Abstract
Tripartite motif (TRIM) protein TRIM5α has been shown to restrict human immunodeficiency virus, type 1 infection in Old World monkey cells at the early post-entry step by poorly understood mechanisms. Currently, the physiological function of TRIM5α is not known. In this study, we showed that transiently overexpressed TRIM5α causes a morphological change in HEK293T cells. A proteomics analysis of the protein complexes that were pulled down with hemagglutinin-tagged TRIM5α suggested that the heat shock protein 70 (Hsp70) may serve as a TRIM5α-binding partner. The interaction between Hsp70 and TRIM5α was confirmed by co-localization and co-immunoprecipitation assays. Co-expression of Hsp70 reversed the TRIM5α-induced morphological change in HEK293T cells. Another heat shock protein Hsc70 also bound to TRIM5α, but unlike Hsp70, Hsc70 was not able to reverse the TRIM5α-induced morphological change, suggesting that Hsp70 specifically reverses the morphological change caused by TRIM5α. Studies using a series of TRIM5α deletion mutants demonstrate that, although the PRYSPRY domain is critical for binding to Hsp70, the entire TRIM5α structure is necessary to induce the morphological change of cells. When the ATPase domain of Hsp70 was mutated, the mutated Hsp70 could not counteract the morphological change induced by TRIM5α, indicating that the catalytic activity of Hsp70 protein is important for this function. Co-expression of Hsp70 elevated the levels of TRIM5α in the detergent-soluble fraction with a concomitant decrease in the detergent-insoluble fraction. Together these results suggest that Hsp70 plays critical roles in the cellular management against the TRIM5α-induced cellular insults.
Keywords: Apoptosis, E3 Ubiquitin Ligase, ER Stress, HIV, Protein Folding, Ubiquitination, Chaperone, Hsp70, TRIM
Introduction
Retroviruses encounter dominant post-entry restrictions in the cells of particular species. Human immunodeficiency virus type 1 (HIV-1)2 infection is blocked in the cells of Old World monkeys and simian immunodeficiency virus (SIVmac) infection is blocked in most New World monkey cells (1–3). These retroviral restrictions are largely mediated by a host protein, tripartite motif 5α (TRIM5α) (4).
TRIM5α is a member of the large family of tripartite motif proteins (TRIMs) (5). Members of this family commonly have a RING domain, one or two B-box domains and coiled-coil (CC) domain, and thus are also called RBCC proteins (5). The RING domain of many TRIM proteins has been shown to have E3 ubiquitin ligase activity, whereas the B box and CC domains may be involved in protein-protein interactions and homo/heterodimerization (5). The alpha isoforms of many cytoplasmic TRIM proteins, including TRIM5α, contain the PRYSPRY domain whose function is unknown. To date, about 70 TRIM proteins have been identified in the human genome and some of their homologs have been found in primates and other species. TRIM proteins arose with the metazoans and have expanded in number during vertebrate evolution (5). Many TRIM proteins, including TRIM19, TRIM21, TRIM22, TRIM34, and TRIM5α, can be up-regulated by interferon, supporting their potential role as effectors in the antiviral cellular response (6–10). However, the natural function of most TRIM proteins is unknown yet.
TRIM5α exhibits species-specific variation in primates, which may account for the species-specific blocking activity of TRIM5α against the retroviral infection (11, 12). In most cases, TRIM5α proteins impair retroviral infection early after the entry of virus into target cells and block the accumulation of viral reverse transcription products (13–15). The viral determinant of susceptibility to TRIM5α-mediated restriction is the capsid protein (16–20). Nevertheless, the detailed mechanism by which TRIM5α restricts retroviral infection is not clear, and the cellular function of TRIM5α is to be determined.
Previously, we reported that heat shock proteins Hsp70 and Hsp90 co-localize with TRIM5α protein (21). Hsp70 and Hsp90 are members of a large group of heat shock proteins that function as molecular chaperones by selectively binding to denatured or partially unfolded domains in polypeptides (22, 23). As nascent polypeptides emerge from the ribosome, most of them first associate with Hsp70 and then with other chaperones, such as Hsp90, Hsp104, Hsp40 (DnaJ), and small Hsp (Hsp27, α-crystallins). This association promotes proper folding of polypeptides and prevents unwanted interactions or aggregation (24). Nonetheless, a significant fraction of newly synthesized polypeptides normally fail to achieve their correct conformation and are rapidly digested (25). Certain combinations of chaperones prevent aggregation of damaged polypeptides, catalyze refolding of denatured molecules, disassemble intracellular protein aggregates, and refold the insoluble molecules into soluble, native species (26). TRIM5α, like other TRIM proteins, self-associates to form cytoplasmic bodies when overexpressed in cells (5, 27, 28). The nature and function of cytoplasmic bodies are unknown, except that they are not required for the antiviral activity (14, 27, 29). Here we investigated the impacts of TRIM5α overexpression on cellular physiology and functions of Hsp70 as a molecular chaperon to reverse the TRIM5α-induced effects.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa (human cervical cancer) and HEK293T (human embryonic kidney) cells were obtained from the American Type Culture Collection, and were grown with 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium (HyClone) containing 10% fetal bovine serum (HyClone), 100 units/ml penicillin (HyClone), and 0.1 mg/ml streptomycin (HyClone). HeLa cells stably expressing human, rhesus, or African green monkey TRIM5α with a HA tag at the N terminus were established using the retroviral vector pLPCX system (Clontech) (12). HEK293T and HeLa cells were transfected using a calcium phosphate method and Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions, respectively. Transfection efficiency was over 50%.
Plasmid Constructs
The cDNAs encoding the amino acid-coding sequence of human (Hu), rhesus monkey (Rh), and African green monkey (AGM) TRIM5α genes were previously described (12). The TRIM5α cDNA with HA tag sequence at the N terminus was initially cloned into pLPCX (Clontech) and subcloned into pcDNA3.1(−) (Invitrogen) by standard subcloning methods.
To delete the RING, B-box, or CC domain of rhesus TRIM5α, the two-step PCR strategy was applied using two external primers and two internal primers. The two external primers are complementary to the vector sequences flanking the HA-TRIM5α sequence. The two internal primers are complementary to the sequences flanking the deleted region and contain 10 nucleotide overlapping sequences at the 5′-end of the primer. The two external primers are pcDNA3.1-for and pcDNA3.1-rev (Table 1). The internal primers used to delete the RING, B-box, and CC domains are as followed: RING-for, RING-rev, B-box-for, B-box-rev, CC-for, and CC-rev (Table 1). To create TRIM5α mutants with deletion of the PRYSPRY domain, a stop codon was introduced upstream of the PRYSPRY domain using the QuikChange site-directed mutagenesis kit (Stratagene) with the primers, PRYSPRY-for and PRYSPRY-rev (Table 1).
TABLE 1.
The oligonucleotide primers used to generate the variants of TRIM5α and Hsp70
| Primer | Nucleotide sequence (5′ to 3′) |
|---|---|
| pcDNA3.1-for | AAGCAGAGCTCTCTGGCTAAC |
| pcDNA3.1-rev | TCCAGGGTCAAGGAAGGCACG |
| RING-for | ACTGATCCGGGTCACCTCCTCCTTTACATTAAG |
| RING-rev | GGTGACCCGGATCAGTTACCAGCCTGAG |
| B-box-for | GGAAAGTGTGATCAACCTTCTGTCCCTC |
| B-box-rev | TTGATCACACTTTCCTCATGGAGGAGGTTG |
| CC-for | CTGAGATGAAAGTGTGGTGACCACGGTGC |
| CC-rev | CACTTTCATCTCAGAACTGGAGCATCG |
| PRYSPRY-fora | GAGAGCTAACAGATGCCTGAATTCGCTACTGGGTTGATGTGAC |
| PRYSPRY-rev | GTCACATCAACCCAGTAGCGAATTCAGGCATCTGTTAGCTCTC |
| Hsp70-for | GCATGCGGCCGCGATGGCCAAAGCCGCGGC |
| Hsp70-rev | GACTTCTAGACTAATCTACCTCCTCAATGGTGGGGC |
| Hsc70-for | GCATGCGGCCGCGATGTCCAAGGGACCTGCAGTTGG |
| Hsc70-rev | GACTTCTAGATTAATCAACCTCTTCAATGGTGGGCC |
| K71E-for | CGCAGAACACCGTGTTTGACGCGGAGCGGCTGATCGGCCGCAAGTTCGG |
| K71E-rev | CCGAACTTGCGGCCGATCAGCCGCTCCGCGTCAAACACGGTGTTCTGCG |
| K71M-for | CCGCAGAACACCGTGTTTGACGCGATGCGGCTGATCGGCCGCAAGTTCGG |
| K71M-rev | CCGAACTTGCGGCCGATCAGCCGCATCGCGTCAAACACGGTGTTCTGCGG |
a The stop codon (bold case) and EcoRI restriction site (italic) introduced are indicated.
The plasmid expressing the FLAG-tagged ubiquitin was kindly provided by Dr. Garrison Fathman (Stanford University, Stanford, CA). Hsp70 expression vector pCMV70, a vector expressing HA-tagged Hsc70 (HA-Hsc70), and a vector expressing FLAG-tagged Hsp90 (FLAG-Hsp90) were provided by Dr. Harm Kampinga (University of Groningen, The Netherlands), Dr. Danny Manor (Case Western Reserve University, Cleveland, OH), and Dr. Len Neckers (NCI, National Institutes of Health, Bethesda, MD), respectively.
To generate FLAG-tagged Hsp70 and Hsc70, the cDNA sequences encoding Hsp70 and Hsc70 were PCR-amplified using pCMV70 and the HA-Hsc70 vector as templates, respectively, and cloned into p3×FLAG-CMV-10 (Sigma) using NotI and XbaI restriction sites. The primers used to amplify Hsp70 and Hsc70 were as followed: Hsp70-for, Hsp70-rev, Hsc70-for, and Hsc70-rev (Table 1). To generate substitute mutants of the lysine residue at the position 71 in the ATPase domain of Hsp70 and Hsc70 to glutamate or methionine, the QuikChange site-directed mutagenesis kit (Stratagene) was employed using the following primers: K71E-for, K71E-rev, K71M-for, and K71M-rev (Table 1).
Cell Viability
HEK293T cells were seeded in 6-well plate and transfected with vectors expressing TRIM5α variants or with an empty vector pcDNA3.1 (Invitrogen) as a control. Forty-eight hours after transfection, cells were stained with trypan blue, and live cells were counted. HEK293T cells were seeded in 96-well plate and transfected for 48 h, and the cell viability was assed using a Vybrant MTT Cell Proliferation Assay Kit (Invitrogen) according to the manufacturer's instructions. The levels of total protein were determined by using the DC Protein Assay System (Bio-Rad).
Cell Death Assays
HEK293T cells were transfected with vectors expressing TRIM5α variants or with an empty vector pcDNA3.1 (Invitrogen) as a control. Forty-eight hours after transfection, cells were assessed for apoptotic cell death using an Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) or Caspase-3/CPP32 Colorimetric Assay Kit (MBL International) according to the manufacturers' instructions. HEK293T cells were also treated with staurosporine (Sigma) at 10 μm for 18 h or with camptothecin (Sigma) at 12 μm for 5 h to induce apoptosis.
Flow Cytometer Analysis
HEK293T cells were transfected with pEGFP-C1 (Clontech), human TRIM5α-GFP, or rhesus TRIM5α-GFP. Forty-eight hours after transfection, GFP-positive cells were analyzed for cell size and cell complexity by forward scatter (FSC) and side scatter (SSC), respectively, using FACSCanto II flow cytometer (BD Biosciences). The cells transfected with pEGFP-C1 were gated as a control so that each quadrant contains ∼25% of the GFP-positive cells, and then the percentage of cells in Q1, Q2, Q3, and Q4 quadrants were determined for the cells expressing human TRIM5α-GFP and rhesus TRIM5α-GFP: Q1 (low FSC, high SSC), Q2 (high FSC, high SSC), Q3 (low FSC, low SSC), and Q4 (high FSC, low SSC).
In-gel Digestion and Mass Spectrometry
Cell lysates were prepared from HEK293T cells transfected with a vector expressing HA-tagged rhesus TRIM5α. HA-TRIM5α complexes were pulled down using anti-HA-agarose (Sigma, A2095), resolved on a SDS-polyacrylamide gel, and stained with Coomassie Blue. Selected gel regions were excised and subjected to the in-gel digestion. The resulting peptides were analyzed by the reversed-phase liquid chromatography coupled with the tandem mass spectrometry using an LTQ-Orbitrap mass spectrometer (ThermoFinnigan, San Jose, CA) at the Emory Proteomics Core Service Center. Peptide masses were matched against the theoretical peptide masses of all proteins in the National Center for Biotechnology Information or SWISS-PROT databases.
Immunofluorescence and Confocal Microscopy
HeLa cells grown in 6-well plates were transfected with 1 μg of plasmids expressing GFP-tagged TRIM5α variants for 24 h using Lipofectamine 2000 (Invitrogen) and then subcultured on the Lab-TekII 8-well chamber slides (Nunc International). HeLa cells stably expressing HA-tagged TRIM5α variants were similarly cultured on the 8-well chamber slides. HEK293T cells were plated onto glass coverslips at a density of 7 × 104/ml in 6-well dishes and transfected with 0.5 μg of wild-type Hsp70 or K71M mutant Hsp70 and a vector expressing HA-tagged rhesus TRIM5α or a control vector using the calcium phosphate procedure. At 40 h after transfection, cells were fixed with 4% para-formaldehyde, permeabilized with 0.2% Triton X-100, and incubated with primary antibody for 1 h. Then, cells were incubated with FITC-conjugated or tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibody (Santa Cruz Biotechnology) plus 4′,6-diamidino-2-phenylindole. Coverslips were mounted in 10% glycerol and examined under a fluorescence microscope. Anti-HA antibody was from Roche Applied Science (3F10), and anti-Hsp70 antibody was from Stressgen (C92F3A-5).
Immunoprecipitation and Immunoblotting Analyses
To prepare cell lysates, cells were washed in phosphate-buffered saline, lysed in Lysis buffer (20 mm HEPES, 150 mm NaCl, 0.5% Triton X-100, 10% glycerol, 1 mm NaF, 0.1 mm Na3VO4, protease inhibitors (Roche Applied Science)) and centrifuged to remove insoluble fraction. Cell lysates (300–400 μg) were incubated with 10 μl of resuspended anti-HA-agarose (Sigma) or anti-FLAG M2 affinity gel (Sigma) for 4 h at 4 °C. The immobilized proteins were collected by centrifugation, washed three times with the Lysis buffer, and solubilized by boiling for 5 min in SDS-PAGE sample buffer (10 mm Tris-HCl (pH 6.8), 1 mm EDTA, 10% glycerol, 0.0004% bromphenol blue, 4% SDS, 20 mm dithiothreitol). After separation on SDS-polyacrylamide gel, proteins were transferred onto a nitrocellulose membrane (Whatman), blocked with 5% nonfat dry milk, washed briefly, and incubated with specific antibodies. Blots were washed three times with TTBS buffer (20 mm Tris (pH 7.4), 150 mm NaCl, and 0.05% Tween 20) and incubated with horseradish peroxidase-conjugated anti-mouse (Santa Cruz Biotechnology) or anti-rabbit (Santa Cruz Biotechnology) antibody, and then developed using a chemiluminescence detection system (Pierce). Total cell lysates (20–30 μg) were also analyzed by immunoblotting analysis to determine the expression level of protein of interest. Mouse monoclonal anti-HA antibody (from Covance (MMS-101R) or Sigma (H6533)) was used to detect HA-tagged TRIM5α variants. Mouse monoclonal anti-FLAG antibody (Sigma (F3165 or F7425)) was used to detect FLAG-tagged heat shock proteins. Ubiquitinated proteins were detected using a rabbit anti-ubiquitin antibody from Santa Cruz Biotechnology (sc-9133). Hsp70 was detected using a mouse monoclonal antibody from Stressgen (SPA-810). The β-actin expression level was served as an internal control for immunoblotting and detected using a mouse monoclonal anti-β-actin antibody from Sigma (A5441). The apoptosis marker proteins (caspase-9, caspase-3, and PARP) and ER stress marker proteins (calnexin, BiP, PDI, and IRE1α) were detected using the antibodies provided in the Apoptosis Antibody Sampler Kit (Cell Signaling) and the ER Stress Antibody Sampler Kit (Cell Signaling), respectively.
RESULTS
Transient Overexpression of TRIM5α Induces a Morphological Change in HEK293T Cells
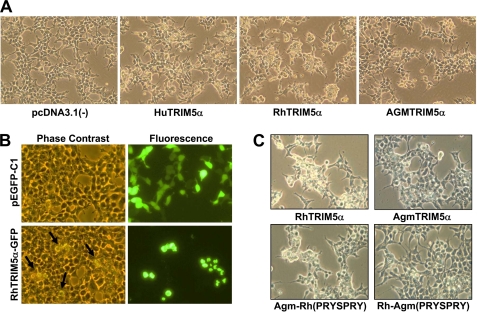
It was previously reported that HA-tagged or fluorescently tagged TRIM5α proteins restrict HIV-1 infection similar to unfused TRIM5α variants (12, 15, 30), and both N- and C-terminally HA-tagged TRIM5α variants efficiently restrict HIV-1 infection (12, 15). To examine the effects of TRIM5α expression on the morphology of cells, we expressed TRIM5α protein tagged with HA or GFP at the N terminus in human HEK293T cells. Interestingly, when transiently overexpressed, HA-tagged human (Hu-) or rhesus (Rh-) TRIM5α caused an alteration in cell morphology with retraction of cellular borders and rounding of cells whereas African green monkey (AGM) TRIM5α caused a minor change in cell morphology compared with human or rhesus TRIM5α (Fig. 1A; herein called “morphological change”). Cells transfected with the empty control vector pcDNA3.1(−) did not show the alteration (Fig. 1A). This is the first demonstration showing that TRIM5α has other cellular function in addition to the restrictive roles against viral infection. To verify that the morphological change observed in the cells transfected with TRIM5α is due to the expression of TRIM5α protein, we expressed GFP-tagged rhesus TRIM5α or pEGFP-C1 as a control in HEK293T cells. Indeed, the cells showing the morphological change corresponded to the cells expressing GFP-tagged rhesus TRIM5α (Fig. 1B, bottom panels), whereas the cells expressing pEGFP-C1 did not cause any change in cell morphology (Fig. 1B, top panels).
FIGURE 1.
Overexpression of TRIM5α induces a morphological change in HEK293T cells. A, the effect of TRIM5α overexpression was evaluated in HEK293T cells transfected with HA-tagged human (Hu), rhesus monkey (Rh), or African green monkey (AGM) TRIM5α or an empty vector pcDNA3.1(−) as a control. Morphological changes were observed under microscope at 48 h post-transfection. B, the effect of TRIM5α overexpression was evaluated in HEK293T cells transfected with GFP-tagged rhesus TRIM5α or pEGFP-C1 as a control. Phase contrast (left panels) and GFP fluorescence (right panels) were analyzed at 48 h post-transfection. The arrows in the phase-contrast image indicate the morphological change (retraction of cellular borders and rounding of cells) induced by TRIM5α overexpression. C, HEK293T cells were transfected with HA-tagged rhesus TRIM5α, AGM TRIM5α, AGM TRIM5α containing the PRYSPRY domain of rhesus TRIM5α, or rhesus TRIM5α containing the PRYSPRY domain of AGM TRIM5α for 48 h and analyzed by phase-contrast microscopy.
The PRYSPRY domain of TRIM5α exhibits the most variation from species to species and determines the restriction specificity against different retroviral infection (11, 12). Because rhesus TRIM5α caused a significant change in cell morphology and AGM TRIM5α caused only a minor change in cell morphology (Fig. 1A), we generated two types of the PRYSPRY domain-swapped TRIM5α mutants. First, Agm-Rh (PRYSPRY) contains the PRYSPRY domain of rhesus TRIM5α in the backbone of AGM TRIM5α. Second, Rh-Agm (PRYSPRY) contains the PRYSPRY domain of AGM TRIM5α in the backbone of rhesus TRIM5α. Agm-Rh (PRYSPRY) was as efficient as the wild-type rhesus TRIM5α in inducing the morphological change (Fig. 1C). In contrast, Rh-Agm (PRYSPRY) was less efficient in inducing the morphological change compared with the wild-type rhesus TRIM5α (Fig. 1C). These results suggest that the PRYSPRY domain of rhesus TRIM5α is critical for inducing the morphological change.
We also examined the deletion mutants of rhesus TRIM5α in which the RING domain, B-box domain, CC domain, or PRYSPRY domain is deleted. None of the deletion mutants was able to induce the morphological change (supplemental Fig. S1 and data not shown). Therefore, we conclude that the overall structure of TRIM5α is necessary for TRIM5α to elicit the morphological change. Finally, we wanted to determine whether the ability to induce an alteration in cell morphology is a general feature of the TRIM family proteins or specific to a subset of TRIM family proteins. Among the TRIM proteins examined, TRIM5α, TRIM14, TRIM32, and TRIM54 caused a morphological change, whereas TRIM4, TRIM25, TRIM28, and TRIM29 did not induce the morphological change (supplemental Fig. S1).
Heat Shock Protein Hsp70 Interacts with TRIM5α
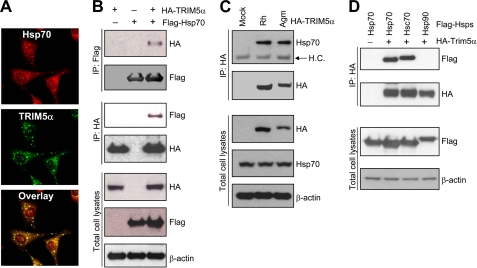
TRIM5 forms cytoplasmic bodies when overexpressed (5, 27, 28), raising the possibility that TRIM5 may form multiprotein complexes in cells. Using a proteomics approach, we analyzed the HA-tagged rhesus TRIM5α complexes that were pulled down with HA antibody, and identified the heat shock proteins Hsp70 and Hsc70 as potential TRIM5α-binding proteins (data not shown). To verify interaction between Hsp70 and TRIM5α, we first examined the intracellular localization of Hsp70 and TRIM5α by immunostaining assay. Fig. 2A shows that Hsp70 and TRIM5α co-localize in cells (Fig. 2A). Furthermore, a co-immunoprecipitation assay confirmed that FLAG-Hsp70 and HA-TRIM5α are in the same complex, or the two proteins directly interact with each other in HEK293T cells (Fig. 2B). The protein bands representing the interaction of FLAG-Hsp70 and HA-TRIM5α were present when the two expression plasmids were co-expressed (Fig. 2B, lane 3), whereas there was no protein band detected when either one of the expression plasmids was omitted from the transfection (Fig. 2B, lanes 1 and 2). Fig. 2C shows that endogenous Hsp70 also interacts with both rhesus TRIM5α and AGM TRIM5α.
FIGURE 2.
Hsp70 interacts with TRIM5α. A, the subcellular localization of endogenous Hsp70 and HA-tagged rhesus TRIM5α was evaluated by double labeling with anti-Hsp70 (red) and anti-HA (green) antibodies in HeLa cells stably expressing HA-tagged rhesus TRIM5α. B, co-immunoprecipitation of FLAG-Hsp70 with HA-TRIM5α from the cell lysates of HEK293T cells transfected with FLAG-Hsp70 and HA-tagged rhesus TRIM5α. Immunoprecipitates of FLAG antibody or HA antibody were analyzed for HA or FLAG by Western blotting. Total cell lysates were also analyzed for HA-TRIM5α, FLAG-Hsp70, or β-actin. C, co-immunoprecipitation of endogenous Hsp70 with HA-TRIM5α from the cell lysates of HeLa cells stably expressing HA-tagged rhesus TRIM5α or AGM TRIM5α. Immunoprecipitates of HA antibody were analyzed for Hsp70 by Western blotting. The arrow indicates the band of IgG heavy chain (labeled as H.C.). Total cell lysates were also analyzed for HA-TRIM5α, Hsp70, or β-actin. D, HEK293T cells were transfected with HA-tagged rhesus TRIM5α and FLAG-tagged Hsp70, Hsc70, or Hsp90, and the cell lysates were subjected to immunoprecipitation and Western blotting. Immunoprecipitates of HA antibody were analyzed for FLAG-Hsp and HA-TRIM5α. Total cell lysates were also analyzed for FLAG-Hsp and β-actin.
To test whether other heat shock proteins also interact with TRIM5α, we examined Hsp70, Hsc70, and Hsp90 for their ability to interact with TRIM5α by co-immunoprecipitation assay. As shown in Fig. 2D, both Hsp70 and Hsc70 interact with TRIM5α, while Hsp90 does not, suggesting that the molecular chaperones Hsp70 and Hsc70 may be involved in regulating the cellular function of TRIM5α.
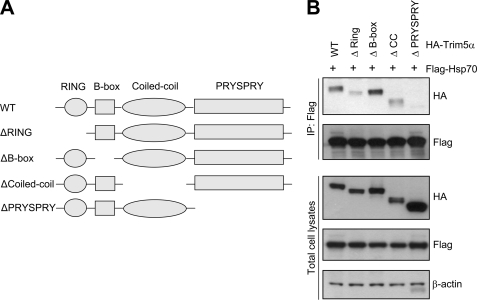
The PRYSPRY Domain Is the TRIM5α Determinant of Hsp70 Binding
To define the domain in TRIM5α that is critical for interaction with Hsp70, we generated a series of truncation mutants of rhesus TRIM5α (Fig. 3A). These TRIM5α mutants, each with deletion of the RING domain, B-box domain, CC domain, or PRYSPRY domain, respectively, have a HA tag at the N terminus. Interestingly, the TRIM5α mutant with the deletion of the PRYSPRY domain could not interact with Hsp70, whereas other TRIM5α variants with the RING, B-box, or CC domain deletion were able to interact with Hsp70 (Fig. 3B). It is noteworthy that the TRIM5α variants with the RING or CC domain deletion showed weak interaction with Hsp70 compared with the wild-type TRIM5α. These results suggest that the PRYSPRY domain is the determinant of TRIM5α for Hsp70 binding. It was previously reported that TRIM5α recognizes the targeted retroviral capsid, and this interaction depends on the PRYSPRY domain (31). Together, these findings indicate that the PRYSPRY domain of TRIM5α may serve as a binding domain for multiple proteins, including the retroviral capsid and molecular chaperone Hsp70.
FIGURE 3.
The PRYSPRY domain of TRIM5α is required for Hsp70 binding. A, schematic representation of wild-type (WT) rhesus TRIM5α and TRIM5α variants with deletion of the RING domain, B-box domain, CC domain, or PRYSPRY domain is shown. B, co-immunoprecipitation of FLAG-Hsp70 with HA-tagged rhesus TRIM5α variants. HEK293T cells were transfected with FLAG-Hsp70 and HA-tagged wild-type TRIM5α or TRIM5α variants containing the deletion of each domain, and the cell lysates were subjected to immunoprecipitation and Western blotting as described in Fig. 2.
Co-expression of Hsp70 Reverses the Cellular Morphological Change Induced by TRIM5α
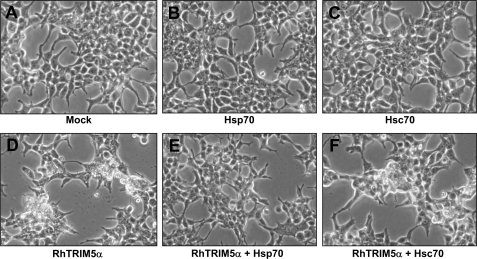
The interaction of Hsp70 and Hsc70 with TRIM5α raised a possibility that the molecular chaperone function of the heat shock proteins may ameliorate the morphological change induced by transiently overexpressed TRIM5α. To test this possibility, we examined the morphology of HEK293T cells transfected with TRIM5α alone or together with Hsp70, Hsc70, or Hsp90 using phase-contrast microscopy. Transient expression of Hsp70 (Fig. 4B) or Hsc70 (Fig. 4C) alone did not cause a morphological change compared with the mock transfected cells (Fig. 4A). Transient expression of TRIM5α induced a morphological change with retraction of cellular borders and rounding of cells (Fig. 4D), as previously shown in Fig. 1. Interestingly, co-expression of Hsp70 alleviated the morphological alteration induced by TRIM5α (Fig. 4E), whereas co-expression of Hsc70 (Fig. 4F) or Hsp90 (data not shown) did not. These results suggest that Hsp70 specifically antagonize the cellular effects of overexpressed TRIM5α.
FIGURE 4.
Co-expression of Hsp70 reverses TRIM5α-induced morphological change. HEK293T cells were transfected with HA-tagged rhesus TRIM5α, FLAG-tagged Hsp, or both. Forty-eight hours after transfection, phase-contrast images were obtained. Cells were transfected with: an empty vector pcDNA3.1 (A), FLAG-Hsp70 (B), FLAG-Hsc70 (C), HA-rhesus TRIM5α (D), HA-rhesus TRIM5α and FLAG-Hsp70 (E), and HA-rhesus TRIM5α and FLAG-Hsc70 (F).
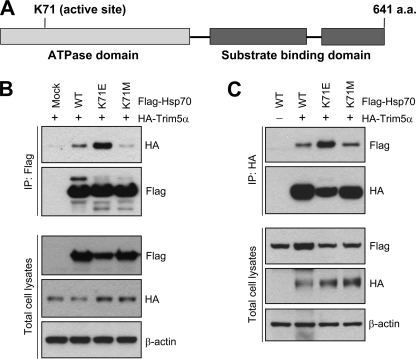
The ATPase Function of Hsp70 Protein Is Important for Reversing TRIM5α-induced Morphological Change
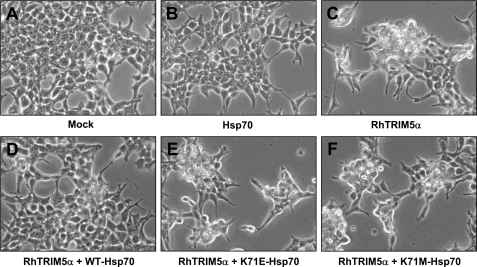
Hsp70 has the ATPase domain and substrate binding domain at the N and C termini, respectively (Fig. 5A) (32). We tested whether the ATPase function of Hsp70 is important for binding to TRIM5α and/or preventing TRIM5α from inducing the morphological change. The lysine residue at position 71 is critical for the ATPase function of Hsp70 protein. The Lys-71 was changed to Glu or Met, and the ability of the Hsp70 mutants to bind TRIM5α was examined. As shown in Fig. 5 (B and C), Hsp70 K71E binds to TRIM5α more efficiently than the wild-type Hsp70, whereas Hsp70 K71M mutant interacted with TRIM5α less efficiently than the wild-type Hsp70. To analyze the impact of the mutation in Hsp70 ATPase domain on the ability of Hsp70 to reverse TRIM5α-mediated morphological change, we examined HEK293T cells transfected with TRIM5α alone or together with the Hsp70 variants by microscopy (Fig. 6). As described previously in Fig. 1, the transient expression of TRIM5α induced a morphological change (Fig. 6C), whereas the transient expression of Hsp70 did not cause the morphological change (Fig. 6B) compared with the mock transfected cells (Fig. 6A). Although the wild-type Hsp70 reversed the TRIM5α-induced morphological change (Fig. 6D), Hsp70 mutants K71E (Fig. 6E) or K71M (Fig. 6F) were not able to reverse TRIM5α-induced morphological change, suggesting that the ATPase function of Hsp70 is important for Hsp70 to protect cells from the TRIM5α-induced phenotype.
FIGURE 5.
The ATPase function of Hsp70 is not required for binding to TRIM5α. A, schematic diagram of Hsp70 with the ATPase domain and substrate binding domain is shown. The lysine residue at the position 71 critical for the ATPase function of Hsp70 was changed into glutamic acid (K71E) or methionine (K71M) by site-directed mutagenesis. B and C, co-immunoprecipitation of HA-TRIM5α with FLAG-tagged Hsp70 variants. HEK293T cells were transfected with HA-tagged rhesus TRIM5α and FLAG-Hsp70 variants, and the cell lysates were subjected to immunoprecipitation and Western blotting. Immune precipitates of FLAG antibody were analyzed for HA-TRIM5α by Western blotting in B, or immune precipitates of HA antibody were analyzed for FLAG-Hsp70 by Western blotting in C. Total cell lysates were also analyzed for FLAG-Hsp70, HA-TRIM5α, or β-actin by Western blotting.
FIGURE 6.
The ATPase function of Hsp70 is critical for reversing the TRIM5α-induced morphological change. HEK293T cells were transfected with: an empty vector pcDNA3.1 (A), FLAG-Hsp70 (B), HA-rhesus TRIM5α (C), HA-rhesus TRIM5α and FLAG-WT-Hsp70 (D), HA-rhesus TRIM5α and FLAG-K71E-Hsp70 (E), and HA-rhesus TRIM5α and FLAG-K71M-Hsp70 (F). Forty-eight hours after transfection, phase-contrast images were obtained.
TRIM5α Does Not Affect the Steady-state Level of Hsp70
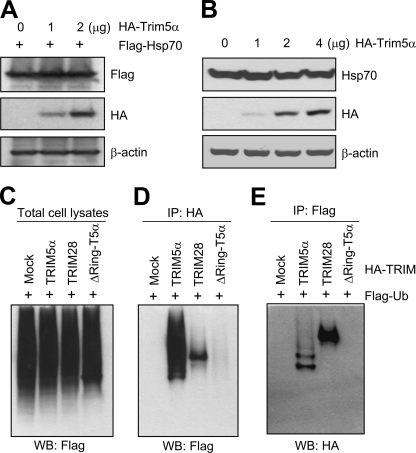
A recent study showed that TRIM5α has a RING domain-dependent self-ubiquitination activity (33), although the cellular substrates of TRIM5α E3 ligase are not known. The E3 ubiquitin ligase activity in general leads to polyubiquitination of target proteins and subsequent degradation through the proteasome pathway. Because TRIM5α interacted with Hsp70, we tested whether overexpression of TRIM5α causes a reduction in the steady-state level of Hsp70 protein through the TRIM5α-mediated ubiquitination and proteasomal degradation of Hsp70. Intriguingly, overexpression of rhesus TRIM5α protein did not cause a reduction in the steady-state levels of the exogenous (Fig. 7A) as well as endogenous Hsp70 protein (Fig. 7B).
FIGURE 7.
Overexpression of TRIM5α does not alter the steady-state level of Hsp70. A, HEK293T cells were transfected with FLAG-Hsp70 and increasing amounts of HA-rhesus TRIM5α. Forty-eight hours after transfection, the cell lysates were evaluated for FLAG, HA, and β-actin by Western blotting. B, HEK293T cells were transfected with increasing amounts of HA-rhesus TRIM5α. Forty-eight hours after transfection, the cell lysates were evaluated for endogenous Hsp70 level by Western blotting. C, HEK293T cells were transfected with FLAG-Ubiquitin and HA-tagged rhesus TRIM5α, human TRIM28, or rhesus TRIM5α mutant with the deletion of RING domain. Forty-eight hours after transfection, the cell lysates were evaluated for FLAG by Western blotting. D, the cell lysates prepared in C were immunoprecipitated using HA antibody and evaluated for FLAG by Western blotting. E, the cell lysates prepared in C were immunoprecipitated using FLAG antibody and evaluated for HA by Western blotting.
Next, we examined the E3 ubiquitin ligase activity of TRIM5α in HEK293T cells following transfection with a vector expressing FLAG-ubiquitin and a vector expressing HA-tagged TRIM5α or TRIM28. TRIM28 was originally identified as a factor important for the transcription repression function of Kruppel-associated box domain (37), and recent studies showed that TRIM28 restricts murine leukemia viruses in embryonic cells by primer binding site-targeted silencing of the integrated proviruses (34–36). The overall pattern and levels of ubiquitination of cellular proteins were indistinguishable in the mock transfected cells and in the cells transfected with TRIM5α variants or TRIM28 protein (Fig. 7C). There was robust ubiquitination of the wild-type TRIM5α, including mono- and polyubiquitinated products, whereas the ubiquitinated TRIM5α products were not detected in the cells expressing the TRIM5α mutant with the deletion of the RING domain (Fig. 7, D and E). In the cells expressing TRIM28 protein, only monoubiquitinated species were detected (Fig. 7, D and E). These results indicate that TRIM5α has self-ubiquitination function, which is dependent on the RING domain. Taken together, these results indicate that, although TRIM5α interacts with Hsp70, this interaction does not lead to degradation of Hsp70. In support of this idea, we could not observe the TRIM5α-mediated ubiquitination of Hsp70 (data not shown).
Co-expression of Hsp70 Elevates the Level of TRIM5α in the Detergent-soluble Fraction
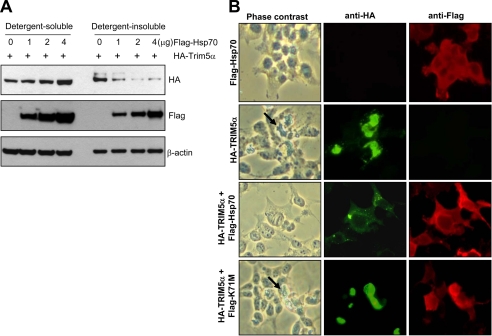
It is possible that the molecular chaperone function of Hsp70 may promote the folding of nascent TRIM5α to prevent the formation of misfolded aggregates. To test this possibility, HEK293T cells were transfected with a vector expressing HA-TRIM5α with increasing amounts of a vector expressing FLAG-tagged Hsp70, and the cell lysates were separated into the detergent-soluble and detergent-insoluble fractions by centrifugation and analyzed by immunoblotting assay. As shown in Fig. 8A, as the level of Hsp70 increased, the level of TRIM5α in the detergent-soluble fraction increased with a concomitant decrease in the level of TRIM5α in the detergent-insoluble fraction.
FIGURE 8.
Co-expression of Hsp70 elevates the level of TRIM5α in the detergent-soluble fraction. A, HEK293T cells were transfected with HA-tagged rhesus TRIM5α and increasing amounts of FLAG-Hsp70. Forty-eight hours after transfection, the cell lysates were separated into the detergent-soluble and detergent-insoluble fractions by centrifugation and evaluated for HA, FLAG, and β-actin by Western blotting. B, HEK293T cells were transfected with FLAG-Hsp70, HA-tagged rhesus TRIM5α, or both. Forth-eight hours after transfection, cells were fixed and labeled with HA antibody (green) and/or FLAG antibody (red). The arrows in the phase-contrast images indicate the morphological change (retraction of cellular borders and rounding of cells) induced by TRIM5α overexpression.
Intracellular localization of HA-TRIM5α and FLAG-Hsp70 was analyzed in HEK293T cells following transient transfection (Fig. 8B). Overexpression of TRIM5α resulted in protein aggregation and induced the morphological change (Fig. 8B, second panel), whereas overexpression of Hsp70 alone did not show any phenotype (Fig. 8B, top panel). When the wild-type Hsp70 was co-expressed with TRIM5α, large TRIM5α aggregates were replaced by smaller puncta, and the TRIM5α-induced morphological alteration was reversed (Fig. 8B, third panel). Co-expression of the Hsp70-K71M neither altered the expression pattern of TRIM5α aggregates nor reversed the morphological change induced by TRIM5α (Fig. 8B, bottom panel). These results indicate the strong correlation between the formation of TRIM5α aggregates and induction of morphological alteration. However, it is not clear whether the TRIM5α aggregates are the direct cause of the morphological alteration or they represent a cellular response to defend the cells from the insults caused by overexpression of TRIM5α.
TRIM5α Overexpression Does Not Induce an Apoptosis in HEK293T Cells
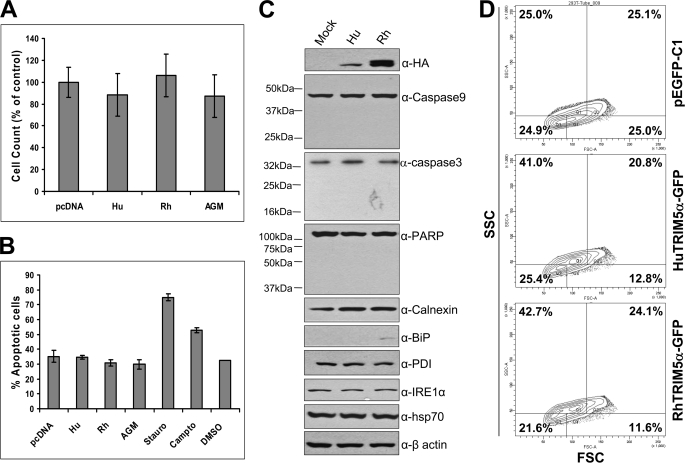
It was possible that the protein aggregates and/or the morphological changes induced by TRIM5α overexpression could cause an ER stress or apoptotic cell death. We first determined whether TRIM5α overexpression can affect the cell viability in HEK293T cells. Overexpression of human, rhesus, or AGM TRIM5α did not significantly affect the viability of HEK293T cells measured by trypan blue staining/cell counting (Fig. 9A) or MTT assay (supplemental Fig. S2A). The levels of total protein in the cells expressing human, rhesus, or AGM TRIM5α were not reduced compared with those in the cells transfected with an empty vector pcDNA3.1 (supplemental Fig. S2B).
FIGURE 9.
Overexpression of TRIM5α does not induce an apoptosis. A, HEK293T cells were transfected with HA-tagged human, rhesus, or AGM TRIM5α, or an empty vector pcDNA3.1, and 48 h later the number of live cells was counted after trypan blue staining. The values are shown as % of the control. Data are expressed as the mean ± S.D. of three independent experiments. B, HEK293T cells were transfected with HA-tagged human, rhesus, or AGM TRIM5α, or an empty vector pcDNA3.1, and 48 h later the percentage of apoptotic cells were determined by using the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences). When indicated, HEK293T cells were treated with staurosporine at 10 μm for 18 h or with camptothecin at 12 μm for 5 h to induce apoptosis. Data are expressed as the mean ± S.D. of three independent experiments. C, HEK293T cells were transfected with HA-tagged human or rhesus TRIM5α, or an empty vector pcDNA3.1, and 48 h later the cell lysates were subjected to Western blot analysis to detect HA-TRIM5α proteins, apoptosis marker proteins (caspase-9, caspase-3, and PARP), ER stress marker proteins (calnexin, BiP, PDI, and IRE1α), Hsp70, or β-actin as an internal control. D, HEK293T cells were transfected with pEGFP-C1, human TRIM5α-GFP, or rhesus TRIM5α-GFP, and 48 h later the forward scatter (FSC) and side scatter (SSC) patterns of GFP-positive cells were analyzed by a flow cytometer: Q1 (low FSC and high SSC), Q2 (high FSC and high SSC), Q3 (low FSC and low SSC), and Q4 (high FSC and low SSC). Data shown here is one representative data of three independent experiments.
We investigated the possibility that TRIM5α overexpression may induce a programmed cell death or apoptosis. Apoptosis is a regulated physiological process leading to cell death, and caspases, a family of cysteine acid proteases, are central regulators of apoptosis, which cleave cytoskeletal and nuclear proteins like PARP, α-fodrin, DEF, and lamin A (38). The expression of human, rhesus, or AGM TRIM5α in HEK293T cells did not cause an increase in the number of apoptotic cells compared with the cells transfected with an empty vector pcDNA3.1 as measured by either annexin V staining (Fig. 9B) or caspase-3 enzyme assay (supplemental Fig. S2C), whereas staurosporine or camptothecin treatment increased the number of apoptotic cells compared with the cells treated with DMSO as a control (Fig. 9B and supplemental Fig. S2C). These results were confirmed by Western blot analysis, which showed that expression of human or rhesus TRIM5α did not affect the levels of full-length caspase-9, caspase-3, or PARP nor induce the production of their cleavage products (Fig. 9C). Only the full-length proteins of caspase-9 (47 kDa), caspase-3 (35 kDa), and PARP (116 kDa) were detected and the cleavage products of caspase-9 (17 kDa, 35 kDa, and 37 kDa), caspase-3 (17 kDa and 19 kDa), and PARP (24 kDa and 89 kDa) were not detected in the cells transfected with a vector expressing TRIM5α or an empty vector (Fig. 9C). Taken together, these results indicate that TRIM5α overexpression does not induce an apoptosis in HEK293T cells.
Disruptions of ER homeostasis leads to the accumulations of unfolded proteins, and the ER has developed an adaptive mechanism called the unfolded protein response or ER stress response, which is mediated by the ER chaperone or regulatory proteins, including calnexin, BiP, protein disulfide isomerase (PDI), and IRE1α (39). We explored the possibility that the overexpression of TRIM5α proteins may induce the ER stress response (Fig. 9C). The levels of ER stress marker proteins calnexin, PDI, and IRE1α were not affected by the expression of TRIM5α protein (Fig. 9C). We observed a very faint band of the ER chaperone protein BiP only in the cells overexpressing rhesus TRIM5α (Fig. 9C), but it was not reproduced in a repeat experiment. The overexpression of TRIM5α did not affect the levels of the cytoplasmic molecular chaperone Hsp70 or β-actin protein used as an internal control (Fig. 9C). Therefore, we conclude that TRIM5α overexpression does not induce the ER stress response in HEK293T cells.
To determine the impact of TRIM5α overexpression on cell size and cell complexity, we analyzed the forward scatter (FSC) and side scatter (SSC) patterns of HEK293T cells expressing GFP-tagged TRIM5α variants using a flow cytometer and quantified the cell populations in each of four quadrants: Q1 (low FSC and high SSC), Q2 (high FSC and high SSC), Q3 (low FSC and low SSC), and Q4 (high FSC and low SSC). Interestingly, the percentage of cells in Q1 and Q2 quadrants (cells with complex or granular structure) was 61.8% for the cells expressing human TRIM5α and 66.8% for the cells expressing rhesus TRIM5α, compared with 50.1% for the cells expressing an empty vector pEGFP-C1 (Fig. 9D). These results indicate that the expression of human or rhesus TRIM5α causes an increase in the number of cells with granular structure (Q1 plus Q2). The functional significance of the increase of cells with granular structure in the cells expressing TRIM5α is currently under investigation.
DISCUSSION
Antiretroviral activity of TRIM5α is well documented, yet the physiological function of TRIM5α is not known (4). In this study, we report that transient overexpression of TRIM5α induces aggregate formation and morphological change in HEK293T cells, and the molecular chaperone Hsp70 counteracts the TRIM5α-induced cellular change. We propose that transient overexpression of TRIM5α induces misfolded structure that may interfere with normal cell functions, and the molecular chaperone Hsp70 relieves this adverse effect by eliminating the misfolded proteins.
This study also demonstrates that TRIM5α has a RING domain-dependent self-ubiquitination activity, which is consistent with the previous report by Yamauchi et al. (33). It was reported that TRIM5α has a tendency to oligomerize (40). It is therefore possible that the ability of TRIM5α to oligomerize may facilitate the self-ubiquitination of TRIM5α protein. We explored the possibility that Hsp70, a TRIM5α-interaction partner, might be the substrate for TRIM5α E3 ligase. However, overexpression of TRIM5α did not affect the ubiquitination status and the steady-state level of Hsp70. The cellular target of TRIM5α remains to be identified. We previously reported that TRIM5α restricts retroviral infection by specifically recognizing the capsid and promoting its rapid, premature disassembly (31). It is therefore possible that TRIM5α may ubiquitinate the capsid protein of incoming viral complex, contributing to accelerated disassembly of viral capsid and inhibition of reverse transcription.
Cytosolic protein aggregates are formed when the quality control mechanisms of the cell fail to ensure the folding and/or degradation of proteins (41). Accumulation of such aggregates is a hallmark of several neurodegenerative disorders, such as Huntington disease and Parkinson disease (26). Molecular chaperones, such as Hsp70, improve cell viability (42–44) and facilitate the elimination of poly(Q) proteins in the cellular models of poly(Q) expansion diseases and ameliorate disease phenotype in transgenic Drosophila models (42, 45–48). Hsp70 also improves the phenotype in a Drosophila Parkinson disease model in which human α-synuclein is overexpressed (49). Upon transient overexpression, TRIM5α forms large cytoplasmic bodies or aggregates, induces a morphological change, including retraction of cellular borders and rounding of cells, and causes an increase in the number of cells with granular structure. Interestingly, co-expression of Hsp70 led to a reduction in the level of TRIM5α in the aggregate or detergent-insoluble fraction and reversed the morphological change induced by TRIM5α overexpression. There is a correlation between the formation of TRIM5α aggregates and the extent of morphological change. Some evidence supports the idea that protein aggregates cause cytopathological effects in cells. For example, it has been reported that aggregated proteins directly inhibit the 26 S proteasome by choking the proteases with degradation-resistant protein aggregates (50–52). It is also possible that accumulation of misfolded protein aggregates may overwhelm and impairs the function of molecular chaperones. However, it remains unclear whether the soluble oligomers, which form during the early stages of assembly, or the large insoluble assemblies are more toxic to the cells.
It will be interesting to define the functional significance of Hsp70-TRIM5α interaction in TRIM5α-mediated restriction of HIV-1 infection. It is possible that the molecular chaperone activity of Hsp70 may be involved in several steps during the HIV-1 life cycle, including the uncoating of the incoming viral capsid, intracellular trafficking of viral complexes, and assembly of new virions. It has been reported that Hsp70 facilitates the nuclear import of the HIV-1 preintegration complexes (53), and Hsp70 is incorporated into the HIV-1 virion (54). Dissecting the functional significance of the Hsp70-TRIM5α interaction in the TRIM5α-mediated antiretroviral activity will be challenging due to the diverse functions of Hsp70. One possibility is that Hsp70 binding to TRIM5α may help TRIM5α maintain its proper conformation and allow TRIM5α to efficiently recognize the incoming viral capsid, consequently enhancing the restriction activity of TRIM5α. An alternative possibility is that Hsp70 may compete with the incoming viral capsid for TRIM5α binding and may interfere with the restriction function of TRIM5α.
TRIM5α overexpression was not linked to the induction of apoptosis or ER stress response, but it induced an alteration in cell morphology with retraction of cellular borders and rounding of cells and caused an increase in the number of cells with complex or granular structure. It is possible that TRIM5α overexpression may not directly induce an apoptosis, but it may sensitize cells to an apoptosis mediated by other apoptosis-inducing agents or ligands. This possibility is currently under investigation. Overexpression of proteins can lead to cellular insults in a variety of ways. Hsp70 as a molecular chaperone may be required for the reversal of these insults, without the interaction being a normal part of the function of the overexpressed protein. Once an antibody specifically recognizing endogenous TRIM5α is available, we will be able to test whether this interaction takes place under the usual levels of expression of both TRIM5α and Hsp70, in primary cells or other cell types.
Further studies will be necessary to understand how TRIM5α overexpression induces the cellular insults, how Hsp70 alleviates the TRIM5α-mediated effect, and what the roles of Hsp70 are in the TRIM5α-mediated restriction of HIV-1 infection. We hope that an approach for examining the cellular insults of TRIM5α protein may provide a clue to prevent a further accumulation of potentially toxic misfolded protein complexes in the cells.
Supplementary Material
Acknowledgments
We thank Dr. Garrison Fathman (Stanford University, Stanford, CA), Dr. Harm Kampinga (University of Groningen, The Netherlands), Dr. Danny Manor (Case Western Reserve University, Cleveland, OH), and Dr. Len Neckers (NCI, National Institutes of Health, Bethesda, MD) for the generous gift of expression vectors for ubiquitin, Hsp70, Hsc70, and Hsp90, respectively. We are grateful to the Emory Proteomics Core Service Center for mass spectral analyses. We thank Hamish Young for helpful discussions, advice, and critically reading the manuscript.
This work was supported by the Campbell Foundation Award, Emory University Research Committee Award, Emory Egleston Children's Research Center Award, and Emory Center for AIDS Research Award P30AI050409 (to B. S.), and partially supported by the Korea Research Council of Fundamental Science and Technology Award M10642040003 and National Research Foundation of Korea Award M10503010002 (to K.-S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HIV-1
- human immunodeficiency virus type 1
- TRIM
- tripartite motif
- AGM
- African green monkey
- Hsp70
- heat shock protein 70
- Hsc70
- heat shock cognate protein 70
- Hsp90
- heat shock protein 90
- EGFP
- enhanced green fluorescence protein
- Ub
- ubiquitin
- PARP
- poly(ADP-ribose) polymerase
- BiP
- Ig binding protein
- PDI
- protein disulfide isomerase
- IRE1α
- inositol requiring enzyme 1α
- MTT
- 1-(4,5-demethyldiazol-2-yl)-2,5-diphenyltetrazolium bromide
- FITC
- fluorescein isothiocyanate
- E3
- ubiquitin-protein isopeptide ligase
- HA
- hemagglutinin
- CMV
- cytomegalovirus
- FSC
- forward scatter
- SSC
- side scatter
- TRITC
- tetramethylrhodamine isothiocyanate
- ER
- endoplasmic reticulum
- CC
- coiled-coil domain
- Hu
- human
- Rh
- rhesus monkey.
REFERENCES
- 1.Himathongkham S., Luciw P. A. (1996) Virology 219, 485–488 [DOI] [PubMed] [Google Scholar]
- 2.Hofmann W., Schubert D., LaBonte J., Munson L., Gibson S., Scammell J., Ferrigno P., Sodroski J. (1999) J. Virol. 73, 10020–10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata R., Sakai H., Kawamura M., Tokunaga K., Adachi A. (1995) J. Gen. Virol. 76, 2723–2730 [DOI] [PubMed] [Google Scholar]
- 4.Nisole S., Stoye J. P., Saïb A. (2005) Nat. Rev. Microbiol. 3, 799–808 [DOI] [PubMed] [Google Scholar]
- 5.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., Ballabio A. (2001) EMBO J. 20, 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asaoka K., Ikeda K., Hishinuma T., Horie-Inoue K., Takeda S., Inoue S. (2005) Biochem. Biophys. Res. Commun. 338, 1950–1956 [DOI] [PubMed] [Google Scholar]
- 7.Chelbi-Alix M. K., Pelicano L., Quignon F., Koken M. H., Venturini L., Stadler M., Pavlovic J., Degos L., de Thé H. (1995) Leukemia 9, 2027–2033 [PubMed] [Google Scholar]
- 8.Gongora C., Tissot C., Cerdan C., Mechti N. (2000) J. Interferon Cytokine Res. 20, 955–961 [DOI] [PubMed] [Google Scholar]
- 9.Orimo A., Tominaga N., Yoshimura K., Yamauchi Y., Nomura M., Sato M., Nogi Y., Suzuki M., Suzuki H., Ikeda K., Inoue S., Muramatsu M. (2000) Genomics 69, 143–149 [DOI] [PubMed] [Google Scholar]
- 10.Tissot C., Mechti N. (1995) J. Biol. Chem. 270, 14891–14898 [DOI] [PubMed] [Google Scholar]
- 11.Song B., Gold B., O'Huigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. (2005) J. Virol. 79, 6111–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song B., Javanbakht H., Perron M., Park D. H., Stremlau M., Sodroski J. (2005) J. Virol. 79, 3930–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keckesova Z., Ylinen L. M., Towers G. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10780–10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Caballero D., Hatziioannou T., Zhang F., Cowan S., Bieniasz P. D. (2005) J. Virol. 79, 15567–15572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. (2004) Nature 427, 848–853 [DOI] [PubMed] [Google Scholar]
- 16.Cowan S., Hatziioannou T., Cunningham T., Muesing M. A., Gottlinger H. G., Bieniasz P. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11914–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatziioannou T., Cowan S., Von Schwedler U. K., Sundquist W. I., Bieniasz P. D. (2004) J. Virol. 78, 6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owens C. M., Song B., Perron M. J., Yang P. C., Stremlau M., Sodroski J. (2004) J. Virol. 78, 5423–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owens C. M., Yang P. C., Göttlinger H., Sodroski J. (2003) J. Virol. 77, 726–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towers G., Bock M., Martin S., Takeuchi Y., Stoye J. P., Danos O. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12295–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Griffero F., Li X., Javanbakht H., Song B., Welikala S., Stremlau M., Sodroski J. (2006) Virology 349, 300–315 [DOI] [PubMed] [Google Scholar]
- 22.Liberek K., Lewandowska A., Zietkiewicz S. (2008) EMBO J. 27, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 24.Frydman J., Hartl F. U. (1996) Science 272, 1497–1502 [DOI] [PubMed] [Google Scholar]
- 25.Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000) Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 26.Sherman M. Y., Goldberg A. L. (2001) Neuron 29, 15–32 [DOI] [PubMed] [Google Scholar]
- 27.Song B., Diaz-Griffero F., Park D. H., Rogers T., Stremlau M., Sodroski J. (2005) Virology 343, 201–211 [DOI] [PubMed] [Google Scholar]
- 28.Xu L., Yang L., Moitra P. K., Hashimoto K., Rallabhandi P., Kaul S., Meroni G., Jensen J. P., Weissman A. M., D'Arpa P. (2003) Exp. Cell Res. 288, 84–93 [DOI] [PubMed] [Google Scholar]
- 29.Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. (2005) J. Virol. 79, 8969–8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell E. M., Dodding M. P., Yap M. W., Wu X., Gallois-Montbrun S., Malim M. H., Stoye J. P., Hope T. J. (2007) Mol. Biol. Cell 18, 2102–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D. J., Sundquist W. I., Sodroski J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bukau B., Horwich A. L. (1998) Cell 92, 351–366 [DOI] [PubMed] [Google Scholar]
- 33.Yamauchi K., Wada K., Tanji K., Tanaka M., Kamitani T. (2008) FEBS J. 275, 1540–1555 [DOI] [PubMed] [Google Scholar]
- 34.Wolf D., Cammas F., Losson R., Goff S. P. (2008) J. Virol. 82, 4675–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf D., Goff S. P. (2007) Cell 131, 46–57 [DOI] [PubMed] [Google Scholar]
- 36.Wolf D., Hug K., Goff S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12521–12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd (1996) Genes Dev. 10, 2067–2078 [DOI] [PubMed] [Google Scholar]
- 38.Strasser A., O'Connor L., Dixit V. M. (2000) Annu. Rev. Biochem. 69, 217–245 [DOI] [PubMed] [Google Scholar]
- 39.Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 40.Mische C. C., Javanbakht H., Song B., Diaz-Griffero F., Stremlau M., Strack B., Si Z., Sodroski J. (2005) J. Virol. 79, 14446–14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 42.Cummings C. J., Mancini M. A., Antalffy B., DeFranco D. B., Orr H. T., Zoghbi H. Y. (1998) Nat. Genet. 19, 148–154 [DOI] [PubMed] [Google Scholar]
- 43.Jana N. R., Tanaka M., Wang G., Nukina N. (2000) Hum. Mol. Genet. 9, 2009–2018 [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi Y., Kume A., Li M., Doyu M., Hata M., Ohtsuka K., Sobue G. (2000) J. Biol. Chem. 275, 8772–8778 [DOI] [PubMed] [Google Scholar]
- 45.Bailey C. K., Andriola I. F., Kampinga H. H., Merry D. E. (2002) Hum. Mol. Genet. 11, 515–523 [DOI] [PubMed] [Google Scholar]
- 46.Kazemi-Esfarjani P., Benzer S. (2000) Science 287, 1837–1840 [DOI] [PubMed] [Google Scholar]
- 47.Marsh J. L., Walker H., Theisen H., Zhu Y. Z., Fielder T., Purcell J., Thompson L. M. (2000) Hum. Mol. Genet. 9, 13–25 [DOI] [PubMed] [Google Scholar]
- 48.Warrick J. M., Chan H. Y., Gray-Board G. L., Chai Y., Paulson H. L., Bonini N. M. (1999) Nat. Genet. 23, 425–428 [DOI] [PubMed] [Google Scholar]
- 49.Auluck P. K., Chan H. Y., Trojanowski J. Q., Lee V. M., Bonini N. M. (2002) Science 295, 865–868 [DOI] [PubMed] [Google Scholar]
- 50.Donaldson K. M., Li W., Ching K. A., Batalov S., Tsai C. C., Joazeiro C. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8892–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holmberg C. I., Staniszewski K. E., Mensah K. N., Matouschek A., Morimoto R. I. (2004) EMBO J. 23, 4307–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A. L. (2004) Mol. Cell 14, 95–104 [DOI] [PubMed] [Google Scholar]
- 53.Agostini I., Popov S., Li J., Dubrovsky L., Hao T., Bukrinsky M. (2000) Exp. Cell Res. 259, 398–403 [DOI] [PubMed] [Google Scholar]
- 54.Gurer C., Cimarelli A., Luban J. (2002) J. Virol. 76, 4666–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.