Abstract
The Hermansky-Pudlak syndrome (HPS) is a genetic hypopigmentation and bleeding disorder caused by defective biogenesis of lysosome-related organelles (LROs) such as melanosomes and platelet dense bodies. HPS arises from mutations in any of 8 genes in humans and 16 genes in mice. Two of these genes, HPS1 and HPS4, encode components of the biogenesis of lysosome-related organelles complex-3 (BLOC-3). Herein we show that recombinant HPS1-HPS4 produced in insect cells can be efficiently isolated as a 1:1 heterodimer. Analytical ultracentrifugation reveals that this complex has a molecular mass of 146 kDa, equivalent to that of the native complex and to the sum of the predicted molecular masses of HPS1 and HPS4. This indicates that HPS1 and HPS4 interact directly in the absence of any other protein as part of BLOC-3. Limited proteolysis and deletion analyses show that both subunits interact with one another throughout most of their lengths with the sole exception of a long, unstructured loop in the central part of HPS4. An interaction screen reveals a specific and strong interaction of BLOC-3 with the GTP-bound form of the endosomal GTPase, Rab9. This interaction is mediated by HPS4 and the switch I and II regions of Rab9. These characteristics indicate that BLOC-3 might function as a Rab9 effector in the biogenesis of LROs.
Keywords: Cell/trafficking, Diseases/Genetic, Membrane/Trafficking, Protein/Assembly, Protein/Protein-Protein Interactions, Subcellular Organelles/Endosomes
Introduction
The Hermansky-Pudlak syndrome (HPS)3 is a group of rare autosomal recessive diseases (OMIM 203300) that are characterized by reduced pigmentation of the eyes and skin (e.g. oculocutaneous albinism) in combination with prolonged bleeding (e.g. bleeding diathesis) (Ref. 1, reviewed in Ref. 2). These symptoms result from respective defects in melanosomes and platelet dense bodies, which are members of the diverse family of “lysosome-related organelles” (LROs) (2–4). Some HPS patients present additional symptoms, including pulmonary fibrosis and granulomatous colitis, which probably result from dysfunction of other LROs (2). Genetic studies have shown that HPS is caused by mutations in any of 8 genes in humans and 16 genes in mouse (2, 5, 6). Some of these HPS genes encode subunits of well-characterized cytosolic complexes (e.g. AP-3, HOPS) that participate in vesicular transport steps in the endosomal-lysosomal system (7). Other HPS genes, however, encode novel proteins of unknown structure and function (7). Biochemical analyses have shown that a subset of these proteins are components of three additional cytosolic complexes named biogenesis of lysosome-related organelles complex-1, -2, and -3 (BLOC-1, -2, and -3) (7). Like AP-3 and HOPS, the BLOCs are expected to mediate vesicular transport events in the biogenesis of LROs. However, their precise molecular functions remain poorly understood.
The simplest of these complexes, BLOC-3, comprises two cytosolic proteins named HPS1 and HPS4 (8–10). Mutations in HPS1 (11) are the most frequent cause of HPS (2). This is particularly the case in Puerto Rico, where a founder effect makes HPS1 the most common single gene inherited disorder in the general population (2, 11, 12). HPS1 and HPS4 are the only types of HPS that present with lung fibrosis, a symptom that often leads to mortality in the fourth or fifth decade of life (2). Although the most visible manifestations of HPS1 or HPS4 deficiency derive from LRO defects, both proteins are expressed in all cell types, including those that normally lack LROs (11, 13, 14). Human HPS1 has 700 amino acids and a predicted molecular mass of 79 kDa (11), whereas human HPS4 has 708 amino acids and a predicted molecular mass of 77 kDa (14). Their similar sizes notwithstanding, HPS1 and HPS4 exhibit no significant sequence homology to each other (18% overall identity with 39.5% gaps, according to the EMBOSS Needle program). Recent bioinformatics analyses have suggested that the N-terminal ∼200 amino acids of both proteins might encompass a CHiPS (15) or longin (16) domain, but this finding remains to be verified experimentally.
The biochemical evidence that HPS1 and HPS4 are part of BLOC-3 comes mainly from experiments involving co-immunoprecipitation from whole cell extracts (8–10). In addition, combined sedimentation velocity and gel filtration analyses of whole cell extracts showed that both proteins migrate together as part of a 140–175-kDa species (9, 10, 17), roughly equivalent to the sum of the HPS1 and HPS4 molecular masses. However, only a small fraction of epitope-tagged forms of HPS1 and HPS4 expressed by transfection into cells occur as a complex, the rest remaining as unassembled proteins (9). Moreover, yeast two-hybrid (Y2H) assays failed to show interaction of HPS1 with HPS4 (8–10). These observations led to speculation that an additional small subunit might be required for assembly of these proteins. Adding to the uncertainty as to the oligomerization state of these proteins, Y2H assays showed an interaction of HPS4 with itself (8), suggesting that BLOC-3 could have more than copy of this protein. Finally, additional species of ∼200 kDa containing only HPS1 and ∼500 kDa containing both HPS1 and HPS4 were also detected in gel filtration analyses (8, 18). To date, no other proteins have been found to interact with HPS1 and HPS4 (6). Thus, the exact composition of BLOC-3 and its connection to other cellular components remain to be established.
To determine the composition and properties of BLOC-3, we produced the recombinant complex by co-expression of human HPS1 and HPS4 in insect cells using a baculovirus system. We found that this complex can be efficiently isolated as a monodisperse assembly of HPS1 and HPS4 in the absence of any other protein. Analytical ultracentrifugation analyses showed that this recombinant complex has a molecular mass of 146 ± 5 kDa, similar to that of the native complex (9, 10, 17). This size is consistent with BLOC-3 being a 1:1 heterodimer of HPS1 and HPS4. Deletion analyses showed that assembly involves the entire HPS1 and HPS4 sequences, with the sole exception of a long central loop in HPS4. Both subunits are thus intimately connected with one another throughout most of their length. An interaction screen revealed that both the recombinant and native BLOC-3 specifically interact with the GTP-bound form of Rab9, a small GTPase localized to endosomes (19). These observations raise the possibility that BLOC-3 could function as a Rab9 effector.
EXPERIMENTAL PROCEDURES
Expression and Reconstitution of BLOC-3
For expression of BLOC-3 using the baculovirus system, the genes for full-length human HPS4 carrying an N-terminal GST tag followed by a TEV site and untagged human HPS1 were ligated into the PH and p10 cassettes of pFastBacDual (Invitrogen, Carlsbad, CA), respectively. Recombinant baculovirus was generated using Escherichia coli DH10multibac cells, which contain a viral genome lacking chitinase and cathepsin genes for increased expression (20). Subsequent steps of baculovirus amplification were performed following the procedure in the Bac-to-Bac system manual (Invitrogen). For protein production, Sf9 cells in Sf-900 III serum-free medium (Invitrogen) were infected at a density of 2 × 106 cells/ml and incubated for 72 h at 27 °C before harvesting. Cells were washed in phosphate-buffered saline containing 5 mm benzamidine and frozen at −80 °C. The frozen cell pellets were lysed by resuspension in buffer A (50 mm HEPES, 150 mm NaCl, 50 mm KCl, 1 mm TCEP, 5% (w/v) glycerol, and 1 mm EDTA pH 7.3) containing DNase I (Sigma) and protease inhibitor mixture (Sigma). After lysis, cell debris was removed by centrifugation (43,000 × g, 40 min) and the supernatant applied to glutathione-Sepharose 4B (GE Healthcare, Piscataway, NJ) equilibrated in buffer A. After 2 h, the resin was washed twice in lysis buffer and twice in buffer B (50 mm HEPES, 250 mm NaCl, 1 mm EDTA, 1 mm TCEP, 0.6% (w/v) β-octylglucopyranoside (β-OG) pH 7.3). Bound proteins were eluted in buffer B containing 20 mm reduced glutathione. The GST tag was removed with TEV protease and BLOC-3 purified further by gel filtration on a Superose 6 column (GE Healthcare) equilibrated in buffer B.
BLOC3(ΔHPS4275–533) was produced by replacing residues 269–275 and 528–534 of HPS4 with TEV recognition sites using site-directed mutagenesis, allowing residues 275–533 of HPS4 to be removed together with the N-terminal GST tag during TEV cleavage.
For E. coli expression of truncated forms of HPS4, the corresponding DNA fragments were ligated into pGST2 (21) and transformed into E. coli BL21(DE3). Cells were grown at 20 °C in ZYM-5052 autoinduction medium (22) containing 100 μg/ml ampicillin and harvested by centrifugation when the culture reached saturation. Cell pellets were resuspended in buffer A, lysed using an Emulsiflex homogenizer (Avestin, Ottawa, Canada) and cleared by centrifugation (40 min, 43,000 × g). Soluble GST fusion proteins were purified further as described for the full-length complex.
BLOC-3 Antibody
A rabbit polyclonal antibody to BLOC-3 was generated using the purified full-length recombinant complex for immunization (Covance, Denver, PA). The antibody was subsequently affinity purified using recombinant BLOC-3 immobilized on NHS-Sepharose (GE Healthcare) following the manufacturer's instructions.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted at 4.0 °C on a Beckman Optima XL-A analytical ultracentrifuge. 400 μl of a 2.7 μm solution of BLOC-3 in buffer B was loaded in a double sector centerpiece cell. 130 scans were acquired at 5-min intervals and rotor speeds of 50 krpm as single absorbance measurements at 280 nm using a radial spacing of 0.003 cm. Data were analyzed in SEDFIT version 11.8 (20) in terms of a continuous c(s) distribution. Solution densities ρ and viscosities η were calculated using the program SEDNTERP 1.2 (John Philo, Software Consultant, Thousand Oaks, CA) (23), as were values for the partial specific volume v of the protein. c(s) analyses were carried out using an s20,w range of 2 to 12 with a linear resolution of 100 and a confidence level (F-ratio) of 0.68; excellent fits were observed with a root mean square deviation of 0.0037 absorbance units. Sedimentation coefficients were corrected to s20,w.
Expression and Purification of Rab Proteins
The genes for human Rab4a, canine Rab5a, and human Rab27a were ligated into pGEX expression vectors (GE Healthcare) and human Rab7a and Rab9a (residues 1–177) into pGST2. GST-Rab fusion proteins were produced in E. coli BL21(DE3) cells in ZYM-5052 autoinduction medium at 20 °C as described above. Cells were lysed in buffer C (50 mm Tris, 200 mm NaCl, 1 mm MgCl2, 1 mm TCEP pH 8.0) containing 50 μg/ml AEBSF. GST-Rab proteins were then purified by glutathione affinity chromatography and eluted in buffer C containing 20 mm reduced glutathione. Eluted proteins were subsequently purified by gel filtration on a Superdex 75 column (GE Healthcare). Fractions containing pure GST-Rab fusion protein were pooled, concentrated, supplemented with 10% (w/v) glycerol, and snap frozen in liquid nitrogen.
Guanine Nucleotide Exchange on Rab Proteins
Nucleotide exchange of wild-type GST-Rab fusion proteins was performed as described (24). Briefly, GST-Rabs were first immobilized on glutathione-Sepharose 4B resin and washed with NS buffer (20 mm HEPES, 100 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol (DTT), pH 7.2). The bound nucleotide was exchanged by washing the resin in NE buffer (20 mm HEPES, 100 mm NaCl, 5 mm MgCl2, 1 mm DTT pH 7.2) containing 20 μm of either GDP, GMP-PNP, GTPγS, or GTP followed by a 30-min incubation in the same buffer containing 1 mm nucleotide. After repeating the exchange procedure, bound nucleotide was stabilized by washing the resin in NS buffer containing 20 μm nucleotide followed by a 30-min incubation with 1 mm nucleotide. All exchange steps were carried out at room temperature.
GST Pulldown Assays
For GST pulldown experiments with recombinant BLOC-3, 50 pmol of GST-Rab fusion protein was immobilized on glutathione-Sepharose 4B and washed in NS buffer containing 0.5% β-OG (assay buffer) and 1 mm GDP, GMP-PNP, or GTPγS. Recombinant BLOC-3 was added, and the volume adjusted to 500 μl with assay buffer. After incubation for 2 h at 4 °C, the resin was washed four times with 500 μl of assay buffer. Bound proteins were eluted with Laemmli sample buffer and separated by SDS-PAGE. Gels were stained with colloidal Coomassie Blue G250 dye and quantified by measuring Coomassie Blue fluorescence on a Typhoon Trio scanner (GE Healthcare) using an excitation wavelength of 532 nm and no emission filter. Integration of band intensities was performed using ImageQuant TL image-analysis software (GE Healthcare). The intensities of BLOC-3 bands were normalized to the intensity of the GST-Rab band in each lane to account for loading variations. Binding constants were obtained by non-linear curve fitting in Microcal Origin (Northampton, MA).
For pulldown from cytosol, pig spleen cytosol was prepared as described (25). Immobilized GST-Rab proteins charged with GMP-PNP or GDP were incubated with cytosol at 4 °C by end-over-end rotation. After 2 h, the beads were washed (26), and bound protein was eluted by boiling for 5 min in Laemmli sample buffer. Bound protein was then detected by immunoblotting with an antibody to HPS4 (9).
For pulldown from cell lysates, COS-7 cells were transfected with pCIneo-HA-HPS1, and/or pCIneo-Myc-HPS4 (9) using Lipofectamine-2000 (Invitrogen). After 24 h, the cells were washed with ice-cold phosphate-buffered saline and lysed in 20 mm HEPES, pH 7.5, 100 mm NaCl, 5 mm MgCl2, and 0.2% (w/v) Triton X-100 for 10 min on ice. Lysates were spun for 10 min at 15,700 × g in a refrigerated benchtop centrifuge (Eppendorf, Nijmegen, The Netherlands). Supernatants were collected and incubated with GST-Rab9a-GMP-PNP (or GST or GST-Rab9a-GDP as negative controls) bound to glutathione-agarose beads for 2 h by end-over-end rotation. Samples were subsequently washed six times with lysis buffer; the excess buffer was drained with a 30.5-gauge needle, and bound proteins were eluted by boiling in Laemmli sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting with rabbit polyclonal antibodies to the Myc (Upstate Cell Signaling, Huissen, The Netherlands) and HA tags (Sigma).
Yeast Two-hybrid Assays
The pGBKT7-Rab9aWT or -Rab9bWT constructs were generated by PCR amplification of cDNAs encoding wild-type human Rab9a or Rab9b, followed by in-frame cloning into the EcoRI-SmaI sites or the EcoRI-SalI sites of the pGBKT7 (TRP1) vector (Clontech, Palo Alto, CA), respectively. Mutagenesis of these constructs was performed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All the constructs were sequenced to confirm their identity and the presence of the desired mutations. Cloning of full-length human HPS4 and HPS1 into the pGADT7 (LEU2) vector (Clontech) has been described before (9). The pGADT7-pallidin construct was generated by PCR amplification of a cDNA-encoding human pallidin, followed by in-frame cloning into the EcoRI-SalI sites of the pGADT7 vector. The Saccharomyces cerevisiae strain AH109 (Clontech) was co-transformed with pGBKT7-Rab9aWT (or pGBKT7-Rab9a(Q66L)) and pGADT7-HPS4, pGADT7-HPS1 or pGADT7-pallidin by the lithium acetate method as described in the instructions for the MATCHMAKER two-hybrid kit (Clontech). As negative controls, AH109 cells were co-transformed with pGBKT7-Rab9aWT (or Rab9a(Q66L)) and pGADT7-SV40, or the empty vector pGADT7. AH109 cells were also co-transformed with pGADT-HPS4 and pGBKT7-p53, and the empty vector pGBKT7. Similar control experiments were performed with the pGBKT7-Rab9b and pGBKT7-Rab9b(Q66L) constructs. AH109 transformants were resuspended in water to 0.2 A600/ml, and 5 μl were spotted on plates in the presence or absence of histidine (His) with or without the histidine biosynthesis inhibitor 3-amino-1,2,4-triazole (3AT). Plates were incubated at 30 °C for 3–4 days. As a positive control for growth on -His (with or without 3AT) plates, AH109 cells were co-transformed with pGBKT7-p53 and pGADT7-SV40.
RESULTS
BLOC-3 Is a 1:1 Complex of HPS1 and HPS4
To gain insight into the assembly of BLOC-3, we co-expressed human HPS1 and HPS4 in Sf9 insect cells using a baculovirus expression system. A GST tag and TEV protease cleavage site were added to the N terminus of HPS4 to aid purification. After capture on glutathione-Sepharose, HPS1 remained tightly associated with GST-HPS4 (data not shown, see below). Upon removal of the GST tag with TEV protease, BLOC-3 remained soluble at low ionic strength. However, aggregates began to form at concentrations of the complex above 0.5 mg/ml, and most of it eluted in the void volume during gel filtration on Superose 6. The solubility of BLOC-3 decreased further with increasing ionic strength, indicating exposure of hydrophobic surface patches as a possible cause of aggregation. Therefore, we screened detergents and other buffer additives for their ability to stabilize the complex after removal of the GST tag. Among the detergents tested, β-OG at 0.6% (w/v), roughly twice the critical micelle concentration, proved the most effective at stabilizing BLOC-3 in its native state, allowing it to be concentrated to several mg/ml without aggregation.
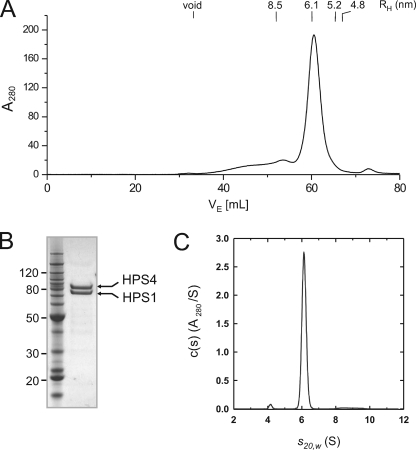
Gel filtration analysis showed that most of BLOC-3 eluted as a species with a hydrodynamic radius (RH) of 6.1 nm, corresponding to an apparent molecular mass of 440 kDa assuming a globular shape (Fig. 1A). SDS-PAGE and Coomassie Blue staining of the peak fractions revealed the presence of both HPS1 and HPS4 in roughly equimolar quantities (Fig. 1B). Given the predicted molecular masses of 79 and 77 kDa for the HPS1 and HPS4 polypeptides, respectively, the apparent molecular mass of 440 kDa suggests either a multimeric assembly or an elongated 1:1 complex.
FIGURE 1.
Hydrodynamic properties of BLOC-3. A, gel filtration profile of recombinant BLOC-3 on a Superose 6 column in 0.6% (w/v) β-OG. The hydrodynamic radii of standard proteins are indicated. B, Coomassie Blue-stained gel of purified BLOC-3 complex after gel filtration. Molecular masses are indicated in kDa. C, analytical ultracentrifugation of BLOC-3 at 4 °C in 0.6% (w/v) β-OG. Analysis using one species for the c(s) distribution results in a molecular mass of 146 kDa and an f/f0 ratio of 1.65.
To determine more accurately the stoichiometry of the complex, we further analyzed BLOC-3 by analytical ultracentrifugation. Sedimentation velocity experiments were carried out at a single loading concentration of 2.7 μm in the presence of β-OG. Data analysis in terms of a continuous c(s) distribution returned excellent fits, showing the presence of 96% of the loading absorbance as a monodisperse species with a sedimentation coefficient (s20,w) of 6.11 ± 0.03 S and a frictional ratio (f/fo) of 1.65 ± 0.05 (Fig. 1C). From these values, we calculated a molecular mass of 146 ± 5 kDa for the recombinant complex. Taken together, these experiments demonstrated that HPS1 and HPS4 interact directly to form a 1:1 heterodimer with an asymmetric shape. The hydrodynamic parameters of the recombinant BLOC-3 are in good agreement with those previously reported for the endogenous complex in various cell lines (9, 17), making it unlikely that this complex contains a third subunit.
The Central Part of HPS4 Contains a Large Unstructured Region Dispensable for Complex Formation
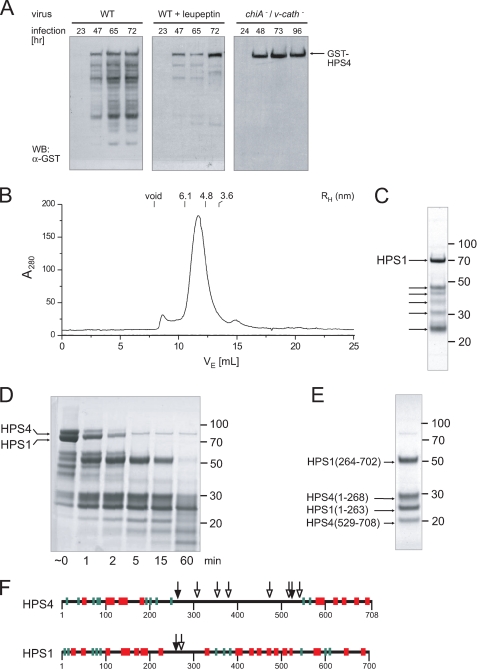
In the course of purification, BLOC-3, and in particular the HPS4 subunit, was found to be prone to degradation. HPS4 degradation was already observed during baculovirus infection, and could be reduced by adding leupeptin to the culture medium or using a baculovirus lacking v-cath, the gene for viral cathepsin protease (Fig. 2A). Therefore, we attempted to map the protease-sensitive regions in BLOC-3 and determine whether their removal might produce a stable subcomplex.
FIGURE 2.
Protease sensitivity and mapping of disordered regions. A, Sf9 cells were infected with either wild-type or a cathepsin/chitinase (v-cath/chiA)-deficient baculovirus expressing GST-HPS4 and HPS1. Total cell lysates taken at the indicated time points after infection were analyzed by immunoblotting using an antibody to GST. B, gel filtration profile of recombinant BLOC-3 after autoproteolytic digestion. The hydrodynamic radii of standard proteins are indicated. C, Coomassie Blue-stained SDS-PAGE of the peak fraction from the gel filtration shown in B. In addition to full-length HPS1, several smaller HPS1 and HPS4 fragments remain tightly associated in the complex (arrows). D, limited proteolysis with trypsin. Full-length recombinant BLOC-3 was incubated with 2 μg/ml trypsin at room temperature and aliquots taken at the indicated time points. E, BLOC-3 was proteolysed with trypsin for 15 min and loaded onto a Superdex 200 gel filtration column. The four major bands in the 15-min digest (D) remain tightly associated and co-elute in the peak fraction. However, the central region in HPS4 between 268 and 529 dissociates from the rest of the complex and is not present in the peak fraction. The gels in C through E were stained with Coomassie Blue, molecular masses are indicated in kDa. F, all protease cleavage sites in BLOC-3 are located in regions lacking predicted secondary structure. Mapped sites from autoproteolysis (open arrowheads) and limited trypsin proteolysis (closed) are shown together with predicted secondary structure elements for HPS1 and HPS4 (α-helices, red; β-strands, green).
First, a sample of purified BLOC-3 was incubated at 4 °C without protease inhibitors. Proteolytic degradation was already apparent after a few days, and after 1 week, HPS4 had been completely cleaved into fragments, whereas some HPS1 remained intact. Surprisingly, the degraded complex gave rise to a good gel filtration profile (Fig. 2B), and all but one of the fragments still co-eluted as one complex in the peak fractions (Fig. 2C). The fragments were analyzed by N-terminal sequencing and MALDI-MS, revealing five cleavage sites in the middle of HPS4, a region predicted to lack secondary structure (Fig. 2F, open arrowheads). The one fragment that did dissociate during gel filtration corresponded to a smaller piece of the same region (residues 387–470). HPS1 was largely stable under these conditions, and we detected only one minor cleavage site at residue 275, also in a region predicted to lack secondary structure (Fig. 2F, open arrowhead).
Next, we confirmed the major protease-sensitive regions using limited trypsin proteolysis. Incubation of BLOC-3 with 2 μg/ml trypsin for different times at room temperature resulted in the production of a series of fragments (Fig. 2D). Gel filtration of the 15-min digest revealed four tightly associated fragments (Fig. 2E). Under the conditions used, HPS1 was cleaved once at residue 263, a position similar to that found in the autoproteolysis experiment. However, HPS4 was cleaved at positions 268 and 529, with the entire region in between dissociating from the complex (Fig. 2F, closed arrowheads).
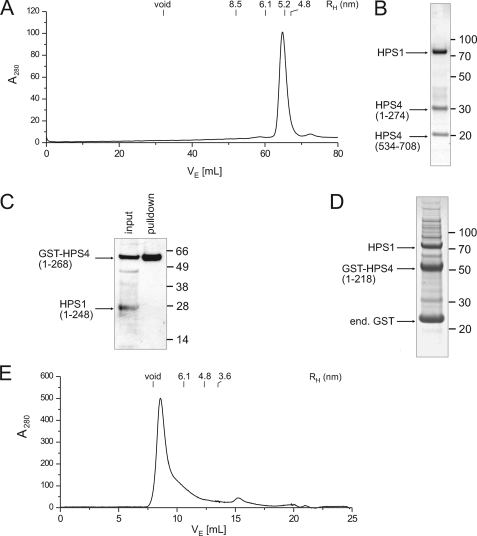
Finally, to test whether the entire central region of HPS4 might be dispensable for complex formation, we generated a construct in which residues 269–275 and 528–534 of HPS4 were replaced with TEV cleavage sites, allowing the region in between to be removed. After TEV cleavage, the HPS4 fragment between the two TEV sites dissociated as expected, and the resulting HPS1-HPS4(1–274)-HPS4(534–708) complex could be purified by gel filtration without dissociating (Fig. 3, A and B). The hydrodynamic radius of the truncated complex was smaller than that of full-length BLOC-3 (5.2 versus 6.1 nm), consistent with the loss of 260 amino acids. Like the full-length complex, the ΔHPS4275–533 variant was only stably soluble in the presence of β-OG. Thus, the putative solvent-exposed hydrophobic patches that cause aggregation in the absence of detergent, possibly a membrane interaction surface, reside in the main bulk of the complex and not in the central region of HPS4.
FIGURE 3.
Assembly of the BLOC-3 complex. A, gel filtration profile of the BLOC-3 complex after removal of HPS4 residues 275–533 using flanking TEV sites. B, Coomassie Blue-stained peak fraction from the gel filtration shown in A. The N-terminal (1–274) and C-terminal (534–708) fragments of HPS4 remain tightly associated with HPS1, whereas the central region dissociates from the complex, showing that it is not needed for complex stability. C, N-terminal domains of HPS1 and HPS4 do not form a stable complex. GST-HPS4-(1–268) and HPS1-(1–248) were co-expressed, captured on glutathione resin, and detected by immunoblot using a rabbit polyclonal antibody against the full-length BLOC-3 complex. D, HPS4 N-terminal domain (1–218) interacts with full-length HPS1. Co-expression of GST-HPS4-(1–218) and full-length HPS1 followed by capture on glutathione resin and Coomassie Blue staining. Molecular masses in C and D are indicated in kDa. E, N-terminal domain of HPS4 forms soluble aggregates. Elution profile of HPS4-(1–218) on a Superdex 200 column after removal of the GST tag and ion exchange chromatography.
Taken together, these results show that the central region of HPS4 encompassing residues 275–535 does not play a role in complex assembly. The functional significance of this region remains unclear. There is little sequence conservation between HPS4 orthologs, and the length of the region itself varies widely, ranging from 260 residues in human HPS4 to 110 residues in the chicken ortholog. This region exhibits an abundance of low complexity sequence, which is a typical feature of a disordered loop. Indeed, the entire region is predicted to lack secondary structure (Fig. 2F), which is supported by our observation of several highly protease-sensitive sites close together.
Assembly of BLOC-3 Depends on Multiple Sites of Interaction between HPS1 and HPS4
Recent bioinformatics studies have suggested the presence of an evolutionarily conserved longin (16) or CHiPS (15) domain at the N terminus of both HPS1 and HPS4, which might interact to form a stable core complex. Because our proteolysis experiments revealed a complex set of interactions, we tried to gain insight into BLOC-3 assembly by co-expressing different fragments.
For reconstitution experiments, we used either the domain boundaries previously mapped by proteolysis, or the shorter, predicted longin domain boundaries (1–268 versus 1–218 for HPS4 and 1–248 versus 1–220 for HPS1). First, we expressed the N-terminal domain of HPS4-(1–268) as a GST fusion together with untagged HPS1-(1–248). Although GST-HPS4-(1–268) was soluble and could be captured on glutathione resin, it did not pull down HPS1-(1–248), showing that the N-terminal domains of HPS1 and HPS4 are not sufficient to form a complex (Fig. 3C). However, GST-HPS4-(1–218) did pull down full-length HPS1 (Fig. 3D), indicating the presence of an HPS4 interaction site in the HPS1 C-terminal region that is necessary for complex formation.
Despite being soluble, the GST-HPS4-(1–218)-HPS1 complex was subsequently found to be completely aggregated, preventing further characterization (data not shown). We observed the same behavior when we produced HPS4-(1–218) by itself (Fig. 3E), or a GST fusion comprising residues 1–268 (data not shown). Apparently, the putative N-terminal longin-like domain of HPS4 is not stable by itself, and association with HPS1 does not prevent the formation of non-native contacts that cause aggregation. However, when we added the C-terminal domain of HPS4 to the assembly, in the form of the aforementioned BLOC-3(ΔHPS4275–533) complex consisting of HPS1, HPS4-(1–274), and HPS4-(534–708), there was no sign of aggregation, and the complex was well-behaved in solution (Fig. 3A). Because the only difference between the two complexes was the presence of the HPS4 C-terminal domain, it must have a stabilizing effect on the N-terminal domain. This indicates a strong interaction between the N- and C-terminal domains despite the very long loop connecting them. The possibility that the N- and C-terminal parts of HPS4 may belong to a single, non-contiguous domain cannot be excluded. Indeed, it is tempting to speculate that the large, poorly conserved unstructured region in the middle of HPS4 might have arisen through an insertion event early in metazoan evolution. Because this region is not required for assembly and only weakly interacts with the rest of the complex, if at all, there would have been few evolutionary constraints on its length and sequence. In summary, our results show that BLOC-3 has no discernible core or peripheral domains. Instead, it appears to be an elongated assembly in which both HPS1 and HPS4 form multiple inter- and intrasubunit interactions.
BLOC-3 Interacts with Rab9
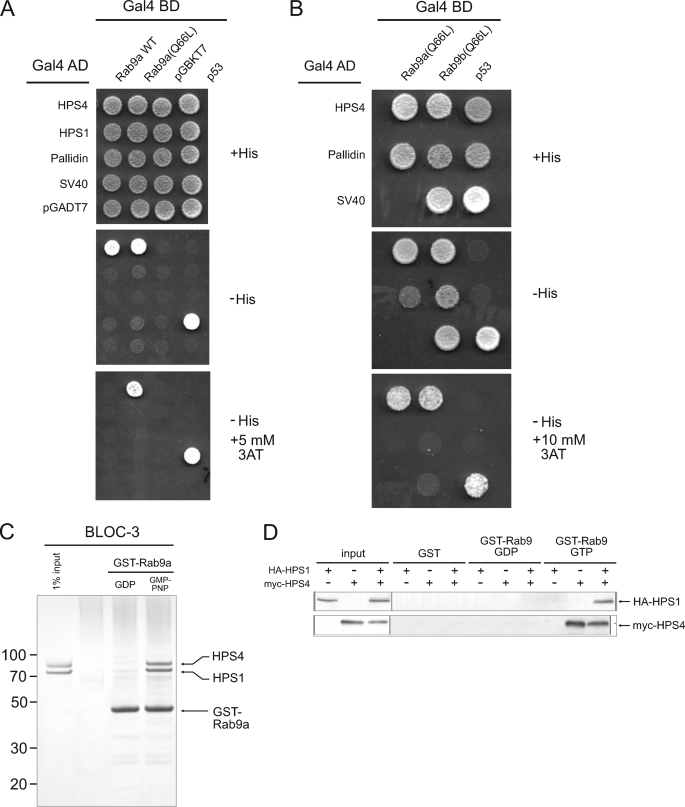
Because several Rab GTPases are involved in melanosome and platelet dense body biogenesis (27, 28, 29, 30, 31, 32, 33, 34), we tested for interactions of various Rab proteins (i.e. Rab7, Rab9a, Rab29, Rab32, and Rab38) with HPS4 and HPS1 using the Y2H system (data not shown). We found a robust interaction between HPS4 and Rab9a (19), an endosomal Rab that had not been previously implicated in LRO biogenesis. HPS4 was found to interact with both wild-type Rab9a and its Q66L variant, a GTP hydrolysis-deficient mutant that remains in the GTP-bound active state (Fig. 4A). Increasing the stringency of the assay by addition of 5 mm 3-amino-1,2,4-triazole (3AT), however, showed interaction with Rab9a-Q66L but not wild-type Rab9a (Fig. 4A), indicating that the interaction is nucleotide-dependent. In contrast, we found no interaction of HPS1 with Rab9a, and Rab9a with the BLOC-1 subunit pallidin. We also found an interaction of HPS4 with the Q66L mutant of Rab9b (Fig. 4B), a second Rab9 isoform in humans that shares 76% sequence identity with Rab9a (35).
FIGURE 4.
GTP-dependent interaction of BLOC-3 with Rab9. A, Y2H assay of HPS1 and HPS4 binding to wild-type and GTP-locked (Q66L) Rab9a. B, Y2H assay of HPS4 binding to GTP-locked (Q66L) Rab9a and Rab9b. Negative and positive controls for interactions are described under “Experimental Procedures.” C, GST pulldown using purified recombinant BLOC-3 (200 nm) and immobilized wild-type GST-Rab9a (100 nm) loaded with either GDP or GMP-PNP. Coomassie Blue-stained gel. Molecular masses are indicated in kDa. D, GST pulldown using lysates from COS-7 cells expressing HA-HPS1, Myc-HPS4, or both. Lysates were passed over GST or GST-Rab9a loaded with GTP or GDP and bound proteins detected by immunoblotting with anti-HA and anti-Myc antibodies.
We confirmed the interaction with Rab9a in vitro using pulldown assays. Wild-type GST-Rab9a was immobilized on glutathione-Sepharose 4B, the bound nucleotide exchanged for GDP or the non-hydrolyzable GTP analog GMP-PNP, and purified recombinant BLOC-3 added. We observed that BLOC-3 bound to Rab9a-GMP-PNP, but not Rab9a-GDP (Fig. 4C). HPS1 and HPS4 bound in equal amounts, showing that the complex remained intact when bound to Rab9a. In another pulldown experiment, we transfected COS-7 cells with constructs expressing HA-HPS1, Myc-HPS4, or both, and incubated the cell lysates with GST-Rab9a loaded with GTP or GDP (Fig. 4D). Rab9a-GTP was able to bind Myc-HPS4 or Myc-HPS4 together with HA-HPS1, but not HA-HPS1 alone. No interaction was observed with Rab9a-GDP or with the GST control (Fig. 4D), confirming that the interaction between BLOC-3 and Rab9a is nucleotide specific.
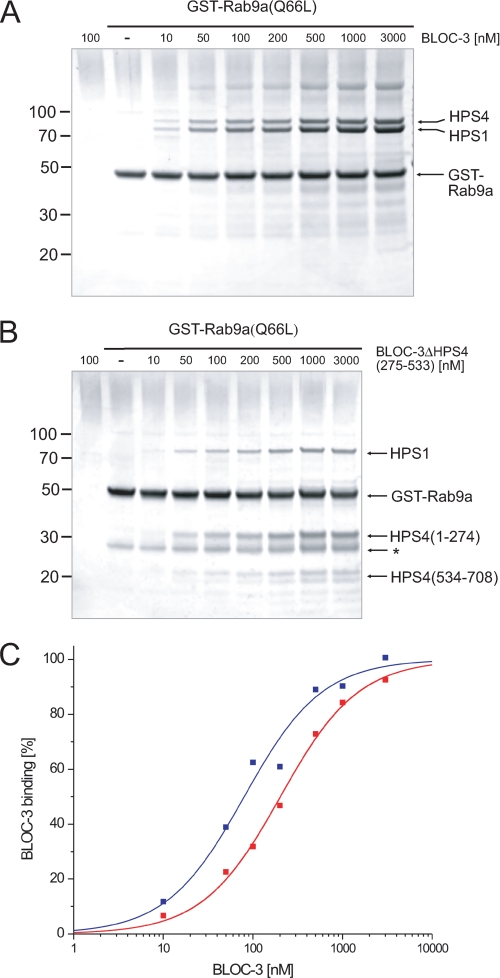
We measured the binding affinity of this interaction by titrating GST-Rab9a(Q66L) with increasing amounts of recombinant BLOC-3 (Fig. 5A). A fit of the binding curve assuming 1:1 stoichiometry between BLOC-3 and Rab9a resulted in a KD of 82 ± 11 nm (Fig. 5C). This is an exceptionally strong interaction for a Rab-binding protein, as most Rab effectors show affinities in the low micromolar range (36). To determine whether the unstructured central loop region of HPS4 is important for the Rab9a-BLOC-3 interaction, we repeated the titration using BLOC-3(ΔHPS4275–533), and obtained a KD of 203 ± 15 nm (Fig. 5B), an affinity that, although lower than that of full-length BLOC-3, is on the same order of magnitude, showing that the flexible loop in HPS4 does not significantly contribute to Rab9a binding.
FIGURE 5.
Deleting the central region of HPS4 does not impair Rab9 binding. A, GST pulldown with different concentrations of recombinant BLOC-3 and GTP-locked GST-Rab9a at 100 nm. B, GST pulldown with different concentrations of recombinant BLOC-3(ΔHPS4275–533) and GTP-locked GST-Rab9a at 100 nm. A GST contaminant is marked by an asterisk. Molecular masses in A and B are indicated in kDa. C, quantification of the titrations in A and B. The amount of bound BLOC-3 was determined by Coomassie Blue G250 fluorescence, normalized to the amount of GST-Rab9 in each lane and plotted against BLOC-3 concentration. Fitting the data assuming 1:1 binding stoichiometry results in a KD of 82 nm for full-length BLOC-3 (blue) and 202 nm for BLOC-3(ΔHPS4275–533) (red).
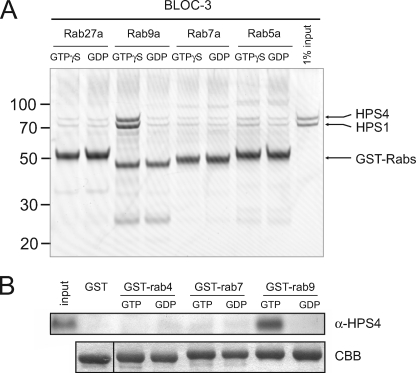
To assess the specificity of the Rab9a-BLOC-3 interaction, we performed pulldown assays with other endosomal Rab proteins as controls. In addition to GST-Rab9a, we tested GST-Rab5a (early endosome), GST-Rab7a (early-late endosome), and GST-Rab27a (melanosome/LRO) after exchanging their bound nucleotide by GDP or another non-hydrolyzable GTP analog, GTPγS. Recombinant BLOC-3 only bound to GST-Rab9a-GTPγS, showing that the interaction is highly specific (Fig. 6A). We then tested whether endogenous BLOC-3 showed the same binding properties as the recombinant protein. GST-Rab4a, -Rab7a, and Rab9a were incubated with mouse spleen cytosol and bound endogenous HPS4 detected by immunoblotting (Fig. 6B). Again, BLOC-3 only bound to Rab9a-GTP, but not Rab9a-GDP or the other Rabs.
FIGURE 6.
The interaction of BLOC-3 with Rab9 is Rab-specific. A, GST pulldown using purified recombinant BLOC-3 (200 nm) and immobilized wild-type GST-Rab5a, -Rab7a, -Rab9a, and -Rab27a (100 nm) loaded with either GDP or GTPγS. Coomassie Blue-stained gel. Molecular masses are indicated in kDa. B, GST pulldown using mouse spleen cytosol and GST-Rab4a, -Rab7a and -Rab9a loaded with GTP or GDP as bait. Endogenous HPS4 protein was detected using an affinity-purified polyclonal antibody.
BLOC-3 Binds to a Canonical Effector-binding Surface on Rab9
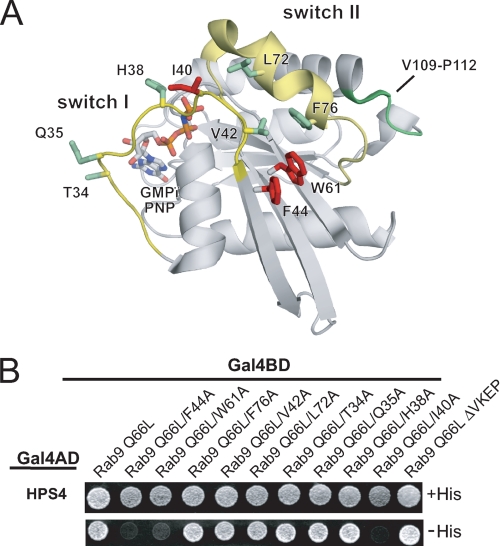
The presence of the γ-phosphate of GTP is sensed by Rab proteins through backbone interactions with a flexible loop, which in turn induces a series of conformational changes in two regions termed the switch I and switch II regions (37). Because the switch conformation serves as a readout for the nucleotide status, potential effector proteins must bind to these regions to discriminate between active and inactive forms of the GTPase. An important determinant is the so-called hydrophobic triad (38), a triplet of aromatic residues close to the switch I and II regions whose side chains form a stacking interaction (Phe-44, Trp-61, and Phe-76 in Rab9a) (Fig. 7A). Because of this interaction, GTP hydrolysis, which induces a shift in the backbone that in turn alters the side chain conformation of one of the residues, will lead to a conformational change in the other two as well, amplifying the effect of the backbone movement.
FIGURE 7.
Mapping the binding site on Rab9. A, scheme of Rab9-GMP-PNP in the active conformation (PDB 1YZL) with the residues mutated to alanine in B shown in stick representation. The switch I and II regions are colored yellow. Point mutations causing loss of HPS4 binding are highlighted in red and those with no effect in green. Deletion of the Val-109-Pro-112 loop (green backbone) also had no effect. B, Y2H assay of Rab9a(Q66L) binding to HPS4. Residues on the Rab9a surface were mutated to alanine as indicated. The ΔVKEP mutation (far right) refers to the Val-109—Pro-112 loop, which only occurs in Rab7, Rab9, Rab32, and Rab38.
We hypothesized that the hydrophobic triad, and perhaps other residues in the switch I and II regions, may be part of the surface epitope that BLOC-3 recognizes. Indeed, mutating either Phe-44 or Trp-61 to alanine caused a complete loss of HPS4 binding in the Y2H assay (Fig. 7B). The same was observed for Ile-40, which lies at the end of the switch I loop and is part of the switch I/switch II interface. However, mutating the third residue in the hydrophobic triad, Phe-76, and two nearby residues, Leu-72 and Val-42, had no effect. Three other alanine mutations directly at the nucleotide-binding site, Thr-34, Gln-35, and His-38, also had no effect, showing that the distal part of the switch I loop is not part of the BLOC-3 interaction surface.
In addition to the point mutations, we also considered a short loop formed by residues Val-109 to Pro-112 that is unique to Rab7, Rab9, Rab32, and Rab38, and that might be important for specific recognition of Rab9 by BLOC-3. However, deletion of this loop had no effect on HPS4 binding (Fig. 7B, ΔVKEP mutant). Taken together, our data are consistent with BLOC-3 binding to the switch I/II region of Rab9a, a canonical effector interaction site.
DISCUSSION
Subunit Composition and Hydrodynamic Properties of BLOC-3
Results presented here shed light on aspects of BLOC-3 that were unclear from previous work. Although both HPS1 and HPS4 had been shown to be components of BLOC-3, these two proteins did not interact in Y2H assays and assembled poorly when expressed as epitope-tagged proteins in transfected cells (8–10). These observations had suggested the possible existence of a third subunit that was necessary for assembly of HPS1 and HPS4 into the complex. This hypothetical third subunit had to be very small because the molecular mass of BLOC-3, as calculated from hydrodynamic analyses, approached the sum of the predicted molecular masses of HPS1 and HPS4 (9, 10, 17). Our characterization of the recombinant BLOC-3, however, shows that HPS1 and HPS4 co-purify as a 1:1 heterodimer without a requirement for a third subunit. The interaction between HPS1 and HPS4 is therefore direct and independent of any other cellular protein. It is unclear why previous analyses failed to show this stoichiometric interaction. In the Y2H system, both proteins were expressed as chimeras in which their N termini were fused to the C terminus of the Gal4 binding and activation domains (8–10). Similarly, in cellular studies, triple repeats of the HA and Myc epitope tags were appended to the N termini of HPS1 and HPS4, respectively (9). In contrast, in the present study we only fused GST to the N terminus of HPS4, leaving HPS1 untagged. It is thus possible that blocking the N terminus of HPS1 hinders assembly, an effect that would not be surprising given the compact nature of the complex. A more trivial explanation would be that HPS1-HPS4 assembly depends on the particular moiety used to tag each subunit. In any event, the notion that BLOC-3 comprises just HPS1 and HPS4 is in line with the identical phenotypes of humans with mutations in the homonymous genes and of the corresponding pale ear (ep) and light ear (le) mice, and with the absence of any other mutants with the same phenotype among the many that have been characterized in both humans and mice (5).
Our results also clarify the issue of the size of BLOC-3. Although previous hydrodynamic analyses of the endogenous complex had indicated that the complex has a molecular mass in the range of 140–175 kDa (9, 17), other analyses limited to gel filtration had revealed larger species of 200 and 500 kDa (8, 18). The size of the recombinant complex determined here (∼146 kDa) is similar to that from the former studies, in which no evidence for larges species was found (9, 10, 17). The appearance of larger species in the other studies might be due to the use of a detergent-free, 10,000 × g cytoplasmic fraction containing particulate materials and to the calculation of molecular masses from gel filtration analysis assuming a globular shape (8, 18). We find that BLOC-3 is not globular but elongated, necessitating the use of ultracentrifugation techniques for accurate molecular mass calculation.
Assembly of BLOC-3
Many components of the machinery involved in the biogenesis of endosomal-lysosomal organelles have a modular structure consisting of tandemly arranged folded domains that are often separated by disordered linker regions (39). A cursory interpretation of secondary structure predictions would indicate that this is also the case for HPS1 and HPS4, as these analyses show folded N- and C-terminal domains connected by long disordered segments for both proteins (Fig. 2F). Moreover, bioinformatics analyses have predicted the occurrence of potentially autonomous CHiPS (15) or longin (16) domains in the N-terminal portions of HPS1 and HPS4. However, limited proteolysis of the complete complex showed that, despite the presence of a long disordered loop in the central part of HPS4, the N- and C-terminal parts of both subunits remain noncovalently attached as part of a complex. Moreover, expression of protein fragments showed that the HPS4 N-terminal domain did not assemble with the HPS1 N-terminal domain of HPS1 and was aggregated. The HPS4 N-terminal domain did assemble with full-length HPS1, but this complex was also aggregated. Finally, the only well-behaved partial complex that we could find comprised the N- and C-terminal domains of HPS4 (without the central loop) plus full-length HPS1. These results indicated that the N- and C-terminal domains of both proteins are intimately associated through multiple interdomain interactions. Thus, unlike other components of the endosomal-lysosomal trafficking machinery, BLOC-3 lacks defined “hinge” regions linking independent folded domains.
The compact nature of BLOC-3 probably explains the severe phenotypes of some mutations in HPS patients. For example, the HPS1(L668P) mutation found in a Japanese patient prevented assembly of the mutant protein into the BLOC-3 complex (40). This mutation, located in a predicted α-helix, presumably disrupts the tertiary structure in the C-terminal region of HPS1, leading to loss of HPS4 binding. Likewise, the most frequent mutation found in HPS patients of Puerto Rican origin, a 16-bp duplication leading to a frameshift at residue 496 and termination of the polypeptide at 586 (11), would be expected to have the same effect, although this has not been shown by in vitro reconstitution. Thirteen other HPS1 mutations have been reported in HPS patients that generate stop codons at positions ranging from 97 to 666 (41).
For HPS4, eight frameshift mutations leading to premature termination have been described in HPS patients from various backgrounds (41). Notably, two of these generate stop codons at positions 701 (42) and 703 (14), leading to a loss of only seven and five residues from the C terminus, respectively. Both are associated with a severe disease phenotype and underscore the importance of the HPS4 C-terminal domain for BLOC-3 assembly, which is in agreement with our in vitro reconstitution experiments.
Interaction with Rab9
Of all the complexes that are defective in HPS, BLOC-3 is the only one for which no interacting partners had been identified (6). Here we present evidence for a strong (i.e. nanomolar affinity) and specific interaction of BLOC-3 with the Rab9a and Rab9b isoforms. This interaction is GTP-dependent and involves binding of HPS4 to the switch I and II regions of Rab9a. These properties indicate that BLOC-3 is likely a Rab9 effector. It is noteworthy that both BLOC-3 and Rab9 exhibit the same phylogenetic pattern of expression. Indeed, recognizable orthologs of HPS1, HPS4, and Rab9 can be found in insects (including Drosophila melanogaster), fish, amphibians, and mammals, but not in yeast (including S. cerevisiae) and nematodes (including Caenorhabditis elegans) (our analysis and Refs. 6 and 43). This pattern suggests that BLOC-3 and Rab9 co-evolved to perform a common function.
Rab9a localizes to late endosomes, and regulates retrograde transport from late endosomes to the trans-Golgi network (TGN) (19) through interaction with several effectors (Ref. 44 and references therein). We have found that, in melanosomes, both Rab9a and Rab9b co-localize to structures that label for Hrs and EEA1,4 which are markers for both endosomes and stage I pre-melanosomes (45). It is thus possible that Rab9 regulates BLOC-3 function in LRO biogenesis from an endosomal compartment. We have been unable to examine the localization of endogenous BLOC-3 relative to Rab9 because the levels of BLOC-3 in all cell lines that we have tested are below the level of detection by immunofluorescence microscopy.
Other connections between Rabs and LRO biogenesis have been previously reported. For instance, the gunmetal mouse lacks a functional Rab geranylgeranyl transferase (RGGT), which leads to underprenylation of Rab27a and other Rab proteins (27, 31). Rab27a in turn mediates attachment of melanosomes to myosin Va through its interaction with the adaptor melanophilin, thereby enabling transport to the cell periphery along microtubules (32–34). More recently, Rab38 and the functionally redundant Rab32 were shown to reside on melanosomes, where they regulate sorting of TYRP1 and tyrosinase to melanosomes (29, 30). In light of our results, it will now be of interest to determine whether Rab9 also regulates LRO biogenesis.
Acknowledgments
We thank Layla Saidi, Xiaolin Zhu, and Nora Tsai for expert technical assistance and Christina Schindler for helpful discussions.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Programs of NICHD (to J. S. B.) and NIDDK (to J. H. H.), the Netherlands' Proteomics Center (to P. v. d. S.), and the DFG-NIH program (to D. P. K.).
J. S. Bonifacino, unpublished observations.
- HPS
- Hermansky-Pudlak syndrome
- GST
- glutathione S-transferase
- MALDI-MS
- matrix-assisted laser desorption/ionization mass spectrometry
- TEV
- tobacco etch virus
- BLOC
- biogenesis of lysosome-related organelles complex
- GMP-PNP
- guanosine 5′-(β,γ-imino)triphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- HA
- hemagglutinin
- LRO
- lysosome-related organelles
- TCEP
- Tris(2-carboxyethyl)phosphine
- β-OG
- β-octylglucopyranoside.
REFERENCES
- 1.Hermansky F., Pudlak P. (1959) Blood 14, 162–169 [PubMed] [Google Scholar]
- 2.Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., Gahl W. A. (2008) Annu. Rev. Genomics Hum. Genet. 9, 359–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dell'Angelica E. C., Mullins C., Caplan S., Bonifacino J. S. (2000) FASEB J. 14, 1265–1278 [DOI] [PubMed] [Google Scholar]
- 4.Raposo G., Marks M. S., Cutler D. F. (2007) Curr. Opin. Cell Biol. 19, 394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W., Rusiniak M. E., Chintala S., Gautam R., Novak E. K., Swank R. T. (2004) Bioessays 26, 616–628 [DOI] [PubMed] [Google Scholar]
- 6.Li W., Feng Y., Hao C., Guo X., Cui Y., He M., He X. (2007) J. Genet. Genomics 34, 669–682 [DOI] [PubMed] [Google Scholar]
- 7.Dell'Angelica E. C. (2004) Curr. Opin. Cell Biol. 16, 458–464 [DOI] [PubMed] [Google Scholar]
- 8.Chiang P. W., Oiso N., Gautam R., Suzuki T., Swank R. T., Spritz R. A. (2003) J. Biol. Chem. 278, 20332–20337 [DOI] [PubMed] [Google Scholar]
- 9.Martina J. A., Moriyama K., Bonifacino J. S. (2003) J. Biol. Chem. 278, 29376–29384 [DOI] [PubMed] [Google Scholar]
- 10.Nazarian R., Falcón-Pérez J. M., Dell'Angelica E. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8770–8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh J., Bailin T., Fukai K., Feng G. H., Ho L., Mao J. I., Frenk E., Tamura N., Spritz R. A. (1996) Nat. Genet. 14, 300–306 [DOI] [PubMed] [Google Scholar]
- 12.Oh J., Ho L., Ala-Mello S., Amato D., Armstrong L., Bellucci S., Carakushansky G., Ellis J. P., Fong C. T., Green J. S., Heon E., Legius E., Levin A. V., Nieuwenhuis H. K., Pinckers A., Tamura N., Whiteford M. L., Yamasaki H., Spritz R. A. (1998) Am. J. Hum. Genet. 62, 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dell'Angelica E. C., Aguilar R. C., Wolins N., Hazelwood S., Gahl W. A., Bonifacino J. S. (2000) J. Biol. Chem. 275, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T., Li W., Zhang Q., Karim A., Novak E. K., Sviderskaya E. V., Hill S. P., Bennett D. C., Levin A. V., Nieuwenhuis H. K., Fong C. T., Castellan C., Miterski B., Swank R. T., Spritz R. A. (2002) Nat. Genet. 30, 321–324 [DOI] [PubMed] [Google Scholar]
- 15.Hoffman-Sommer M., Grynberg M., Kucharczyk R., Rytka J. (2005) Traffic 6, 534–538 [DOI] [PubMed] [Google Scholar]
- 16.Kinch L. N., Grishin N. V. (2006) Protein Sci. 15, 2669–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nazarian R., Huizing M., Helip-Wooley A., Starcevic M., Gahl W. A., Dell'Angelica E. C. (2008) Mol. Genet. Metab. 93, 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh J., Liu Z. X., Feng G. H., Raposo G., Spritz R. A. (2000) Hum. Mol. Genet. 9, 375–385 [DOI] [PubMed] [Google Scholar]
- 19.Lombardi D., Soldati T., Riederer M. A., Goda Y., Zerial M., Pfeffer S. R. (1993) EMBO J. 12, 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger I., Fitzgerald D. J., Richmond T. J. (2004) Nat. Biotechnol. 22, 1583–1587 [DOI] [PubMed] [Google Scholar]
- 21.Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 22.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 23.Cole J. L., Lary J. W., Moody T. P., Laue T. M. (2008) Methods Cell Biol. 84, 143–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christoforidis S., Zerial M. (2000) Methods 20, 403–410 [DOI] [PubMed] [Google Scholar]
- 25.van der Sluijs P., Neeft M., van Vlijmen T., Elstak E., Wieffer M. (2008) Methods Enzymol. 438, 185–201 [DOI] [PubMed] [Google Scholar]
- 26.Rojas R., van Vlijmen T., Mardones G. A., Prabhu Y., Rojas A. L., Mohammed S., Heck A. J., Raposo G., van der Sluijs P., Bonifacino J. S. (2008) J. Cell Biol. 183, 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Detter J. C., Zhang Q., Mules E. H., Novak E. K., Mishra V. S., Li W., McMurtrie E. B., Tchernev V. T., Wallace M. R., Seabra M. C., Swank R. T., Kingsmore S. F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ménasché G., Pastural E., Feldmann J., Certain S., Ersoy F., Dupuis S., Wulffraat N., Bianchi D., Fischer A., Le Deist F., de Saint Basile G. (2000) Nat. Genet. 25, 173–176 [DOI] [PubMed] [Google Scholar]
- 29.Loftus S. K., Larson D. M., Baxter L. L., Antonellis A., Chen Y., Wu X., Jiang Y., Bittner M., Hammer J. A., 3rd, Pavan W. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4471–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasmeier C., Romao M., Plowright L., Bennett D. C., Raposo G., Seabra M. C. (2006) J. Cell Biol. 175, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q., Zhen L., Li W., Novak E. K., Collinson L. M., Jang E. K., Haslam R. J., Elliott R. W., Swank R. T. (2002) Br. J. Haematol. 117, 414–423 [DOI] [PubMed] [Google Scholar]
- 32.Wu X., Rao K., Bowers M. B., Copeland N. G., Jenkins N. A., Hammer J. A., 3rd (2001) J. Cell Sci. 114, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 33.Wu X. S., Rao K., Zhang H., Wang F., Sellers J. R., Matesic L. E., Copeland N. G., Jenkins N. A., Hammer J. A., III (2002) Nat. Cell Biol. 4, 271–278 [DOI] [PubMed] [Google Scholar]
- 34.Hume A. N., Collinson L. M., Hopkins C. R., Strom M., Barral D. C., Bossi G., Griffiths G. M., Seabra M. C. (2002) Traffic 3, 193–202 [DOI] [PubMed] [Google Scholar]
- 35.Seki N., Azuma T., Yoshikawa T., Masuho Y., Muramatsu M., Saito T. (2000) J. Hum. Genet. 45, 318–322 [DOI] [PubMed] [Google Scholar]
- 36.Eathiraj S., Pan X., Ritacco C., Lambright D. G. (2005) Nature 436, 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabe Lee M. T., Mishra A., Lambright D. G. (2009) Traffic 10, 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merithew E., Hatherly S., Dumas J. J., Lawe D. C., Heller-Harrison R., Lambright D. G. (2001) J. Biol. Chem. 276, 13982–13988 [DOI] [PubMed] [Google Scholar]
- 39.Evans P. R., Owen D. J. (2002) Curr. Opin. Struct. Biol. 12, 814–821 [DOI] [PubMed] [Google Scholar]
- 40.Ito S., Suzuki T., Inagaki K., Suzuki N., Takamori K., Yamada T., Nakazawa M., Hatano M., Takiwaki H., Kakuta Y., Spritz R. A., Tomita Y. (2005) J. Invest. Dermatol. 125, 715–720 [DOI] [PubMed] [Google Scholar]
- 41.Wei M. L. (2006) Pigment Cell Res. 19, 19–42 [DOI] [PubMed] [Google Scholar]
- 42.Bachli E. B., Brack T., Eppler E., Stallmach T., Trüeb R. M., Huizing M., Gahl W. A. (2004) Am. J. Med. Genet. A 127A, 201–207 [DOI] [PubMed] [Google Scholar]
- 43.Pereira-Leal J. B., Seabra M. C. (2001) J. Mol. Biol. 313, 889–901 [DOI] [PubMed] [Google Scholar]
- 44.Espinosa E. J., Calero M., Sridevi K., Pfeffer S. R. (2009) Cell 137, 938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raposo G., Tenza D., Murphy D. M., Berson J. F., Marks M. S. (2001) J. Cell Biol. 152, 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]