Abstract
Cancer stem cells represent a novel therapeutic target. The major challenge in targeting leukemic stem cells (LSCs) is finding therapies that largely spare normal hematopoietic stems cells (HSCs) while eradicating quiescent LSCs. We present a mathematical model to predict how selective a therapy must be to ensure that enough HSCs survive when LSCs have been eradicated. Stem cell population size is modeled as a birth-death process. This permits comparison of LSC and HSC eradication times under therapy and calculation of the number of HSCs at the time of LSC eradication for varied initial population sizes and stem cell death rates. We further investigate the effects of LSC quiescence and resistance mutations on our predictions. From a clinical point of view, our models suggest criteria by which cancer stem cell therapy safety can be assessed. We anticipate that in conjunction with experimental observation of cancer stem cells killing rates, our results will be useful in screening targeted therapies for both hematologic and solid tumor malignancies.
Keywords: cancer stem cell, chronic myelogenous leukemia, recurrence, mutation, quiescence, rational drug design
Introduction
Cancer stem cells play roles in both solid tumors (1–10) and hematopoietic malignancies (11–15). Cancer stem cells are involved in disease pathogenesis and progression as well as in relapse and resistance to therapy (8, 9, 14, 15). Therapies proposed to target cancer stem cells include: monoclonal antibodies (16); vaccines (17); apoptosis induction via oxidative stress responses, NF-κB inhibition and p53 activation (18); drugs targeting the Notch and hedgehog (Hh) signaling pathway (19, 20); and other targeted therapies (21–23).
One major concern in targeting leukemic stem cells (LSCs) is the risk of killing large numbers of non-malignant hematopoietic stem cells (HSCs). Therapy designed to completely eradicate cancer stem cells could lead to dangerously low HSC populations, threatening their ability to maintain homeostasis and tissue repair. Recognizing this threat, therapies are being designed to selectively eliminate cancer stem cells in acute myelogenous leukemia (18, 24–26), chronic myelogenous leukemia (CML) (27), and solid malignancies (23). However, it is unclear how selective a therapy must be to ensure that an adequate population of HSCs is present. Other major therapeutic complications include quiescence and heterogeneity in genetic expression and phenotype of LSCs, both across patients with a given leukemia and within a patient over disease course (24). Quiescence is a challenge because quiescent leukemic stem cells may be resistant to targeted therapy (28). Finally, one must consider the role of the stem cell niche, the microenvironment that houses stem cells and regulates their proliferation, quiescence, and differentiation. The HSC niche is partially driven by the Wnt-Hedgehog signaling pathway, a complex protein network with roles in both embryogenesis and carcinogenesis. Therapy inhibiting this pathway can impair CML stem cell self-renewal (20). Combining such therapy with a dasatinib-like drug, which induces apoptosis by binding to and inhibiting the constitutively active bcr-abl tyrosine kinase in CML cells, may lead to LSC eradication.
Mathematical models of both healthy and cancer stem cells provide insights into their biology (29–33). This paper discusses the clinical implications of a quantitative model developed in full mathematical detail elsewhere (34). As an example, we apply our model to CML. The model’s primary purpose is to determine the differential in death rates a therapy must possess to eradicate a LSC population while maintaining an adequate HSC population. This differential is succinctly expressed by selectivity and efficiency indices introduced later. A branching process elaboration of the model highlights the importance of following quiescent as well as active LSCs during therapy. When the model incorporates features such as drug resistant mutations, it becomes difficult to derive explicit analytic results. We therefore briefly describe numerical tools for fast simulation of complicated versions of the model. These tools allow us to explore combination therapies that target both LSCs and the microenvironmental niche signaling supporting their expansion.
Materials and Methods
Quick Guide to Equations and Assumptions
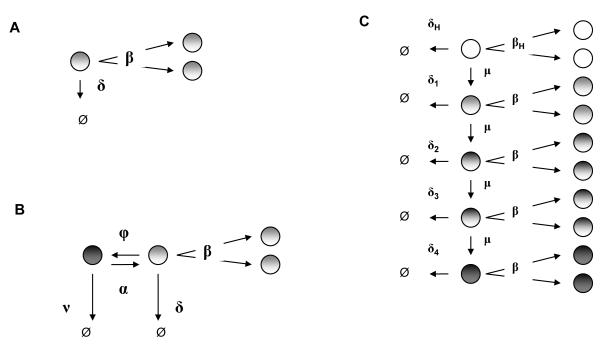
In studying stem cell population dynamics as a linear birth-death process Xt, it is helpful to clearly lay out all assumptions. Our simplest model is diagrammed in Panel A of Figure 1.
Fig. 1.
Modeling stem cell dynamics as a birth-death process. Panel A: Basic model: linear birth-death process. Panel B: Extension of model to include quiescence of LSCs. Panel C: Model including heterogeneity of LSCs.
Equation 1
| (1) |
Several well-known formulas are required for understanding the implications of the birth-death models. The most important formulas cover the average (equation 1) and standard deviation of the number of stem cells at time t, where n is the initial number of cells at the beginning time 0 (35). When the death rate exceeds the birth rate (δ > β), the most crucial feature of these formulas is the exponential decline in both the average and standard deviation with time.
Major Assumptions
All LSCs divide at the same rate β and die at the same rate δ per cell. Thus, in the small time interval from t to t + Δt, a particle dies with approximate probability δΔt and symmetrically divides with approximate probability βΔt. Each stem cell behaves independently and initiates a clan of descendants, which individually behave like the ancestral cell. When the death rate exceeds the birth rate (δ > β), clan extinction is certain. The extinction time for the entire process is the maximum extinction time for the different clans issuing from the various ancestral stem cells. We ignore asymmetric division that leaves a stem cell intact and produces one partially differentiated progenitor cell. Although such events are relevant to the entire organism’s health, they are irrelevant to the ultimate fate of a stem cell population.
Equation 2
The probability that all cells derived from n clans are extinct by time t
| (2) |
is also important (34).
Major Assumptions
Different clans’ independent behaviors allow us to express the extinction probability as the nth power of the extinction probability of a single clan. Because δ > β, this probability tends to 1 as t tends to ∞.
Equation 3
Birth-death process theory permits us to answer questions such as what is the probability that at least one normal stem cell survives after all LSCs have been eradicated? The answer to this and more nuanced questions depends on the initial numbers of the two cell types as well as on the killing differentials. The extinction time Mn over all n clans initiated by the original population of stem cells, leukemic or normal, equals the maximum extinction time for the separate clans. Based on equation (2), one can exploit the asymptotic theory of extreme-value statistics (36) to demonstrate that the standardized random variables (δ – β)(Mn – an) stabilize (34) for the carefully chosen sequence
In fact, the sequence of random variables (δ – β)(Mn – an) tends to the Gumbel distribution with mean γ ≈ 0.57722 and variance π2/6 (37).
These considerations produce the accurate mean and variance approximations
| (3) |
The sequence an is a linear function of ln n. This weak dependence on n suggests the possibility of safely eradicating LSCs even when they sharply outnumber HSCs. In this regard, it is helpful to define therapy selectivity σ as the ratio of the differences between death and birth rates for LSCs versus HSCs, namely
Here rates are subscripted by L for leukemia and H for healthy. For safe eradication one needs σ to be substantially greater than ln nL/ ln nH.
Equation 4
We also solve for the time t that renders the extinction probability (2) equal to a specific value p > 0. To good approximation (34)
| (4) |
Alternating between formulas (2) and (4) allows us to determine the extinction probability for the healthy stem cells at the time when the extinction probability for the LSCs reaches a predetermined level p. When birth rates are unaffected by therapy, the survival probability of the HSC population increases quickly as the difference between δH and δL grows. For example, take δH = 0.08 week−1 and βL = βH = 0.024 week−1. To be 80% certain that at least one ordinary stem cell remains when we are 99.9% certain no LSC remains, δL must be 0.15 week−1 or larger, corresponding to therapy selectivity σ of at least 2.1. We provide more definitive estimates in Results.
Equation 5
Perhaps the most useful measure of success in a targeted therapy is the number of HSCs NH left at the precise moment of LSC extinction. We define the killing efficiency κ of a therapy as the ratio δL/βL of the death rate to the birth rate of LSCs. The average (expected) number of healthy stem cells present at the time when all the LSCs are eradicated (34) depends on the selectivity σ and killing efficiency κ of the therapy through the formula
| (5) |
whereГ(·) is Euler’s gamma function. For an effective therapy, δL > βL, κ > 1, and σ > 1. The presence of the σth root in the denominator of (5) implies that the expected number of HSC is driven more by therapy selectivity and the initial numbers of HSCs and LSCs than by killing efficiency.
Quiescence
Quiescence is a reversible state of not dividing; the analogous state of senescence is not. Quiescent LSCs have been isolated in CML (38). Although some therapies are effective against both active and quiescent cells, other therapies such as dasatinib are effective in inducing apoptosis in active CML stem cells, but do not eliminate quiescent ones (27). In this setting, we predict that the active LSC population will be eradicated first and leave behind a quiescent LSC population, which on awakening causes cancer recurrence.
Panel B of Figure 1 presents a variant of our model that captures both HSC and LSC quiescence and awakening. Here a quiescent stem cell dies with rate ν and awakens to an actively dividing state with rate α. Active stem cells divide or die. We consider the case where a therapy is less effective at killing quiescent LSCs than active LSCs, so that ν < δ. For simplicity, we initially ignore transitions to quiescence by taking ϕ = 0 in Figure 1. These transitions to quiescence only slow LSC eradication and do not detract from the thrust of our arguments.
Modeling LSC heterogeneity
Our simple model and its elaboration are both multitype branching processes (35). This general framework is compatible with even more complicated models, but mathematical analysis is more formidable than for single-type processes such as birth-death processes. If one is satisfied with population means, then considerable progress can be made. Multitype branching particles behave independently according to prescribed stochastic rules. Each particle of type i has death intensity λi. At the end of its life, a type i particle reproduces particles of its own type and particles of other types. Let fij be its expected number of type j daughter particles. The mean behavior of the system of particles is summarized by a matrix M(t) = [mij(t)] whose entry mij(t) equals the mean number of type j particles at time t starting with a single type i particle at time 0. When the process starts with ni type i particles, there are on average type j particles at time t. M(t) satisfies the matrix differential equation , where Ω = (ωij) has diagonal entries ωii = λi(fii – 1) and off diagonal entries ωij = λifij. Because the process starts with a single type i particle, we have initial conditions mii(0) = 1 and mij(0) = 0 for i ≠ j. The matrix exponential solves the differential equation for M(t) subject to the initial conditions.
As an illustration, we model heterogeneity in gene expression of LSCs occuring across patients or within a patient over time. Let a stem cell acquire progressive decreases in susceptibility to therapy through a series of resistance mutations, each occurring at rate μ. For simplicity, we neglect other heterogeneity mechanisms such as epigenetics and niche regulatory signaling.
Panel C of Figure 1 describes 4 LSC types, each with varying therapy susceptibility manifested through different death rates δ1, … , δ4. Each LSC type has the same symmetric division rate β, and any cell of type 0 through 3 can acquire a mutation at rate μ rendering it less susceptible to therapy. As the figure indicates, more susceptible cell types gradually progress to less susceptible types. The model excludes back-mutation to a more susceptible state. The HSC population constitutes a separate type, type 0, which mutates to a type 1 LSC with rate μ. As in our original model, HSCs divide with rate βH and die with rate δH per cell. Mutation is rare. As a guess consider CML with approximately 40 known mutations. If each mutation occurs at a rate of 10−8 per cell division, then μ = 4 × 10−7 (31).
In this extended model, the death intensities are
The matrix F = (fij) has trivial entries except for
Thus the matrix Ω is
Because no back mutation occurs, Ω is upper triangular, and its eigenvalues appear along its diagonal. Along with their corresponding eigenvectors, these determine the system’s asymptotic mean behavior.
Stochastic simulation
Stochastic simulation is useful in modeling stochastic processes where analytic results are not easily obtained. It can also verify complicated analytic results. We apply stochastic simulation to extend our birth-death process model to account for both LSC and HSC quiescence. This permits backflow into quiescence at rate ϕ per active cell (see Figure 1). We also compare the matrix exponentiation and stochastic simulation results for our LSC heterogeneity model.
Stochastic simulation algorithms range from fine-grained exact algorithms in which every reaction is simulated (SSA) (39), to coarse-grained approximation methods (40). The τ-leaping method is intermediate (41). Over time interval (t, t + τ), the number of times a reaction fires is approximated by a Poisson random variable with mean equal to τ times the propensity of the reaction. We apply a variant of the τ-leaping method, the SAL algorithm (42), that includes linear and quadratic changes in reaction propensities. These improvements increase accuracy for larger systems without compromising speed.
Parameter choices
Supplementary Table 1 lists our model parameter choices. Murine experimentation reveals that 0.007% of bone marrow cells are long-term hematopoietic stem cells with life-long myelolymphoid reconstruction ability (43, 44). Extrapolating murine and feline data to humans predicts a total HSC population of 22,000 stem cells (45). We consider two coexisting stem cell populations: LSCs and HSCs, with separate rates of birth, death, quiescence and awakening, and distinct initial numbers. We consider advanced leukemia in which the LSCs outnumber the HSCs at therapy initiation.
In CML, birth and death rates per stem cell occur on a time scale of months. These assumptions are based on a 42 week replication time for human HSCs, extrapolated from feline HSC replication (45), and a death rate of 1 per 71 weeks, extrapolated from CML patients on imatinib therapy (31, 32). Because CML LSCs are insensitive to imatinib therapy (27), these death rates are likely to be close to natural death rates for LSCs. Under therapy directly targeting LSCs, such as dasatinib or farnesyl transferase inhibitors, the death rate should be much higher and we consider effects of varying death rate in our calculations and simulations. Death rates νL and νH for quiescent LSCs and quiescent HSCs, respectively, also vary under therapy and we consider the case where νL << δL and νH << δH.
HSC quiescence and awakening rates are unknown. However, approximately 90% of LSCs are reproductively active (46), while approximately 75% of HSCs are quiescent (47). We fix an awakening rate, of α = 0.07 week−1 and we estimate the backflow to quiescence by assuming equilibrium for the stem cell population. In Results, we consider 2 scenarios: the case in which the HSC and LSC quiescence rates are equal ϕH = ϕL and low, and the case in which ϕL << ϕH. In the first case, both the LSC and HSC populations respond to aberrant microenvironmental niche signaling. In the second case, LSCs fail to respond to normal niche signaling.
Results
Safety depends mainly on selectivity
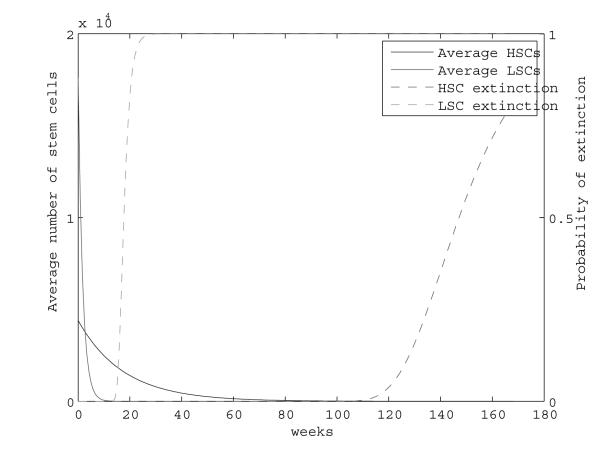
First we ignore quiescence. Figure 2 plots the trajectories of the average number of HSCs and LSCs, calculated using exact analytic results, and their extinction probabilities, calculated using the Gumbel approximation (3). Although LSCs initially outnumber healthy stem cells, the LSCs are much more likely to go extinct first. At the time when the LSC extinction probability approaches 1 (18-20 weeks), approximately 1,000 HSCs remain.
Fig. 2.
Mean population size and extinction probability of LSCs and HSCs during therapy. Here δL = 0.59, δH = 0.08, and βL = βH = 0.024.
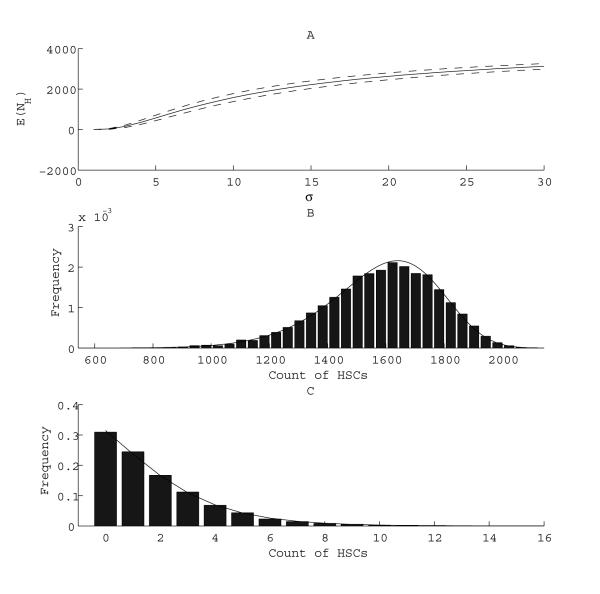
Panel A of Figure 3 plots as a function of therapy selectivity σ the average number of HSCs left at the moment when LSCs reach extinction. As discussed, this average number is insensitive to changes in the killing efficiency κ. (See Materials and Methods for definitions of σ and κ.) Clearly, higher selectivity entails higher average numbers of HSCs at LSC eradication. For example with κ = 25 and σ = 10 or greater, we expect more than 1,000 HSCs to survive. The selectivity required to ensure that the mean number of surviving HSCs is over 1,000 at the time of LSC extinction (σ = 10) is much higher than the selectivity required to be 80% sure that at least one healthy stem cell survives (σ = 2.2). Although both results assess safety in theory, predicting is more valuable in practice, because a handful of HSCs is unlikely to be sufficient for tissue repair and maintenance.
Fig. 3.
Dependence of the mean (Panel A) and distribution (Panels B and C) of the number of HSCs at LSC extinction time on the selectivity (σ) of therapy. Panel A: The mean is shown (solid line) plus or minus 1 standard deviation (dashed lines). δH = 0.08. Panel B: δH = 0.08, σ = 10. Panel C: δH = 0.48, and σ = 1.2. In Panels B and C, solid bars represent the distribution obtained using stochastic simulation, while lines represent the distribution obtained from analytic results.
We also examine the full distribution of HSCs at the precise moment when LSCs become extinct. This distribution is more helpful than the average in predicting the rarity of the event that HSC numbers become dangerously low under targeted therapy. Results obtained using analytic methods (34) and stochastic simulation show nice agreement. Panels B and C of Figure 3 depict distributions obtained for varying selectivities. With decreasing selectivity, HSCs will very likely be wiped out by the time of LSC eradication.
It is interesting to compare the dependence of the mean number of HSCs at the time of LSC extinction on the selectivity, killing efficiency, and the initial proportion nL/(nL + nH) of LSCs. We start with σ = 10, κ = 25, and nL/(nL + nH) = 0.8, and change one variable at a time, holding the others constant. Based on equation (5), Table 1 demonstrates that is more sensitive to selectivity than to killing efficiency or the initial fraction of CSCs. When σ = 1.25, approaches dangerously low numbers. also falls as the initial proportion of nL increases, but less dramatically than with σ. When the proportion of LSCs is 99%, there are still 77 HSCs at the time of LSC eradication.
Table 1.
The average number of HSCs remaining at LSC eradication depends on σ, but the mean time to lSC extinction depends on κ. We start with σ = 10, κ = 25, and nL/(nL + nH) = 0:8, varying each parameter in turn while holding the others constant at their initial values.
| Variable | Value | (weeks) | |
|---|---|---|---|
| σ | 10 | 1581 | * |
| 5 | 577 | * | |
| 2.5 | 80 | * | |
| 1.25 | 2 | * | |
|
| |||
| κ | 25 | 1581 | 18 |
| 12.5 | 1588 | 37.5 | |
| 6.25 | 1603 | 81.4 | |
| 3.125 | 1637 | 197 | |
|
| |||
| nL/(nL + nH) | 0.80 | 1581 | 18 |
| 0.90 | 781 | 18.25 | |
| 0.95 | 389 | 18.35 | |
| 0.99 | 77 | 18.42 | |
When βL and κ are held constant, is not a function of σ.
Although κ has little effect on , it determines the LSC mean extinction time. This fact becomes evident when we rewrite (equation 3) in terms of κ as
For example, κ = 25 leads to LSC eradication in 18 weeks on average, whereas κ = 3.125 increases mean time to eradication to almost 4 years (Table 1). Higher killing efficiency should lead to faster cures, which are important in reducing morbidity and mortality from side effects.
Safe therapy must target quiescent LSCs
When the active LSC elimination rate, δL – βL, is greater than the quiescent LSC elimination rate α + ν, recurrence is likely. To highlight the danger in targeting only active LSCs in the parameter regime α + ν < δL – βL, we derived (34) the approximation
| (6) |
to the probability that a quiescent LSC and all of its descendants are extinct by time t. Here o(1) is an error term that tends to 0 exponentially fast as t tends to ∞, and c is a positive constant that is well approximated by
when δL >> βL. Thus, the behavior of G(t) is determined by the effective elimination rate α + ν of quiescent LSCs and to a lesser extent by the difference (δ – β) – (α + ν).
If we start with nQL quiescent LSCs, then asymptotic extreme value theory entails a mean extinction time of
| (7) |
for the quiescent LSCs and their descendants. In contrast the mean extinction time for the nL active LSCs is
| (8) |
Because α + ν < δL – βL, the multipliers of ln nQ and ln nL in these formulas satisfy (δL–βL)−1 < (α+ν)−1. Thus our analytical results show that when α+ν < δL–βL, the mean extinction time of the nQL quiescent LSCs and their descendents exceeds the mean extinction time of the nL active LSCs unless nL is much larger than nQL. Therefore oncologists potentially face two dangers: (a) driving the active healthy stem cell population to a low level or (b) prematurely stopping therapy shortly after actively dividing LSCs are eradicated, and risking quiescent LSCs awakening to revive the cancer.
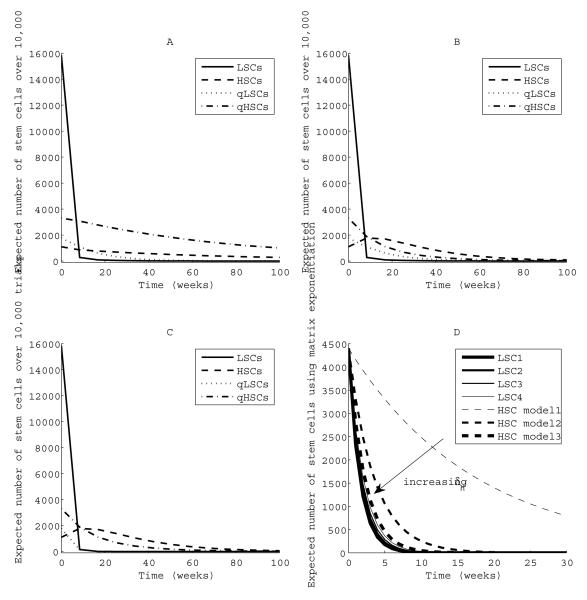
When we consider quiescence in the HSC population, the relevant population to preserve includes both the active and quiescent HSCs. Stochastic simulation helps us understand the effects of incorporating backflow to quiescence and the effects of letting the quiescent LSC death rate νL approach the active LSC death rate δL. Panels A, B and C of Figure 4 show the results of stochastic simulation when we incorporate both quiescence and awakening of LSCs and HSCs. Simulations were predominately implemented by SAL with SSA steps when needed and run for fixed time periods with increments of 8.4 weeks. Mean population counts over 10,000 trials are plotted for each run. Negative population counts were avoided by rejecting any leap taking a population below zero. We considered three scenarios by varying death rates of quiescent leukemic stem cells (qLSCs) and quiescence rates of LSCs. For σ = 10 and a lower quiescence rate for LSCs than for HSCs (Panel A), there are adequate HSC population counts (quiescent and active) at the time of LSC eradication. The LSC extinction time is far longer when quiescence is involved (around 100 weeks versus 18 weeks).
Fig. 4.
Effects of quiescence and heterogeneity of LSCs on population sizes during therapy. Panel A: ϕL = 0.007 << ϕH = 0.21, and νL = νH = 0.00024 week−1. The coefficient of variation of population size increases as stem cell counts approach very small numbers, with ranges from 0.17 to 1.2 for LSCs, 0.03 to 0.11 for HSCs, 0.04 to 0.44 for qLSCs, and 0.03 to 0.17 for qHSCs. Panel B: ϕL = ϕH = 0.007, and νL = νH = 0.00024 week−1. Panel C: ϕL = ϕH = 0.007, νL = 0.25, and νH = 0.00024 week−1. In panels A to C, δL = 0.59, δH = 0.08, βL = βH = 0.024, and αL = αH = 0.07 week−1. Panel D: Arrow represents increasing δH. In all cases, δ4 = 0.52, δ3 = 0.57, δ2 = 0.62, δ1 = 0.67. Model 1: δH = 0.08 week−1. Model 2: δH = 0.31 week−1. Model 3: δH = 0.48 week−1.
When LSC and HSC quiescence rates are equal, it is difficult to eradicate LSCs before active HSCs reach a critically low number at around 100 weeks (Panel B). Finally, when we increase the death rate of quiescent LSCs to about half the death rate of active LSCs, there are still adequate numbers of HSCs at the time of LSC eradication (Panel C) and a faster time to cure is observed (around 30 weeks). We conclude that targeted LSC therapy must kill quiescent LSCs in order to be safe. Our results highlight the need for experimental determination of quiescence and awakening rates for both LSCs and HSCs.
LSC heterogeneity
Panel D of Figure 4 plots the mean trajectories of HSCs and a heterogeneous populations of LSCs using matrix exponentiation. Results from stochastic simulation closely match our analytic results. The arrow in Figure 4 denotes gradually increasing HSC death rates. Not surprisingly, the least susceptible LSC population drives the overall LSC eradication dynamic. Here we made the simplifying assumption of equal initial numbers (4400) of each LSC type. Changing the initial numbers of each LSC type may shorten the overall extinction time. For example, less susceptible LSCs may go extinct before more susceptible LSCs if there are initially only a few less susceptible LSCs.
As we decrease the difference between δH and the most susceptible death rate δ1, there are fewer expected HSCs at the time of LSC eradication. With δH = 0.48, Panel D of Figure 4 shows that HSCs are nearly extinct by the time all LSCs are eradicated. Therapy selectivity ranges from 1.1 for the least susceptible LSC to 1.4 for the most susceptible LSC. Based on these results, it seems reasonable to define a heterogenous LSC population’s selectivity using the least susceptible population’s death rate.
Role of the stem cell niche
Stem cell microenvironmental signaling may play a large role in cancer stem cell dynamics. Hedgehog (Hh) signaling inhibition slows LSC self-renewal, suggesting a potential benefit of combining this therapy with standard tyrosine kinase inhibitors such as imatinib or dasatinib. Estimated death rates of LSCs on imatinib therapy are low (31). On imatinib therapy alone, we predict using equation (3) that extinction would take years. However, if Hh inhibition is added to imatinib therapy, thus decreasing the birth rate βL relative to the death rate δL, the killing efficiency may become sufficient to eradicate a LSC population in a reasonable amount of time.
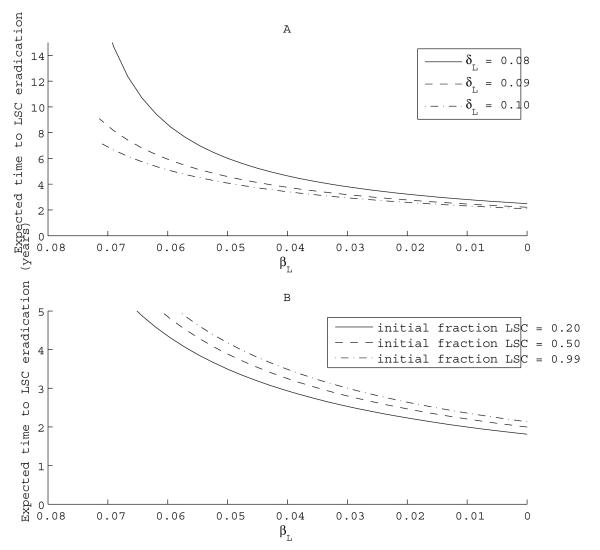
Suppose at therapy initiation, the LSC birth rate is higher than the HSC birth rate, say βH = 0.024 week−1 and βL = 0.07 week−1. Before initiating therapy, δL = δH = 0.03 week−1. Tyrosine kinase inhibition results in a very modest selective increase in the LSC death rate to δL = 0.10 week−1. Under these assumptions, σ = 5, assuring a safe eradication of LSCs with the average remaining HSCs around 577 (Table 1). The killing efficiency is low, and the mean disease eradication time is approximately 6 years. If we start with a more modest effect, δL = 0.08 week−1, then selectivity is 1.7 and time to eradication is 16 years. However, when we add Hh inhibition, causing a decreased self-renewal rate βL, the eradication time falls sharply. Panel A of Figure 5 plots average LSC extinction times for varying βL, starting with different LSC death rates. When δL = 0.10, a βL of 0.02 leads to an expected LSC eradication time of 1 to 2 years rather than 6 years. When δL is smaller, the dependence on βL is more dramatic. These results suggest that with combined therapy, modest increases in apoptosis caused by dasatinib may be augmented by the addition of therapy decreasing βL. However, under our assumptions, even if the birth rate is lowered to zero, it may still take 2 years to eradicate the LSCs. Similar results are obtained for initial LSC proportions ranging from 20% to 99% (Panel B of Figure 5). If Hh signaling inhibition sufficiently lowers βL rates, combined therapy with dasatinib and niche targeted therapy may lead to a cure, even when starting with high proportions of LSCs.
Fig. 5.
Effects of combining therapy inhibiting self-renewal with tyrosine kinase inhibition. Panel A: Effects of varying δL on expected time to eradication. Here we hold the initial proportion of LSCs constant at 80 percent. Panel B: Effects of varying initial proportion of LSCs on expected time to eradication. Here δL is held constant at 0.10.
Discussion
In conclusion, rational therapy is informed by mathematical modeling. The birth-death model gives us hope that attacking LSCs can cure CML. To be successful, targeted therapies must be tuned to spare HSCs and eradicate quiescent as well as active LSCs. This second criterion eliminates therapies that only attack dividing cells.
Highlighting dependence on selectivity is helpful in considering therapies that might differentially affect birth rates rather than death rates. For example, if we start with different birth rates for HSC and LSC and apply a therapy that decreases βL without affecting βH, we can calculate selectivity and in a similar fashion. Thus, our model is equally useful in predicting safety in this setting.
Our mathematical models may prove helpful in predicting therapeutic safety and efficacy as well as time to cure. Predictions will require reasonable estimates of LSC and HSC birth and death rates. Recently developed high throughput techniques allow the identification of agents that selectively target cancer stem cells and spare ordinary cells (23). Of course, in vivo rates may be less than in vitro rates, so these predictions must be confirmed in animal studies. Some therapies will demand a delicate balance between the time to cure and safety. For example, a therapy may have a high killing efficiency with a time to cure of 3 weeks, but may have low selectivity and cause dangerously low levels of HSCs for a transient time period. Another therapy may have high selectivity, ensuring an adequate HSC number when all LSCs have been eradicated, but still have a reasonable expected time to cure, say less than 1 year. From our perspective, slower cures are likely safer because they permit better monitoring of both HSC and LSC levels. On the other hand, the adverse side effects could tip the treatment balance toward quicker cures.
Our results may generalize to other leukemias and solid tumors in which cancer stem cells are being targeted. Their relevance depends on how well the modeling assumptions fit these applications. A limiting assumption in our model is that cancer stem cells behave independently. Although LSC populations occupying separate bone-marrow niches may not influence each other, spatial considerations may be important in solid tumors. Stem cells are influenced by their microenvironment, and their fate is regulated by interactions with cells of the niche, as well as other stem cells, adhesion molecules, extracellular matrix, and gradients in oxygen tension, pH, calcium concentration, growth factors, and cytokines. Furthermore, gradients in blood supply and hence drug delivery to solid tumors may render some stem cells within a tumor more susceptible to therapy. Other mathematical models address vasculature and spatial effects on drug delivery (48). Incorporation of stem cell spatial dependence in our models would help generalize our results to solid tumors.
No one should underestimate the difficulty in developing targeted drugs. For the full spectrum of cancers, this may take decades. Though great improvements are underway (49), the ability to monitor and detect stem cells is currently primitive. The mean extinction time and expected number of remaining HSCs at the time of LSC eradication are predicted by the killing efficiency and selectivity, which in turn are based on our chosen death rates and initial LSC population size. The minimum HSC numbers adequate for tissue survival and maintenance are not known and may vary by tissue type. Therapy dosing and duration must be determined by observations of toxicity in preclinical and early clinical trials. Such trials should also address whether stem cell therapies should be combined with standard chemotherapy regimens targeting the differentiated cells comprising the bulk of the tumor. If available in the future, direct measurement of both the actively dividing and quiescent cancer stem cell populations, used in conjunction with our modeling predictions, may be ideal for making decisions about discontinuation of therapy.
Therapy successes are apt to come first for well characterized cancers such as chronic myelogenous leukemia. Our optimism is not entirely predicated on the stem cell hypothesis of cancer. Recent evidence suggests that the number of cells with tumorigenic potential under various conditions may be several orders of magnitude higher than the number of cancer stem cells (50). Our models are flexible enough to include a wider class of cells. Provided the selectivity and killing efficiency are sufficiently high, nothing in principle prevents a cure. Cancer stem cell targeted therapy is more radical and risky than management of cancer as a chronic disease, and it must be performed with extraordinary care. Finally, it is helpful to keep in mind the potential of targeted therapies for diseases such as HIV. If a therapy could be devised to preferentially kill infected immune cells, it might have a chance of succeeding.
Major Findings.
We apply a birth-death process model to investigate how selective a therapy must be to ensure that an adequate number of normal hematopoietic stem cells survive when leukemic stem cells (LSCs) have been driven extinct. Our models take into account quiescence and genetic heterogeneity, and make specific predictions in terms of two interpretable quantities, therapeutic selectivity and killing efficiency, which are readily calculated for a proposed therapy using in vitro death rates. Major clinical implications include our finding that the number of LSCs at therapy initiation is less important than therapy selectivity and the necessity of directly targeting quiescent LSCs. Finally, we consider the effects of combining targeting LSC therapy with niche signaling inhibitors and conclude that modest increases in apoptosis rates can be augmented by inhibiting stem cell renewal, leading to cure.
Supplementary Material
Acknowledgments
We would like to thank Drs. Richard Gatti, Elliot Landaw, Julian Martinez, Van Savage, and Desmond Smith and Ms. Lindsay Riley for critical discussions and manuscript review. This research is supported in part by USPHS grants GM53275, MH59490, and GM008185 and by a Young Investigator Award from the American Society of Clinical Oncology.
References
- [1].Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- [3].Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell sub-population that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102:6942–7. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- [5].Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Suva M, Riggi N, Stehle J, et al. Identification of cancer stem cells in Ewing’s sarcoma. Cancer Res. 2009;69:1776–81. doi: 10.1158/0008-5472.CAN-08-2242. [DOI] [PubMed] [Google Scholar]
- [7].Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–9. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- [9].Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast cancer cells. N Engl J Med. 2007;356:217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- [10].Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–12. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- [11].Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–48. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- [12].Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- [13].Matsui W, Huff CA, Wang Q, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332–6. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kavalerchik E, Goff D, Jamieson CHM. Chronic myeloid leukemia stem cells. J Clin Oncol. 2008;26:2911–5. doi: 10.1200/JCO.2008.17.5745. [DOI] [PubMed] [Google Scholar]
- [15].Jamieson CH, Ailles LE, Dylla SJ, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- [16].Okamoto OK, Perez JF. Targeting cancer stem cells with monoclonal antibodies: a new perspective in cancer therapy and diagnosis. Expert Rev Mol Diagn. 2008;8:387–93. doi: 10.1586/14737159.8.4.387. [DOI] [PubMed] [Google Scholar]
- [17].El-Shami K, Smith BD. Immunotherapy for myeloid leukemias: current status and future directions. Leukemia. 2008;22:1658–64. doi: 10.1038/leu.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guzman ML, Rossi RM, Neelakantan S. An orally bioavailable parthenolide analog selectively eradicates acute myelogenous leukemia stem and progenitor cells. Blood. 2007;110:4427–35. doi: 10.1182/blood-2007-05-090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shih I, Wang T. Notch signaling, γ-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–82. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- [20].Zhao C, Chen A, Jamieson CH, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–9. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–8. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yilmaz OH, Morrison SJ. The PI-3kinase pathway in hematopoietic stem cells and leukemia-initiating cells: a mechanistic difference between normal and cancer stem cells. Blood Cells Mol Dis. 2008;41:73–6. doi: 10.1016/j.bcmd.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:1–15. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jordan CT. Can we finally target the leukemic stem cells? Best Pract Res Clin Haematol. 2008;21:615–20. doi: 10.1016/j.beha.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Konopleva M, Contractor R, Tsao T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–88. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- [26].Hassane DC, Guzman ML, Corbett C, et al. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public expression data. Blood. 2008;111:5654–62. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Copland M, Hamilton A, Elrick LJ, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–9. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- [28].Graham SM, Jergensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–25. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- [29].Enderling H, Chaplain MAJ, Anderson ARA, Vaidya JS. A mathematical model of breast cancer development, local treatment and recurrence. J Theor Biol. 2007:245–59. doi: 10.1016/j.jtbi.2006.12.010. [DOI] [PubMed] [Google Scholar]
- [30].Ashkenazi R, Gentry SN, Jackson TL. Pathways to tumorigenesis – Modeling mutation acquisition in stem cells and their progeny. Neoplasia. 2008:1170–82. doi: 10.1593/neo.08572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Michor F, Hughes TP, Iwasa Y, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–70. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- [32].Dingli D, Michor F. Successful therapy must eradicate cancer stem cells. Stem Cells. 2006;24:2603–10. doi: 10.1634/stemcells.2006-0136. [DOI] [PubMed] [Google Scholar]
- [33].Wodarz D. Stem cell regulation and the development of blast crisis in chronic myeloid leukemia: Implications for the outcome of Imatinib treatment and discontinuation. Med Hypotheses. 2008;70:128–36. doi: 10.1016/j.mehy.2007.03.040. [DOI] [PubMed] [Google Scholar]
- [34].Sehl ME, Zhou H, Sinsheimer JS, Lange KL. Extinction models for cancer stem cell therapy. SIAM Review. 2009 May 8; doi: 10.1016/j.mbs.2011.09.005. Submitted to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lange KL. Applied Probability. Springer-Verlag; New York: 2003. [Google Scholar]
- [36].Ferguson TS. A Course in Large Sample Theory. Chapman & Hall/CRC; New York: 1996. [Google Scholar]
- [37].Gumbel EJ. Bivariate Logistic Distributions. J Amer Stat Assoc. 1961;56:335–49. [Google Scholar]
- [38].Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent sub-population of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94:2056–64. [PubMed] [Google Scholar]
- [39].Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–61. [Google Scholar]
- [40].Higham DJ. Modeling and simulating chemical reactions. SIAM Rev. 2008;50:347–68. [Google Scholar]
- [41].Gillespie DT. Approximate accelerated stochastic simulation of chemically reacting systems. J Chem Phys. 2001;115:1716–33. [Google Scholar]
- [42].Sehl ME, Alekseyenko AL, Lange KL. Accurate stochastic simulation via the step anticipation τ-leaping (SAL) algorithm. J Comput Biol. doi: 10.1089/cmb.2008.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cheshier SH, Prohaska SS, Weissman IL. The effect of bleeding on hematopoietic stem cell cycling and self-renewal. Stem Cells Dev. 2007;16:707–17. doi: 10.1089/scd.2007.0017. [DOI] [PubMed] [Google Scholar]
- [44].Bonnet D. Haematopoietic stem cells. J Pathol. 2002;197:430–40. doi: 10.1002/path.1153. [DOI] [PubMed] [Google Scholar]
- [45].Abkowitz JL, Catlin S, McCallie MT, Guttorp P. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood. 2002;100:2665–7. doi: 10.1182/blood-2002-03-0822. [DOI] [PubMed] [Google Scholar]
- [46].Holyoake TL, Jiang X, Jorgensen HG, et al. Primitive quiescent leukemic cells from patients with chronic myeloid leukemia spontaneously initiate factor-independent growth in vitro in association with up-regulation of expression of interleukin-3. Blood. 2001;97:720–8. doi: 10.1182/blood.v97.3.720. [DOI] [PubMed] [Google Scholar]
- [47].Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96:3120–5. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jackson TL, Byrne HM. A mathematical model to study the effects of drug resistance and vasculature on the response of solid tumors to chemotherapy. Math Biosci. 2000;164:17–22. doi: 10.1016/s0025-5564(99)00062-0. [DOI] [PubMed] [Google Scholar]
- [49].Wu M, Kwon HY, Rattis F, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–54. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.