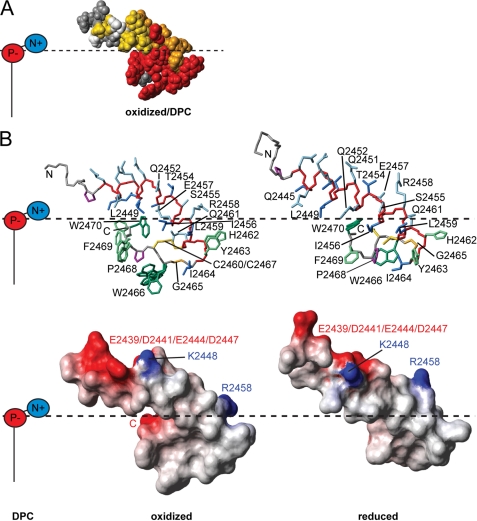
The titrations of oxidized y1fatc with DPC and other lipids (Fig. 2) indicate which residues form the interaction surface with the micelle. This information, together with the information about the distribution of surface charges, can be used to derive a model for the membrane association of y1fatc. The region that is embedded in the micelle and thereby makes contacts with the hydrophobic fatty acid tails consists of several aromatic amino acids (His-2462, Tyr-2463, Trp-2466, Phe-2469, and Trp-2470), some aliphatic residues (Ile-2456, Leu-2459, Ile-2464, and Pro-2468), Gly-2465, and the two cysteines (Cys-2460 and Cys-2467), either oxidized or reduced (Fig. 5B and supplemental Fig. 10). The interface region that is close to the polar headgroups contains two charged (Glu-2457 and Arg-2458) as well as some polar (Ser-2455 and Gln-2461) residues and the C-terminal carboxylate. The N-terminal mostly solvent-exposed part contains many polar and charged amino acids and is overall rather acidic.
FIGURE 5.
Model of y1fatc membrane association. A, for this representation, the results from the titration with DPC (Fig. 2) have been mapped onto a space-filled model of the structure of oxidized y1fatc bound to DPC micelles; the color coding is the same as in Fig. 2B. B, the top shows the lowest energy structure of micelle-bound oxidized and reduced y1fatc with the same color coding as in Fig. 4, A and B, and with the bonds between heavy atoms shown in a neon representation. For the side chains of Trp-2466 and Phe-2469 of micelle-bound oxidized y1fatc several conformers are shown to represent the structural heterogeneity observed in the ensemble (Fig. 4A). Below, the surface charge distribution of both structures was visualized. Positively charged areas are colored blue and negatively charged ones are red. DPC with its negatively charged phosphate and its positively charged choline group is schematically depicted to the left. The solvent-membrane interface that has been estimated based on the titrations with DPC and the distribution of surface charges is indicated by a dotted black line. Views of A and B, where the structures have been rotated by 180° around the vertical axes are given in supplemental Fig. 10, A and B. All structure images were made with the programs MolMol (55) and POV-Ray. Supplemental Fig. 10C shows further structure images of a DPC micelle (56) and oxidized micelle-bound y1fatc at the same scale.
The y1fatc Mutants W2466A and W2466A/W2470A Still Bind DPC Micelles
Tryptophans play an important role for the interaction of many membrane proteins with lipid bilayers and often favor a position at the interface of the aqueous solution and the lipid phase (45, 46). Even very short tryptophan-containing peptides can associate with lipid bilayers (45). The lipid-binding motif of y1fatc contains two conserved, sequentially close tryptophans (Trp-2466 and Trp-2470) that may be critical determinants for the affinity and specificity of membrane association. To test this hypothesis, two y1fatc mutant proteins were prepared, in which one or both tryptophans were replaced by alanine (W2466A and W2466A/W2470A). The 1H,15N HSQC spectra of the free forms indicate that both can still form the disulfide bond and maintain the overall fold of the wild-type protein (supplemental Fig. 11A). The ability of the mutant proteins to bind micelles was analyzed by titrating each protein in the oxidized form with DPC from 0 to ∼100 mm (supplemental Fig. 11B). Both mutant proteins showed as the wild type strong chemical shift changes above the CMC of DPC (1.1 mm) and a spectrum different from the free form with well dispersed peaks at high DPC concentrations. Based on the number of peaks, the spectra of the micelle-bound oxidized forms of the mutant proteins show the resonances for most if not all residues in the membrane-binding C-terminal loop at 298 K and not as the wild type only at higher temperatures (318 K). Reduction of the disulfide bond results, as for the wild type, in stronger resonances for several residues (supplemental Fig. 11, C and D). In summary, replacement of one or both tryptophans by alanine is not sufficient to abrogate micelle binding.
DISCUSSION
Recent studies demonstrated that TOR can associate with different cellular membranes (21–25, 27, 28) and mTOR has even been detected in the nucleus (26). The cellular localization of TOR was suggested to influence the specific outcome of TOR signaling (21), however it is not clear how TOR localization is regulated in response to various stimuli and with respect to the different composition of the two TOR complexes. The presented NMR binding studies of the yeast TOR1 FATC domain (y1fatc) with different lipids (Fig. 2 and supplemental Fig. 6) indicate that it specifically interacts with a membrane-mimetic environment but appears not to recognize a specific lipid headgroup.
It has been observed that mTOR associates with ER membranes tighter than other peripheral membrane proteins (21). It was further shown that the HEAT repeats mediate the localization of yeast TOR2 in the P13 (plasma membrane, ER, vacuole, and mitochondria) and P100 (Golgi, endosomes, and secretory vesicles) fractions. However, a C-terminal fragment encompassing part of the FAT, the FRB, the kinase, and the FATC domain was still able to localize to the P13 fraction. These observations would be consistent with the possibility that TOR cannot only associate with the ER, mitochondrial, and other cellular membranes by protein-protein interactions, but additionally via the here detected lipid-binding motif in the FATC domain.
In the structures of oxidized and reduced y1fatc bound to DPC micelles residues Ile-2456 through Trp-2470 form a hydrophobic bulb that has a rim of charged residues (Glu-2457 and Arg-2458) that can interact with the positively charged choline and the negatively charged phosphate groups of DPC (Fig. 5 and supplemental Fig. 10). The TOR FATC membrane anchor has thereby a similar buildup as the one of the C2 domains of the peripheral membrane protein Lactadherin (47). However, the hydrophobic and aromatic amino acids of the C2 membrane-binding motif are not provided from a single larger loop as is the case for y1fatc, but from three short loops named “spike1–3” (47).
The NMR characterization of the membrane association of TOR was done using the isolated FATC domain, which raises the question of whether this domain is accessible for membrane interactions in the context of the full-length protein. Based on a recent electron microscopy structure of yeast TOR1, the FATC domain was assigned to a region that sticks out like a “finger” and thereby appears amenable to interactions with cellular components (48). The same publication contained also the electron microscopy structure of yeast TOR1 in complex with KOG1, the yeast analog of Raptor (48). The low resolution (∼25 Å) of both electron microscopy structures made it difficult to precisely assign the different functional regions, especially the location of the very small FATC domain in the TOR1-KOG1 complex structure. Based on the provided electron microscopy structures, it can therefore not be clearly determined whether the FATC domain may be accessible for membrane interactions in the complex with KOG1/Raptor.
It has been shown that the interaction with the mTORC1-specific protein Raptor is redox-sensitive and that this may be connected to the redox state of the FATC domain (16). Changing the composition and the ratio of the two TOR complexes may be one possibility to change the cellular localization of TOR. Immunofluorescence experiments on whole fixed cells indicate that mutation of the second of the disulfide bond-forming cysteines did not alter the cellular localization of TOR in yeast (15). However, the C → S mutation lowered its cellular stability. Moreover, yeast TORC1 localizes exclusively to the vacuolar membrane, whereas TORC2 localizes dynamically to a new plasma membrane compartment (24). Cellular redox signals may influence the composition and ratio of the two TOR complexes and thereby also the amount of the yeast TOR1 and -2 at the two localization sites.
The Kd values (Table 4) that were estimated from NMR-diffusion measurements (Table 3) indicate that the TOR FATC domain binds DPC micelles with low millimolar affinity (Table 4). Using a similar approach, the equilibrium constants for the association of different leucine- and methionine-enkephalin peptides (34) and of the tripeptide GHG (33) with SDS micelles had been determined. Calculating the reciprocal and dividing it by the aggregation number for SDS micelles (= 51 (49)) gives the Kd values for the association with SDS micelles, which range for the different leucine-enkephalin peptides from 0.385 to 0.008 mm and for the different methionine-enkephalin peptides from 0.754 to 0.029 mm. Accordingly, the Kd for the GHG tripeptide is 1.153 mm. Better comparable to the FATC domain is the FYVE domain, which also binds membrane-mimetic environments peripherally and with very similar affinity. Based on NMR-titration data the Kd for the association of the FYVE domain with DPC micelles is 7 ± 1.4 mm (40). The presence of phosphatidylinositol 3-phosphate increases the membrane-affinity of the FYVE domain ∼6-fold (Kd = 1.1 mm ± 0.2 mm) (40). Because the cellular localization of TOR varies in response to different stimuli, the TOR FATC domain is not expected to anchor TOR to a specific cellular membrane with constantly high affinity but rather to regulate the membrane association of TOR. Similar to the FYVE domain, the affinity of the TOR FATC domain for specific cellular membranes may be altered by changes in the membrane composition. Alternatively, the affinity of TOR for a specific cellular membrane may be further increased by the presence of certain TOR ligands.
Based on the NMR-derived Kd values (Table 4) the oxidized form binds DPC micelles slightly better than the reduced form. Because, however, both redox states bind micelles with overall similar affinities (Table 4), modulation of the redox state of the membrane-associated form may allow regulation of the interaction of TOR with other membrane proteins and thereby influence its localization at different cellular membranes, e.g. at the “redox-active” mitochondrial outer membrane (17, 18). It has already been shown that several TOR interaction partners and regulators such as lipidated Rheb (50) and Avo1 (24) or phospholipase D2 (51), as well as many TOR target proteins (24), are membrane-associated. The redox state of the membrane-associated TOR FATC domain may further be sensitive to lipid oxidation products, the concentration of which appears to increase upon aging (52).
Replacement of one or both tryptophans in the membrane-binding motif of y1fatc by alanine was not sufficient to abrogate micelle binding. An analysis of the localization of tryptophans in membrane proteins indicates a preference of tryptophan for the interface between the polar aqueous environment and the more apolar interior of the membrane (46). This was further confirmed by binding studies using different membrane-mimetic systems and tryptophan alone or short tryptophan peptides (2–3 residues) (46). Alkylation of the indole nitrogen, which destroys the hydrogen-bonding capability and increases the hydrophobicity, resulted in stronger interactions with lipid bilayers by enabling a deeper insertion into the apolar interior (46). Similar effects were observed with gramicidin A, a 15-residue tryptophan-rich peptide, and its N-methyltryptophan derivatives (45). Therefore, the two tryptophans of the membrane-binding motif of the TOR FATC domain may be important for the exact positioning in the membrane and possibly omit a membrane insertion being too deep.
Mutation of Trp-2545 (= Trp-2466 of yeast TOR1) to glycine or phenylalanine in rapamycin-resistant mTOR resulted in a loss of the autophosphorylation activity and the kinase activity toward two TOR substrates (4EBP1, p70 S6 kinase α) (13). In another study it was observed that deletion of the C-terminal tryptophan (Trp-2549 of mTOR = Trp-2470 of yeast TOR1) in rapamycin-resistant mTOR was enough to destroy the ability of mTOR to protect p70 S6 kinase α from rapamycin-induced dephosphorylation (14). Whether the effects of the mTOR W2545F/G mutants and the Trp-2549 deletion mutant are related to the membrane-binding properties of the TOR FATC domain or whether these mutations affect only the interactions with other domains of TOR or with TOR interaction partners or the cellular stability of TOR cannot be clarified based on the current data (13).
The lipid-binding data of the y1fatc W2466A and W2466A/W2470A mutants indicate that the other aromatic and aliphatic residues in the membrane anchor contribute significantly to the affinity. This is consistent with the observation that most of the respective amino acids show, as tryptophan, a positive free energy contribution for the transfer of a model peptide from a lipid bilayer to water (53). Therefore, more intensive mutagenesis studies in vitro must be performed to find mutations that impair the ability of the FATC domain to bind membrane-mimetic systems and that can be incorporated in full-length TOR to characterize the role of FATC membrane association and its potential redox regulation in vivo.
In conclusion, the presented NMR data suggest that TOR cannot only associate with biological membranes via protein-protein interactions or the association of the FRB domain with phosphatidic acid but, further, by a lipid-binding motif in its C-terminal FATC domain.
Supplementary Material
Acknowledgments
I thank Dr. Gela G. Tevzadze from Prof. Dr. Rochelle Esposito's group at the University of Chicago, for bringing the potential lipid-binding motif in the FATC domain to my attention and for showing me some of his results. I thank Klara Rathgeb-Szabo for help with the protein purification. I thank Prof. Dr. Mike Hall for stimulating discussions and methodological support in the initial stage of the structural characterization of the TOR FATC domain. Finally, I am very grateful to Prof. Dr. Stephan Grzesiek for his continuous support throughout the last years.
This work was supported in part by the Swiss National Science Foundation through a grant (SNF Grant 31-109712) to Stephan Grzesiek and by the German Research Foundation (Deutsche Forschungsgemeinschaft Grant DA 1195/3-1 to S. A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 6–11 and additional Refs. 1–3.
The atomic coordinates and structure factors (codes 2kio and 2kit) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The NMR restraints used have been deposited at the Biological Magnetic Resonance Databank (accession nos. 16284 and 16295 for oxidized y1fatc bound to DPC micelles at 318 K and reduced y1fatc bound to DPC micelles at 298 K, respectively).
G. G. Tevzadze, University of Chicago, personal communication.
- TOR
- target of rapamycin
- mTOR
- mammalian TOR
- CMC
- critical micelle concentration
- Dihex-PIP3
- 1,2-dihexanoyl-sn-glycero-3-(phosphoinositol 3,4,5-trisphosphate)
- DHepPC
- 1,2-diheptanoyl-sn-glycero-3-phosphocholine
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- Dioct-PA
- 1,2-dioctanoyl-sn-glycero-3-phosphate
- DOPA
- 1,2-dioleoyl-sn-glycero-3-phosphate
- DPC
- dodecyl phosphocholine
- FAT
- FRAP, ATM, TTRAP
- HSQC
- heteronuclear single quantum coherence
- NOE
- nuclear Overhauser Effect
- NOESY
- NOE spectroscopy
- r.m.s.d.
- root mean square deviation
- PIP3
- phosphoinositol 3,4,5-trisphosphate
- d38-DPC
- deuterated DPC
- y1fatc
- yeast TOR1 FATC domain
- FRB
- FKBP12-rapamycin binding
- PA
- phosphatidic acid
- SED
- spin-echo difference.
REFERENCES
- 1.Polak P., Hall M. N. (2009) Curr. Opin. Cell. Biol. 21, 209–218 [DOI] [PubMed] [Google Scholar]
- 2.Dunlop E. A., Tee A. R. (2009) Cell. Signal. 21, 827–835 [DOI] [PubMed] [Google Scholar]
- 3.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 4.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 5.Blagosklonny M. V. (2008) Cell Cycle 7, 3344–3354 [DOI] [PubMed] [Google Scholar]
- 6.Hall M. N. (2008) Transplant. Proc. 40, Suppl. 1, S5–S8 [DOI] [PubMed] [Google Scholar]
- 7.Lane H. A., Breuleux M. (2009) Curr. Opin. Cell. Biol. 21, 219–229 [DOI] [PubMed] [Google Scholar]
- 8.Jaworski J., Sheng M. (2006) Mol. Neurobiol. 34, 205–219 [DOI] [PubMed] [Google Scholar]
- 9.Dann S. G., Selvaraj A., Thomas G. (2007) Trends Mol. Med. 13, 252–259 [DOI] [PubMed] [Google Scholar]
- 10.Sabatini D. M. (2006) Nat. Rev. Cancer 6, 729–734 [DOI] [PubMed] [Google Scholar]
- 11.Groves M. R., Barford D. (1999) Curr. Opin. Struct. Biol. 9, 383–389 [DOI] [PubMed] [Google Scholar]
- 12.Bosotti R., Isacchi A., Sonnhammer E. L. (2000) Trends Biochem. Sci. 25, 225–227 [DOI] [PubMed] [Google Scholar]
- 13.Takahashi T., Hara K., Inoue H., Kawa Y., Tokunaga C., Hidayat S., Yoshino K., Kuroda Y., Yonezawa K. (2000) Genes Cells 5, 765–775 [DOI] [PubMed] [Google Scholar]
- 14.Peterson R. T., Beal P. A., Comb M. J., Schreiber S. L. (2000) J. Biol. Chem. 275, 7416–7423 [DOI] [PubMed] [Google Scholar]
- 15.Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S. (2005) J. Biol. Chem. 280, 20558–20564 [DOI] [PubMed] [Google Scholar]
- 16.Sarbassov D. D., Sabatini D. M. (2005) J. Biol. Chem. 280, 39505–39509 [DOI] [PubMed] [Google Scholar]
- 17.Desai B. N., Myers B. R., Schreiber S. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4319–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schieke S. M., Phillips D., McCoy J. P., Jr., Aponte A. M., Shen R. F., Balaban R. S., Finkel T. (2006) J. Biol. Chem. 281, 27643–27652 [DOI] [PubMed] [Google Scholar]
- 19.Cunningham J. T., Rodgers J. T., Arlow D. H., Vazquez F., Mootha V. K., Puigserver P. (2007) Nature 450, 736–740 [DOI] [PubMed] [Google Scholar]
- 20.Asnaghi L., Bruno P., Priulla M., Nicolin A. (2004) Pharmacol. Res. 50, 545–549 [DOI] [PubMed] [Google Scholar]
- 21.Drenan R. M., Liu X., Bertram P. G., Zheng X. F. (2004) J. Biol. Chem. 279, 772–778 [DOI] [PubMed] [Google Scholar]
- 22.Kunz J., Schneider U., Howald I., Schmidt A., Hall M. N. (2000) J. Biol. Chem. 275, 37011–37020 [DOI] [PubMed] [Google Scholar]
- 23.Wedaman K. P., Reinke A., Anderson S., Yates J., 3rd, McCaffery J. M., Powers T. (2003) Mol. Biol. Cell 14, 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berchtold D., Walther T. C. (2009) Mol. Biol. Cell 20, 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Withers D. J., Ouwens D. M., Nave B. T., van der Zon G. C., Alarcon C. M., Cardenas M. E., Heitman J., Maassen J. A., Shepherd P. R. (1997) Biochem. Biophys. Res. Commun. 241, 704–709 [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Shu L., Hosoi H., Murti K. G., Houghton P. J. (2002) J. Biol. Chem. 277, 28127–28134 [DOI] [PubMed] [Google Scholar]
- 27.Partovian C., Ju R., Zhuang Z. W., Martin K. A., Simons M. (2008) Mol. Cell 32, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen I. A., Attardo G. M., Roy S. G., Raikhel A. S. (2005) J. Biol. Chem. 280, 20565–20572 [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 30.Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 31.Grzesiek S., Bax A., Hu J. S., Kaufman J., Palmer I., Stahl S. J., Tjandra N., Wingfield P. T. (1997) Protein Sci. 6, 1248–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas L. H., Larive C. K. (2004) Concepts Magn. Reson. A 20A, 24–41 [Google Scholar]
- 33.Orfi L., Lin M., Larive C. K. (1998) Anal. Chem. 70, 1339–1345 [DOI] [PubMed] [Google Scholar]
- 34.Begotka B. A., Hunsader J. L., Oparaeche C., Vincent J. K., Morris K. F. (2006) Magn. Reson. Chem. 44, 586–593 [DOI] [PubMed] [Google Scholar]
- 35.Lauterwein J., Bösch C., Brown L. R., Wüthrich K. (1979) Biochim. Biophys. Acta 556, 244–264 [DOI] [PubMed] [Google Scholar]
- 36.Lazaridis T., Mallik B., Chen Y. (2005) J. Phys. Chem. B 109, 15098–15106 [DOI] [PubMed] [Google Scholar]
- 37.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 38.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. (2001) Science 294, 1942–1945 [DOI] [PubMed] [Google Scholar]
- 39.Toschi A., Lee E., Xu L., Garcia A., Gadir N., Foster D. A. (2009) Mol. Cell. Biol. 29, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutateladze T. G., Capelluto D. G., Ferguson C. G., Cheever M. L., Kutateladze A. G., Prestwich G. D., Overduin M. (2004) J. Biol. Chem. 279, 3050–3057 [DOI] [PubMed] [Google Scholar]
- 41.Ahn H. C., Juraniæ N., Macura S., Markley J. L. (2006) J. Am. Chem. Soc. 128, 4398–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanga R. P., Brender J. R., Xu J., Hartman K., Subramanian V., Ramamoorthy A. (2009) J. Am. Chem. Soc. 131, 8252–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seewald M. J., Pichumani K., Stowell C., Tibbals B. V., Regan L., Stone M. J. (2000) Protein Sci. 9, 1177–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dosset P., Hus J. C., Blackledge M., Marion D. (2000) J. Biomol. NMR. 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 45.Sun H., Greathouse D. V., Andersen O. S., Koeppe R. E., 2nd. (2008) J. Biol. Chem. 283, 22233–22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W., Caffrey M. (2006) Biochemistry 45, 11713–11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao C., Novakovic V. A., Head J. F., Seaton B. A., Gilbert G. E. (2008) J. Biol. Chem. 283, 7230–7241 [DOI] [PubMed] [Google Scholar]
- 48.Adami A., García-Alvarez B., Arias-Palomo E., Barford D., Llorca O. (2007) Mol. Cell 27, 509–516 [DOI] [PubMed] [Google Scholar]
- 49.Thèvenot C., Grassl B., Bastiat G., Binana W. (2005) Colloids Surf. A: Physicochem. Engineer. Aspects 252, 105–111 [Google Scholar]
- 50.Buerger C., DeVries B., Stambolic V. (2006) Biochem. Biophys. Res. Commun. 344, 869–880 [DOI] [PubMed] [Google Scholar]
- 51.Ha S. H., Kim D. H., Kim I. S., Kim J. H., Lee M. N., Lee H. J., Kim J. H., Jang S. K., Suh P. G., Ryu S. H. (2006) Cell. Signal. 18, 2283–2291 [DOI] [PubMed] [Google Scholar]
- 52.Almeida M., Ambrogini E., Han L., Manolagas S. C., Jilka R. L. (2009) J. Biol. Chem. 284, 27438–27448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wimley W. C., White S. H. (1996) Nat. Struct. Biol. 3, 842–848 [DOI] [PubMed] [Google Scholar]
- 54.Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 56.Tieleman D. P., van der Spoel D., Berendsen H. J. (2000) J. Phys. Chem. B. 104, 6380–6388 [Google Scholar]
- 57.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR. 8, 477–486 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.