Introduction
Epithelial ovarian cancer (EOC) is the 4th leading cause of cancer deaths among USA women and the most common cause of death from gynecologic cancers (1, 2). It arises from ovarian surface epithelial (OSE) cells, and is currently classified by surgical and histological appearance (see Table 1), although the predictive value of this morphologic classification is limited (3).
Table 1.
Current concepts regarding the molecular pathology of specific histologic forms of epithelial ovarian carcinomaa
| Histology | Likely precursor | Molecular features |
|---|---|---|
| High-grade serous carcinoma | “De novo” in epithelial inclusion cysts | p53 mutation and BRCA1 dysfunction (promoter methylation) |
| High-grade endometrioid carcinoma | Epithelial inclusion glands/cysts | p53 mutation and BRCA1 dysfunction (promoter methylation) PIK3CA mutation |
| Low-grade serous carcinoma | Cystadenoma-borderline tumor-carcinoma sequence | Mutations in K-ras and/or b-raf |
| Mucinous carcinoma | Cystadenoma-borderline tumor-carcinoma sequence | Mutations in K-ras; ? p53 mutation associated with transition from borderline tumor to carcinoma |
| Low-grade endometrioid carcinoma | Endometriosis and endometrial-like hyperplasiab | Mutations in CTNNB1 (B-catenin gene) and PTEN with microsatellite instability |
| Clear cell carcinoma | ? endometriosis | ? PTEN mutation/loss of heterozygosity PIK3CA mutation |
PIK3CA is the gene at 3q26 that encodes the p110α subunit of phosphatidylinositol-3-kinase (reproduced from reference 3 with permission)
Endometriosis and adjacent low-grade endometrioid carcinoma share common genetic events such as loss of heterozygosity involving the same allele (i.e., PTEN). In contrast, high-grade and poorly differentiated endometrioid carcinomas are similar to high-grade serous carcinomas
However, while the exact cell of origin and the reasons underlying morphological differences in ovarian tumors are unclear, the histological differentiation of specific ovarian tumors appears to be due to the expression and activity of homeobox genes that regulate differentiation and proliferation of the Mullerian duct (4). The dismal outcome of EOCs is due to the majority of patients presenting with advanced stage III/IV disease. Despite improved median survival in patients with paclitaxel/carboplatin chemotherapy after surgical debulking, relapse occurs in most patients with advanced disease, and only 20% are alive and disease-free at 5 years. Although current efforts to improve outcomes focus on both earlier diagnosis and the incorporation of additional cytotoxic chemotherapy drugs and novel-targeted therapies into treatment, these efforts have not provided significant improvements in patient outcomes in comparison to paclitaxel/carboplatin therapy alone. The results of the ICON5 trial, for example, demonstrate that adding a third cytotoxic agent (gemcitabine, topotecan, or liposomal doxorubicin) to paclitaxel/carboplatin does not prolong progression-free survival in first-line therapy of EOC (5). Current targeted approaches include utilization of epidermal growth factor receptor (EGFR) family inhibitors, or inhibitors of Kit and platelet-derived growth factor receptor (PDGFR), although EGFR and Kit mutations, associated with targeted therapy responsiveness in other tumor types, are rare in EOC.
Like many solid tumors, EOC has a high degree of chromosomal instability, and both total and regional instability are associated with altered patient outcomes (6). Unpublished studies from the Gray group and the Ovarian Cancer SPORE support this contention (personal communication). EOC evolves through the multistep acquisition of genomic and epigenetic aberrations that initially deregulate normal cell growth control, followed by autonomous proliferation, and eventually other “hallmark” features of malignancy (7). Although there are only few “hallmarks”, many genomic aberrations contribute to the underlying process, and many of these events act in concert to generate the tumorigenic phenotype. Indeed, aberrations in single oncogenes frequently result in senescence or oncogene-induced death. Combined with the evidence for multiple different nonoverlapping defects in cancer cells, this suggests a major degree of complexity. An improved understanding of the underlying genomic aberrations driving EOC, and how they disrupt protein function leading to oncogenesis is greatly needed. Thus, we have advocated a comprehensive characterization of genomic anomalies in EOC as a common first step in studies that attempt to advance our understanding of ovarian oncogenesis and to discover promising new markers and targets for its therapy.
The selection of EOC as one of three tumor types for extensive characterization as part of The Cancer Genome Atlas (TCGA) project offers an excellent opportunity to extend our understanding of this disease. Although genomic mutations commonly affect only a limited number of genes in EOC, chromosomal copy number aberrations typically involve relatively large numbers of genes and other innovative techniques are needed in an attempt to determine a more limited list of genes that act as “key drivers” of oncogenesis. Using breast cancer (BC) as an example, the her2/neu oncogene is considered a “key driver” of the her2/neu amplicon on chromosome 17q, although multiple other nearby genes are frequently coamplified with her2/neu. Whether these coamplified genes are “innocent” passengers or act cooperatively, her2/neu remains an unanswered question. Theoretically, genes selected for mutation as well as “key drivers” of chromosomal copy number alterations that play a key role in the oncogenesis process will manifest significant aberrations at the transcription level and/or protein expression/function than other aberrant gene “passengers” at copy number changes. “Key drivers” represent potential markers for early diagnosis and targets for therapy, their identification are critical. Therefore, study of the transcriptional and proteomic effects of genomically aberrant genes in EOC is essential to determine their importance in the oncogenic process and to explore the antitumor efficacy and proteomic effects of therapies designed to specifically target the protein products of these aberrant genes. Alternatively, a systematic down regulation of each gene in an amplicon may expose phenotypic responses in cells where the genes are amplified. However, such approaches may miss critical aberrations with context-dependent effects such as those that manifest only in vivo. Moreover, as there are too many candidates for systematic analysis of all potential drivers at genomic aberrations, we have concentrated our efforts specifically on genomic changes that correlate with patient outcomes. This imperfect filter focuses on genes that are most likely to provide novel targets or molecular markers.
Mutations in Ovarian Cancer
Genomic mutations play a key role in the pathogenesis of multiple forms of cancer. High prevalence mutations (>5%) have been identified only in a limited number of genes in EOC. Genes known to develop mutations at high rate in certain EOC histologies and their frequency are shown in Table 1 (3-10). Current evidence, in particular the subtype-restricted expression patterns, suggests that specific mutations may be important in the pathogenesis of specific EOC subtypes. Moreover, germ-line mutations of BRCA1, BRCA2, and the hereditary non-polyposis coli (Lynch syndrome II) genes are all associated with DNA repair processes, a markedly increased risk of EOC, and account for 5–10% of all EOCs (11). Hereditary BRCA1/2-related EOCs tend to occur at an earlier age than sporadic tumors, possibly by their predisposition to genomic instability, and are usually high-grade serous tumors with p53 dysfunction.
Recent high-throughput approaches such as sequencing of specific functional groups of genes (i.e., the kinome) have not revealed other commonly mutated genes in EOC, although only limited sections of the genome have been probed (12). A large number of “rare” events have been identified that may act coordinately during oncogenesis. Whole genome sequencing has now been reported preliminarily for breast and colorectal cancers but has similarly not uncovered novel unknown mutations occurring at high frequency; rather, it has demonstrated that infrequent mutations affect a significantly higher number of genes in individual tumors than previously suspected (13). Moreover, there is a great diversity in the genes targeted, with few mutations in common. Thus, high-throughput sequencing by the Sanger Center (www.sanger.ac.uk), as proposed by TCGA in EOC, will likely identify a large number of low frequency mutations, although pathway or network analyses as part of a systems biology approach may demonstrate that these “rare” mutations result in coordinate aberrations in common critical processes. Therefore, it will be a challenge to determine specific mutations that play an important role in oncogenesis vs. gene mutations that result as a consequence of genomic instability.
By ectopically manipulating genes in mice OSE cells, it has been possible to induce a very poorly differentiated form of EOC by combining p53 loss with activation/overexpression of two of three oncogenes (myc, K-ras, and AKT1) (14, 15). While the relevance of this model to human EOC remains disputable, it has the potential to allow further confirmation for the role of specific oncogenes in EOC, as well as allowing potential evaluation of targeted therapeutics. Similar studies in human epithelium have demonstrated that introduction of SV40 large and small T antigen (TAg and tag) increase the number of cell divisions that normal OSE undergoes before entering senescence (16). Similarly, introduction of telomerase (hTERT) into these cells results in an extended life span in the absence of a transformed phenotype (17). Disrupting retinoblastoma function replaces TAg in cellular immortalization. Immortalized OSE can be transformed by the introduction of activated ras but not other oncogenes. Whether this represents a specific role in the transformation of OSE or complementation of TAg and hTERT is unknown. OSE cells provide potent reagents to determine mechanisms underlying immortalization and transformation (18). However, these studies, while supportive, do not definitively establish OSE as the EOC cell of origin.
Although the transcriptional and proteomic effects of mutations on protein expression and signaling in EOC as well as the roles played by these proteomic effects in oncogenesis remain only partly understood, mutations do allow identification of a number of logical potential markers and targets to facilitate earlier diagnosis and better treatment. On the other hand, successful targeting of the consequences of mutations in human EOC treatment has not been achieved. In an early example, despite promising preclinical and clinical data leading to the initiation of an international randomized phase II/III trial of p53 gene-therapy using replication-deficient adenoviral vectors in combination with first-line chemotherapy, a therapeutic benefit was not shown (19). However, there were multiple problems including some that potentially affected the successful intratumoral delivery of p53. Only a limited number of targeted therapeutics relevant to the genomic aberrations present in EOC has been assessed. Importantly, a number of therapeutics targeting aberrations relevant to EOC such as mutations in PIK3CA will enter clinical trials over the next year. The successful application of genomic mutations in EOC therapy will require not only effective delivery of the agent to the tumor cells, but a comprehensive understanding of the transcriptional and functional proteomic effects of these mutations, changes induced in response to in vitro and in vivo targeting of the protein product of the mutant gene, and its interaction with other genomic changes. After all, there are multiple genetic changes in EOC and it is possible that these may bypass the tumor dependence on a single mutation/change. Functional studies of the protein signaling and related pathways associated with or affected by mutant genes will facilitate “real time” monitoring of mechanisms associated with the ability of a cell to survive therapy targeted specifically toward an important mutational event.
Comprehensive Genomic Copy Number Screening
Genomic instability is very frequent in EOC and it represents at least two theoretically different processes in ovarian oncogenesis. Of less importance are those genomic areas that are destabilized as a result of the oncogenic process per se. More important are particular areas of the genome that may be targeted and selected specifically due to their functional importance for tumor initiation and progression. Although such anomalous areas of DNA may carry multiple genes, only a limited number of these are “key drivers” of the process. These “key drivers” are perceived as the most critical markers and potential treatment targets. An identification of the “key drivers” of genomic copy number aberrations could greatly improve our understanding of EOC and contribute to improved patient management. It is not clear, however, whether each genomic aberration harbors a single “key driver” or whether mulitple genes within a genomically aberrant region may contribute to “cooperative oncogenesis”. These two alternatives may be reflected in narrow alterations areas in contrast to large regional aberrations. Alternatively, the underlying mechanisms of fragile areas in the genome may determine the aberration type.
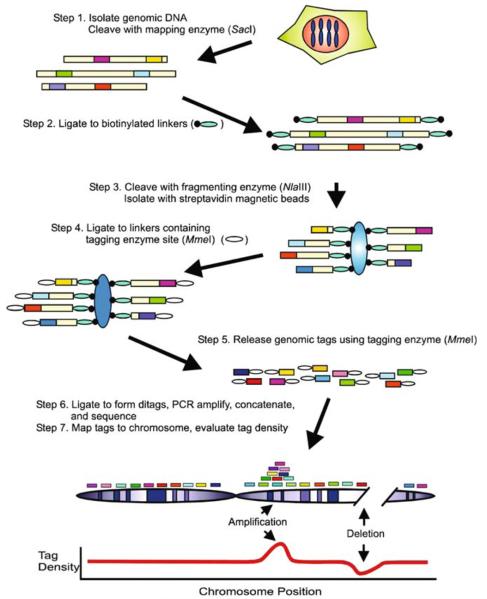
A number of relatively new high throughput technologies are now capable of profiling copy number changes across the entire genome in cancer. Comparative genomic hybridization (CGH) is one such technology and has recently become a conventional first step in the identification of potentially important markers and targets in EOC. The resolution of single nucleotide polymorphism (SNP)-based CGH approaches, and that of other recent methodologies developed to identify and map genome wide copy number changes in EOC, such as digital karyotyping (Fig. 1) or ROMA, has improved rapidly and dramatically (20, 21). A novel emerging molecular inversion probe (MIP) has the potential to supplant previous low resolution methods. Conventionally, strategies to distinguish candidate “drivers” from other less important coamplified or deleted genes in areas of chromosomal copy number change have focused on finding aberrations that induce significant changes at both the RNA and protein levels resulting in alterations in the phenotypic behavior of manipulated cells in vitro, and in significant changes to patient outcome (22). Important induced changes at the transcriptional level generally entail a significant increase (amplified genes) or decrease (deleted genes) in the amount of mRNA, and changes in expression and, more importantly, function at the protein level. For example, her2/neu gene amplification in BC dramatically increases not only HER2 protein expression but, importantly, activation of downstream proteins such as protein kinase B (AKT). These “drivers” may be applicable through small interfering (si)RNA technology or overexpression of candidate genes. The challenge is to determine which strategies and assays will reveal the spectrum of functional candidates within a genomic aberration. For example, knockdown and expression approaches may result in false positive data when a gene may be important for normal cellular function but is not specifically involved in oncogenesis.
Fig. 1.
Digital karyotyping approach: Dark boxes represent genomic tags; Small ovals represent linkers; Larger ovals represent streptavidin-coated magnetic beads (From (20) with permission)
In EOC, frequent chromosomal losses have been observed at 4p11-14, 9p, 11p14-15, 13q22-34, and 18q22-23, while common areas of gain include 1q22, 3q26, 5p15, 7q32-36, 8q23-24, 12p11-12, 17q12-23, 20p, and 20q12-13 (23). Multiple studies have identified candidate “drivers” by exploring genes that impact transcription, protein expression and function, cell behavior in vitro, and patient outcome. Since inhibition of protein function is more likely to occur than restoration of function, a perception reinforced by the clinical failure in EOC to exploit p53 loss, and by the success of the anti-HER2 monoclonal antibody trastuzumab in BC, most studies have focused on areas of chromosomal gain (amplicons) rather than on areas of deletion. Candidate “drivers” at areas of copy number anomaly have been identified. They include the small G protein RAB25 at 1q22, evi1, protein kinase C iota (PKCi), SnoN, BCL6, and PIK3CA [the gene that encodes the p110α protein subunit of phosphatidylinositol-3-kinase (PI3K)] at 3q26.2, myc and PVT1 at 8q24.2, remodeling and spacing factor 1 (rsf-1, HBXAP) and PAK1 at 11q13, her-neu at 17q12, AKT2 at 19q13.2 and ZNF217 and EEF1A2, both at 20q13.2 (21, 24-31).
In addition to mutations and DNA copy number changes, rearrangements, epi-genetic changes, and imprinting also affect cellular function potentially identifying important markers and therapy targets. Several imprinted genes including ARH1 (NOEY2) and LOT1/ZAC function as tumor suppressor genes, and these and other potential tumor suppressors (i.e., WWOX, HIC1, OVCA1, and OVCA2) may be lost in EOC, either by epigenetic mechanisms and/or deletions (32-33). New comprehensive profiling approaches to detect methylation patterns and to explore imprinted genes are likely to identify new candidates (34). To date, however, none of the aforementioned potential targets have had an impact on patient care in the clinic; in fact, clinical studies have not yet even begun to assess therapies that are targeted to the majority of these aberratations. Most current clinical studies in EOC still focus on alternative cytotoxic chemotherapy regimens, in addition to treatments that target EGFR, despite the lack of evidence of genomic aberrations in the gene that encodes EGFR, and vascular endothelial growth factor (VEGF) or its receptors; indeed, it may be VEGF-targeted therapies that will prove most effective of this particular list of treatments (35).
Transcriptional Profiling
Transcriptional profiling approaches have identified expression patterns associated with the different histological types of EOC (36). The similar expression patterns in EOCs and Mullerian tissues and the comparable histological features strengthen concerns regarding the tissue of origin and differentiation patterns in EOC. A number of transcriptional profiles that correlate with patient outcomes have been identified. However, none of these parameters have sufficient power or been confirmed to warrant alterations in patient management. A combination of a platin and taxane remain the primary therapy for EOC. However, since only 70% of patients respond to this chemotheapy and fewer than 20% are cured by it, approaches to identify patients likely to respond and, in particular, unresponsive are desperately needed to allow triage to alternative therapies including clinical trials.
Transcriptional profiling approaches have also identified a number of candidate genes relevant to EOC. The availability of public databases such as www.oncomine.org, and the prerequisite of journals and granting agencies that transcriptional profiling data be deposited in MIAME-compliant databases such as the Gene Expression Ominibus (GEO http://www.ncbi.nlm.nih.gov/geo) will provide a robust resource. This will be greatly strengthened by the efforts of TCGA. Indeed, it behoves the research community to rapidly and efficiently exploit the outcomes of these efforts.
However, transcriptional profiling has major limitations. The absence of the clear identity of the precursor cell of each EOC histotype and the limited population of “relevant stem cells” make interpretation of transcriptional profiles difficult. Is the “pattern” observed in a tumor representative of the underlying defects driving oncogenesis, or is simply a reflection of the tissue of origin of the tumor? Moreover, if there is a “tumor stem cell” population in EOC, does the observed transcriptional profile have relevance to the stem cell population, or does it reflect the irrelevant but predominant differentiated cell population? A potentially critical problem regarding current transcriptional profling efforts is the alternate and aberrant splicing that occurs in cancer and specifically in EOC. Indeed, our recent data indicate that EOC may be a disease of “aberrant” splicing, since a wide variety of splice aberrations are present in multiple critically important genes (37). DNA copy number errors may contribute to EOC by causing aberrant splicing. We have identified functionally relevant splicing aberrations in multiple genes implicated in the inititation and progression of EOC. Therefore, new splicing arrays such as those being incorporated into the TCGA may reveal a whole new level of complexity.
Functional Proteomics
As discussed earlier, novel technologies such as whole genome sequencing and SNP-based CGH have revolutionized our ability to visualize genomic changes in cancer cells. Quantitative polymerase chain reaction (QPCR) and transcriptional profiling are both well established approaches to measure changes in mRNA levels in cancer cells. DNA copy number, mutation, rearrangement, and methylation alterations act in coordination to alter critical cellular functions through changes at the protein level; however, because of posttranslational modification and other events, protein levels and function offer a limited correlation to DNA and RNA changes. Thus, it is necessary to develop and implement functional proteomics approaches that can assay posttranslational and other protein modifications. In terms of our ability to globally assess cancer cells across DNA, RNA, and protein platforms, functional proteomics is significantly less advanced than genomic technologies. An efficient and clinical applicable functional proteomics strategy is needed.
Mass spectroscopy-based approaches represent the emerging most exciting technology, although they are not applicable to patient management. Currently, they are low throughput, in depth analytical strategies for a limited number of specimens. New mass spectroscopy platforms and the efforts of the Clinical Proteome Analysis Technology Consortium provide new strategies and opportunities. ELISA, Western blotting, and immunohistochemistry are still the most robust and, therefore, the standard approaches to assess protein expression and activation. However, these conventional proteomic technologies are semiquantitative and lack high-throughput capability and capacity to comprehensively assess changes in the activation of the multiple signaling pathways and events that may be perturbed by genomic aberrations. Thus, efforts currently focus on mass spectrometry and particularly on protein array platforms in an effort to address these shortcomings.
We have focused our efforts on the development of a novel moderate- throughput and quantitative proteomic technology known as reverse phase tissue lysate array (RPPA) (38-40). Using this approach, cell/tumor protein lysates are arrayed in serial dilutions on nitrocellulose (or other affinity reagent)-coated glass slides and probed with monospecific antibodies that quantify protein expression or a specific posttranslational modification (e.g., phosphorylation), followed by signal amplification using a colorimetric or similar methodology. This approach can be multiplexed with fluorescent, near infrared, or quantum dot methologies; however, we have restricted our approaches to single reagents to decrease the possibility of interference by multiple probes. Quantification involves software-based construction of serial dilution-signal intensity curves for individual samples with logistic fit models that use logarithmic values representative of the intensity of individual spots in serial dilutions of each sample. A representative value of each sample curve on the slide is then generated and used as a relative quantification of the amount of the particular protein in each sample. Absolute quantification can be achieved by comparison to samples with known amounts of protein or of the immunizing peptide. Protein loading is corrected among samples using loading controls of relatively constant protein concentrations or using an average of all proteins assayed. Each slide is stained with a single antibody under optimal conditions to allow comparison and quantification. Slides can be replicated robotically, allowing analysis of many samples (currently up to 1 000 per slide) with multiple antibodies targeting functionally relevant antigens in signaling and other pathways (we have validated almost 100 different antibodies). As the assay is “solid phase” it requires little starting material (nanograms) and is extremely sensitive (femtograms of target). Thus, it is ideal for valuable patient samples and is highly applicable to laser-aided microdis-section or needle biopsies. A major drawback of the RPPA approach is the lack of validated monospecific antibodies able to assess expression and activation of the majority of proteins that make up the proteome. New technologies for the generation of affinity reagents as well as improved validation approaches are ameliorating this challenge. Further, RPPA is primarily a candidate gene approach based on known proteins with high quality antibodies, and, therefore, it is not ideal for discovery. It may, however, be useful for the discovery of proteomic “patterns” indicative of prognosis or predictive of response to therapy.
A major advantage of RPPA is that it can assess activity of many relevant signaling pathways at multiple levels. This allows an integration of information across several pathways. Our initial data suggest that many of the “standard” signaling diagrams may not be applicable to patient samples. Further, genomic aberrations may impact signaling pathways at different levels. The ability to comprehensively analyze particular signaling cascades will provide evidence to identify which pathways are important in tumor progression. RPPA is thus ideally suited to determine a “global” proteomic view of signaling changes induced by genomic aberrations and occurring in response to the targeting of proteins encoded by these aberrations. Its major limitation is that it does not assay spatial organization; thus it is best combined with immunohistochemistry.
RPPA can also play a major role in drug development and implementation. The process from target identification, to drug evaluation and implementation, into patient care is slow and fraught with a high failure rate. RPPA can be used early in drug development as a secondary screen to identify potential “hit” targets from high-throughput screens. It can also be used in structure/function analysis to decrease the likelihood of off-target activity. RPPA may also identify critical feedback loops that lead to unexpected drug effects and rationale therapy combinations. Indeed, we have utilized RPPA to comprehensively map feedback loops driven by inhibitors of mTOR and AKT. Such loops may play a role in resistance to targeted therapeutics or lead to unexpected deleterious effects such as toxicity or increased tumor growth. RPPA, particularly when used early in drug development, may also be capable of identifying patterns of protein expression representative of likelihood of response to targeted therapeutics. It also allows determination of appropriate dosing and scheduling of drugs (Hennessy, et al., in preparation). Despite the difficulty in obtaining serial biopsies for tailoring dose/response in human tumors, RPPA may be applicable to accessible surrogate tissues such as white blood cells and, more importantly, may prove critical to the identification and validation of targets for molecular imaging.
Early Detection of Ovarian Cancer
The great majority of EOC patients are diagnosed with advanced disease (stage III/IV). Early detection implies the diagnosis of EOC at an early stage in its development with resultant expected improvements in patient outcome using conventional treatment strategies. For early diagnosis, technologies must be available that are capable of detecting small tumors at an early stage; in addition, since screening of the entire population with such technologies is often not practical, guidelines as to patient risk assessment are often necessary to appropriately target technologies for the purposes of early cancer diagnosis and prevention. Age, for example, is a major factor used to delineate risk in current population screening approaches to breast and colorectal cancers. As with BC, BRCA1 and BRCA2 mutations are associated with a particularly high risk of EOC in many cases.
Serum or urine markers have clear potential utility in identifying patients with early stage EOC as well as those at high risk who will likely benefit from chemo-prevention. It has become the “holy grail” of most studies aimed at early detection of EOC. Such markers also offer foreseeable means to better facilitate treatment planning and even individualization of care than is currently possible. The most well-known serum marker associated with EOC is CA125. Its routine use in screening is impeded by a lack of sufficient sensitivity and specificity (41). Since EOC prevalence is low and secondary screening approaches such as CAT scans are expensive and inaccurate, an extremely high specificity is necessary to prevent frequent false positive data with resultant morbidity and high financial costs. Combinations of multiple markers will likely be necessary to provide sufficient sensitivity and specificity for early diagnosis. Thus, it has been theorized that there are proteomic panels that can accurately identify tumors at an early stage of development. Indeed, several groups have applied proteomic approaches such as surface-enhanced laser desorption/ionization-time of flight (SELDI-TOF) mass spectrometry (MS) for identification of the theorized serum biomarker panels that will allow early detection of EOC. For example, there is evidence that serum transthyretin (TTR), β-hemoglobin (Hb), apolipoprotein (Apo), AI, and transferrin (TF), when combined with CA125, significantly improve the detection of early stage EOC (42). However, although potentially exciting, these and similar studies suffer from potential drawbacks including impracticality, expense, difficulty in identifying specific candidate proteins at recognized peaks, and lack of prospective validation in large studies. Nevertheless, they have important potential to significantly impact EOC mortality.
RPPA may have a role in the analysis and evaluation of serum markers. With appropriate high quality validated antibodies, RPPA can be developed with single antibodies rather than sandwich approaches. This provides a potential advantage over ELISA or Luminex methodologies. Indeed, with the sensitivity of RPPA, it may also be possible to quantify low levels of markers that cannot be assessed effectively by other methods. Further, the solid phase approach may be less sensitive to “interference” by contaminating compounds. We have successfully quantitated potential markers in as little as one nanoliter of serum when compared with the 100–500 μl required for most ELISA assays. The ability to analyze up to 1000 samples at a time greatly decreases cost when compared with ELISA and Luminex.
Although data regarding the potential usefulness of circulating tumor cells in EOC are currently very limited, these as well as ascitic fluid cells can theoretically provide an easily accessible and abundant source of tumor genome and proteome that may be useful in early cancer detection and in novel molecular studies of markers and therapy targets. Circulating and ascitic tumor cell DNA in EOC patients is thus currently being studied for a potential role in early diagnosis, as a prognostic factor, and in monitoring response to treatment (43). One early study found peripheral blood circulating tumor-specific p53 sequences in only a small proportion (15.3%) of FIGO stage III/IV EOC patients, demonstrating that this approach, as it stands, will not be of benefit for early EOC detection (44). However, current methods for enrichment of circulating tumor cells including immunomagnetic enrichment technologies such as CellSearch™ (Veridex, LLC) or enhanced density gradient systems may increase yield (45, 46). The application of sensitive mass spectrometry approaches such as those used by Sequenom (San Diego, CA) to assess fetal DNA in perinatal diagnosis may also have major advantages. A major challenge remains the ability to detect and amplify, without error, DNA from as little as 1–5 cancer cells per each 10 ml of blood. Expansion of the list of probed genomic changes to more than a limited number of specific p53 sequences may also increase yield. As the sensitivity of these assays improves, the usefulness of circulating tumor cells in guiding early diagnosis and treatment may increase.
Novel high-throughput molecular technologies are adding multiple new possibilities for early EOC detection, particularly with integration of data across DNA, RNA, and protein platforms. Ideally, such markers will not only allow early detection of cancer but will also facilitate prognosis and prediction/treatment planning. In other words, they should ideally provide information about the presence of both current and potentially exploitable molecular targets. These studies may also offer a route to preferentially target treatments guided by specific genomic or other cancer cell changes. However, the application of such a global approach to early detection is still very much in its infancy.
Challenges
Before studies that link genomics to the discovery of new targets and molecular markers are ready for routine clinical implementation, many challenges must be addressed and overcome. As mentioned earlier, potential targets discovered by the approaches described in this chapter have yet to be tested in EOC clinical trials. The “accessibility” of the target itself may be partly responsible in some cases. HER2, for example, is a membrane protein with a large and easily accessed extracellular domain, while many of the potentially important targets already uncovered in EOC are intracellular and more difficult to attain. Indeed, the success of targeting HER2 in BC led to a number of clinical trials in EOC. However, based on those studies, it appears that less than 10% of patients express elevated levels of HER2. This makes the successful completion of targeted therapeutic trials against HER2 technically impractical in a relatively uncommon disease such as EOC. Moreover, while targeted monoclonal antibodies such as trastuzumab are a major part of current targeted therapy armamentarium, they are generally not useful for intracellular targets. More interaction between basic, translational, and clinical investigators along with an improved general understanding of the processes involved, and the events that drive each of these stages will also facilitate more logical and targeted translation of the results of preclinical studies into human clinical trials. At present, while marker and target discovery using high throughput genomic approaches with initial validation using transcriptional and proteomic technologies is becoming easier to implement in preclinical studies, many challenges lie in the way of their practical application to human clinical trials. Therefore, each stage of research must be planned in a fashion that better considers how best to integrate any derive findings and data into subsequent stages of research. Concurrent development of targeted therapeutics and molecular markers capable to direct clinical evaluation (theragnostics) is critically important.
Studies utilizing human tumor cell lines have been criticized for their inability to adequately represent the molecular heterogeneity of human cancers in vivo. In some cases, this may contribute to failure to replicate findings from in vitro studies in human clinical trials. Newly developed targeted treatments have frequently shown promising activity in cell line studies but fail to subsequently demonstrate efficacy in human cancer patients. Multiple reasons have been postulated to explain the difficulty in translating early studies to improved patient outcomes. Most of the reasons remain purely speculative. However, it is quite likely that the use of small numbers of cell lines and the failure to match molecular aberrations (i.e., underlying genetic changes) in cell lines to genomic and proteomic changes in specific human tumors are some of the most important underlying problems. Therefore, it is probable that using baseline characterization of molecular abnormalities in a large number of cell lines to link particular cell lines to populations of patient tumors will allow better in vitro modeling of the molecular heterogeneity of human tumors. This will require development of a large set of EOC cell lines that are comprehensively characterized for genomic, transcriptional, and proteomic aberrations. We have implemented this process with academic/industrial partnerships and philanthropic support to embark on a major genomic, transcriptional, and functional proteomic evaluation of over 40 EOC cell lines and isogenic pairs that reflect known genomic aberrations in EOC patients. These lines will also be characterized for responsiveness to therapeutics with a major emphasis on targeted drugs. The resulting database will be made available to the research community to facilitate improved patient management. Indeed, we request that investigators provide additional cell lines to aid in their characterization. The data and lines will be shared with the community to fulfill our covenant with patients who donated the material and with the public that provides support for EOC research.
Likewise, the development of more appropriate animal tumor models driven by specific genetic aberrations will be necessary. Although current therapies are mainly developed based on the “average” patient and not embracing the concept of patient and tumor heterogeneity, overcoming these aforementioned difficulties may allow identification of effective targeted therapies in specific patient tumors that may otherwise be able to be dismissed as inactive in clinical trials. It will be a challenge, however, to introduce personalized molecular medicine in a relatively uncommon disease such as EOC, particularly because of difficulty in implementing clinical trials. We may need to “borrow” information and strategies derived from more common tumors. Thus, as discussed in the next section, as high throughput approaches become more important to study the genome and the proteome in pre-clinical and clinical research, the creation of widely accessible and user-friendly databases will be essential to maximize their impact and avoid effort duplication. With such resources, characterization of the genomic, transcriptional, and proteomic aberrations of human tumor and cell line panels, along with the responsiveness of each to multiple therapies, will advance our understanding of the heterogeneity of EOC and facilitate logical studies that utilize more targeted cell lines, along with predictive drug activity models in specific populations of EOC patients.
Following the identification of high-quality target tools, including chemical genomics and siRNA, it will be necessary to apply them concurrently with high-throughput genomic and proteomic studies to allow determination of on- and off-target effects of inhibition and pathway crosstalk. Although siRNA validation offers specific and effective knockdown, it may not mimic the effects of drugs that potentially have other, including off-target, functions. Further, because of the slow action of siRNA, the cells studied may represent those that survived the stress of siRNA knockdown of critical targets. These cells may have extensive “pathogenic” rewiring of critical pathways, making interpretation of the data challenging. Early in development, methods that combine novel targeted treatments with standard radiation or chemotherapy should also be sought as this may better facilitate translation of novel targets to clinical trials.
Clinical trials will also need to advance in a fashion that allows clinical drug development to keep pace with preclinical target development (47). Mandated tumor biopsies with well-designed correlative studies in molecular-marker-driven trials will be necessary for the efficient evaluation of novel targeted therapeutics. It will be crucial to distinguish on- from off-target activity to prevent the elimination of a good target because of an incorrect drug. We must begin to employ the concept of “biologically relevant dose” rather than maximum tolerated dose (MTD) to determine drug dosing by correlating biologic effects with clinical efficacy; this open the possibility to merge the three clinical trial phases into one continuous clinical/biological assessment of drug efficacy in patients. The goals of Phase I trials in particular will need to be addressed if the initial drug testing is not driven by MTD. In preclinical studies when no validated mechanism(s) have been discovered, large, randomized Phase III trials are particularly necessary to assess drugs that benefit a small proportion of cancer patients to separate likely responders from nonresponders. After drug approval, studies must continue to improve the identification of likely responders and associated tumor molecular aberrations, to facilitate early identification of nonresponders and to monitor off-target effects of each drug. When emergent resistance in tumors possessing the target is observed, it is important to initiate biological studies to determine molecular mechanism(s) likely to be predictable secondary events in many cases. Indeed, well-designed preclinical studies should uncover potential mechanisms of resistance to targeted therapies before resistance becomes a clinical problem in patients, particularly as functional proteomic technologies improve. Comprehensive characterization of human ovarian tumor genomic and proteomic aberrations, and their similarity to particular groups of EOC cell lines, may ultimately make the preclinical to clinical translation of targets and specific therapies a more logical and predictable process than it is at the present.
Data Integration
As studies characterize EOC cell lines and human EOCs using high-throughput genomic, transcriptional, and ultimately proteomic approaches, it is critical that adequate and centralized computational infrastructure, in addition to bioinformatics and biostatistical support, be developed to allow storage and utilization/integration of the vast and highly heterogeneous data derived from “omics” technologies (48). Such a computational resource should be made available to all investigators in a manner that is easy to use but also protects patient information and confidentiality. This will avoid duplication of efforts. In addition, these resources should facilitate data mining, retrieval, and automatic analysis with statistical software packages such as R or Matlab, thus allowing automatic high-throughput data integration across genomic, transcriptional, and proteomic platforms, between datasets (i.e., to model specific groups of cell lines to human ovarian tumors with similar patterns of molecular aberrations), and analysis of the association of specific aberrations and changes with clinical endpoints, and with the characterized responsiveness of cell lines to multiple targeted and chemotherapies. Repeated updating should be facilitated as novel “omics” technologies are continually introduced and upgraded (49). Such access would also allow and encourage novel bioinformatics and biostatistical approaches that further our ability to work with and integrate large amounts of data across multiple platforms to advance our understanding of the pathogenesis and molecular biology of EOC.
Potential Pitfalls
Overall, we are still limited by poor understanding of the molecular mechanisms underlying the development and selection of genomic aberrations in EOC. Moreover, the complexity of molecular changes raises the very daunting possibility that the “key driver” hypothesis is overly simple. If each of the large number of molecular anomalies plays an individual minor role in most EOCs, indicating that they are truly molecularly heterogeneous, then identification and targeted therapy planning will be considerably more difficult and complicated than is presumed today.
Conclusion
EOC is characterized by a large number of complicated genomic and proteomic changes with functional consequences that converge to create the relatively few “hallmarks” of cancer. Novel high-throughput technologies enable us to comprehensively profile these changes and identify “key drivers” in oncogenesis. Although these “key drivers” are potentially important targets for patient treatment, currently, their clinical application is considerably more difficult than their identification. Thus, basic, translational, and clinical studies must evolve in a reciprocal fashion as part of an integrated and overlapping continuum. As our capacity to investigate EOC molecular changes increases exponentially, more organized and accessible data collection as well as more formal bioinformatics and biostatistical support is becoming necessary. The current focus of the TCGA on EOC offers a major opportunity to the research community that will hopefully be rapidly translated into improved patient outcomes. However, the complexity of genomic changes may preclude a comprehensive understanding of the many interactions of EOC genetic and proteomic aberrations. Indeed, this complexity will likely bypass the ability of current reductionist approaches to deal with the amount and complexity of data. New computational and systems biology approaches may allow generation of usable models that result in testable predictions.
Finally, data from high-throughput approaches must be integrated with the results of other more conventional research studies to allow a comprehensive and real understanding of EOC. EOC pathogenesis involves a number of cellular functions such as DNA repair and intracellular kinase signaling that are not only influenced by genomic aberrations, but potentially, by other processes, as well, including autocrine and membrane-initiated signaling loops involving molecules such as VEGF and phospholipids including lysophosphatidic acid (LPA) (3, 50). Knowledge of these and other processes will expand our knowledge of potential markers and targets, allowing the formulation of approaches that best assimilate all of these data. After a recent important study in EOC pathogenesis identifying a specific role of changes on homeobox gene expression, researchers are now attempting to integrate this novel finding with the presence of EOC histological-specific mutations and other genomic changes. These studies may ultimately uncover the sequence of events underlying the initiation of EOC oncogenesis (50).
References
- 1.Jemal A, et al. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy BT, Mills GB, et al. Ovarian cancer: Homeobox genes, autocrine/paracrine growth, and kinase signaling. Int J Biochem Cell Biol. 2006;38:1450–1456. doi: 10.1016/j.biocel.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Taylor HS, Vanden Heuvel GB, Igarashi P. A conserved Hox axis in the mouse and human female reproductive system: late establishment and persistent adult expression of the Hoxa cluster genes. Biol Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 5.International Collaborative Ovarian Neoplasm Group Paclitaxel plus carboplatin versus standard chemotherapy with either single-agent carboplatin or cyclophosphamide, doxorubicin, and cisplatin in women with ovarian cancer: the ICON3 randomised trial. Lancet. 2002;360:505–515. doi: 10.1016/S0140-6736(02)09738-6. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki S, Moore DH, III, Ginzinger DG, et al. An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 2000;60:5382–5385. [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 8.Levine DA, Bogomolniy F, Yee CJ, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 9.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 10.Shih IeM, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narod SA, Boyd J. Current understanding of the epidemiology and clinical implications of BRCA1 and BRCA2 mutations for ovarian cancer. Curr Opin Obstet Gynecol. 2002;14:19–26. doi: 10.1097/00001703-200202000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Stephens P, Edkins S, Davies H, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 13.Sjoblom T, Jones S, Wood LD, et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Orsulic S, Li Y, Soslow RA, et al. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinulescu DM, Ince TA, Quade BJ, et al. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- 16.Yang G, Rosen DG, Mercado-Uribe I, et al. Knockdown of p53 combined with expression of the catalytic subunit of telomerase is sufficient to immortalize primary human ovarian surface epithelial cells. Carcinogenesis. 2007;28:174–182. doi: 10.1093/carcin/bgl115. [DOI] [PubMed] [Google Scholar]
- 17.Alvero AB, Fishman DA, Qumsiyeh MB, et al. Telomerase prolongs the lifespan of normal human ovarian surface epithelial cells without inducing neoplastic phenotype. J Soc Gynecol Investig. 2004;11:553–561. doi: 10.1016/j.jsgi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Auersperg N, Wong AS, Choi KC, et al. Ovarian surface epithelium: biology, endocrinology and pathology. Endocr Rev. 2001;22:255–288. doi: 10.1210/edrv.22.2.0422. [DOI] [PubMed] [Google Scholar]
- 19.Zeimet AG, Marth C. Why did p53 gene therapy fail in ovarian cancer? Lancet Oncol. 2003;4:415–422. doi: 10.1016/s1470-2045(03)01139-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih IeM, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci USA. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennessy BT, Nanjunden M, Cheng KW, Nolden L, Mills GB. Ovarian cancer: identification of remodeling and spacing factor 1 (rsf-1, HBXAP) at chromosome 11q13 as a putative oncogene in ovarian cancer (News and Commentary) Eur J Hum Genet. 2006;14:381–383. doi: 10.1038/sj.ejhg.5201570. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe T, Imoto I, Kosugi Y, et al. A novel amplification at 17q21-23 in ovarian cancer cell lines detected by comparative genomic hybridization. Gynecol Oncol. 2001;81:172–177. doi: 10.1006/gyno.2001.6132. [DOI] [PubMed] [Google Scholar]
- 24.Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 25.Cheng KW, Lahad JP, Kuo WL, et al. The Rab 25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 26.Eder A, Sui X, Rosen D, et al. Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:12519–12524. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown LA, Irving J, Parker R, et al. Amplification of EMSY, a novel oncogene on 11q13, in high grade ovarian surface epithelial carcinomas. Gynecol Oncol. 2006;100:264–70. doi: 10.1016/j.ygyno.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 28.Hughes-Davies L, Huntsman D, Ruas M, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 29.Anand N, Murthy S, Amann G, et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 30.Schraml P, Schwerdtfeger G, Burkhalter F, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163:985–992. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, Maines-Bandiera S, Kuo WL, et al. Multiple roles of the candidate oncogene znf217 in ovarian epithelial neoplastic progression. Int J Cancer. 2007;120:1863–73. doi: 10.1002/ijc.22300. [DOI] [PubMed] [Google Scholar]
- 32.Kamikihara T, Arima T, Kato K, et al. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 33.Strathdee G, Appleton K, Illand M, et al. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158:1121–1127. doi: 10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei SH, Balch C, Paik HH, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12:2788–2794. doi: 10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- 35.Wright JD, Hagemann A, Rader JS, et al. Bevacizumab combination therapy in recurrent, platinum-refractory, epithelial ovarian carcinoma: a retrospective analysis. Cancer. 2006;107:83–89. doi: 10.1002/cncr.21969. [DOI] [PubMed] [Google Scholar]
- 36.Smith DI. Transcriptional profiling develops molecular signatures for ovarian tumors. Cytometry. 2002;47:60–62. doi: 10.1002/cyto.10042. [DOI] [PubMed] [Google Scholar]
- 37.Nanjundan M, Zhang F, Schmandt R, Smith-McCune K, Mills GB. Identification of a novel splice variant of AML1b in ovarian cancer patients conferring loss of wild-type tumor suppressive functions. Oncogene. 2007;26:2574–84. doi: 10.1038/sj.onc.1210067. [DOI] [PubMed] [Google Scholar]
- 38.Charboneau L, Scott H, Chen T, et al. Utility of reverse phase protein arrays: applications to signalling pathways and human body arrays. Brief Funct Genomic Proteomic. 2002;1:305–315. doi: 10.1093/bfgp/1.3.305. [DOI] [PubMed] [Google Scholar]
- 39.Sheehan KM, Calvert VS, Kay EW, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array (RPPA): validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoetic stem cells (HSC) Mol Cancer Ther. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 41.Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 42.Kozak KR, Su F, Whitelegge JP, et al. Characterization of serum biomarkers for detection of early stage ovarian cancer. Proteomics. 2005;5:4589–4596. doi: 10.1002/pmic.200500093. [DOI] [PubMed] [Google Scholar]
- 43.Judson PL, Geller MA, Bliss RL, et al. Preoperative detection of peripherally circulating cancer cells and its prognostic significance in ovarian cancer. Gynecol Oncol. 2003;91:389–394. doi: 10.1016/j.ygyno.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Swisher EM, Wollan M, Mahtani SM, et al. Specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193:662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 45.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 46.Muller V, Stahmann N, Riethdorf S, et al. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Can Res. 2005;11:3678–3685. doi: 10.1158/1078-0432.CCR-04-2469. [DOI] [PubMed] [Google Scholar]
- 47.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Gorlitsky R, Almeida JS. From XML to RDF: how semantic web technologies will change the design of ‘omic’ standards. Nat Biotechnol. 2005;23:1099–1103. doi: 10.1038/nbt1139. [DOI] [PubMed] [Google Scholar]
- 49.Almeida JS, Chen C, Gorlitsky R, et al. Data integration gets ‘Sloppy’. Nat Biotechnol. 2006;24:1070–1071. doi: 10.1038/nbt0906-1070. [DOI] [PubMed] [Google Scholar]
- 50.Cheng W, Liu J, Yoshida H, et al. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]