Abstract
Apoptosis contributes to the development of diabetic nephropathy (DN), but the mechanisms that lead to diabetes-induced cell death are not fully understood. Here, we combined a functional genomics screen for cDNAs that induce apoptosis in vitro with transcriptional profiling of renal biopsies from patients with DN. Twelve of the 138 full-length cDNAs that induced cell death in human embryonic kidney cells matched upregulated mRNA transcripts in tissue from human DN. Confirmatory screens identified induction of BASP1 in tubular cross sections of human DN tissue. In vitro, apoptosis-inducing conditions such as serum deprivation, high concentrations of glucose, and proinflammatory cytokines increased BASP1 mRNA and protein in human tubular epithelial cells. In normal cells, BASP1 localized to the cytoplasm, but in apoptotic cells, it colocalized with actin in the periphery. Overexpression of BASP1 induced cell death with features of apoptosis; conversely, small interfering RNA (siRNA)-mediated knockdown of BASP1 protected tubular cells from apoptosis. Supporting possible involvement of BASP1 in renal disease other than DN, we also observed significant upregulation of renal BASP1 in spontaneously hypertensive rats and a trend toward increased tubulointerstitial BASP1 mRNA in human hypertensive nephropathy. In summary, a combined functional genomics approach identified BASP1 as a proapoptotic factor in DN and possibly also in hypertensive nephropathy.
Diabetic end-organ damage contributes to at least 233,000 yearly deaths in the United States alone, with diabetic nephropathy (DN) being the most common cause of ESRD.1 The diabetic milieu is known to induce apoptosis in various end-organ systems2,3 and is believed to contribute to the gradual loss of renal function and mass in DN, because apoptosis has been detected in tubular epithelial, endothelial, and interstitial cells of renal biopsy samples from patients with DN.4,5
Cell death via apoptosis is an active response to an altered microenvironment and is characterized by the activation of specific intracellular pathways. The presence of injurious factors and/or the lack of survival factors may lead to engagement of the apoptotic molecular machinery. Apoptosis is modulated by a host of checks and balances with a multitude of positive and negative regulators, many of which are disease and organ specific.6
The combination of unbiased gene expression profiling with focused data mining has proven to be a powerful tool to expand our knowledge of relevant pathways and players in human disease.7,8 We have recently used this approach to find specific changes in apoptosis-related genes in the renal tubulointersitium transcriptome of the human diabetic kidney using gene ontology.6 A key limitation of this method is the need of a priori knowledge of the cell-death-associated function of the regulated transcript. Genome-wide functional assays for the identification of novel cell-death-associated molecules have recently been developed.9 For this study this strategy was adapted to identify apoptosis-inducing full-length reading frames in human embryonic kidney cells as a primary screening step. The in vitro assay was followed by analysis of the regulation of the corresponding mRNA transcripts in human DN. From the genes passing both filter steps, brain acid soluble protein 1/brain-abundant signal protein 1 (BASP1) mRNA and protein was induced in the tubulointerstitial compartment of human DN.
BASP1 (also neuron-enriched acidic protein, having a molecular mass of 22 kD/cortical cytoskeleton-associated protein) is a 23-kDa myristoilated protein originally isolated from brain extracts10,11 that shares 70% homology in human and rat.12 It was initially described as a brain-specific protein,10,11 but later studies revealed that BASP1 is also expressed by human endothelium,13 developing mammary gland, kidney, testis, and lymphoid tissues.12–16 BASP1 is involved in cytoskeletal and lipid raft dynamics, as well as in the nuclear regulation of transcription. However, a role in apoptosis has not been previously reported. BASP1 contains an effector domain (ED) that dynamically binds to the plasma membrane, to calmodulin, and to actin fibrils. Reversible phosphorylation of ED by protein kinase C modulates these interactions.12 In the plasma membrane BASP1 localizes to lipid rafts and may influence the behavior and composition of the membrane.17 In addition, BASP1 promotes actin dynamics, including loss of stress fibers and bleb formation.18 Additionally, BASP1 is present in the developing nephron structures of the embryonic kidney coinciding with the transcriptional regulator Wilm's tumor gene (WT1). In the adult kidney the main site of BASP1 expression is the podocyte, where it behaves as a transcriptional cosuppressor of WT1. Low levels of BASP1 expression are present in the cytoplasm of tubular cells in vivo and in cell lines.15
BASP1 expression is increased in human DN tubulointerstitium and in tubules from experimental DN. BASP1 is also expressed by cultured renal tubular cells, where it is regulated by stimuli that promote cell death and has a role in apoptotic cell death.
Results
Functional Genomics cDNA Screening for Cell-Death-Inducing Genes
In a previous search for genes with a potential tumor suppressor phenotype, a high throughput (HT) functional genomics screen was established to identify novel cell-death-inducing genes.9 In brief, 596,832 independent clones from a human embryo cDNA expression library and 17,680 defined full-length cDNA expression plasmids were screened for cell death induction after transient transfection into human embryonic kidney (HEK293) cells. Of these, 194 unique cDNAs induced cell death and DNA fragmentation: 138 contained full-length open reading frames (Figure 1). Thirty-nine genes were known inducers or associated with apoptosis.
Figure 1.
Integrative functional genomics screen for apoptosis mediators in DN. 138 of 18000 unique cDNA clones containing full-length open reading frames induced cell death upon overexpression in HEK293 cells. Genome-wide expression analysis of renal biopsies from DN and control patients identified 12 of the 138 unique clones to be upregulated in the tubulointerstitium with q ≤ 0.05 (Table 1). Of these, only BASP1 was positive in confirmatory apoptosis screening tests and was chosen for further analysis.
The list of full-length cDNAs whose overexpression induced cell death in the HT functional screen was compared with genome-wide expression profiles from tubulointerstitial compartments of DN biopsies.6,7 Twelve genes were upregulated in DN (q < 0.05) and induced cell death in the HT functional screen as assessed by β-galactosidase/chlorophenolred-β-d-galactopyranoside and DNA fragmentation assays in HEK293 cells (Table 1). Of these genes, only BASP1 overexpression yielded positive results in all three subsequent confirmatory screens for caspase activation, externalization of phosphatidylserin, and loss of mitochondrial potential (Figure 1).
Table 1.
Open reading frames capable of induction of cell death when overexpressed and with concomitant upregulation in the tubulointerstitial compartment in human DN biopsies (q ≤ 0.05)a
| GeneSymbol | FC | LogFC | q |
|---|---|---|---|
| EMP3 | 1.75 | 0.81 | 0.00 |
| BASP1 | 1.62 | 0.70 | 0.00 |
| PLP2 | 1.61 | 0.69 | 0.00 |
| ARL6IP5 | 1.41 | 0.50 | 0.00 |
| PTPLAD1 | 1.39 | 0.48 | 0.00 |
| GNAS | 1.38 | 0.47 | 0.00 |
| CYBA | 1.33 | 0.41 | 0.00 |
| PPIB | 1.29 | 0.36 | 0.00 |
| CLDN4 | 1.58 | 0.66 | 0.03 |
| TMEM59 | 1.22 | 0.29 | 0.03 |
| ITGB2 | 1.84 | 0.88 | 0.04 |
| SSR4 | 1.31 | 0.39 | 0.04 |
FC, fold change over control; EMP3, epithelial membrane protein 3; PLP2, proteolipid protein 2 (colonic epithelium-enriched); ARL6IP5, ADP-ribosylation-like factor 6 interacting protein 5; PTPLAD1, protein tyrosine phosphatase-like A domain containing 1; GNAS, guanine nucleotide binding protein, alpha stimulating complex locus; CYBA, cytochrome b-245, alpha polypeptide; PPIB, peptidylprolyl isomerase B (cyclophilin B); CLDN4, claudin 4; TMEM59, transmembrane protein 59; ITGB2, integrin, beta 2 (complement component 3 receptor 3 and 4 subunit); SSR4, signal sequence receptor, delta.
aData are presented as the fold-change increase in mRNA expression in DN as assessed by Affymetrix GeneChip.
BASP1 Is an Apoptosis Inducer and Is Upregulated in the Tubulointerstitium of DN
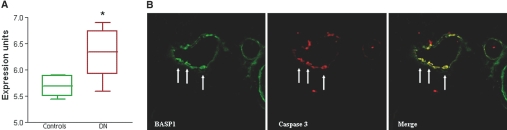
Overexpression of BASP1-induced death as assessed by β-galactosidase/chlorophenolred-β-d-galactopyranoside and DNA fragmentation assays in HEK293 cells. BASP1 was also positive in screening tests for caspase activation, externalization of phosphatidylserine, and of loss of mitochondrial potential. BASP1 mRNA was increased 1.6-fold in the renal tubulointerstitium of DN patients when compared with controls (Figure 2A). No significant changes were observed in the glomerular compartment (1.09-fold change, data not shown). BASP1 mRNA expression correlated with 24-hour proteinuria (R2 = 0.40, P = 0.04) in DN patients but not in minimal change disease (MCD) patients (R2 = 0.14, P = 0.61). In an independent cohort of patients with DN, BASP1 mRNA was evaluated by quantitative real-time reverse-transcriptase PCR (RT-PCR). The BASP1-to-18S rRNA ratio was induced 2.4 times in DN compared with controls [mean ± SD; control 0.5 ± 0.25, DN 1.18 ± 0.6, P = 0.02]. BASP1 expression in nondiabetic renal diseases is summarized in Supplemental Table 1). Immunofluorescence confirmed an increase of tubular expression of BASP1 in human DN compared with minimal detectable levels in control kidneys (Supplemental Figure 1), as well as co-staining for BASP1 and active caspase 3 in human DN (Figure 2B). The percentage of positive tubulointerstitial area was significantly higher in DN (18.25 ± 2.49, versus MCD 7.2 ± 3.11% and control kidney 0%, P < 0.05) and this was associated with an increased apoptosis rate (active caspase 3 staining DN 5.25 ± 2.96% of cells, versus MCD 0.6 ± 0.89 versus control kidney 0.33 ± 0.57%, P < 0.05). In addition, total kidney BASP1 protein was increased in two animal models of chronic kidney disease: diabetes induced by streptozotocin and hypertensive nephropathy of spontaneously hypertensive rats (Figure 3, A and B). Immunohistochemistry localized increased BASP1 expression to tubular epithelia in these models (Figure 3C). Lectin binding experiments identified proximal tubules as the main source of tubular BASP1 in DN (Supplemental Figure 2). As previously reported in rodent models of DN, an increased tubular apoptosis rate was observed in diabetic kidneys (Supplemental Figure 3A).4,19–21 Apoptotic cells tended to be grouped in certain tubular epithelia where colocalization of active caspase 3 and BASP1 expression was noted (Supplemental Figure 3B).
Figure 2.
Increased BASP1 expression in human DN. (A) BASP1 mRNA was increased in the renal tubulointerstitium of DN patients (n = 11) when compared with the controls (n = 7) (*q = 0.001). There was no difference in BASP1 expression between living donors and MCD biopsies, thus they were grouped as controls. The array data are expressed as normalized log-transformed expression units. The box extends from 25th percentile to the 75th percentile and the line gives the median at the 50th percentile. (B) Most cells in a tubule from a type II DN biopsy co-stain for BASP1 (green) and active caspase 3 (red) in DN tubulointerstitium. Note co-stained cells in the merged image (yellow, arrows). Original magnification 400×.
Figure 3.
BASP1 expression in experimental DN and hypertensive nephropathy. (A) Representative image of Western blot of whole kidney extracts (SHR, spontaneously hypertensive rats). (B) Quantification of Western blot results. *P < 0.05 versus Wistar control rats. Mean ± SD of 5 rats. (C) Immunohistochemistry localized increased BASP1 expression to tubular cells. Detail: Note an apoptotic BASP1-stained cell in the tubular lumen. Original magnification 200× and 1000×.
BASP1 Regulation in Cultured Human Tubular Epithelial Cells
Cultured human proximal tubular epithelial cells constitutively express BASP1 mRNA and protein (Figure 4). Deprivation of the survival factors from serum increased BASP1 mRNA (Figure 4A) and protein (Figure 4, B and C) expression. Confocal fluorescence localized the protein to the perinuclear area (Figure 4D). This is consistent with the known localization of BASP1 in human umbilical vascular endothelial cells and COS-1 cells.13,15 The increased protein expression found in serum-deprived cells was mainly localized to the perinuclear area. A peripheral location near the cell membrane was more conspicuous in apoptotic cells, where it colocalized with actin and α-tubulin in the cell periphery (Figure 4E). BASP1 expression was also increased by a high glucose concentration and by proinflammatory cytokines (Figure 5A and B), but not by mannitol used as an osmotic control (<5% variation versus control). Deprivation of survival factors and exposure to a high glucose environment and proinflammatory cytokines are known inducers of apoptosis in tubular cells.22–24
Figure 4.
Serum deprivation increases BASP1 expression in human proximal tubular epithelial HK2 cells. (A) Real-time RT-PCR analyses of BASP1 mRNA expression in cells cultured in the presence or absence of 10% FBS. Mean ± SD of three independent experiments. *P < 0.05 versus 10% FBS for 24 hours. (B) Quantification of Western blot results. Mean ± SD of three independent experiments. *P < 0.05 versus 10% FBS for 24 hours. **P < 0.01 versus 10% FBS for 48 and 72 hours. (C) Representative Western blot of BASP1. Cells were cultured in the presence or absence of 10% FBS for 72 hours. (D) Confocal microscopy confirmed increased BASP1 expression in cells deprived of the survival factors from serum for 24 hours, although in the presence and absence of serum BASP1 was mainly cytoplasmic and perinuclear in live cells. By contrast, in apoptotic cells found among serum-deprived cells BASP1 displayed a peripheral distribution consistent with its presence in the cell cortex. BASP1, green; propidium iodide, red. Original magnification 320×. (E) Colocalization of BASP1 was further explored in cells undergoing apoptosis upon serum deprivation. BASP1 localized to the cell periphery of apoptotic cells with pyknotic nuclei, where it appeared to colocalize with tubulin (arrows, upper panel, conventional fluorescence microscopy). Confocal microscopy confirmed colocalization with tubulin (middle panel, confocal microcopy) and actin (arrow, lower panel, confocal microscopy) in the periphery of apoptotic cells. In the two lower panels the image is focused on the apoptotic cells. Nevertheless, the different pattern of actin filament distribution between the apoptotic cell (arrow) and the living cells is clearly observed in the lower panel.
Figure 5.
BASP1 expression is increased by apoptosis-inducing stimuli. Cells exposed to high glucose concentration or proinflammatory cytokines (100 ng/ml TWEAK, 30 ng/ml TNFα, and INFγ 30 U/ml) for 48 hours. (A) Representative Western blot. (B) Quantification of Western blot results. Mean ± SD of three independent experiments *P < 0.05 versus control, **P < 0.01 versus control.
BASP1 Overexpression Induces Apoptosis in Tubular Epithelial Cells
Once we had demonstrated that BASP1 expression is regulated by an adverse microenvironment in tubular epithelia in vivo and in cell culture, we approached the consequences of BASP1 overexpression. As predicted by the screening assay, overexpression of BASP1 decreased cell survival in a dose-dependent manner (Figure 6A). Inhibitors of specific caspases, such as caspase 3 [Z-Asp(OMe)-Glu(OMe)-Val-DL-Asp(OMe)-fluoromethylketone (DEVD)], caspase 8 [Z-Ile-Glu(OMe)-Thr-Asp(OMe)-fluoromethylketone (IETD)], and caspase 9 [Z-Leu-Glu(OMe)-His-Asp(OMe)-fluoromethylketone (LEHD)], as well as multiple caspases [Z-Val-Ala-DL-Asp-fluoromethylketone (zVAD)] prevented the decrease in cell viability associated with BASP1 overexpression (Figure 6B). In addition P5, a KU-70-derived Bax inhibitory peptide, also prevented cell death (Figure 6B). Overexpression of BASP1 also increased the percentage of hypodiploid apoptotic cells (Figure 7A) and of apoptotic cells staining for caspase-generated cytokeratin fragments (Figure 7B), as well as caspase 3 activity (Figure 7C). Cotransfection with a green fluorescent protein (GFP) plasmid allowed quantification of apoptotic cells with the typical nuclear morphology among transfected cells and showed that BASP1 overexpression promoted apoptosis that was prevented by zVAD or P5 (Figure 8). Taken together, these data suggest the involvement of caspases and mitochondria in the cell death process activated by BASP1 overexpression.
Figure 6.
Overexpression of BASP1 induces caspase-dependent death in human tubular epithelial cells. (A) Dose-response curve: Cells were co-transfected with a β-galactosidase expression plasmid and different amounts of BASP1 expression vector. β-galactosidase activity was quantified at 24 hours. Mean ± SD of three independent experiments. *P < 0.05 versus control. (B) Protective effect of caspase inhibition and Bax antagonism. Cells were transfected with BASP1 expression vector and treated with 200 μM DEVD, LEDH, P5, or zVAD. Cell survival was assessed at 24 hours. Mean ± SD of three independent experiments. *P < 0.05 versus control, #P < 0.005 versus BASP1 alone.
Figure 7.
Cell death induced by BASP1 has features of apoptosis. (A) BASP1 induces apoptosis in tubular cells. Quantification of hypodiploid cells by flow cytometry of DNA content. Note the increase in the hypodiploid, apoptotic population in cultured cells transfected with 5 μg BASP1 for 24 hours. Mean ± SD for three independent experiments. *P < 0.05 versus control. (B) BASP1 overexpression increases the percentage of apoptotic cells stained with caspase-specific cytokeratin fragments (cytodeath), as assessed by flow cytometry. Mean ± SD for three independent experiments. *P < 0.05 versus control. (C) Caspase 3 activity is increased in tubular cells overexpressing BASP1. Mean ± SD for three independent experiments #P < 0.005 versus control. Cells were transfected with 5 μg BASP1 for 20 hours.
Figure 8.
BASP1 overexpression promotes human tubular epithelial cell apoptosis. (A) Tubular cells were cotransfected with GFP plasmid and BASP1 or empty vector for 24 hours. Note the presence of apoptotic nuclei (shrunk, bright) among cells transfected with BASP1 (arrows). (B) Quantification of apoptotic cells expressed as percentage of cells with apoptotic nuclei among green cells. Mean ± SD for three independent experiments. #P < 0.005 versus control, **P < 0.01 versus BASP1, ###P < 0.004 versus BASP1.
Inhibition of BASP1 Expression Protects against Apoptosis
A siRNA approach was used to assess the requirement of endogenous BASP1 for cell death induction in human kidney epithelial cells. Western blot confirmed efficient gene silencing (Figure 9A). Deprivation of survival factors is a classical inducer of apoptosis that also upregulates BASP1 expression in tubular cells. HK2 cells underwent apoptosis when deprived of serum that was prevented by targeting of BASP1 (Figure 9B). BASP1 targeting also protected from high-glucose-induced apoptosis but not from cytokine-induced death (Figure 9, B and C). Surviving cells looked healthy by contrast-phase microscopy (not shown).
Figure 9.
Inhibition of BASP1 expression by siRNA prevents apoptosis in human tubular epithelial cells. (A) BASP1 siRNA decreased the expression of BASP1 protein. (B) Cells transfected with BASP1 siRNA were protected from serum-deprivation and high glucose-induced apoptosis but not from cytokine-induced apoptosis. Cell death was assessed by flow cytometry of DNA content (hypodiploid cells) after culture in the absence of FBS (control, 5 mM glucose) and in the presence of high glucose (25 mM) or TWEAK/TNFα/IFNγ for 48 hours. Mean ± SD of three independent experiments. *P < 0.05 versus 0% FBS control, **P < 0.01 versus high glucose.
Discussion
Recent studies suggest a role for cell death in the progression of human DN.4,25 Gene ontology identified 112 cell-death-related genes that were significantly differentially regulated in the tubulointerstitium of renal biopsies from DN patients.24 To search for further apoptosis regulatory genes not previously identified as such, we took advantage of HT screening for genes for which overexpression induced cell death. BASP1 was increased in the tubulointerstitium of human DN samples, induced apoptosis upon overexpression, and was thereby identified as a novel death-related molecule. Animal experiments confirmed that BASP1 expression is increased in tubular cells in progressive nephropathies and cell culture demonstrated induction of apoptosis upon BASP1 overexpression and protection from apoptosis when BASP1 expression was suppressed.
BASP1 is widely expressed. Sites of expression include the developing and adult kidney15 and mammary gland.14 The localization of the protein is cell-type-specific. Although its nuclear expression in podocytes was recently stressed, the protein was also found in the cytoplasm of tubular and interstitial cells in vivo.15 BASP1 contains a nuclear localization sequence, and BASP1 is exclusively nuclear in some cell lines.26 BASP1 associates with WT1 in the nucleus of cells that naturally express both proteins (e.g., podocytes).15 Despite its wide expression, the functions of BASP1 have only been characterized in neurons and podocytes while remaining poorly understood in other cell types. Available data suggest that BASP1 is a multifunctional protein that may behave as a transcription factor and may have lipid raft and cytoskeletal organizational properties15,18,27; either of these properties could contribute to apoptosis. BASP1 regulates transcription by acting as a cosuppressor for WT1 in glomerular epithelial cells. The subcellular localization of BASP1 in the course of tubular epithelial cellapoptosis points to a relationship between BASP1 actions and actin and/or lipid raft organization. Lipid rafts and the actin cytoskeleton are key determinants of the cell susceptibility to apoptosis and there is an intimate relationship between them.28–30 Cytoskeletal remodeling is a hallmark of apoptosis. Concomitantly with rounding of the apoptotic cell, actin rearranges into a peripheral cortical ring.31,32 Appropriate remodeling of the actin cytoskeleton is required for cell survival, because disassembly and excessive crosslinking adversely affect survival.33,34 In tubular epithelia BASP1 overexpression induces apoptosis and endogenous BASP1 expression is increased by stimuli that promote apoptosis. Conversely, BASP1 downregulation protects from serum deprivation and high-glucose-induced apoptosis. Lack of protection from cytokine-induced cell death suggests that BASP1 may be critical for some, but not all, forms of apoptotic cell death and supports a role in DN.35 In addition, in apoptotic cells BASP1 redistributes to the cell cortex where it colocalizes with the apoptotic actin ring. BASP1 also colocalizes at this location with tubulin in a pattern similar to the tubulin cocoon of apoptotic cells.36 Taken together, these data point to a previously undescribed proapoptotic role for BASP1.
The precise localization of BASP1 in the chain of events that leads to apoptosis is not clear. Prevention of BASP1-induced apoptosis by Bax antagonists and caspase inhibitors suggests that it has a role upstream of mitochondrial injury and caspase activation or that, at least, participates in the positive feedback loop, leading to further mitochondrial injury and caspase activation.35 Recent reports have implicated changes in the dynamics of the actin cytoskeleton in mitochondrial injury and subsequent cell death.32,34 Thus, actin binding proteins such as gelsolin and cofilin regulate apoptosis at the mitochondrial injury and caspase activation levels.37–39
BASP1/cortical cytoskeleton-associated protein 23, growth-associated protein 43 (GAP43), and myristoylated alanine-rich C-kinase substrate (MARCKS) define a novel class of functionally related proteins (termed “GMC proteins”) that share the ED domain and certain properties (e.g., acylation-mediated targeting to the cell membrane and association with the cortical cytoskeleton).18 BASP1, GAP43 and MARCKS bind to and can regulate key actin binding proteins such as gelsolin and cofilin.40,41 Although a role for BASP1 in apoptosis is novel, it has been suggested for other GMC proteins, although the molecular pathways are not well understood. GAP43 and a novel xenopus MARCKS-related protein induce apoptosis.42–44 The ED domain is essential for the proapoptotic function of the Xenopus MARCKS-like protein.44 Phosphorylation of the ED in adducin precedes caspase activation, actin cytoskeleton changes, and apoptosis in tubular epithelia.45–47
In summary, we have identified BASP1 as a protein for which expression is increased in tubular cells in the course of tubulointerstitial injury in DN and in cultured cells stressed by a proapoptotic microenvironment. BASP1 may contribute to the apoptotic loss of tubular cells induced by an inadequate supply of survival factors or by high glucose concentrations in DN. BASP1 expression may be increased in other nephropathies, suggesting a potential role in kidney disease of different etiologies.
Concise Methods
Functional Genomics cDNA Screening for Cell-Death-Inducing Genes
The HT functional genomics approach has been previously detailed.9 In brief, a cDNA expression library generated from human embryo tissue (Scinet, Braunschweig, Germany) and a full-length cDNA expression plasmid collection have been propagated in E. Coli SURE. In a robot cascade, individual colonies were picked and automatically transferred for cDNA plasmid preparation in a 96-well format followed by transient transfection into HEK293 cells. The cells were grown in DMEM medium/5% FCS in 96-well plates and transfected by calcium phosphate coprecipitation. Cotransfection with CRE-lacZ reporter plasmid encoding for β-galactosidase enabled assessment of growth and viability 48 hours later by stimulation with forskolin and measuring of β-galactosidase activity (chlorophenolred-β-d-galactopyranoside, Roche). In total 596,832 independent clones from the human embryo cDNA library and all 17,680 defined full-length cDNA expression plasmids were screened. Positive clones inducing cell death by overexpression were verified after transient transfection by DNA-fragmentation ELISA (CDD+, Roche) and were bioinformatically annotated after sequencing. Flow cytometric determinations of active caspases in HeLa cells (CaspaTag assay with carboxyfluorescein and propidium iodide, Intergen, Oxford, United Kingdom), apoptotic membrane alterations (Annexin V, externalization of phosphatidylserine), and measurements of loss of mitochondrial potential with the fluorochrome DiOC6 were performed on selected genes.
Human Renal Biopsy Samples for Oligonucleotide-Array-Based Gene Expression Profiling
Human renal biopsies were collected in a multicenter study, the European Renal cDNA Bank (ERCB; see Acknowlegdments). The biopsies were performed according to the local ethical committees after informed consent was obtained from the patients and the samples processed according to the ERCB protocol.8 For oligonucleotide-array-based gene expression profiling, a total of 18 kidney biopsies from individual patients were included: 3 pretransplantation kidney biopsies from living donors [n = 3, age 39 ± 7 years, female:male ratio (F:M) 1:2, serum creatinine <1 mg/dl, proteinuria absent], 4 biopsies with histologic diagnosis of MCD (n = 4, age 25 ± 4.1 years, F:M 1:3, serum creatinine 1.0 ± 0.14 mg/dl, proteinuria 4.9 ± 2.3 g/24 h), and 11 biopsies with histologic diagnosis of DN (n = 11, age 58.3 ± 3.2 years, F:M 3:8, diabetes duration 9.8 ± 3.3 years, hemoglobin A1C levels 7.1% ± 0.3%, serum creatinine 1.98 ± 0.37 mg/dl, proteinuria 2.95 ± 0.8 g/24 h). Detailed clinical and histologic patient characteristics have been recently published.48
For validation quantitative, real-time RT-PCR analysis of biopsies from progressive DN samples (n = 7, age 57.83 ± 3.15 years, F:M 2:5, serum creatinine 2.48 ± 0.79 mg/dl, proteinuria 3.68 ± 0.97 g/24 h), pretransplantation kidney biopsies from living donors (n = 6, age 63.3 ± 8.44 years, F:M 1:5, serum creatinine 1.02 ± 0.09 mg/dl, proteinuria absent) using ABI assay on demand for BASP1 and 18S rRNA and the relative standard curve method for BASP1 quantification were performed as described.48
Cortical tissue segments were manually microdissected in glomerular and tubulointerstitial compartments.7 Total RNA was isolated from the microdissected tubulointerstitial tissue. HG-U133A Affymetrics microarray chips displayed 22,283 probe sets. Image files were initially obtained through Affymetrix GeneChip software (MAS5). Subsequently, robust multichip analysis was applied using RMAexpress (http://statwww.berkeley.edu/users/bolstad/RMAExpress) for the preprocessing of data, and significance analysis of microarrays (http://www-stat.stanford.edu/∼tibs/SAM/) was used to identify differentially expressed genes.7
Immunofluorescence in Human Samples
Stainings were performed by indirect immunofluorescence. Briefly, 5-μm thick acetone-fixed frozen sections from human control and diabetic kidneys were incubated with the primary antibody (rabbit polyclonal anti-BASP1, AbCAM, Cambridge, United Kingdom), followed by fluorescence-labeled secondary antibody (Alexa Fluor 546 goat anti-rabbit IgG (H+L) (Molecular Probes, Invitrogen, S. Giuliano Milanese, Italy). Double-staining sections were sequentially incubated with the primary antibodies (rabbit polyclonal anti-BASP1, Abcam; mouse anti-active caspase 3, eBioscience Ltd., Hatfield, United Kingdom), followed by proper fluorescence-labeled secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG, Alexa Fluor 546 goat anti-mouse IgG, Invitrogen). Specificity of antibody labeling was demonstrated by the lack of staining after substituting proper control immunoglobulins (rabbit primary antibody isotype control, Zymed, Invitrogen) for the primary antibody. Slides were mounted with Fluorsave aqueous mounting medium (Calbiochem, VWR International, Milan, Italy) and observed under a Zeiss Axioscope 40FL microscope equipped with an AxioCam MRc5 digital videocamera and immunofluorescence apparatus (Carl Zeiss SpA). A total of 16 biopsies were studied: DN samples (n = 8, serum creatinine 1.55 ± 0.33 mg/dl, proteinuria 1.82 ± 0.77 g/24 h), MCD samples (n = 5, serum creatinine 0.84 ± 0.11 mg/dl, proteinuria 5.72 ± 1.66 g/24 h), and control kidney biopsy samples (n = 3, serum creatinine 0.80 ± 0.10 mg/dl, proteinuria 0.02 ± 0.01 g/24 h).
Animal Model
Diabetes was induced by a single intraperitoneal injection of streptozotocin (Sigma, St. Louis, MO; 50 mg/kg in 0.01 M citrate buffer, pH 4.5)19 in ten 10-week-old Wistar Kyoto rats (Criffa, Barcelona, Spain). Ten control rats received the vehicle. Rats were sacrificed 7 months after induction of diabetes. Additionally, ten 10-week-old spontaneously hypertensive rats were followed for 7 months. Insulin (1 to 4 IU subcutaneously, Insulatard NPH, Novo Nordisk, Denmark) was administered weekly to prevent death from 7 days after administration of streptozotocin, having checked that all of the animals had blood glucose levels >400 mg/dl. Systolic blood pressure was measured monthly in conscious, restrained rats by a tail-cuff sphygmomanometer (NARCO, Biosystems, CO). The average of three separate measurements was calculated at each time point. Albuminuria was assessed by ELISA every 24 hours (Celltrend, Luckenwalde, Germany). Table 2 presents animal model data. All studies were performed in accordance with the European Union normative.
Table 2.
Animal modela
| Animal | Weight Gain (g) | SBP (mmHg) | Glycemia (mg/dl) | Albuminuria (μg/24 h) |
|---|---|---|---|---|
| WKY | 222 ± 15 | 138.1 ± 6.7 | 87 ± 3 | 390 ± 70 |
| WKY-D | 102 ± 32c | 136.2 ± 7.6 | 422 ± 4c | 1071 ± 247b |
| SHR | 160 ± 11b | 205.7 ± 2d | 90 ± 6 | 1800 ± 464c |
WKY, Wistar-Kyoto rats; WKY-D, diabetic WKY rats; SHR, spontaneously hypertensive rats; SBP, systolic blood pressure.
aData at latest follow-up (month 7), except for glycemia, which represents mean values throughout the study. Mean ± SD.
bP < 0.02.
cP < 0.002 versus WKY.
dP < 0.05 versus WKY.
Immunostaining of Rat Tissue Samples
Immunohistochemistry was carried out in 5-μm thick paraffin-embedded rat tissue sections.49 The primary antibody was a rabbit polyclonal anti-BASP1 (1:200). Sections were counterstained with Carazzi's hematoxylin. Negative controls included incubation with a nonspecific Ig of the same isotype as the primary antibody. Some sections were subsequently incubated with the proximal tubule marker, fluorescein-conjugated Tetranogolobus lotus (diluted 1:33) (Sigma) or the collecting tubule marker, fluorescein-conjugated Dolichos biflorum (diluted 1:100) (Sigma) lectins. These sections were mounted in 90% glycerol/PBS and photographed immediately.50
Immunofluorescence was used to co-stain with anti-BASP1 antibody (1:100) and anti-active caspase 3 (1:100, Promega). Secondary antibodies were anti-goat Alexa Fluor 488 and anti-rabbit Alexa Fluor 633 (1:300, Invitrogen).
Apoptotic cells were stained using the terminal deoxynucleotidyl-transferase-mediated dUTP nick-end labeling (In Situ Cell Death Detection Kit; Promega) as described previously.51
Cell Lines and Reagents
HK-2 human proximal tubular epithelial cells (ATCC, Rockville, MD) were grown on RPMI 1640 (Life Technologies, Grand Island, NY) with 10% heat-inactivated FBS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite, and 5 ng/ml hydrocortisone in 5% carbon dioxide at 37°C. For experiments cells were cultured in serum-free media 24 hours previous to the addition of the stimuli and throughout the experiment.
The pancaspase inhibitor zVAD (Bachem, Bubendorf, Switzerland), the caspase 3 inhibitor DEVD, and the caspase 9 inhibitor LEHD (Calbiochem, San Diego, CA) were dissolved in DMSO and used at 200 μM.23 Final concentration of DMSO in culture was 0.05% and it did not influence apoptosis. Bax inhibitor peptide P5 (Tocris, Ellisville, MO) was used at 200 μM.52,53
Transient Transfection
For transient transfection, cells were plated at a density of 8 × 104 cells/plate in six-well plates (Costar, Cambridge, MA) with RPMI 1640 (10% FBS) 24 hours before transfection. Cells were transfected with 5 μg of BASP1 containing plasmid (unless otherwise indicated) or empty vector and 1 μg of reporter vector (pCMVβ) or GFP (kindly donated by Miguel Angel del Pozo, CNIC, Madrid, Spain) using FuGENE 6 (Roche, Indianapolis, IN) according to the manufacturer's protocol. The efficacy of transfection was visualized by confocal microscopy or galactosidase staining. Real-time RT-PCR analyses of BASP1 mRNA in cells transfected with BASP1 confirmed a 300-fold BASP1 overexpression. BASP1 was inserted into modified pcDNA3 and sequenced to confirm its identity.
Cell Death and Apoptosis
Cells were cultured to subconfluence in 12-well plates and transfected with BASP1 plasmid (5 μg) for 24 hours. Apoptosis was assessed by functional and morphologic studies. Flow cytometry of DNA content was used to quantitate apoptosis. Adherent cells were pooled with spontaneously detached cells; stained in 100 μg/ml propidium iodide, 0.05% NP-40, and 10 μg/ml RNAse A in PBS; and incubated at 4°C for >1 hour. This assay permeabilizes the cells. Permeabilization allows entry of propidium iodide in all cells, dead and alive. A lower DNA content (hypodiploid cells) due to nuclear fragmentation characterizes apoptotic cells; thus, this assay is not based on the known ability of propidium iodide to enter dead cells. The percentage of apoptotic cells with decreased DNA content (A0) was counted.54 Several proapoptotic stimuli were studied, such as deprivation of growth factors from serum, exposure to high (450 mg/dl) glucose concentration, or exposure to a lethal cytokine cocktail (30 ng/ml TNFα, 100 ng/ml TWEAK, and 30 U/ml IFNγ) for 24 hours. In addition, flow cytometry was used to quantify the appearance of a specific epitope generated by caspase cleavage of cytokeratin 18 and identified by the M30 cytodeath antibody (Roche Biochemicals, Mannheim, Germany).
To assess the typical nuclear changes seen in apoptosis, cells were cultured in chamber slides (Labtek, Nunc, Naperville, IL), transfected with GFP and BASP1 plasmid, fixed with methanol:acetone (1:1), and stained with 4′,6-diamidino-2-phenylindole (Sigma) or FITC-M30 cytodeath stain (1:250 Roche) and BASP1. Thus, the typical condensed, shrunk, and fragmented nuclei of apoptotic cells were identified by a laser scanning confocal microscope (Leika).
Caspase 3 activity was assessed in cells transfected with BASP1 plasmid (5 μg) or vehicle for 24 hours. Caspase 3 activity (MBL, Nagoya, Japan) was measured following the manufacturer's instructions.23 In brief, cell extracts (100 μg of protein) were incubated in half-area 96-well plates at 37°C with 200 μM DEVD-pNA peptide in a total volume of 100 μl. The pNA light emission was quantified using a spectrophotometer plate reader at 405 nm. Comparison of the absorbance of pNA from an apoptotic sample with an uninduced control allows determination of the fold increase in caspase activity.
Survival Assays
To provide an objective assessment of cell loss, the β-Galactosidase Enzyme Assay System (Promega) was used to assay β-galactosidase activity in lysates prepared from cells transfected with β-galactosidase reporter vector and BASP1 vector or empty vector. Cell lysates were prepared 24 hours after transfection and the absorbance was read at 420 nm with a Microplate Absorbance Reader Anthos 2020 (Anthos Labtec Instruments, Wals, Austria). Absorbance for the control vector (empty vector) was considered to be 100% and percent survival refers only to the transfected population.55
To observe the morphology of β-galactosidase-transfected cells, 24 hours after transfection cells were fixed with 2% paraformaldehyde/0.2% glutaraldehyde in PBS (5 minutes, 4°C), washed with PBS twice, and stained (1 mg/ml X-gal, 5 mM potassium ferricyanide, 2 mM MgCl2, 0.02% NP-40, and 0.01% SDS) at room temperature until cells turn blue.
Western Blot
Western blots were performed as described previously.23 Membranes were incubated overnight at 4°C with anti-BASP1 antibody (1:2000, AbCAM) or mouse anti-tubulin monoclonal antibody (1:5000, Sigma, St. Louis, MO) followed by incubation with horseradish-peroxidase-conjugated secondary antibody (1:2000, Amersham, Aylesbury, United Kingdom). Blots were developed with the enhanced chemiluminescence method following the manufacturer's instructions (Amersham).
Confocal Microscopy
Cells plated onto Labtek slides were fixed in 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 in PBS for 10 minutes each. After washing in PBS cells were incubated overnight at 4°C with rabbit polyclonal anti-BASP1 antibody (1:100) (AbCAM), followed by 1-hour incubation with FITC secondary antibody (1:200, Sigma). Cell nuclei were counterstained with propidium iodide. After washing, cells were mounted in 70% glycerol in PBS and analyzed with a DM-IRB confocal microscope (Leica DM, Bannockburn, IL).49 F-actin was stained with 800 μM TRITC-phalloidin (Sigma) in the dark for 30 minutes at room temperature and anti-α-tubulin antibodies (1:200, Sigma) followed by a 1-hour incubation with TRITC secondary antibody (1:200, Sigma). Apoptosis was characterized by morphologic and functional criteria. Nuclei of formalin-fixed cells were stained with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Inc., Burlingame, CA) to observe the typical morphologic changes.
Transfection of siRNA
Cells were grown in six-well plates (Costar, Cambridge, MA) and transfected with a mixture composed of 10 nmol/ml siRNA (Ambion, Applied Biosystems, Foster City, CA), Opti-MEM I Reduced Serum Medium, and siPORT Amine Transfection Agent (Ambion, Applied Biosystems). After 18 hours cells were washed and cultured for 48 hours in complete medium and serum-depleted for 24 to 72 hours. These time points were selected from a time course of BASP1 protein expression in response to siRNA. A negative control scrambled siRNA provided by the manufacturer did not reduce BASP1 protein.
Statistical Analysis
Results are expressed as mean ± SD of at least three independent experiments. Significance at the P < 0.05 level was assessed by nonparametric Mann–Whitney test for two groups of data and Kruskal–Wallis or one-way analysis (ANOVA) with a Bonferroni post hoc correction for analysis of differences between three or more groups by SPSS software (SPSS Inc., Chicago, IL). Correlation of BASP1 tubular expression to proteinuria in DN (n = 11) was estimated using Pearson correlation method by GraphPad Prism version 3.00 (GraphPad Software, San Diego CA).
Disclosures
None.
Acknowledgments
We thank all members of the ERCB and their patients for their support and cooperation. C.L. was funded by Instituto de Salud Carlos III (ISCIII), A.B.S. by ISCIII, and M.D.S.N. by Ministerio de Ciencia y Tecnología-MCYT. This study was supported by the following grants: EU Framework V Program “Progressive Renal Disease” QLRT-2001-01215, Comunidad de Madrid (08.2/0030.1/2003, S-BIO-0283–2006) EU project DIALOK: LSHB-CT-2007-036644, MCYT SAF 2003/884, FIS 06/0046 y PI081564, FIS PS09/00447 ISCIII-RETICS: RedinREN 06/0016, and Programa de Intensificación de la Actividad Investigadora in the Sistema Nacional de Salud of the ISCIII and the Agencia “Pedro Laín Entralgo.” We are indebted to Dr. Almut Nitsche and Dr. Bodo Brunner (Sanofi-Aventis Deutschland GmbH) for DNA chip hybridizations and delivery of the data set of hybridization results. Mar Gonzalez García-Parreño helped with confocal microscopy and Susana Carrasco with histologic studies.
Members of ERCB-Kroener Fresenius Bank at the time of this study: J.P. Rougier, P. Ronco, Paris; M. P. Rastaldi, G. D'Amico, Milano; F. Mampaso, Madrid; P. Doran, H.R. Brady, Dublin; D. Mönks, C. Wanner, Würzburg; A.J. Rees, Aberdeen; F. Strutz, G. Müller, Göttingen; P. Mertens, J. Floege, Aachen; T. Risler, Tübingen; L. Gesualdo, F.P. Schena, Bari; J. Gerth, U. Ott, G. Wolf, Jena; R. Oberbauer, D. Kerjaschki, Vienna; B. Banas, B. Krämer, Regensburg; W. Samtleben, Munich; H. Peters, H.H. Neumayer, Berlin; K. Ivens, B. Grabensee, Düsseldorf; M. Zeier, H.J. Gröne, Heidelberg; M. Merta, V. Tesar, Prague; and C.D. Cohen, M. Kretzler, D. Schlöndorff, Munich.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1. Engelgau MM, Geiss LS, Saaddine JB, Boyle JP, Benjamin SM, Gregg EW, Tierney EF, Rios-Burrows N, Mokdad AH, Ford ES, Imperatore G, Narayan KM: The evolving diabetes burden in the United States. Ann Intern Med 140: 945– 950, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Adeghate E: Molecular and cellular basis of the aetiology and management of diabetic cardiomyopathy: A short review. Mol Cell Biochem 261: 187– 191, 2004 [DOI] [PubMed] [Google Scholar]
- 3. Kowluru RA: Diabetic retinopathy: Mitochondrial dysfunction and retinal capillary cell death. Antioxid Redox Signal 7: 1581– 1587, 2005 [DOI] [PubMed] [Google Scholar]
- 4. Kumar D, Robertson S, Burns KD: Evidence of apoptosis in human diabetic kidney. Mol Cell Biochem 259: 67– 70, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Susztak K, Ciccone E, McCue P, Sharma K, Bottinger EP: Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med 2: e45, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lorz C, Benito-Martin A, Justo P, Sanz AB, Sanchez-Nino MD, Santamaria B, Egido J, Ortiz A: Modulation of renal tubular cell survival: Where is the evidence? Curr Med Chem 13: 449– 454, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Grone HJ, Nelson PJ, Schlondorff D, Cohen CD, Kretzler M: Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993– 3003, 2006 [DOI] [PubMed] [Google Scholar]
- 8. Cohen CD, Frach K, Schlondorff D, Kretzler M: Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int 61: 133– 140, 2002 [DOI] [PubMed] [Google Scholar]
- 9. Koenig-Hoffmann K, Bonin-Debs AL, Boche I, Gawin B, Gnirke A, Hergersberg C, Madeo F, Kazinski M, Klein M, Korherr C, Link D, Rohrig S, Schafer R, Brinkmann U: High throughput functional genomics: Identification of novel genes with tumor suppressor phenotypes. Int J Cancer 113: 434– 439, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Maekawa S, Maekawa M, Hattori S, Nakamura S: Purification and molecular cloning of a novel acidic calmodulin binding protein from rat brain. J Biol Chem 268: 13703– 13709, 1993 [PubMed] [Google Scholar]
- 11. Iino S, Taguchi K, Maekawa S, Nojyo Y: Motor, sensory and autonomic nerve terminals containing NAP-22 immunoreactivity in the rat muscle. Brain Res 1002: 142– 150, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Mosevitsky MI, Capony JP, Skladchikova GY, Novitskaya VA, Plekhanov AY, Zakharov VV: The BASP1 family of myristoylated proteins abundant in axonal termini. Primary structure analysis and physico-chemical properties. Biochimie 79: 373– 384, 1997 [DOI] [PubMed] [Google Scholar]
- 13. Sprenger RR, Fontijn RD, van MJ, Pannekoek H, Horrevoets AJ: Spatial segregation of transport and signaling functions between human endothelial caveolae and lipid raft proteomes. Biochem J 400: 401– 410, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morris JS, Stein T, Pringle MA, Davies CR, Weber-Hall S, Ferrier RK, Bell AK, Heath VJ, Gusterson BA: Involvement of axonal guidance proteins and their signaling partners in the developing mouse mammary gland. J Cell Physiol 206: 16– 24, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Carpenter B, Hill KJ, Charalambous M, Wagner KJ, Lahiri D, James DI, Andersen JS, Schumacher V, Royer-Pokora B, Mann M, Ward A, Roberts SG: BASP1 is a transcriptional cosuppressor for the Wilms' tumor suppressor protein WT1. Mol Cell Biol 24: 537– 549, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lund TC, Anderson LB, McCullar V, Higgins L, Yun GH, Grzywacz B, Verneris MR, Miller JS: iTRAQ is a useful method to screen for membrane-bound proteins differentially expressed in human natural killer cell types. J Proteome Res 6: 644– 653, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Shaw JE, Epand RF, Sinnathamby K, Li Z, Bittman R, Epand RM, Yip CM: Tracking peptide-membrane interactions: Insights from in situ coupled confocal-atomic force microscopy imaging of NAP-22 peptide insertion and assembly. J Struct Biol 155: 458– 469, 2006 [DOI] [PubMed] [Google Scholar]
- 18. Wiederkehr A, Staple J, Caroni P: The motility-associated proteins GAP-43, MARCKS, and CAP-23 share unique targeting and surface activity-inducing properties. Exp Cell Res 236: 103– 116, 1997 [DOI] [PubMed] [Google Scholar]
- 19. Ortiz A, Ziyadeh FN, Neilson EG: Expression of apoptosis-regulatory genes in renal proximal tubular epithelial cells exposed to high ambient glucose and in diabetic kidneys. J Investig Med 45: 50– 56, 1997 [PubMed] [Google Scholar]
- 20. de Haan JB, Stefanovic N, Nikolic-Paterson D, Scurr LL, Croft KD, Mori TA, Hertzog P, Kola I, Atkins RC, Tesch GH: Kidney expression of glutathione peroxidase-1 is not protective against streptozotocin-induced diabetic nephropathy. Am J Physiol Renal Physiol 289: F544– F551, 2005 [DOI] [PubMed] [Google Scholar]
- 21. Kelly DJ, Cox AJ, Tolcos M, Cooper ME, Wilkinson-Berka JL, Gilbert RE: Attenuation of tubular apoptosis by blockade of the renin-angiotensin system in diabetic Ren-2 rats. Kidney Int 61: 31– 39, 2002 [DOI] [PubMed] [Google Scholar]
- 22. Ortiz A, Lorz C, Catalan MP, Danoff TM, Yamasaki Y, Egido J, Neilson EG: Expression of apoptosis regulatory proteins in tubular epithelium stressed in culture or following acute renal failure. Kidney Int 57: 969– 981, 2000 [DOI] [PubMed] [Google Scholar]
- 23. Justo P, Sanz AB, Sanchez-Nino MD, Winkles JA, Lorz C, Egido J, Ortiz A: Cytokine cooperation in renal tubular cell injury: The role of TWEAK. Kidney Int 70: 1750– 1758, 2006 [DOI] [PubMed] [Google Scholar]
- 24. Lorz C, Benito-Martin A, Boucherot A, Ucero AC, Rastaldi MP, Henger A, Armelloni S, Santamaria B, Berthier CC, Kretzler M, Egido J, Ortiz A: The death ligand TRAIL in diabetic nephropathy. J Am Soc Nephrol 19: 904– 914, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar D, Zimpelmann J, Robertson S, Burns KD: Tubular and interstitial cell apoptosis in the streptozotocin-diabetic rat kidney. Nephron Exp Nephrol 96: e77– e88, 2004 [DOI] [PubMed] [Google Scholar]
- 26. Wagner KJ, Roberts SG: Transcriptional regulation by the Wilms' tumour suppressor protein WT1. Biochem Soc Trans 32: 932– 935, 2004 [DOI] [PubMed] [Google Scholar]
- 27. Park S, Kim YI, Kim B, Seong C, Oh Y, Baek K, Yoon J: Characterization of bovine and human cDNAs encoding NAP-22 (22 kDa neuronal tissue-enriched acidic protein) homologs. Mol Cells 8: 471– 477, 1998 [PubMed] [Google Scholar]
- 28. Malorni W, Giammarioli AM, Garofalo T, Sorice M: Dynamics of lipid raft components during lymphocyte apoptosis: The paradigmatic role of GD3. Apoptosis 12: 941– 949, 2007 [DOI] [PubMed] [Google Scholar]
- 29. Yang B, Oo TN, Rizzo V: Lipid rafts mediate H2O2 prosurvival effects in cultured endothelial cells. FASEB J 20: 1501– 1503, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Gajate C, Mollinedo F: Cytoskeleton-mediated death receptor and ligand concentration in lipid rafts forms apoptosis-promoting clusters in cancer chemotherapy. J Biol Chem 280: 11641– 11647, 2005 [DOI] [PubMed] [Google Scholar]
- 31. Mills JC, Stone NL, Pittman RN: Extranuclear apoptosis. The role of the cytoplasm in the execution phase. J Cell Biol 146: 703– 708, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boldogh IR, Pon LA: Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta 1763: 450– 462, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Bijian K, Takano T, Papillon J, Le BL, Michaud JL, Kennedy CR, Cybulsky AV: Actin cytoskeleton regulates extracellular matrix-dependent survival signals in glomerular epithelial cells. Am J Physiol Renal Physiol 289: F1313– F1323, 2005 [DOI] [PubMed] [Google Scholar]
- 34. Gourlay CW, Ayscough KR: The actin cytoskeleton: A key regulator of apoptosis and ageing? Nat Rev Mol Cell Biol 6: 583– 589, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Sanz AB, Santamaria B, Ruiz-Ortega M, Egido J, Ortiz A: Mechanisms of renal apoptosis in health and disease. J Am Soc Nephrol 19: 1634– 1642, 2008 [DOI] [PubMed] [Google Scholar]
- 36. Sanchez-Alcazar JA, Rodriguez-Hernandez A, Cordero MD, Fernandez-Ayala DJ, Brea-Calvo G, Garcia K, Navas P: The apoptotic microtubule network preserves plasma membrane integrity during the execution phase of apoptosis. Apoptosis 12: 1195– 1208, 2007 [DOI] [PubMed] [Google Scholar]
- 37. Koya RC, Fujita H, Shimizu S, Ohtsu M, Takimoto M, Tsujimoto Y, Kuzumaki N: Gelsolin inhibits apoptosis by blocking mitochondrial membrane potential loss and cytochrome C release. J Biol Chem 275: 15343– 15349, 2000 [DOI] [PubMed] [Google Scholar]
- 38. Harms C, Bosel J, Lautenschlager M, Harms U, Braun JS, Hortnagl H, Dirnagl U, Kwiatkowski DJ, Fink K, Endres M: Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol Cell Neurosci 25: 69– 82, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Chua BT, Volbracht C, Tan KO, Li R, Yu VC, Li P: Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol 5: 1083– 1089, 2003 [DOI] [PubMed] [Google Scholar]
- 40. Frey D, Laux T, Xu L, Schneider C, Caroni P: Shared and unique roles of CAP23 and GAP43 in actin regulation, neurite outgrowth, and anatomical plasticity. J Cell Biol 149: 1443– 1454, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laux T, Fukami K, Thelen M, Golub T, Frey D, Caroni P: GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol 149: 1455– 1472, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gagliardini V, Dusart I, Fankhauser C: Absence of GAP-43 can protect neurons from death. Mol Cell Neurosci 16: 27– 33, 2000 [DOI] [PubMed] [Google Scholar]
- 43. Stocker KM, Ciment G, Baizer L: GAP-43 in non-neuronal cells of the embryonic chick limb: Clues to function. Perspect Dev Neurobiol 1: 53– 62, 1992 [PubMed] [Google Scholar]
- 44. Zhao H, Cao Y, Grunz H: Isolation and characterization of a Xenopus gene (XMLP) encoding a MARCKS-like protein. Int J Dev Biol 45: 817– 826, 2001 [PubMed] [Google Scholar]
- 45. Matsuoka Y, Li X, Bennett V: Adducin: structure, function and regulation. Cell Mol Life Sci 57: 884– 895, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imamdi R, de GM, van de WB: Protein kinase C mediates cisplatin-induced loss of adherens junctions followed by apoptosis of renal proximal tubular epithelial cells. J Pharmacol Exp Ther 311: 892– 903, 2004 [DOI] [PubMed] [Google Scholar]
- 47. van de WB, Tijdens IB, Verbrugge A, Huigsloot M, Dihal AA, Stevens JL, Jaken S, Mulder GJ: Cleavage of the actin-capping protein alpha -adducin at Asp-Asp-Ser-Asp633-Ala by caspase-3 is preceded by its phosphorylation on serine 726 in cisplatin-induced apoptosis of renal epithelial cells. J Biol Chem 275: 25805– 25813, 2000 [DOI] [PubMed] [Google Scholar]
- 48. Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, Boucherot A, Neusser MA, Cohen CD, Carter-Su C, Argetsinger LS, Rastaldi MP, Brosius FC, Kretzler M: Enhanced expression of Janus kinase-signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes 58: 469– 477, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanz AB, Justo P, Sanchez-Nino MD, Blanco-Colio LM, Winkles JA, Kreztler M, Jakubowski A, Blanco J, Egido J, Ruiz-Ortega M, Ortiz A: The cytokine TWEAK modulates renal tubulointerstitial inflammation. J Am Soc Nephrol 19: 695– 703, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lorz C, Ortiz A, Justo P, Gonzalez-Cuadrado S, Duque N, Gomez-Guerrero C, Egido J: Proapoptotic Fas ligand is expressed by normal kidney tubular epithelium and injured glomeruli. J Am Soc Nephrol 11: 1266– 1277, 2000 [DOI] [PubMed] [Google Scholar]
- 51. Sanz AB, Sanchez-Nino MD, Izquierdo MC, Jakubowski A, Justo P, Blanco-Colio LM, Ruiz-Ortega M, Egido J, Ortiz A: TWEAK induces proliferation in renal tubular epithelium: A role in uninephrectomy-induced renal hyperplasia. J Cell Mol Med 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Justo P, Sanz AB, Egido J, Ortiz A: 3,4-Dideoxyglucosone-3-ene induces apoptosis in renal tubular epithelial cells. Diabetes 54: 2424– 2429, 2005 [DOI] [PubMed] [Google Scholar]
- 53. Sawada M, Hayes P, Matsuyama S: Cytoprotective membrane-permeable peptides designed from the Bax-binding domain of Ku70. Nat Cell Biol 5: 352– 357, 2003 [DOI] [PubMed] [Google Scholar]
- 54. Catalan MP, Reyero A, Egido J, Ortiz A: Acceleration of neutrophil apoptosis by glucose-containing peritoneal dialysis solutions: Role of caspases. J Am Soc Nephrol 12: 2442– 2449, 2001 [DOI] [PubMed] [Google Scholar]
- 55. Justo P, Sanz AB, Lorz C, Egido J, Ortiz A: Lethal activity of FADD death domain in renal tubular epithelial cells. Kidney Int 69: 2205– 2211, 2006 [DOI] [PubMed] [Google Scholar]