Abstract
The neuronal adhesion protein Dragon acts as a bone morphogenetic protein (BMP) coreceptor that enhances BMP signaling. Given the importance of BMP signaling in nephrogenesis and its putative role in the response to injury in the adult kidney, we studied the localization and function of Dragon in the kidney. We observed that Dragon localized predominantly to the apical surfaces of tubular epithelial cells in the thick ascending limbs, distal convoluted tubules, and collecting ducts of mice. Dragon expression was weak in the proximal tubules and glomeruli. In mouse inner medullary collecting duct (mIMCD3) cells, Dragon generated BMP signals in a ligand-dependent manner, and BMP4 is the predominant endogenous ligand for the Dragon coreceptor. In mIMCD3 cells, BMP4 normally signaled through BMPRII, but Dragon enhanced its signaling through the BMP type II receptor ActRIIA. Dragon and BMP4 increased transepithelial resistance (TER) through the Smad1/5/8 pathway. In epithelial cells isolated from the proximal tubule and intercalated cells of collecting ducts, we observed coexpression of ActRIIA, Dragon, and BMP4 but not BMPRII. Taken together, these results suggest that Dragon may enhance BMP signaling in renal tubular epithelial cells and maintain normal renal physiology.
Bone morphogenetic proteins (BMPs) represent a large subfamily of the transforming growth factor β (TGF-β) superfamily of ligands that play roles in numerous physiologic and pathologic processes including cell proliferation, differentiation, apoptosis, and specification of developmental fate during embryogenesis and in adult tissues.1 In the kidney, BMPs play an important role in nephrogenesis. During normal development, BMPs are expressed in the metanephric mesenchyme and the ureteric bud and play a key role in the epithelialization of the metanephric mesenchyme and reciprocal induction of collecting duct differentiation. Loss of BMP expression has profound effects on kidney development.2–11 The role of BMPs in the adult kidney is less well understood. The expression of multiple BMP ligands, including BMP4, BMP6, and BMP7, and BMP receptors persists in the adult kidney,12 supporting the notion that the adult kidney can respond to BMP stimulation.13 Response to injury and repair frequently recapitulates development, and given the function of BMPs during nephrogenesis, it has been hypothesized that BMPs may have a similar role in the adult kidney as epithelial differentiation and survival factors that protect against damage and promote recovery in response to injury14,15.
Functional tight junctions are essential for the establishment and maintenance of the polarized architecture of the epithelial cells,16–18 a process that occurs during kidney development and in response to injury and repair.19–22 Tight junctions also provide a barrier that is involved in regulation of paracellular transport of small molecules.23 Transepithelial resistance (TER) reflects paracellular ionic conductance and it is a measure of tight junction complexity and function.24,25 In the kidney, TER varies dramatically across different nephron tubules and changes in response to physiologic and pathologic conditions.26 Interestingly, it has been shown that BMP signals enhance TER in some epithelial cells,27 suggesting that this could be one function of BMP signaling in the kidney.
BMP signaling is initiated by ligand binding to combinations of two type II and two type I serine/threonine kinase receptors. Upon ligand binding, the type I receptor is phosphorylated by the type II receptor. Type I receptors then act downstream, determining the specificity of the signal via phosphorylation of the receptor-activated Smads (R-Smads). The BMP subfamily signals via one set of R-Smads (Smad1, Smad5, and Smad8), whereas the TGF-β subfamily signals via another set of R-Smads (Smad2, Smad3). All R-Smads then form heteromeric complexes with the common mediator (co-Smad), Smad4. The activated Smad complexes then move from the cytoplasm to the nucleus where they act as transcriptional regulators to modulate gene expression.1
Recently, we identified the three repulsive guidance molecule (RGM) proteins including RGMa, RGMb (Dragon), and RGMc (hemojuvelin) as coreceptors for BMP signaling.28–30 RGM proteins share 50% to 60% sequence homology and have similar structural features including a signal sequence, conserved proteolytic cleavage site, partial von Willebrand factor type D domain, and glycophosphatidylinositol (GPI) anchor. RGM proteins are retained on the outer layer of the plasma membrane through the GPI anchor motif,31,32 although they can be shed from the cell membrane through cleavage at the GPI anchor by phospholipases.31 We have shown that all three RGM proteins physically interact with BMP receptors and specific BMP ligands and increase intracellular Smad phosphorylation in response to BMP ligands.28–30,33 We have also revealed a mechanism shared by RGMa and hemojuvelin in increasing BMP signaling (i.e., facilitating the use of ActRIIA by endogenous BMP-2 and BMP-4 ligands that normally prefer signaling through BMPRII).33,34 However, the precise molecular mechanisms of Dragon's action in regulating BMP signaling remain to be investigated.
Dragon is expressed in the central nervous system, where it is involved in neuronal cell adhesion through homophilic interactions.31 However, a detailed examination of Dragon expression in other tissues and a physiologic role for the BMP signaling function of Dragon has yet to be determined.
Here, we show that Dragon is expressed in the kidney epithelial cells in the thick ascending limbs, distal tubules, and collecting ducts. We demonstrate that Dragon can act in a mouse inner medullary collecting duct (mIMCD3) cell line to generate BMP signals as a ligand-dependent coreceptor. Furthermore, Dragon enhances utilization of a BMP type II receptor, ActRIIA, by BMP4 in mIMCD3 cells. Finally, we show that Dragon and BMP4 increase TER in mIMCD3 cells, and that the small-molecule BMP inhibitor LDN-193189 can block this increase. Thus Dragon could potentially generate BMP signals in tubule cells in the kidney and may play a role in regulating the functions of tight junctions in the epithelia of kidney tubules.
Results
Dragon mRNA and Protein Are Expressed in the Kidney
To investigate the presence of Dragon mRNA in mouse tissues, Northern blots containing 10 μg of total RNAs prepared from various mouse tissues (whole embryos, kidney, heart, liver, muscle, and brain) were hybridized with a specific probe for Dragon. As shown in Supplemental Figure 1A, there are two message sizes for Dragon—a major band at approximately 4 kb in size and a smaller band at 2.4 kb. Both are expressed in the kidney and in other tissues such as brain, heart, liver, muscle, and whole embryo.
To further confirm Dragon expression in the kidney, we performed Western blot analyses of proteins from adult mouse whole kidney lysates separated by reducing SDS polyacrylamide gel electrophoresis and probed with a previously characterized anti-Dragon antibody.31 A band of approximately 55 kD was seen in the kidney lysates (Supplemental Figure 1B, lane 2). This correlates well with the previously described molecular weight of Dragon in Western blots of protein extracts from neonatal and adult dorsal root ganglia.31 The 55-kD band was eliminated when Dragon antibody was preincubated with competing immunizing peptide (Supplemental Figure 1B, lane 1). A band of approximately the same size was detected by Western blot analysis of lysates generated from HEK293 cells transfected with Dragon cDNA but not in untransfected cells (Supplemental Figure 1C); thus, Dragon protein is expressed in the kidney.
Cellular Localization of Dragon in the Kidney
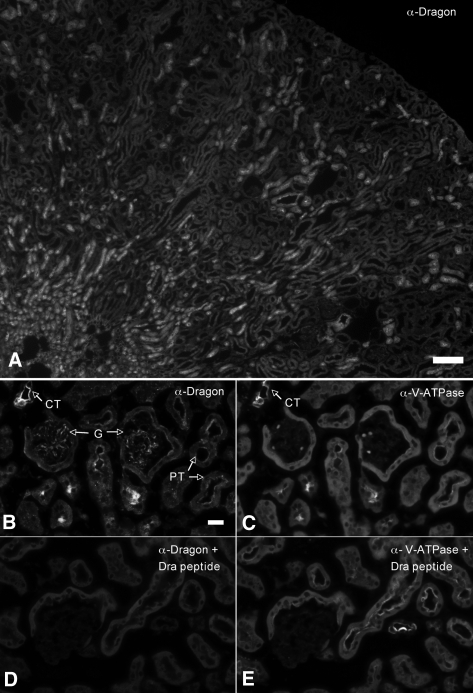
We then investigated the localization of Dragon protein in the adult mouse kidney by indirect immunofluorescence. As shown in the photograph at low magnification (Figure 1A), Dragon is expressed in various tubules. In photographs at high magnification, Dragon protein was weakly expressed in the glomeruli and Bowman's capsules (Figure 1B) compared with Dragon expression in collecting ducts (Figure 1B) identified by V-ATPase staining (Figure 1C). Weak staining was also detected in the brush border of proximal tubules (Figure 1B). The staining appears to be specific for Dragon, because it could be completely blocked by preincubation of the Dragon antibody with the competing immunizing peptide (Figure 1D) whereas the V-ATPase staining was not affected by the presence of the Dragon peptide (Figure 1, C and E). The expression of Dragon in proximal tubules is supported by the detection of Dragon mRNA in isolated epithelial cells of proximal tubules from mice obtained through BSA-Alexa555 labeling of proximal tubule cells followed by laser capture microdissection (Figure 7A).
Figure 1.
Dragon is localized in various tubules in (A) the whole mouse kidney and (B through E) is localized in glomeruli and proximal tubules in the cortex. Adult mouse kidney sections were co-immunostained with (A, B) rabbit anti-Dragon antibody and (C, D) chicken anti-V-ATPase E1 subunit antibody. The Dragon and V-ATPase were visualized using Cy3-donkey anti-rabbit and FITC-donkey anti-chicken secondary antibodies, respectively. Photographs were taken at (A, 4×) low and (B through E, 40×) high magnifications. (D) The staining of Dragon in the cortex was completely blocked in the presence of its competing immunizing peptide (Dra peptide, 200 ng/μl), whereas (E) V-ATPase staining was not affected by the Dragon peptide. Dragon staining was seen in glomeruli, Bowman's capsules, and in epithelial cells of proximal tubules, which show moderate apical V-ATPase staining. (B) The strongest Dragon staining was found in cortical tubules that contain cells with abundant V-ATPase staining, suggesting that collecting ducts express the Dragon protein. G, glomerulus; PT, proximal tubules; CT, collecting ducts. Scale bars = 80 μm for (A) low magnification and 20 μm for (B through E) high magnification.
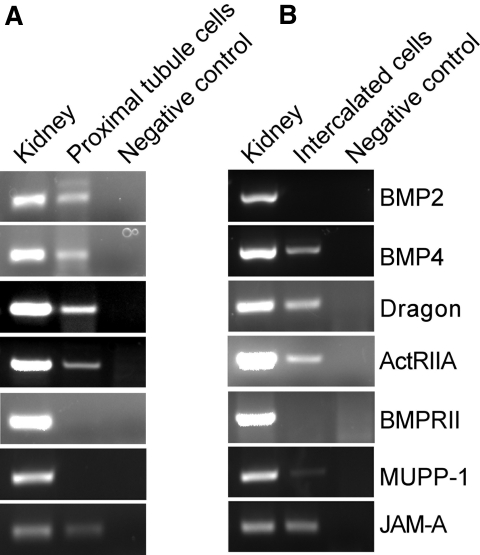
Figure 7.
Expression of mRNAs for BMP2, BMP4, Dragon, ActRIIA, BMPRII, and tight junction proteins MUPP-1 and JAM-A in isolated proximal tubule cells and intercalated cells. RT-PCR analyses were performed for (A) isolated proximal tubule cells and (B) intercalated cells. cDNA from the adult kidney was used as positive control and replacement of the template with water was used as negative control.
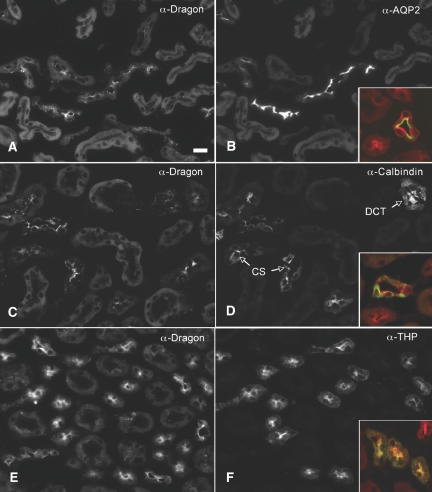
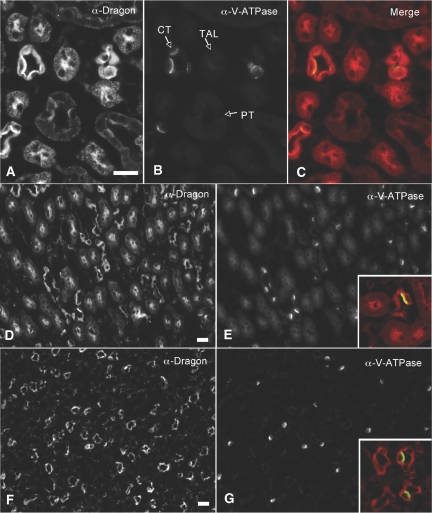
To further characterize the cellular distribution of Dragon in collecting ducts, we performed dual staining of Dragon protein with aquaporin 2 (AQP2), a marker for principal cells,35 or V-ATPase, a proton pump that is localized to intercalated cells.36 Dragon (red) was expressed in the AQP2-containing cells (green, inset, Figure 2B) and V-ATPase-containing cells (Figures 1 and 3). These results suggest that Dragon is expressed in principal and intercalated cells in collecting ducts. Of note, Dragon expression was polarized in collecting ducts in the cortex (Figures 1B and 2A) and in the outer stripe (Figure 3A), with Dragon staining being concentrated toward the apical pole of the cells, although weak staining was observed throughout the cytoplasm and at the basal membrane. In contrast, Dragon staining appears to be diffuse in collecting ducts in the inner stripe (Figure 3D) and papilla (Figure 3F).
Figure 2.
Dragon is localized in collecting ducts, distal convoluted tubules, connecting segments, and thick ascending limbs in the (A through D) cortex and (E, F) outer stripe of the mouse kidney. Mouse kidney sections were co-immunostained with (A, C and E) rabbit anti-Dragon antibody and (B) goat anti-AQP2, (D) goat anti-calbindin 28, or (F) goat anti-Tamm-Horsfall glycoprotein. Sections were then stained with Cy3-donkey anti-rabbit (red) and FITC-donkey anti-goat (green) secondary antibodies to visualize Dragon, AQP2, calbindin 28, or THP. The merged images for Dragon and AQP2, calbindin 28, or THP are shown in the respective insets. Specific staining for Dragon was seen in (A, B) principal cells in collecting ducts, (C, D) epithelial cells in distal convoluted tubules and connecting segments, and (E, F) thick ascending limbs. DCT, distal convoluted tubules; CS, connecting segments; anti-Tamm-Horsfall glycoprotein (α-THP); THP, Tamm-Horsfall glycoprotein. Scale bar = 20 μm.
Figure 3.
Dragon is localized in collecting ducts and thick ascending limbs in the (A through C) outer and (D, E) inner stripes and (F, G) collecting ducts and thin loops of Henle in the papilla of the mouse kidney. Mouse kidney sections were co-immunostained with (A, D, E) rabbit anti-Dragon antibody and (B, E, F) chicken anti-V-ATPase E1 antibody. The Dragon and V-ATPase were visualized using Cy3-donkey anti-rabbit (red) and FITC-donkey anti-chicken secondary antibodies (green), respectively. The merged images for Dragon and V-ATPase are shown in panel C for the outer stripe or in the insets in panels E and F for the inner stripe and papilla. Strong staining for Dragon was seen in intercalated cells of collecting ducts (panel C and the inset in panel E), but adjacent principal cells were also positive. (A, B) Thick ascending limbs (TAL) of the outer stripe were also strongly staining for Dragon. Staining for Dragon was seen in collecting ducts and thin limbs of Henle's loop (inset, panel G) in the papilla. Scale bars = 30 μm for the outer stripe (A through C) and 20 μm for the inner stripe and papilla (D through G).
Apical staining of Dragon was also seen in the epithelial cells positive for calbindin 28, a calcium-binding protein specifically expressed in the mouse distal convoluted tubules and connecting segments37 (Figure 2, C and D), and in epithelial cells positive for Tamm-Horsfall glycoprotein, a marker for thick ascending limbs38 (Figure 2, E and F). Dragon was also detected in thin limbs of Henle's loop in the papilla (Figure 3F). Taken together, the data show that Dragon protein is expressed in the glomerulus and in epithelial cell types in various tubules in the kidney.
Dragon Mediates BMP4 Signaling in the Kidney Collecting Duct Cell Line mIMCD3
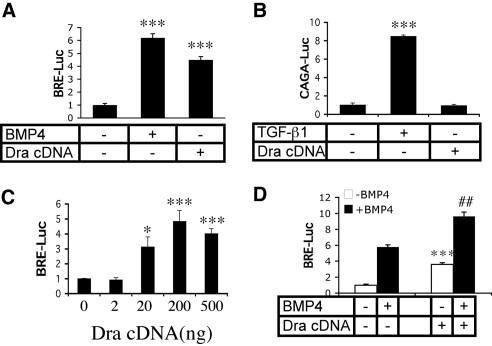
Because Dragon is highly expressed in several kidney tubule epithelial cell types, including collecting duct cells, we tested if Dragon can mediate BMP signaling in mIMCD3 cells, a well established kidney inner medullary collecting duct cell line known to respond to BMP ligands. mIMCD3 cells were transfected with a BMP-responsive luciferase reporter gene construct (BRE-Luc)39 (Figure 4A) or a TGF-β-responsive luciferase reporter gene construct [(CAGA)12MLP-Luc]40 (Figure 4B) alone or in combination with Dragon cDNA. Transfected cells were then incubated with or without 40 ng/ml BMP4 or 2 ng/ml TGF-β1. In the absence of Dragon, stimulation with BMP4 and TGF-β increased the relative luciferase activity for their respective reporters compared with unstimulated cells (Figure 4, A and B). Transfection with Dragon increased BRE luciferase activity even in the absence of BMP stimulation (Figure 4A) but did not alter (CAGA)12 MLP-Luc activity compared with the baseline (Figure 4B). The increase of Dragon signaling is dose-dependent, and Dragon signaling reached peak at 200 ng (Figure 4C). These results demonstrate that Dragon can stimulate BMP signaling but not TGF-β signaling in mIMCD3 cells.
Figure 4.
Dragon increases BMP but not TGF-β signaling in kidney tubule cells. (A) Dragon increases BMP signaling. mIMCD3 cells were transfected with BRE-Luc construct in the absence (bars 1 and 2), or presence (bar 3) of Dragon cDNA. Cells were then starved and incubated for 16 hours in the absence (bars 1 and 3) or presence of 40 ng/ml BMP4 (bar 2), and luciferase activity was measured the next day from cell extracts. ***P < 0.001 versus the control. (B) Dragon does not affect TGF-β signaling. mIMCD3 cells were transfected with the (CAGA)12MLP-Luc construct in the absence (bars 1 and 2) or presence (bar 3) of Dragon cDNA. Cells were incubated for 16 hours in the absence (bars 1 and 3) or presence of 2 ng/ml TGF-β1 (bar 2), and luciferase activity was measured the next day. ***P < 0.001 versus the control. (C) Dragon increases BMP signaling in a dose dependent manner. mIMCD3 cells were transfected with BRE-Luc construct with increasing amounts of Dragon cDNA (0, 2, 20, 200, and 500 ng). Relative luciferase activity was measured from cell extracts. *P < 0.05 and ***P < 0.001 versus the control. (D) BMP4 and Dragon signals are additive. mIMCD3 cells were transfected with the BRE-Luc construct in the absence or presence of Dragon cDNA. Cells were then incubated without (open bars) or with BMP4 (filled bars) and relative luciferase activity was measured from cells extracts. ***P < 0.001, bar 3 versus bar 1; ###P < 0.05, bar 4 versus bar 2.
To determine whether Dragon and BMP ligands are additive in their BMP signaling effects, mIMCD3 cells were transfected with the BRE-Luc construct in the absence or presence of Dragon cDNA. Cells were then incubated in the absence or presence of BMP4. The presence of Dragon augmented BMP4 stimulation of BRE-Luc activity (Figure 4D). Similar results were obtained when BMP4 was replaced with BMP2 (Supplemental Figure 2). Thus, Dragon behaves in a manner consistent with a role as an accessory receptor for BMPs.
We then explored whether Dragon expression in vivo correlates with its hypothesized role as a coreceptor for BMP signaling. We therefore determined whether Dragon-expressing cells in the kidney showed evidence of BMP signaling (i.e., nuclear accumulation of p-Smad1/5/8). As shown in Supplemental Figure 3, nuclear staining of phospho-Smad1/5/8 was detected in glomeruli, principal and intercalated cells of collecting ducts, distal convoluted tubules and connecting segments, and thick ascending limbs. Thus, BMP signaling occurs in Dragon-expressing cells in the kidney, consistent with a role for Dragon as a BMP coreceptor in vivo.
BMP4 Is the Endogenous Ligand for Dragon in mIMCD3 Cells
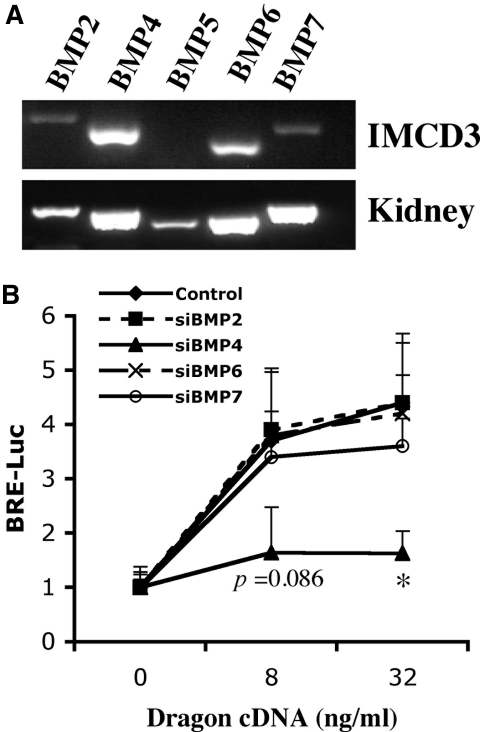
Dragon has been shown to signal in a ligand-dependent manner.28 We have previously shown that Dragon binds BMP2 and BMP4 but not BMP7 and TGF-β1.28 Therefore, it is possible that BMP2, BMP4, or both are endogenous ligands for Dragon. To investigate endogenous ligands for the Dragon coreceptor in mIMCD3 cells, we screened these cells by reverse-transcriptase PCR (RT-PCR) for expression of mRNAs for BMP2; BMP4; and the closely related members BMP5, BMP6, and BMP7 (Figure 5A). Among these ligands, BMP4 and BMP6 mRNA were readily detected in mIMCD3 cells, whereas BMP2 and BMP7 were barely detectable and BMP5 was not detected, although all of these ligands were expressed in the native mouse kidney (Figure 5A), which contains many different cell types. We then tested whether Dragon-induced BMP signaling is affected by small interfering RNA (siRNA)-mediated specific inhibition of BMP2, BMP4, BMP6, or BMP7. As shown in Supplemental Figure 4, BMP2, BMP4, BMP6, or BMP7 expression was specifically inhibited by 60% to 70% by the respective gene-specific siRNA duplexes (60 nM), with minimal effect on the expression of the other ligands. mIMCD3 cells were transfected with BRE-Luc in combination with control, BMP2, BMP4, BMP6, or BMP7 siRNA (60 nM) (Figure 5B). Inhibition of BMP4 expression abolished Dragon-mediated BMP signaling. In contrast, inhibition of BMP2, BMP6, and BMP7 expression did not change BRE-Luc activity induced by Dragon compared with control siRNA (Figure 5B). These results demonstrate that BMP4 is the predominant endogenous ligand for Dragon in mIMCD3 cells.
Figure 5.
BMP4 is the endogenous ligand for Dragon in mIMCD3 cells. (A) Expression of BMP ligands in mIMCD3 cells. Total RNA from IMCD3 cells was extracted for RT-PCR to determine the expression of BMP2 and BMP4 through BMP7. Total RNA from mouse kidney was used in PCR analyses as positive controls. BMP4 and BMP6 mRNAs were readily detected, BMP2 and BMP7 mRNAs were weak, and BMP5 mRNA was undetectable in mIMCD3 cells. (B) Effect of siRNA targeting of BMP2, BMP4, BMP6, and BMP7 on Dragon-mediated BMP signaling. mIMCD3 cells were transfected with BRE luciferase reporter alone or with increasing amounts of Dragon cDNA and in combination with control, BMP2 (siBMP2), BMP4 (siBMP4), BMP6 (siBMP6), or BMP7 (siBMP7) siRNA (60 nM) for 46 hours before measurement of luciferase activity. Inhibition of BMP4 expression significantly reduced Dragon-mediated BMP signaling, whereas inhibition of BMP2, BMP6, or BMP7 had little or no effect. *P < 0.05, siBMP4 versus control at 32 ng/ml Dragon cDNA. The exact P value was shown for siBMP4 versus control at 8 ng/ml Dragon cDNA.
Dragon Enhances Utilization of ActRIIA by BMP4 in mIMCD3 Cells
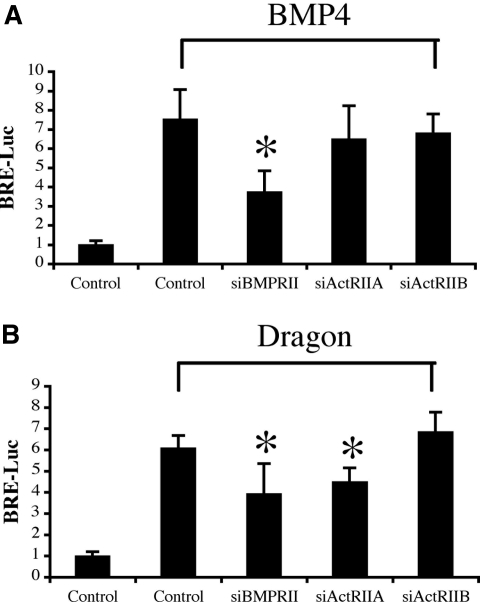
We previously demonstrated that RGMa and RGMc/hemojuvelin increase BMP signaling by increased utilization of BMP type II receptor ActRIIA by BMP2 and BMP4, which normally signal through BMPRII.33,34 To test whether Dragon also alters utilization of BMP type II receptors, we examined the effect of siRNA-mediated specific inhibition of BMP type II receptors on Dragon-induced BMP signaling in mIMCD3 cells. To this end, we screened mIMCD3 cells for expression of BMPRII, ActRIIA, and ActRIIB mRNA by RT-PCR. As shown in Supplemental Figure 5A, all three type II receptors were expressed. The expression of the three receptors was selectively reduced by more than 50% with the introduction of specific siRNAs (60 nM) (Supplemental Figure 5B). We examined whether or not inhibition of endogenous type II receptors affects Dragon-mediated stimulation of BRE-Luc activity. As expected, treatment of mIMCD3 cells with BMP4 (20 ng/ml) increased BRE-Luc activity to 7.5-fold above the baseline (Figure 6A). This stimulation was dramatically reduced by BMPRII-specific siRNA to 3.7-fold above the baseline (P < 0.05) but was not reduced by ActRIIA- or ActRIIB-specific siRNA. These results suggest that in mIMCD3 cells BMP4 signaling is primarily transduced by BMPRII but not by ActRIIA or ActRIIB.
Figure 6.
Dragon enhances utilization of ActRIIA by BMP4. (A) Effect of siRNA targeting of BMPRII, ActRIIA, and ActRIIB on BMP4 signaling. mIMCD3 cells were transfected with the BRE luciferase reporter in combination with control siRNA or siRNA specific for BMPRII (siBMPRII), ActRIIA (siActRIIA), or ActRIIB (siActRIIB) (60 nM). Transfected cells were then incubated in the absence or presence of 20 ng/ml BMP4. Luciferase activity was measured in cell extracts. Only inhibition of BMPRII expression reduced BMP4 signaling. (B) Effect of siRNA targeting of BMPRII, ActRIIA, and ActRIIB on Dragon-mediated BMP signaling. mIMCD3 cells were transfected with BRE luciferase reporter in combination with control siRNA or siRNA specific for BMPRII, ActRIIA, or ActRIIB (60 nM) and in the absence or presence of Dragon cDNA for 46 hours before measurement of luciferase activity. In the presence of Dragon, inhibition of BMPRII and ActRIIA reduced endogenous BMP4 signaling. *P < 0.05 versus controls with treatments.
Transfection of mIMCD3 cells with Dragon cDNA increased BRE-Luc activity to 6.1-fold above the baseline (Figure 6B). This stimulation was reduced to 3.9- and 4.5-fold above the baseline by BMPRII and ActRIIA-specific siRNAs, respectively (P < 0.05 for both). Dragon-mediated BRE-Luc activity was not altered by inhibition of ActRIIB expression. These results suggest that BMPRII and ActRIIA are both utilized to transduce endogenous BMP4 signal in the presence of the Dragon coreceptor, indicating that Dragon enhances the utilization of ActRIIA by BMP4 in mIMCD3 cells.
ActRIIA is Coexpressed with Dragon in Proximal Tubule Cells and Intercalated Cells
To examine whether BMP4/Dragon/ActRIIA signaling occurs in the kidney, we analyzed expression of these genes along with BMPRII by RT-PCR in proximal tubule cells and intercalated cells isolated from adult kidneys (Figure 7). Consistent with the Dragon immunostaining, Dragon mRNA was detected in proximal tubule cells (Figure 7A) and intercalated cells (Figure 7B). BMP4 and ActRIIA mRNAs were also detected in both cell types, whereas BMPRII mRNA was not detected in either cell type. BMP2 was detected in proximal tubule cells but not in intercalated cells. These results suggest that Dragon may form a functional signaling unit with BMP4 and ActRIIA in vivo to enhance BMP signaling.
Dragon Increases TER
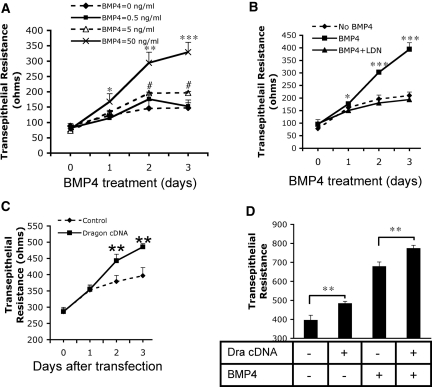
It has been shown that BMP ligands enhance TER in colonic epithelial cells.27 We therefore examined whether BMP4 and Dragon played a role in the establishment of TER of kidney epithelial cells. mIMCD3 cells were plated on Transwells (25,000 cells/well) in 12-well plates. Three days later TER was measured daily. As shown in Figure 8A, untreated mIMCD3 cells showed a small increase in TER with time. The addition of BMP4 (5 ng/ml) to these cells significantly increased TER on days 2 and 3 compared with untreated control cells. Increased concentration of BMP4 (50 ng/ml) caused greater increases in TER on days 1, 2, and 3. This increase in TER was not due to an increase in cell proliferation because cell numbers decreased in a dose-dependent manner after a 3-day exposure to BMP4 (Supplemental Figure 6A).
Figure 8.
BMP4 and Dragon increase TER in mIMCD3 cells. (A) Exogenous BMP4 increased TER. mIMCD3 cells were cultured in Transwells and different concentrations of BMP4 (0, 0.5, 5, and 50 ng/ml) were added. TER was measured daily. *P < 0.05, **P < 0.01, ***P < 0.001, 50 ng/ml BMP4 versus controls; #P < 0.05, 5 ng/ml BMP4 versus controls. (B) LDN-193189 inhibition of BMP4 signaling reduced its stimulatory effect on TER. mIMCD3 cells were cultured in Tranwells and were incubated with BMP4 (40 ng/ml) in the presence or absence of LDN-193189 (40 nM). TER was measure daily. *P < 0.05, ***P < 0.001, BMP4 versus BMP4 + LDN-193189. (C) Transfection with Dragon cDNA increased TER. mIMCD3 cells cultured in Transwells were transfected with Dragon cDNA and TER was measured daily. **P < 0.01 versus controls. (D) Stimulatory effects of BMP4 and Dragon on TER are additive. mIMCD3 cells cultured in Transwells were transfected with (bars 2 and 4) or without (bars 1 and 3) Dragon cDNA and were incubated with (bars 3 and 4) or without (bars 1 and 2) 10 ng/ml BMP4. TER was measured on day 3 after transfection. **P < 0.01.
To test whether the effect of BMP4 on TER is mediated through the Smad1/5/8 pathway or through the mitogen-activated protein kinase (MAPK) pathway, we incubated mIMCD3 cells with BMP4 in the presence or absence of LDN-193189 (DM-3189), a small-molecule derivative of the BMP inhibitor Dorsomorphin.41,42 Consistent with previous findings,43 LDN-193189 inhibited Smad1/5/8 phosphorylation induced by BMP4 but did not affect p38 MAPK phosphorylation induced by BMP4 (Supplemental Figure 6B). As shown in Figure 8B, LDN-193189 (40 nM) completely abolished stimulation of TER by BMP4. These results suggest that BMP4 signals through the Smad1/5/8 pathway but not through the MAPK pathway to regulate TER.
To examine the effect of Dragon on TER, mIMCD3 cells at 90% confluency in Transwells were transfected with Dragon cDNA, and TER was measured daily. Dragon overexpression significantly increased TER on days 2 and 3 after transfection (Figure 8C), whereas cell numbers were significantly reduced as counted on day 3 after transfection (Supplemental Figure 6C). Transfection of Dragon in the presence of BMP4 further increased TER measured on day 3 compared with Dragon transfection or BMP4 treatment alone (Figure 8D). These results suggest that BMP and Dragon stimulate TER in mIMCD3 cells.
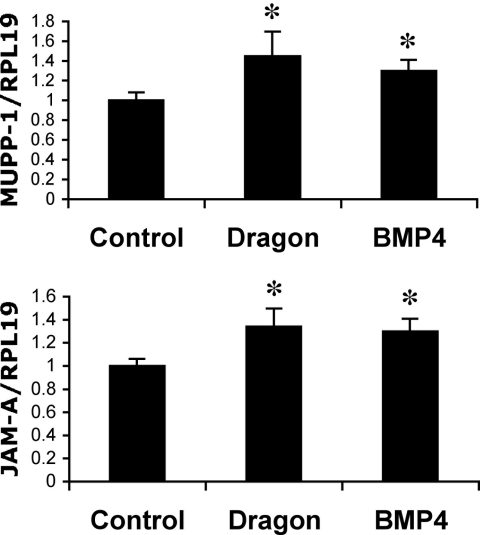
To determine the potential mechanism of BMP4 and Dragon action, we examined the mRNA expression levels of tight junction proteins claudins 1 to 16; occludin; ZO-1, ZO-2, and ZO-3; MUPP-1; JAM-A, JAM-B, and JAM-C; and cingulin in mIMCD3 cells treated with BMP4. We found that MUPP-1 and JAM-A were significantly upregulated by BMP4. The expression of MUPP-1 and JAM-A was also increased by transfection of Dragon (Figure 9). MUPP-1 and JAM-A were expressed in intercalated cells, and JAM-A but not MUPP-1 were detected in proximal tubule cells (Figure 7). These results suggest that Dragon may regulate expression of tight junction components such as JAM-A and MUPP-1 in the kidney.
Figure 9.
BMP4 and Dragon increase expression of MUPP-1 and JAM-A mRNAs. mIMCD3 cells cultured in Transwells were transfected with Dragon cDNA for 46 hours or treated with BMP4 for 18 hours. MUPP-1 or JAM-A relative to RPL19 mRNA levels were quantified by real-time RT-PCR. *P < 0.05.
Discussion
The RGMs RGMa, RGMb (Dragon), and RGMc (hemojuvelin) are GPI-anchored membrane proteins. Our studies have shown that the three proteins are BMP coreceptors that enhance BMP signaling. Although RGMc/hemojuvelin has been shown to regulate hepcidin expression and iron homeostasis through the BMP pathway,30,44 the biologic roles of RGMa and Dragon as BMP coreceptors remain largely unknown.
To investigate the potential biologic functions of Dragon in the kidney, we examined the cellular distribution of Dragon in this organ. We found that Dragon is expressed in epithelial cells of various tubules, including thick ascending limbs, distal tubules, and collecting ducts in the kidney. Similar to other GPI-anchored proteins,45 Dragon is concentrated most heavily on the apical surfaces of tubular epithelial cells in the kidney, although basolateral staining is also present. These results suggest that Dragon may play a role in transport or signaling in kidney tubular epithelial cells.
BMPs play important roles in numerous physiologic and pathologic processes during kidney development and in response to injury and repair. Here we found that Dragon can mediate BMP signaling but not TGF-β signaling in mIMCD3 cells. Interestingly, Dragon-expressing cells in the kidney (e.g., intercalated cells and principal cells of collecting ducts, epithelial cells of distal convoluted tubules and connecting segments, and thick ascending limbs) exhibited active BMP signaling as shown by the accumulation of nuclear phospho-Smad1/5/8 proteins in the nucleus. These results are consistent with our previous findings with other cell types,28,32 suggesting that Dragon also functions as a BMP coreceptor in epithelial cells in the kidney.
We previously showed that Dragon-mediated BMP signaling is ligand-dependent and that Dragon.Fc binds radiolabeled BMP2 and BMP4 but not BMP7 using solid phase binding assays.28 We have also shown that Dragon.Fc inhibits the biologic activity of BMP2 and BMP4 ligands much more potently than other BMP ligands, including BMP5, BMP6, BMP7, and BMP9, presumably by binding these ligands and preventing their interaction with cell surface signaling receptors. BMP2 and BMP4 are expressed in the kidney, suggesting that BMP2 and BMP4 are very likely to be the key endogenous ligands used by the Dragon coreceptor in the kidney. In mIMCD3 cells, Dragon-induced BMP signaling was blocked by siRNA inhibition of BMP4 expression but was not affected by siRNA inhibition of BMP2, BMP6, and BMP7. This suggests that BMP4 is likely the sole endogenous ligand for the Dragon BMP coreceptor in mIMCD3 cells. The failure of BMP2 inhibition to affect Dragon-mediated BMP signaling in mIMCD3 cells was unexpected, but it is most likely due to the low levels of endogenous BMP2 expression. However, this result does not rule out the possibility that BMP2 is also an endogenous ligand for the Dragon coreceptor in the native kidney. The failure of BMP6 inhibition to affect Dragon-mediated BMP signaling suggests that endogenous BMP6 did not play a significant role in Dragon action in this system, presumably because of the lower affinity of Dragon for BMP6 compared with that for BMP4. These data are consistent with our bioinhibition data showing that Dragon.Fc was significantly less potent at inhibiting BMP6 compared with BMP4. Interestingly, this contrasts to hemojuvelin, which uses BMP6 as a ligand and appears to have higher affinity for BMP6 compared with BMP4.34,46
We also tested the utilization of BMP type II receptors used by the BMP4 ligand in mIMCD3 cells in the presence or absence of transfected Dragon. siRNA-mediated inhibition of BMPRII or ActRIIA expression significantly reduced BMP4 signaling mediated by Dragon. This indicates that, in the presence of transfected Dragon, endogenous BMP4 ligand signals through BMPRII and ActRIIA. This is consistent with prior data showing that Dragon physically interacts with BMPRII and ActRIIA when overexpressed in HEK293 cells.28 In contrast, in the absence of Dragon overexpression, exogenous BMP4 signals were inhibited by BMPRII siRNA but not ActRIIA siRNA, suggesting that BMP4 normally signals through BMPRII in mIMCD3 cells. These results suggest that Dragon enhances utilization of ActRIIA by BMP4 in mIMCD3 cells, similar to our previously reported finding for RGMa33 and hemojuvelin.34 This ability to enhance utilization of ActRIIA by BMP ligands appears to be a common feature of the entire RGM family and may be one mechanism by which these coreceptors enhance cellular responses to BMP ligands.
In the isolated proximal tubule cells and intercalated cells, mRNAs for Dragon and ActRIIA were both detected, whereas BMPRII was not detected. These results are consistent with our general hypothesis that Dragon or other RGM family members may play an important role in regulating BMP4 (or BMP2) signaling in BMPRII-null, BMPRII-low, or BMPRII-deficient cells,33 which would not respond well to BMP4 or BMP2 in the absence of the RGM family of coreceptors. Indeed, several other cell types have been identified where RGM family members are expressed, and ActRIIA is the predominant BMP type II receptor whereas BMPRII is not or is barely detectable, including epithelial cells of ureteric branches in embryonic kidneys,47,48 human liver cells,34 and oocytes in the ovary.49,50 Although it remains to be investigated whether ActRIIA is expressed in other Dragon-expressing cells within the kidney, the expression of the BMP4/Dragon/ActRIIA system in the proximal tubule cells and intercalated cells supports our hypothesis that Dragon plays a role in adult kidney function.
A study by Peiris et al. showed that BMP2 modulated epithelial barrier maturation as assessed by the increases in TER in colonic epithelial cells.27 Consistent with this result, we found that BMP4 dramatically increased TER in mIMCD3 cells cultured in Transwell inserts through the Smad1/5/8 pathway, and Dragon further enhanced the BMP4 effect on TER. Furthermore, Dragon and BMP4 significantly increased the mRNA expression of tight junction components MUPP-1 and JAM-A, providing a potential mechanism by which Dragon and BMP increase TER.
Functional tight junctions are essential for the establishment and maintenance of the polarized architecture of the epithelial cells.16–18 Therefore, modulation of tight junction functions by BMP signaling may be a mechanism by which BMPs regulate kidney recovery in response to injury, and Dragon may play an important role in these processes by sensitizing renal epithelial cells to BMP signals. In addition, tight junctions provide a barrier that regulates paracellular transport. In renal tubules, the paracellular pathway is recognized to play an important role in the epithelial permeability of ions (e.g., sodium, chloride, calcium, and magnesium), and variations in TER among different nephron segments may be involved in regulation of selectivity for these transported ions.26,51–53 Thus, our results raise the possibility that BMPs/Dragon may play a role in paracellular transport in tubular epithelia in the kidney.
In summary, we found that Dragon is strongly expressed in epithelial cells of thick ascending limbs, distal tubules, and collecting ducts of the kidney and is weakly expressed in proximal tubules, glomeruli, and Bowman's capsules. In mIMCD3 cells, BMP4 is the predominant endogenous ligand used by Dragon to mediate BMP signaling. Similar to RGMa and RGMc/hemojuvelin, Dragon enhances utilization of ActRIIA by BMP4, which normally signals through BMPRII. ActRIIA is expressed in proximal tubule cells and intercalated cells, which also express Dragon. Dragon and BMP4 increase tight junction protein expression and TER of cultured mIMCD3 cells. These results suggest that Dragon may play an important role in kidney function by enhancing BMP signaling.
Concise Methods
Western Blot Analysis
Extracts from mIMCD3 cells treated with BMP4 in the presence or absence of LDN-193189 were subjected to Western blotting with anti-phospho-SMAD1/5/8 (Cell Signaling), anti-total-SMAD1 (Cell Signaling), anti-phospho-MAPK p38 (Cell Signaling), or anti-total MAPK p38 (Cell Signaling).
Immunohistochemistry Analysis
Sixty-day-old mice were perfused with paraformaldehyde-lysine-periodate fixative (4% paraformaldehyde, 75 mM lysine-HCl, 10 mM sodium periodate, and 0.15 M sucrose in 37.5 mM sodium phosphate). Frozen sections (5 μm) were collected onto Superfrost Plus precleaned charged microscope slides (Fisher Scientific, Pittsburgh, PA).
Sections were treated with 1% SDS for 4 minutes for retrieval of antigenic sites.54 The sections were incubated with a previously characterized rabbit anti-Dragon antibody31,32 diluted at 1:5000 in Dako antibody diluent (Dako, Carpinteria, CA) in combination with chicken anti-V-ATPase E1 subunit (1:10),55 goat anti-AQP2 (0.4 μg/ml, Santa Crutz Biotechnology), mouse anti-calbindin 28 (1: 1600, Sigma-Aldrich, St. Louis, MO), or mouse anti-Tamm Horsfall glycoprotein (1 μg/ml, Cappel Laboratories, Inc., Cochranville, PA). The Dragon staining specificity was assessed by preincubating the Dragon antibody with immunizing peptide (200 ng/μl) before applying to tissue sections. For phospho-Smad1/5/8 staining, immersion-fixed kidneys were used; the sections were treated with 1% SDS for 12 minutes.
Luciferase Assays
mIMCD3 cells (ATCC CRL-2123) were cultured in DME medium (Cellgro, Mediatech., VA) supplemented with 10% FBS. All transfections were performed with Lipofectamine-2000 (Invitrogen Life Technologies, Carlsbad, CA).
To test the effect of Dragon on BMP signaling, mIMCD3 cells were transiently transfected with Bre-Luc39 or (CAGA)12MLP-Luc)40 alone or in combination with cDNA encoding full-length Dragon. A control pRL-TK Renilla luciferase reporter (Promega, Madison, NY) was included to control for transfection efficiency. Cells were then serum starved for 6 hours before treatment with varying amounts of BMP2, BMP4, or TGF-β1 ligands (R&D systems, Minneapolis, MN) for 16 hours. Cells were lysed and luciferase activity was determined with the dual reporter assay (Promega, Madison, NY). Relative light units were calculated as ratios of Firefly (reporter) and Renilla (transfection control) values. Results from luciferase assay experiments are expressed as mean ± SD of triplicates from representative experiments. Two to three independent experiments were performed in each experimental setting.
Isolation of Proximal Tubule Cells and Intercalated Cells
The epithelial cells of proximal tubules were isolated as described previously.56 Briefly, mice were injected with BSA-Alexa555 via tail veins to label proximal tubules and 15 minutes later kidneys were collected and frozen. Sections (5 μm) were used to isolate proximal tubule cells using laser capture microdissection.
To isolate intercalated cells, we used ATP6V1B1-EGFP mice, which express enhanced green fluorescent protein (EGFP) specifically in intercalated cells within the kidney.57 Populations of EGFP-positive cells from kidney preparations were isolated by FACS on the basis of their green fluorescence intensity. A fraction of each sample was reanalyzed by flow cytometry to estimate the purity (>95%). EGFP-positive cell samples were collected in nuclease-free PBS (N. Da Silva, unpublished material).
RT-PCR
Total RNA was isolated from mIMCD3 cells, intercalated cells, and mouse kidneys using an RNeasy mini kit (Qiagen Inc.) or PicoPure RNA isolation kit (Molecular Devices) according to the manufacturer's instructions. First-strand cDNA synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad). For samples from isolated proximal tubule cells captured by laser microdissection, total RNAs were amplified using RiboAmp Plus KIT0521 (Molecular Devices) before reverse transcription. Transcripts of mouse BMP2, BMP4, RGMb, BMPRII, ActRIIA, and JAM-A in isolated proximal tubule cells and intercalated cells were amplified using the primers summarized in Supplemental Table 1, whereas MUPP-1 was amplified using primers previously described.58 Transcripts of mouse BMP2, BMP4, BMP5-7 in mIMCD3 cells were amplified using the primers summarized in Supplemental Table 2, and BMPRII, ActRIIA, and ActRIIB in mIMCD3 cells were amplified using the primers previously described.33
siRNA Targeting
Mouse BMPRII, ActRIIA, and ActRIIB siRNA sequences were described previously,59 and siRNA duplexes in annealed and purified form were obtained from Invitrogen. SMARTpool siRNAs against mouse BMP2, BMP4, BMP6, and BMP7 were purchased from Dharmacon. siRNA duplexes were added at the concentrations indicated along with plasmids to subconfluent mIMCD3 cells using Lipofectamine-2000 (Invitrogen). Assays to measure target mRNA levels or luciferase activity were performed 46 hours after transfection.
Measurement of Gene Expression
Real-time quantification of mRNA transcripts was performed as described previously. First-strand cDNA was amplified with the primers as in Supplemental Tables 1 and 2 (mouse BMP2, BMP4, BMP6, and BMP7; JAM-A, JAM-B, and JAM-C; ZO-1, ZO-2 and ZO-3) or as described previously (mouse BMPRII, ActRIIA, ActRIIB, and RPL1933; claudins 1 to 1660; occludin61; cingulin62; and MUPP-158). Results are expressed as a ratio of the gene of interest to RPL19.
TER
mIMCD3 cells were seeded to form monolayers on clear Transwells (0.4 μm pore size) in 12-well plates (Corning). TER was measured using the Millicell electrical resistance system (Millipore) at 0-, 24-, 48-, and 72 hours in the presence of BMP4 or after Dragon cDNA transfection. LDN-193189, a specific inhibitor for BMP signaling,41–43 was used at 40 nM to inhibit BMP4 activity. LDN-193189 was custom synthesized by Shanghai United Pharmatech Company, Shanghai, China.
Disclosures
None.
Supplementary Material
Acknowledgments
H.Y.L. was supported by National Institutes of Health (NIH) grant RO1 DK-071837 and RO1 DK-069533. D.B. was supported by NIH grant DK-42956. Y.X. was supported by NIH grant R03HD60641. J.L.B. was supported by NIH grant K08 DK-075846. The authors thank David A. Fabizio and Jason Campagna for technical assistance.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
References
- 1. Shi Y, Massague J: Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685– 700, 2003 [DOI] [PubMed] [Google Scholar]
- 2. Green MC: Mechanism of the pleiotropic effects of the short-ear mutant gene in the mouse. J Exp Zool 167: 129– 150, 1968 [DOI] [PubMed] [Google Scholar]
- 3. Kingsley DM, Bland AE, Grubber JM, Marker PC, Russell LB, Copeland NG, Jenkins NA: The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell 71: 399– 410, 1992 [DOI] [PubMed] [Google Scholar]
- 4. Dudley AT, Lyons KM, Robertson EJ: A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev 9: 2795– 2807, 1995 [DOI] [PubMed] [Google Scholar]
- 5. Luo G, Hofmann C, Bronckers AL, Sohocki M, Bradley A, Karsenty G: BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev 9: 2808– 2820, 1995 [DOI] [PubMed] [Google Scholar]
- 6. Dunn NR, Winnier GE, Hargett LK, Schrick JJ, Fogo AB, Hogan BL: Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev Biol 188: 235– 247, 1997 [DOI] [PubMed] [Google Scholar]
- 7. Martinez G, Mishina Y, Bertram JF: BMPs and BMP receptors in mouse metanephric development: In vivo and in vitro studies. Int J Dev Biol 46: 525– 533, 2002 [PubMed] [Google Scholar]
- 8. Miyazaki Y, Oshima K, Fogo A, Hogan BL, Ichikawa I: Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J Clin Invest 105: 863– 873, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miyazaki Y, Oshima K, Fogo A, Ichikawa I: Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int 63: 835– 844, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Bush KT, Sakurai H, Steer DL, Leonard MO, Sampogna RV, Meyer TN, Schwesinger C, Qiao J, Nigam SK: TGF-beta superfamily members modulate growth, branching, shaping, and patterning of the ureteric bud. Dev Biol 266: 285– 298, 2004 [DOI] [PubMed] [Google Scholar]
- 11. Hartwig S, Hu MC, Cella C, Piscione T, Filmus J, Rosenblum ND: Glypican-3 modulates inhibitory Bmp2-Smad signaling to control renal development in vivo. Mech Dev 122: 928– 938, 2005 [DOI] [PubMed] [Google Scholar]
- 12. Ozkaynak E, Schnegelsberg PN, Jin DF, Clifford GM, Warren FD, Drier EA, Oppermann H: Osteogenic protein-2. A new member of the transforming growth factor-beta superfamily expressed early in embryogenesis. J Biol Chem 267: 25220– 25227, 1992 [PubMed] [Google Scholar]
- 13. Gould SE, Day M, Jones SS, Dorai H: BMP-7 regulates chemokine, cytokine, and hemodynamic gene expression in proximal tubule cells. Kidney Int 61: 51– 60, 2002 [DOI] [PubMed] [Google Scholar]
- 14. Vukicevic S, Basic V, Rogic D, Basic N, Shih MS, Shepard A, Jin D, Dattatreyamurty B, Jones W, Dorai H, Ryan S, Griffiths D, Maliakal J, Jelic M, Pastorcic M, Stavljenic A, Sampath TK: Osteogenic protein-1 (bone morphogenetic protein-7) reduces severity of injury after ischemic acute renal failure in rat. J Clin Invest 102: 202– 214, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simon M, Maresh JG, Harris SE, Hernandez JD, Arar M, Olson MS, Abboud HE: Expression of bone morphogenetic protein-7 mRNA in normal and ischemic adult rat kidney. Am J Physiol 276: F382– F389, 1999 [DOI] [PubMed] [Google Scholar]
- 16. Brown D, Orci L: Junctional complexes and cell polarity in the urinary tubule. J Electron Microsc Tech 9: 145– 170, 1988 [DOI] [PubMed] [Google Scholar]
- 17. Brown D, Stow JL: Protein trafficking and polarity in kidney epithelium: From cell biology to physiology. Physiol Rev 76: 245– 297, 1996 [DOI] [PubMed] [Google Scholar]
- 18. Shin K, Fogg VC, Margolis B: Tight junctions and cell polarity. Annu Rev Cell Dev Biol 22: 207– 235, 2006 [DOI] [PubMed] [Google Scholar]
- 19. Bonventre JV: Mechanisms of ischemic acute renal failure. Kidney Int 43: 1160– 1178, 1993 [DOI] [PubMed] [Google Scholar]
- 20. Thadhani R, Pascual M, Bonventre JV: Acute renal failure. N Engl J Med 334: 1448– 1460, 1996 [DOI] [PubMed] [Google Scholar]
- 21. Humes HD, Liu S: Cellular and molecular basis of renal repair in acute renal failure. J Lab Clin Med 124: 749– 754, 1994 [PubMed] [Google Scholar]
- 22. Witzgall R, Brown D, Schwarz C, Bonventre JV: Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J Clin Invest 93: 2175– 2188, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Itallie CM, Anderson JM: The molecular physiology of tight junction pores. Physiology 19: 331– 338, 2004 [DOI] [PubMed] [Google Scholar]
- 24. Claude P: Morphological factors influencing transepithelial permeability: A model for the resistance of the zonula occludens. J Membr Biol 39: 219– 232, 1978 [DOI] [PubMed] [Google Scholar]
- 25. Schneeberger EE, Lynch RD: The tight junction: A multifunctional complex. Am J Physiol Cell Physiol 286: C1213– C1228, 2004 [DOI] [PubMed] [Google Scholar]
- 26. Reyes JL, Lamas M, Martin D, del Carmen Namorado M, Islas S, Luna J, Tauc M, González-Mariscal L: The renal segmental distribution of claudins changes with development. Kidney Int 62: 476– 487, 2002 [DOI] [PubMed] [Google Scholar]
- 27. Peiris D, Pacheco I, Spencer C, MacLeod RJ: The extracellular calcium-sensing receptor reciprocally regulates the secretion of BMP-2 and the BMP antagonist Noggin in colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol 292: G753– G766, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, Lin HY, Brivanlou AH, Attisano L, Woolf CJ: DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem 280: 14122– 14129, 2005 [DOI] [PubMed] [Google Scholar]
- 29. Babbit JL, Zhang Y, Samad T, Xia Y, Tang J, Schneyer A, Woolf CJ, Lin HY: Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem 280: 29820– 29827, 2005 [DOI] [PubMed] [Google Scholar]
- 30. Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY: Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 38: 531– 539, 2006 [DOI] [PubMed] [Google Scholar]
- 31. Samad TA, Srinivasan A, Karchewski LA, Jeong SJ, Campagna JA, Ji RR, Fabrizio DA, Zhang Y, Lin HY, Bell E, Woolf CJ: DRAGON: A member of the repulsive guidance molecule-related family of neuronal- and muscle-expressed membrane proteins is regulated by DRG11 and has neuronal adhesive properties. J Neurosci 24: 2027– 2036, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xia Y, Sidis Y, Mukherjee A, Samad T, Brenner G, Woolf CJ, Lin HY, Schneyer A: Localization and action of Dragon (repulsive guidance molecule b), a novel bone morphogenetic protein coreceptor, throughout the reproductive axis. Endocrinology 146: 3614– 3621, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xia Y, Yu PB, Sidis Y, Beppu H, Bloch KD, Schneyer AL, Lin HY: Repulsive guidance molecule (RGMa) alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J Biol Chem 282: 18129– 18140, 2007 [DOI] [PubMed] [Google Scholar]
- 34. Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY: Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood 111: 5195– 5204, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown D, Katsura T, Kawashima M, Verkman AS, Sabolic I: Cellular distribution of the aquaporins: A family of water channel proteins. Histochem Cell Biol 104: 1– 9, 1995 [DOI] [PubMed] [Google Scholar]
- 36. Paunescu TG, Da Silva N, Marshansky V, McKee M, Breton S, Brown D: Expression of the 56-kDa B2 subunit isoform of the vacuolar H(+)-ATPase in proton-secreting cells of the kidney and epididymis. Am J Physiol Cell Physiol 287: C149– C162, 2004 [DOI] [PubMed] [Google Scholar]
- 37. Lai LW, Yong KC, Lien YH: Site-specific expression of IQGAP1, a key mediator of cytoskeleton, in mouse renal tubules. J Histochem Cytochem 56: 659– 666, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sikri KL, Foster CL, MacHugh N, Marshall RD: Localization of Tamm-Horsfall glycoprotein in the human kidney using immuno-fluorescence and immuno-electron microscopical techniques. J Anat 133: 425– 442, 1981 [PMC free article] [PubMed] [Google Scholar]
- 39. Korchynskyi O, ten Dijke P: Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem 277: 4883– 4891, 2002 [DOI] [PubMed] [Google Scholar]
- 40. Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM: Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091– 3100, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT: Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 4: 33– 41, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT: Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett 18: 4388– 4392, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, Kamiya N, Fukuda T, Mishina Y, Peterson RT, Bloch KD: BMP type I receptor inhibition reduces heterotopic ossification. Nat Med 14: 1363– 1369, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY: Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest 117: 1933– 1939, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown D, Waneck GL: Glycosyl-phosphatidylinositol-anchored membrane proteins. J Am Soc Nephrol 3: 895– 906, 1992 [DOI] [PubMed] [Google Scholar]
- 46. Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietragelo A, Vukicevic S, Lin HY, Babitt JL: BMP-6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet 41: 482– 487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martinez G, Loveland KL, Clark AT, Dziadek M, Bertram JF: Expression of bone morphogenetic protein receptors in the developing mouse metanephros. Exp Nephrol 9: 372– 379, 2001 [DOI] [PubMed] [Google Scholar]
- 48. Maeshima A, Yamashita S, Maeshima K, Kojima I, Nojima Y: Activin a produced by ureteric bud is a differentiation factor for metanephric mesenchyme. J Am Soc Nephrol 14: 1523– 1534, 2003 [DOI] [PubMed] [Google Scholar]
- 49. Erickson GF, Shimasaki S: The spatiotemporal expression pattern of the bone morphogenetic protein family in rat ovary cell types during the estrous cycle. Reprod Biol Endocrinol 1: 9, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cameron VA, Nishimura E, Mathews LS, Lewis KA, Sawchenko PE, Vale WW: Hybridization histochemical localization of activin receptor subtypes in rat brain, pituitary, ovary, and testis. Endocrinology 134: 799– 808, 1994 [DOI] [PubMed] [Google Scholar]
- 51. Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP: Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103– 106, 1999 [DOI] [PubMed] [Google Scholar]
- 52. Yamauchi K, Rai T, Kobayashi K, Sohara E, Suzuki T, Itoh T, Suda S, Hayama A, Sasaki S, Uchida S: Disease-causing mutant WNK4 increases paracellular chloride permeability and phosphorylates claudins. Proc Natl Acad Sci U S A 101: 4690– 4694, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu ASL, Enck AH, Lencer WI, Schneeberger EE: Claudin-8 expression in Madin-Darby canine kidney cells augments the paracellular barrier to cation permeation. J Biol Chem 278: 17350– 17359, 2003 [DOI] [PubMed] [Google Scholar]
- 54. Brown D, Lydon J, McLaughlin M, Stuart-Tilley A, Tyszkowski R, Alper S: Antigen retrieval in cryostat tissue sections and cultured cells by treatment with sodium dodecyl sulfate (SDS). Histochem Cell Biol 105: 261– 267, 1996 [DOI] [PubMed] [Google Scholar]
- 55. Breton S, Wiederhold T, Marshansky V, Nsumu NN, Ramesh V, Brown D: The B1 subunit of the H+ATPase is a PDZ domain-binding protein. Colocalization with NHE-RF in renal B-intercalated cells. J Biol Chem 275: 18219– 18224, 2000 [DOI] [PubMed] [Google Scholar]
- 56. Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, Brown D, Marshansky V: V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol 8: 124– 136, 2006 [DOI] [PubMed] [Google Scholar]
- 57. Miller RL, Zhang P, Smith M, Beaulieu V, Paunescu TG, Brown D, Breton S, Nelson RD: V-ATPase B1-subunit promoter drives expression of EGFP in intercalated cells of kidney, clear cells of epididymis and airway cells of lung in transgenic mice. Am J Physiol Cell Physiol 288: C1134– C1144, 2005 [DOI] [PubMed] [Google Scholar]
- 58. Lanaspa MA, Almeida NE, Andres-Hernando A, Rivard CJ, Capasso JM, Berl T: The tight junction protein, MUPP1, is up-regulated by hypertonicity and is important in the osmotic stress response in kidney cells. Proc Natl Acad Sci U S A 104: 13672– 13677, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu PB, Deng DY, Beppu H, Hong CC, Lai C, Hoyng SA, Kawai N, Bloch KD: Bone morphogenetic protein (BMP) type II receptor is required for BMP-mediated growth arrest and differentiation in pulmonary artery smooth muscle cells. J Biol Chem 280: 24443– 24450, 2005 [DOI] [PubMed] [Google Scholar]
- 60. Abuazza G, Becker A, Williams SS, Chakravarty S, Truong HT, Lin F, Baum M: Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am J Physiol Renal Physiol 291: F1132– F1141, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang RS, Yeh S, Chen LM, Lin HY, Zhang C, Ni J, Wu CC, di Sant'Agnese PA, DeMesy-Bentley KL, Tzeng CR, Chang C: Androgen receptor in Sertoli cell is essential for germ cell nursery and junctional complex formation in mouse testes. Endocrinology 147: 5624– 5633, 2006 [DOI] [PubMed] [Google Scholar]
- 62. Guillemot L, Hammar E, Kaister C, Ritz J, Caille D, Jond L, Bauer C, Meda P, Citi S: Disruption of the cingulin gene does not prevent tight junction formation but alters gene expression. J Cell Sci 117: 5245– 5256, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.