Abstract
Noninvasive methods to diagnose rejection of renal allografts are unavailable. Mass spectrometry followed by multiple-reaction monitoring provides a unique approach to identify disease-specific urine peptide biomarkers. Here, we performed urine peptidomic analysis of 70 unique samples from 50 renal transplant patients and 20 controls (n = 20), identifying a specific panel of 40 peptides for acute rejection (AR). Peptide sequencing revealed suggestive mechanisms of graft injury with roles for proteolytic degradation of uromodulin (UMOD) and several collagens, including COL1A2 and COL3A1. The 40-peptide panel discriminated AR in training (n = 46) and test (n = 24) sets (area under ROC curve >0.96). Integrative analysis of transcriptional signals from paired renal transplant biopsies, matched with the urine samples, revealed coordinated transcriptional changes for the corresponding genes in addition to dysregulation of extracellular matrix proteins in AR (MMP-7, SERPING1, and TIMP1). Quantitative PCR on an independent set of 34 transplant biopsies with and without AR validated coordinated changes in expression for the corresponding genes in rejection tissue. A six-gene biomarker panel (COL1A2, COL3A1, UMOD, MMP-7, SERPING1, TIMP1) classified AR with high specificity and sensitivity (area under ROC curve = 0.98). These data suggest that changes in collagen remodeling characterize AR and that detection of the corresponding proteolytic degradation products in urine provides a noninvasive diagnostic approach.
Despite an improvement in renal allograft survival reflecting advances in immunosuppressive medications,1,2 a critical unmet need in patient care is the requirement for sensitive and graft-etiology-specific, noninvasive methodologies for monitoring transplant recipients.3 Expression analyses of urine immune mediators,4 peripheral blood samples, and transplant biopsies5,6 support that distinct molecular pathways can define the injury of acute rejection (AR). Some of the concerns relating to biomarker discovery in urine lie with the confounding effect of proteinuria and high-abundance plasma proteins from nonspecific injury (which also occurs in AR). In this study, we have chosen to only analyze naturally occurring peptides in urine samples from transplant patients for three reasons: (1) because the roughly equal mass of protein and peptide in urine translates into at least a ten-fold greater molar abundance of peptides, urinary peptides provide a fertile ground for biomarker discovery; (2) urinary peptide analysis, unlike intact urinary proteomics analysis, is not hampered by the presence of highly abundant urinary proteins that can obscure the discovery of more informative lower abundance biomarker proteins7; and (3) analysis of urinary peptides is relatively easier than the analysis of complex tissues such as biopsy and blood because one-dimensional HPLC separation is sufficient for the analysis of >21,000 urine peptides.7
An additional important confounder for AR diagnosis and management is BK nephritis. To address these issues, this study performed noninvasive, urine peptidomic analysis of 70 unique urine samples, collected from renal transplant patients and controls, by liquid chromatography and mass spectrometry (LC-MS), followed by multiple reaction monitoring (MRM) verification, on five different cohorts, including samples with nonspecific proteinuria, BK nephritis, and vyuria.
To explore the relevance of altered urinary peptide abundance, we also performed integrated transcriptomic analysis on matching biopsy microarrays, paired with the urine samples, available in the Sarwal Lab (GEO, GSE14328). Quantitative real-time PCR (Q-PCR) verified significant overlapping genes in an independent set of 34 biopsy samples.
Our results indicate that disease-specific alteration of proteolytic and antiproteolytic activities is the underlying mechanism by which these urine peptide biomarkers are generated in graft rejection. To our knowledge, this study represents the first study that analyzed urinary peptidomic and matching renal biopsy transcriptomic analyses, which will help in elucidating the pathophysiological relationships between our nested urine peptide biomarkers and allograft proteolytic networks in vivo in renal allograft diseases.
Results
Sample Characteristics
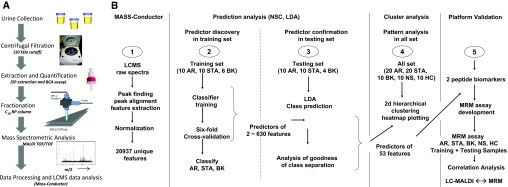
The overall study design for the peptidomic urine analysis is shown in Figure 1. Seventy unique urine samples were analyzed from the following five cohorts: pediatric kidney transplant patients with biopsy-proven acute allograft rejection (AR, n = 20), stable allograft with normal protocol biopsies (STA, n = 20), BK virus nephropathy with vyurina (BK, n = 10), nonspecific proteinuria with native renal disease (biopsy-proven nephrotic syndrome; NS, n = 10), and healthy age-matched volunteers (HC, n = 10). Samples were split into training sets (n = 46) for urine peptide discovery, and test sets (n = 24) (sample demographics in Supplementary Table 1) for urine peptide prediction and verification.
Figure 1.
Peptidomics approach for biomarker discovery. (A) Schematics for peptidomic analysis of naturally occurring urinary peptides. (B) Study design for the urine peptide biomarker discovery.
Discovery of a Urine Peptide Panel for AR by LC-Matrix-Assisted Laser Desorption/Ionization
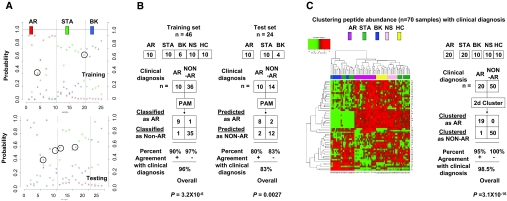
A total of 20,937 unique peptide peaks with distinct m/z and HPLC fractions were resolved in the 900- to 4000-Da range. Prediction analysis by a nearest shrunken centroid (NSC) algorithm8 was performed, and 6-fold internal crossvalidation analysis led to the discovery of a set of 630 peptide features with the lowest classification error (Supplementary Figure 1). Discriminant class probabilities and Gaussian linear discriminant analysis (LDA) were performed for each sample8 (Supplementary Figure 2) in both sample sets and resulted in misclassification of only 2 of the 24 samples in the test set. To find a predictive biomarker panel of optimal feature number, various classifiers were tested for their spread of distribution and goodness of the separation (Figure 1B and Supplementary Figure 3). Linear discriminant probabilities of a biomarker panel of 53 peptide peaks was sufficient for goodness of separation of the clinically relevant transplant categories (AR, STA, and BK) in the training and the test sample sets (Figure 2, A and B). This biomarker panel classified the AR samples with 96% overall agreement with clinical diagnosis of AR in the training set (P = 3.2 × 10−6 by Fisher exact test) and 83% agreement with clinical diagnosis of AR in the test set (P = of 0.0027 by Fisher exact test). When all 70 samples were clustered by unsupervised analysis of their peptide abundance across the 53 peak features, all AR samples, save one, co-clustered, and importantly, all of the non-AR samples (STA, BK, NS, and HC) clustered disparate from the AR sample cluster (Figure 2C). Interestingly, the STA samples separated into two clusters, suggesting that STA samples might harbor two subclasses at the urine peptide level. On the basis of the discriminant-analysis-derived prediction scores for each sample, a receiver operating characteristic (ROC) curve was constructed to evaluate the testing performance of our peptide biomarker panel9,10 and resulted in area under the curve (AUC) values of 0.97 and 0.96 for the training and the test set, respectively (Figure 3A).
Figure 2.
Statistical analyses of the peptide biomarker panel. (A) The discriminant of the peptide biomarker panel for the training (upper) and testing data (lower) probabilities for all transplant samples were calculated from the LDA. The maximum estimated probability for each of the wrongly classified samples is marked with a circle. Two of the 46 samples in the training set and 4 of the 24 samples in the test set were misclassified, giving a correct classification rate of 96% in the training set and 83% in the test set. (B) Left panel: Modified 2 × 2 contingency tables were used to calculate the percentage of classification that agreed with clinical diagnosis for the biomarker panel. P values were calculated with Fisher's exact test. Right panel: A prediction of AR from the non-AR phenotype (a so-called “two-class” prediction) was used to assess the performance of the biomarker panel in the classification of unknown samples. STA and BK were combined into one group as “NON-AR.” Fisher exact test was to compute the P value for the blind test. (C) Unsupervised clustering based on the peptide biomarker panel was used to construct a heat map in which the colors indicate the intensity of peptide concentration by LC-MALDI: red indicates high peptide abundance and green indicates low peptide abundance in the comparative analysis. It can be seen that by unsupervised analysis, the AR samples, save one, all co-cluster together and all of the non-AR samples cluster together. Modified 2 × 2 contingency tables were used to calculate the percentage of unsupervised clustering that agreed with clinical diagnosis for the biomarker panel. P values were calculated with Fisher's exact test.
Figure 3.
Discovery and verification of AR-specific peptides. (A) Discovery of the 40-peptide biomarker panel and their performance on the training set (top panel) and the test set (bottom panel) using ROC analysis. (B) MRM analyses of the two UMOD peptide biomarkers (top panels). The distribution of MRM signals were analyzed by box-whisker graphs according to the sample categories. The boxes are bound by 75th and 25th percentiles of the data, and the whiskers extend to the minimum and maximum values. ROC analysis (bottom panel) of the classification performance of the two UMOD peptide biomarkers. When ROC analysis was performed to test the diagnostic accuracy of the two UMOD peptide biomarkers for AR, the AUCs were computed as 0.83 for the UMOD 1679.98-Da peptide and 0.74 for the UMOD 1911.07-Da peptide.
Identification of AR-Specific Urine Peptides
Manual review of the biomarker panel and associated mass spectrometry (MS) spectra interpreted and de-isotoped the 53 MS peak features, which could be mapped to 40 unique urine peptides and were further identified by matrix-assisted laser desorption/ionization (MALDI)-time-of-flight (TOF)/TOF and LTQ Orbitrap MS/MS analysis. In general, the naturally occurring peptides are more difficult to study with current standard mass spectrometric search engines because of the difficulty in complexity reduction in the search space with the knowledge of peptide-ending amino acids. For those peptides unable to be identified by MS/MS analysis, we are currently following up to scale up the purification of these peptides to have sufficient quantity for protein identification using the Edman sequencing approach. We grouped the identified peptides according to their common protein precursor and computed the medians of LC-MS measurements according to sample categories. The peptides were found to map to nine different proteins, eight of which belonged to the collagen family (COL1A1, COL1A2, COL3A1, COL4A3, COL4A4, COL4A5, COL7A1, COL18A1) and uromodulin (UMOD). When MS/MS analysis was extended to the original 630-peptide feature set, 142 urine peptides were identified, again with predominant presence of collagen peptides (n = 47) and UMOD peptides (n = 16) (Figure 4, A and B). The UMOD peptide biomarker cluster discovered in this study spans from serine residue 589, following arginine residue 588, and to lysine residue 607 (Figure 4C). Little is known about the metabolic pathway of this C-terminal peptide and its biologic role after UMOD is shed from the apical plasma membrane into the tubule lumen. UMOD, the most abundant urinary protein in mammals, has recently been shown to be significantly lower in abundance in urine samples from patients with renal transplant rejection.11 UMOD peptides analyzed in pooled urine samples have also been found to be significantly reduced in patients with transplant rejection compared with patients without rejection.7 This study confirms the results that UMOD peptides are much lower in individual urine samples taken from patients when the filtering kidney has ongoing AR. Although the significance of these findings is unclear at present, a recent genome-wide association study has identified significant single-nucleotide polymorphism associations with chronic kidney disease at the UMOD locus.12
Figure 4.
Mapping of collagen and UMOD peptides in the urine. Identified urine peptide biomarkers yielded clusters of overlapping (A) collagen and (B) UMOD peptides (mass/charge ratio, MH+). “P” in red indicates 4-hydroxyproline. Peptides in brackets derive from the same region of the same precursor proteins. Because the genes labeled in red were significantly regulated in microarray data, we tested them by Q-PCR. (C) Human UMOD precursor. Recent MS analyses50 proved that C-terminal cleavage of the precursor, which has 640 amino acids, occurred after phenylalanine residue 587. Because part of the C-terminal peptide cleaved from the UMOD precursor, the UMOD peptide biomarker cluster (colored in red) discovered in this study spans from serine residue 589, following arginine residue 588, and to lysine residue 607.
Interestingly, all of the identified UMOD and collagen urine peptides showed much lower abundance during AR when compared with other samples, with overall lower abundance in transplant patients when compared with nontransplanted patients (NS) and healthy controls (Supplementary Figure 4). Sequence alignment analysis of the collagen and UMOD peptides were found to line up by forming clusters within either the C- or N-terminal end with ladder like truncations at the opposite ends, suggesting that there is likely disease-specific proteolytic degradation of the parent protein. Similar to the proteolytic degradation of urine proteins in AR, serum proteins have also been found to show differences in degradation in cancer.13
MRM Verification of Selected Urine Peptides
To verify the presence and quantify differences in peptides between AR and non-AR groups, MRM was performed on two selected peptides14 [UMOD1 (1679.98 Da) and UMOD2 (1911.07 Da); Figure 3] on all 70 samples. The box-whisker graphs in Figure 3B illustrate the spread of the distribution of the MRM measurements in AR (n = 20), STA (n = 20), BK (n = 10), NS (n = 10), and HC (n = 10) sample categories for peptides with UMOD 1680.98 and 1912.07 Da, respectively. As seen in Figure 3B (upper panel, left-hand side), similar to the results obtained by LC-MALDI, the abundance of UMOD peptide 1679.98 was significantly lower in AR (P = 0.0003), and as seen in Figure 3B (upper panel, right-hand side), the abundance of UMOD 1911 was also significantly lower in AR (P = 0.0006) when compared with all other non-AR categories. ROC analysis to test the diagnostic ability of the two UMOD peptide biomarkers for AR was seen in terms of AUC. AUCs for UMOD1 and UMOD2 were 0.83 and 0.74, respectively.
Integrated Analysis of Matched Samples: Transcriptional Analysis of Biopsy AR and Peptidomic Analysis of Urine AR
Because urine is an ultrafiltrate of the kidney, we hypothesized that the alteration of the urinary proteins and peptides in urine may relate to processes occurring directly in the kidney. To address this we analyzed archived microarray data in the Sarwal Lab (GSE14328) on matched kidney biopsies (20 AR and 20 STA; taken at the time of urine collection, before any treatment intensification for AR) for expression differences between AR and STA samples for the corresponding UMOD and the collagen genes. We also looked for any expression differences in extracellular matrix proteins in AR, because some of these have been previously demonstrated to be differentially expressed in AR.15 We observed that whereas UMOD gene expression in AR biopsy was significantly lower in AR [false discovery rate (FDR) = 0.02%; similar results to the low UMOD peptide abundance in AR urine], the three collagen genes (COL1A2, FDR = 0.18%; COL3A1, FDR = 0.67%; COL4A1, FDR = 1.82%) were upregulated in AR (different from low collagen peptide abundance in AR urine). Gene expression for matrix metalloproteinase-7 (MMP-7; FDR = 0.03%), tissue inhibitor of metalloproteinase 1 (TIMP1; FDR = 24%), and the serpin peptidase inhibitor (SERPING1; FDR = 33%) was higher in AR when compared with STA biopsies, although only MMP7 expression was significant.
We performed Q-PCR in biopsies from a separate set of 34 kidney biopsies (14 AR, 10 STA, and 10 healthy kidney donor biopsies) for UMOD; the most significant collagen genes in rejection, namely COL1A2 and COL3A1; as well as all MMP7, SERPING1, and TIMP1 (Figure 5A). The Q-PCR results validated that the six genes had statistically significant expression differences in AR, with similar results between the microarray and Q-PCR; lower gene expression for UMOD in AR (P = 0.011); and higher gene expression for COL1A2 (P = 0.027), COL3A1 (P = 0.013), MMP7 (P = 0.013), SERPING1 (P = 0.005), and TIMP1 (P = 0.013) in AR when compared with samples without AR (Figure 5A). The importance of these pathways is underscored by the finding that LDA can also use the gene expression values of the six genes in biopsy AR tissue (ROC curve value of 0.98; Figure 5B) to accurately classify a rejection episode similar to the results obtained from analysis of the corresponding urine peptides (Figure 3B, lower panel). Interestingly, irrespective of the confounder of BK virus, biopsy UMOD gene expression and urinary peptide abundance are significantly lower in AR, whereas biopsy collagen gene expression is significantly higher in AR and collagen peptide abundance in rejecting urine is significantly lower. The dysregulation of collagen expression in the rejecting graft and altered proteolysis of collagens in the urine may provide novel insight into the cascade of events that prime a graft for chronic injury and fibrosis after an AR episode (Figure 6).
Figure 5.
A gene panel specific for AR. (A) The distribution of COL1A2, COL3A1, MMP-7, SERPING1, TIMP1, and UMOD genes' Q-PCR measurements in kidney biopsy were analyzed by box-whisker graphs. (B) ROC analysis was performed to evaluate the performance of the six-member RNA biomarker panel classifying AR from STA. The plotted ROC curve is the vertical average of the 500 bootstrapping runs, and the boxes and whiskers plot the vertical spread around the average.
Figure 6.
A proposed mechanism of fibrosis caused by AR as indicated by the observations of increased collagen gene transcription in the rejection biopsy and reduced collagen peptides in the urine during graft rejection.
Discussion
Proteomic and peptidomic analysis of urine collected from healthy individuals (22 mg peptides in urine per day)16and patients with renal disease have identified more than 1500 different proteins11,17,18 and over 100,000 different peptide biomarkers19 in health and disease.20 This is the first study of an integrated analysis of the urine peptidome and the biopsy transcriptome in graft rejection that uncovers that overlapping key gene and peptide pathways can be jointly dysregulated in AR. The resultant alterations in the abundance of selected genes and the peptide products of the corresponding proteins can highlight potential mechanisms of graft injury in rejection. Disease-specific alterations of gene transcription in the tissue (by array and Q-PCR) and a change in the balance of proteolytic and antiproteolytic activities in urine appear to be important mechanisms resulting in an altered pattern of a specific panel of urinary peptides in AR.
There are at least 28 different human collagens that represent approximately 25% of the total protein content of mammals,21 but in the kidney type I and III collagen are most abundant, whereas type IV collagen is a major component of basement membranes.22 The increase in the aminoterminal and carboxy terminal propeptides from the procollagen of types I, III, and IV during collagen anabolism and later decrease in the collagen-derived urinary naturally occurring peptides during collagen catabolism suggest that increased turnover of renal collagens23–26 may be valuable biomarkers for noninvasive diagnosis of the rejection process in the kidney. The upregulation of extracellular matrix regulators (MMP-7, SERPING1, and TIMP1) also supports the hypothesis of tissue remodeling at the time of AR. The observance of high MMP-7 expression in the kidney at the time of AR has also been previously reported in chronic kidney rejection,27 human kidney aging,28 and a rat renal AR model.29 MMP-7 is a collagenase-related connective-tissue-degrading metalloproteinase and plays a role in the breakdown of extracellular matrix in normal physiologic processes, tissue remodeling during injury,30 and neutrophil influx to sites of injury.31 SERPRING1 regulates leukocyte trafficking and complement (inactivating C1r, C1s, MASP2, and C3b proteases),32 which is also locally regulated in the kidney during ischemia reperfusion injury.33 Similar to the finding in this study, SERPING1 has previously been shown to be regulated in the graft during AR.34 Tissue-specific inhibitors of metalloproteinases are endogenous, specific inhibitors that bind and inhibit matrix metalloproteinases.35 TIMP1 is a physiological inhibitor of the matrix-degrading enzymes, collagenases, gelatinase, and stromelysin and plays a major role in the inhibition of matrix degradation. Upregulation of TIMP1 mRNA and protein has been previously reported in different models of renal disease36–41 and in human sclerotic glomeruli.42 The increased expression of TIMP1, a collagenase inhibitor, may be a reason for the reduced activity of collagenases and subsequent reduced breakdown of tissue collagen, leading to the observance of increased graft collagen expression and reduced collagen urine peptides in graft rejection. Thus, altered collagen and extracellular matrix turnover in graft rejection with altered regulation of collagenases in the graft, as seen in independent data sets by microarray and Q-PCR, may be critical pathways that link AR injury with the observed increased downstream clinical risk of chronic injury and graft fibrosis.43,44
In conclusion, the analysis of the naturally occurring urinary peptides using the LC-MS method in kidney transplant rejection is a novel approach because it provides AR injury-specific peptide biomarkers and also highlights local injury mechanisms in the inflamed tissue relating to a cascade of collagen proteins, which may be important harbingers of chronic graft injury. Future prospective studies of these urine peptide biomarkers by antibody-based or quantitative MS-based approaches are needed to optimize this approach for clinical application and to test the validity of these urinary peptides for prediction of acute and chronic graft injury.
Concise Methods
Urine Samples
Seventy unique urine samples from 50 pediatric renal transplant recipients (20 biopsy-proven AR, 20 STA, 10 BK), 10 age-matched healthy controls (HC), and 10 pediatric patients with nonspecific proteinuria from native renal disease due to nephrotic syndrome (NS; to control for nonspecific renal injury) were collected at Lucile Packard Children's Hospital at Stanford University from 2004 to 2006. Details on patient age, gender, and other transplantation-related clinical indicators are given in Supplementary Table 1. Informed consent was obtained from all patients and the Stanford University Institutional Review Board approved the study.
Urine Collection, Storage, and Processing
Second-morning void midstream urine samples (50 to 100 ml) were collected in sterile containers and were centrifuged at 2000 × g for 20 minutes at room temperature within 1 hour of collection. The details of urine processing and preparation of peptide extraction and fraction are reported elsewhere.7
Peptidomic Data Analysis
We used the approach of ion mapping,45,46 in which biomarker candidate MS peaks are selected on the basis of discriminant analysis and then targeted for MS/MS sequencing analysis to obtain protein identification. We have developed an informatics platform, “MASS-Conductor,”7 which contains an integrated suite of algorithms, statistical methods, and computer applications to allow for signal processing and statistical analysis in LC-MS-based urine peptide profiling. The peaks are located in the raw spectra of the MALDI data by an algorithm that looks for sites (m/z values) for which the intensity is higher than the estimated average background and the approximately 100 surrounding sites, with peak widths approximately 0.5% of the corresponding m/z value. The binned LC-MALDI MS peak data (20,937 m/z values) obtained for all 70 samples were analyzed separately for the training sample set (n = 46) for discovery of discriminant biomarkers using algorithms8 of NSC, 6-fold crossvalidation analyses, and Gaussian LDA. The predictive capabilities of the 53 most discriminant peptide peaks were used to blindly test for differentiating AR, STA, and BK samples in the test set (n = 24). To control the number of false significant features found during NSC mining, we permutated the data set 500 times to calculate the global FDR.47
MRM Assay for Peptide Marker Verification
Stable isotope-labeled peptides (with a 13C-labeled amino acid) were synthesized and used as internal standard peptides. Each urine peptide sample, prepared as described above, was diluted 10-fold with 10% acetonitrile/0.1% formic acid and spiked with the internal standard peptide to a final concentration 0.1 μM. Peptides were resolved in an HPLC equipped with a Polaris C18 column (50 × 20 mm, 3 μM, 6-minute gradient elution; Buffer A: 0.1% formic acid in water; Buffer B: 0.1% formic acid in acetonitrile; flow rate of 200 μl/min). A triple quadrupole mass spectrometer was used. The data were assessed and visualized by an ROC curve ROCR package.10
Integrated Analysis of Peptidomic Data in Urine and Microarray Data from Matched Transplant Biopsies
Affymetirx HU133 plus 2 GeneChips on matched kidney transplant biopsies (20 AR and 20 STA) have been previously performed in the Sarwal Lab (National Center for Biotechnology Information GEO database GSE14328). Raw expression data were preprocessed and normalized using dChip software.48 Supervised, two-class unpaired Significance Analysis of Microarray49 analyses were applied to calculate FDR for differences in expression of the corresponding UMOD and the collagen genes in rejection. Additionally, we searched for any differences in the expression of extracellular matrix proteins (TIMP1, SERPING1, and MMP-7) in the rejecting graft.
RNA Preparation and Q-PCR
Total RNA was extracted from kidney biopsy samples using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA) and later was DnaseI treated and purified using the RNeasy mini kit according to the manufacturer's protocol (Qiagen, Valencia, CA). cDNA was synthesized from 250 ng of RNA using the RT2 First Strand Kit (SABioscience Corporation, Frederick, MD). Q-PCR reactions were performed on 5 ng of cDNA using RT2 SYBR Green/ROX PCR master mix and commercially available primers: PPH12000A-200 for UMOD, PPH00771A-200 for TIMP1, PPH18747E-200 for SERPING1, PPH00809E-200 for MMP-7, PPH01918B-200 for COL1A2, PPH00439E-200 for COL3A1, PPH20687A-200 for COL4A1, and PPH05666E-200 for 18S rRNA (SuperArray Bioscience Corporation, Frederick, MD). All RNA samples were analyzed in duplicate and normalized relative to 18S rRNA levels.
Disclosures
None.
Supplementary Material
Acknowledgments
The work was supported by National Institutes of Health grant RO1-AI-061739 (M.S.), the Deans Fellowship, and the Child Health Research Program (TS, MS). The authors thank Karolina Krasinska at the Stanford University Mass Spectrometry Center for the MRM assay development and applications, Tonya Pekar at Thermo for LTQ Orbitrap MS, and the Stanford University IT group for excellence in Linux cluster support.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1. Nankivell BJ, Borrows RJ, Fung CL, O'Connell PJ, Allen RD, Chapman JR: The natural history of chronic allograft nephropathy. N Engl J Med 349: 2326– 2333, 2003 [DOI] [PubMed] [Google Scholar]
- 2. Opelz G: Influence of treatment with cyclosporine, azathioprine and steroids on chronic allograft failure. The Collaborative Transplant Study. Kidney Int Suppl 52: S89– S92, 1995 [PubMed] [Google Scholar]
- 3. Marsden PA: Predicting outcomes after renal transplantation—New tools and old tools. N Engl J Med 349: 182– 184, 2003 [DOI] [PubMed] [Google Scholar]
- 4. Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, Qian B, Serur D, Mouradian J, Schwartz JE, Suthanthiran M: Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med 344: 947– 954, 2001 [DOI] [PubMed] [Google Scholar]
- 5. Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra O, Jr: Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med 349: 125– 138, 2003 [DOI] [PubMed] [Google Scholar]
- 6. Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, Chismar JD, Horvath S, Mondala T, Gilmartin T, Cook DJ, Kay SA, Walker JR, Salomon DR: Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant 4: 1475– 1489, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sigdel TK, Ling XB, Lau K, Li L, Sarwal MM, Schilling J, Sarwal M: Urinary peptidomic analysis identifies potential biomarkers for acute rejection of renal. Clin Proteom 5: 103– 113, 2009 [Google Scholar]
- 8. Tibshirani R, Hastie T, Narasimhan B, Chu G: Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 99: 6567– 6572, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zweig MH, Campbell G: Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin Chem 39: 561– 577, 1993 [PubMed] [Google Scholar]
- 10. Sing T, Sander O, Beerenwinkel N, Lengauer T: ROCR: Visualizing classifier performance in R. Bioinformatics 21: 3940– 3941, 2005 [DOI] [PubMed] [Google Scholar]
- 11. Sigdel TK, Kaushal A, Gritsenko M, Norbeck AD, Qian W, Xiao W, Camp DG, II, Smith RD, Sarwal MM: Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin Appl 2010, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kottgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Ida Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Pare G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS: Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 2009. [ Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, Sanchez-Carbayo M, Holland EC, Cordon-Cardo C, Scher HI, Tempst P: Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest 116: 271– 284, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai SS, Bunk DM, White ET, Welch MJ: Development and evaluation of a reference measurement procedure for the determination of total 3,3`,5-triiodothyronine in human serum using isotope-dilution liquid chromatography-tandem mass spectrometry. Anal Chem 76: 5092– 5096, 2004 [DOI] [PubMed] [Google Scholar]
- 15. Kuyvenhoven JP, Verspaget HW, Gao Q, Ringers J, Smit VT, Lamers CB, van Hoek B: Assessment of serum matrix metalloproteinases MMP-2 and MMP-9 after human liver transplantation: Increased serum MMP-9 level in acute rejection. Transplantation 77: 1646– 1652, 2004 [DOI] [PubMed] [Google Scholar]
- 16. Strong KJO, Comper TM, Wayne D: Urinary-peptide excretion by patients with and volunteers without diabetes. J Lab Clin Med 145: 239– 246, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M: The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol 7: R80, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA: Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20: 363– 379, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coon JJ, Zurbig P, Dakna M, Dominiczak AF, Decramer S, Fliser D, Frommberger M, Golovko I, Good DM, Herget-Rosenthal S, Jankowski J, Julian BA, Kellmann M, Kolch W, Massy Z, Novak J, Rossing K, Schanstra JP, Schiffer E, Theodorescu D, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P: CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin Appl 2: 964– 973, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Decramer S, Z Uuml Rbig P, Wittke S, Mischak H, Bascands JL, Schanstra JP: Identification of urinary biomarkers by proteomics in newborns: Use in obstructive nephropathy. Contrib Nephrol 160: 127– 141, 2008 [DOI] [PubMed] [Google Scholar]
- 21. Myllyharju J, Kivirikko KI: Collagens and collagen-related diseases. Ann Med 33: 7– 21, 2001 [DOI] [PubMed] [Google Scholar]
- 22. Miner JH: Renal basement membrane components. Kidney Int 56: 2016– 2024, 1999 [DOI] [PubMed] [Google Scholar]
- 23. Querejeta R, Varo N, Lopez B, Larman M, Artinano E, Etayo JC, Martinez Ubago JL, Gutierrez-Stampa M, Emparanza JI, Gil MJ, Monreal I, Mindan JP, Diez J: Serum carboxy-terminal propeptide of procollagen type I is a marker of myocardial fibrosis in hypertensive heart disease. Circulation 101: 1729– 1735, 2000 [DOI] [PubMed] [Google Scholar]
- 24. Keller F, Rehbein C, Schwarz A, Fleck M, Hayasaka A, Schuppan D, Offermann G, Hahn EG: Increased procollagen III production in patients with kidney disease. Nephron 50: 332– 337, 1988 [DOI] [PubMed] [Google Scholar]
- 25. Keller F, Lyreal Ser Y, Schuppan D: Raised concentrations of the carboxy terminal propeptide of type IV (basement membrane) procollagen (NC1) in serum and urine of patients with glomerulonephritis. Eur J Clin Invest 22: 175– 181, 1992 [DOI] [PubMed] [Google Scholar]
- 26. Heickendorff L, Frost L, Madsen JK, Pedersen EB: Serum propeptides of type I and III procollagens in renal transplant recipients. A comparison of cyclosporine and azathioprine treatment. Nephron 67: 203– 208, 1994 [DOI] [PubMed] [Google Scholar]
- 27. Berthier CC, Lods N, Joosten SA, van Kooten C, Leppert D, Lindberg RL, Kappeler A, Raulf F, Sterchi EE, Lottaz D, Marti HP: Differential regulation of metzincins in experimental chronic renal allograft rejection: Potential markers and novel therapeutic targets. Kidney Int 69: 358– 368, 2006 [DOI] [PubMed] [Google Scholar]
- 28. Melk A, Mansfield ES, Hsieh SC, Hernandez-Boussard T, Grimm P, Rayner DC, Halloran PF, Sarwal MM: Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int 68: 2667– 2679, 2005 [DOI] [PubMed] [Google Scholar]
- 29. Edemir B, Kurian SM, Eisenacher M, Lang D, Muller-Tidow C, Gabriels G, Salomon DR, Schlatter E: Activation of counter-regulatory mechanisms in a rat renal acute rejection model. BMC Genomics 9: 71, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, Sheppard D, Pardo A, Selman M, Heller RA: Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci U S A 99: 6292– 6297, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Q, Park PW, Wilson CL, Parks WC: Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 111: 635– 646, 2002 [DOI] [PubMed] [Google Scholar]
- 32. Davis JE, Moss DJ: Treatment options for post-transplant lymphoproliferative disorder and other Epstein-Barr virus-associated malignancies. Tissue Antigens 63: 285– 292, 2004 [DOI] [PubMed] [Google Scholar]
- 33. Naesens M, Li L, Ying L, Sansanwal P, Sigdel TK, Hsieh SC, Kambham N, Lerut E, Salvatierra O, Butte AJ, Sarwal MM: Expression of complement components differs between kidney allografts from living and deceased donors. J Am Soc Nephrol 20: 1839– 1851, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cai S, Dole VS, Bergmeier W, Scafidi J, Feng H, Wagner DD, Davis AE, III: A direct role for C1 inhibitor in regulation of leukocyte adhesion. J Immunol 174: 6462– 6466, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Woessner JF: MMPs and TIMPs: An historical perspective. Mol Biotechnol 22: 1073– 6085, 2002 [DOI] [PubMed] [Google Scholar]
- 36. Engelmyer E, van Goor H, Edwards DR, Diamond JR: Differential mRNA expression of renal cortical tissue inhibitor of metalloproteinase-1, -2, and -3 in experimental hydronephrosis. J Am Soc Nephrol 5: 1675– 1683, 1995 [DOI] [PubMed] [Google Scholar]
- 37. Sharma AK, Mauer SM, Kim Y, Michael AF: Altered expression of matrix metalloproteinase-2, TIMP, and TIMP-2 in obstructive nephropathy. J Lab Clin Med 125: 754– 761, 1995 [PubMed] [Google Scholar]
- 38. Nakamura T, Takahashi T, Fukui M, Ebihara I, Osada S, Tomino Y, Koide H: Enalapril attenuates increased gene expression of extracellular matrix components in diabetic rats. J Am Soc Nephrol 5: 1492– 1497, 1995 [DOI] [PubMed] [Google Scholar]
- 39. Jones CL, Buch S, Post M, McCulloch L, Liu E, Eddy AA: Pathogenesis of interstitial fibrosis in chronic purine aminonucleoside nephrosis. Kidney Int 40: 1020– 1031, 1991 [DOI] [PubMed] [Google Scholar]
- 40. Jones CL, Buch S, Post M, McCulloch L, Liu E, Eddy AA: Renal extracellular matrix accumulation in acute puromycin aminonucleoside nephrosis in rats. Am J Pathol 141: 1381– 1396, 1992 [PMC free article] [PubMed] [Google Scholar]
- 41. Eddy AA, Giachelli CM: Renal expression of genes that promote interstitial inflammation and fibrosis in rats with protein-overload proteinuria. Kidney Int 47: 1546– 1557, 1995 [DOI] [PubMed] [Google Scholar]
- 42. Carome MA, Striker LJ, Peten EP, Moore J, Yang CW, Stetler-Stevenson WG, Striker GE: Human glomeruli express TIMP-1 mRNA and TIMP-2 protein and mRNA. Am J Physiol 264: F923– F929, 1993 [DOI] [PubMed] [Google Scholar]
- 43. Weber KT: Monitoring tissue repair and fibrosis from a distance. Circulation 96: 2488– 2492, 1997 [PubMed] [Google Scholar]
- 44. Paul LC: Chronic allograft nephropathy: An update. Kidney Int 56: 783– 793, 1999 [DOI] [PubMed] [Google Scholar]
- 45. Fach EM, Garulacan LA, Gao J, Xiao Q, Storm SM, Dubaquie YP, Hefta SA, Opiteck GJ: In vitro biomarker discovery for atherosclerosis by proteomics. Mol Cell Proteomics 3: 1200– 1210, 2004 [DOI] [PubMed] [Google Scholar]
- 46. Gao J, Opiteck GJ, Friedrichs MS, Dongre AR, Hefta SA: Changes in the protein expression of yeast as a function of carbon source. J Proteome Res 2: 643– 649, 2003 [DOI] [PubMed] [Google Scholar]
- 47. Ling XB, Cohen H, Jin J, Lau I, Schilling J: FDR made easy in differential feature discovery and correlation analyses. Bioinformatics 25: 1461– 1462, 2009 [DOI] [PubMed] [Google Scholar]
- 48. Li C: Automating dChip: toward reproducible sharing of microarray data analysis. BMC Bioinformatics 9: 231, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Efron B, Tibshirani R: Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol 23: 70– 86, 2002 [DOI] [PubMed] [Google Scholar]
- 50. Santambrogio S, Cattaneo A, Bernascone I, Schwend T, Jovine L, Bachi A, Rampoldi L: Urinary uromodulin carries an intact ZP domain generated by a conserved C-terminal proteolytic cleavage. Biochem Biophys Res Commun 370: 410– 413, 2008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.