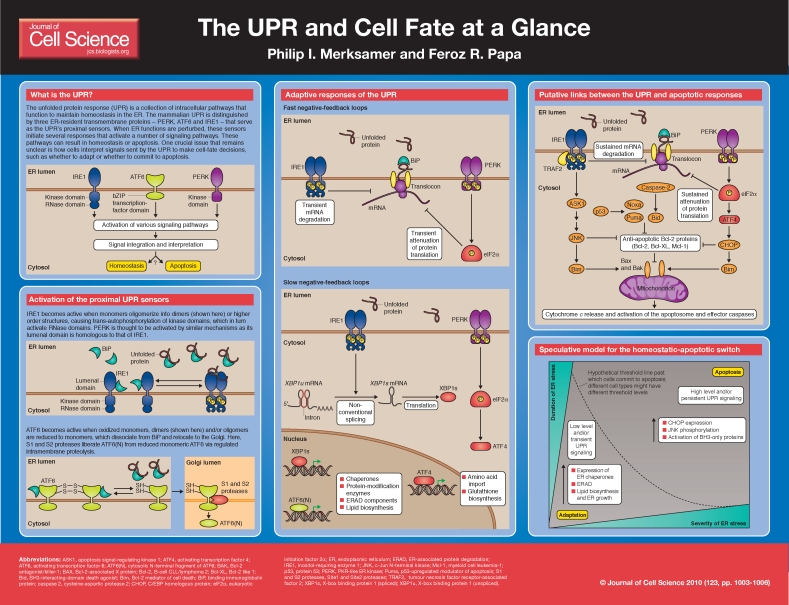
In eukaryotic cells, secreted and resident proteins of the endomembrane system fold into their native structures within the endoplasmic reticulum (ER). The ER is a network of membranous tubules and sheets whose lumenal environment is crowded with molecular chaperones and protein-modification enzymes that are specialized to fold proteins. In addition, the ER contains stringent quality-control systems that selectively export correctly folded proteins and selectively extract terminally misfolded proteins for ubiquitin-dependent proteolytic degradation in the cytosol, a process known as ER-associated protein degradation (ERAD) (Vembar and Brodsky, 2008). The ER is a dynamic organelle, and its capacity to fold proteins can be adjusted in response to changes in cellular protein-folding requirements through several intracellular signaling pathways that are collectively known as the unfolded protein response (UPR) (Ron and Walter, 2007). Dysregulation of the UPR contributes to the pathology of several important human diseases, including diabetes, neurodegeneration and cancer (Kim et al., 2008). In this article and its accompanying poster, we summarize how the mammalian UPR influences cell fate by promoting either cell adaptation or apoptosis when protein folding homeostasis is perturbed.
Activation of the proximal UPR sensors
Mammalian UPR signaling is initiated by three ER-resident transmembrane proteins: protein kinase RNA (PKR)-like ER kinase, (PERK), activating transcription factor-6 (ATF6) and inositol-requiring enzyme-1 (IRE1). The presence of unfolded ER proteins is thought to activate each of these three proximal detectors; however, the ‘sensing’ mechanism remains unclear (Kohno, 2007). IRE1 was the first characterized sensor and its mechanism of activation has been the most thoroughly studied. IRE1 is a type-I transmembrane protein containing three domains: an N-terminal lumenal domain, a cytosolic kinase domain and a cytosolic RNase domain (Tirasophon et al., 1998; Wang et al., 1998). IRE1 becomes active when monomers oligomerize into either dimers or higher order structures, causing trans-autophosphorylation of the kinase domains, which in turn activate the RNase domains. Two
models have been proposed to explain IRE1 oligomerization. In the first model, the ER-resident chaperone immunoglobulin-binding protein (BiP) functions as a master regulator by binding to IRE1 and inhibiting its oligomerization under basal conditions. In this scheme, when unfolded proteins accumulate, BiP dissociates from IRE1 to preferentially interact with them, thus allowing IRE1 to oligomerize (Bertolotti et al., 2000). The second model proposes that unfolded proteins bind directly to the lumenal domain of IRE1, which induces its oligomerization (Credle et al., 2005). More work is needed to resolve the relative contributions of BiP dissociation and direct binding of unfolded proteins to IRE1 activation.
PERK is also a type-I transmembrane protein that has a cytosolic kinase domain and an N-terminal lumenal domain that is homologous to that of IRE1. As a consequence, it has been postulated that PERK is activated by similar mechanisms to those involved in IRE1 activation (Bertolotti et al., 2000; Liu et al., 2000).
ATF6 is an ER-resident type-II transmembrane protein that exists as an oxidized monomer, dimer, and/or oligomer and that associates with BiP under basal conditions. When unfolded proteins accumulate, ATF6 dissociates from BiP and conserved intra- and/or intermolecular disulfide bonds in the lumenal domain of ATF6 are reduced, creating ATF6 monomers (Nadanaka et al., 2006; Shen et al., 2005). Reduced monomeric ATF6 translocates to the Golgi and becomes a substrate for the Site-1 and Site-2 proteases, which liberate the N-terminal cytosolic fragrament of ATF6 [ATF6(N), a basic leucine zipper (bZiP) transcription factor] via regulated intramembrane proteolysis (Haze et al., 1999). More work is needed to elucidate the mechanisms governing ATF6 disulfide bond reduction, BiP dissociation and regulation of ATF6 translocation to the Golgi.
Adaptive responses of the UPR
When the proximal UPR sensors become activated, they initiate a response to restore protein-folding homeostasis in the ER. This adaptive response involves several outputs and can be conceptualized as two negative-feedback loops acting on two different time scales: a fast negative-feedback loop that decreases the influx of proteins into the ER, and a slow negative-feedback loop that requires de novo mRNA and protein synthesis to increase the folding capacity of the ER (Trusina et al., 2008).
The kinase activity of activated PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α), which impedes subsequent rounds of translation initiation (Harding et al., 1999). In addition, IRE1 is responsible for the rapid degradation of several ER-localized mRNAs (Hollien and Weissman, 2006). Transient translation attenuation and mRNA decay constitute the fast negative-feedback loops because they rapidly reduce the protein load on the ER. This provides the ER folding machinery an extended opportunity to fold existing unfolded proteins and the ERAD machinery an extended period of time to degrade them.
IRE1 also catalyzes the non-conventional splicing of XBP1u mRNA into XBP1s mRNA, which encodes the bZiP transcription factor X-box-binding protein 1 (XBP1s) (Calfon et al., 2002; Yoshida et al., 2001). The slower phase of the adaptive response is controlled by XBP1s together with ATF6(N). ATF6(N) and XBP1s increase transcription rates of genes encoding ER-resident chaperones, protein-modification enzymes, ERAD components and lipid biosynthetic enzymes to augment the size and folding and degradation activities of the ER (Yamamoto et al., 2007). Translation of the activating transcription factor-4 (ATF4) also increases when eIF2α is phosphorylated by PERK, causing increased transcription of many genes that promote survival under many types of cellular stress (Harding et al., 2003). Additional transcription factors might also contribute to the transcriptional UPR program in certain cell types; for example, CREBH (cyclic AMP response element-binding protein H) appears to be involved in hepatocytes (Zhang et al., 2006). Together, these negative-feedback loops reduce the concentration of unfolded proteins in the ER to maintain cellular homeostasis in the face of changing metabolic and protein-folding requirements. As the concentration of unfolded proteins decreases, the UPR shuts off, although the molecular details of UPR attenuation remain unclear.
ER stress
The term ‘ER stress’ is often used to describe a condition in which ER homeostasis is lost because of an overload on the ER's protein-folding capacity. In practice, however, ER stress is often used operationally to describe any condition in which cells have activated the UPR. This operational definition has evolved because it is difficult to directly measure the ER unfolded proteins that are thought to be the activating signals of the UPR. However, solely monitoring UPR signaling does not necessarily provide information about the functional state of protein folding in the ER. Therefore, it is important to consider additional physiological end-points, such as ER distention, or changes in the secretion, glycosylation or oxidation of ER proteins (Merksamer et al., 2008). In experimental settings, ER stress is generally induced by treating cells with toxic chemicals that severely comprise ER protein folding or trafficking. Under these non-physiological conditions, the adaptive mechanisms of the UPR are insufficient to maintain homeostasis in the ER and cells ultimately die, typically through apoptosis.
Putative links between the UPR and apoptotic responses
Cells experiencing irremediable ER stress commit to apoptosis when the outer mitochondrial membrane (OMM) is permeabilized and cytochrome c is released to activate executioner caspases. This intrinsic (mitochondrial) apoptotic pathway, which is typically triggered in response to intracellular stresses including DNA damage and viral infections, is regulated by the Bcl-2 protein family (Youle and Strasser, 2008). The Bcl-2 family can be divided into three groups: multi-domain proapoptotic proteins (e.g. Bax, Bak), anti-apoptotic proteins (e.g. Bcl-2, Bcl-XL) and proapoptotic BH3-only proteins (e.g. Bid, Bad, Bim, Noxa, Puma) (Brunelle and Letai, 2009). In response to ER stress, the proapoptotic BH3-only proteins are transcriptionally or post-translationally activated to stimulate proapoptotic Bax and Bak either directly or indirectly through antagonizing anti-apoptotic members. Once activated, Bax and/or Bak form homo-oligomers in the OMM to initiate mitochondrial permeabilization (Wei et al., 2001). At this time, it is unclear whether and how UPR signaling components communicate with the Bcl-2 family members or other apoptotic signaling molecules to initiate apoptosis. In the following section, we summarize some of the more compelling data supporting such a link.
Of the 11 members of the BH3-only family, Puma, Noxa, Bid and Bim have been described to mediate apoptosis triggered by ER stress (Li et al., 2006; Puthalakath et al., 2007; Upton et al., 2008). However, it remains possible that other BH3-only proteins serve important roles, with the relative contribution(s) of each BH3-only member varying in different tissues. Recently, the transcription factor C/EBP homologous protein (CHOP) was found to increase the rate of Bim transcription during ER stress, marking an important connection between a UPR signaling component and a BH3-only protein (Puthalakath et al., 2007). CHOP mRNA levels increase sharply during ER stress, an effect that is mediated primarily through the upstream transcription factor ATF4. In addition to regulating Bim expression, CHOP has been reported to antagonize the expression of anti-apoptotic Bcl-2. Although CHOP is clearly an important mediator between the UPR and the apoptotic machinery, CHOP−/− cells are only partially resistant to ER-stress-induced apoptosis. In addition, PERK−/− cells readily undergo apoptosis despite minimal CHOP expression (Oyadomari and Mori, 2004). Therefore, parallel signaling pathways might compensate for the loss of CHOP and possibly other proapoptotic components upstream of Bax and Bak.
Another signaling pathway that operates in parallel with CHOP is mediated by the mitogen-activated protein kinase (MAPK) c-Jun NH2-terminal kinase (JNK). JNK is activated by cytokines and several cellular stresses, and JNK signaling can promote protective or apoptotic responses, depending on cellular context (Weston and Davis, 2007). JNK signaling increases during ER stress in a manner that depends on IRE1 and the MAPK kinase kinase (MAP3K) apoptosis signal-regulating kinase 1 (ASK1). Activated IRE1 associates with tumor necrosis factor receptor-associated factor 2 (TRAF2), leading to the activation of ASK1, which in turn initiates a phosphorylation cascade resulting in JNK phosphorylation and activation (Nishitoh et al., 2002; Urano et al., 2000). How TRAF2 is recruited to IRE1 and how this complex activates ASK1 remains unclear. JNK is thought to promote apoptosis under these conditions through several interactions with Bcl-2 family members: there is evidence that JNK can phosphorylate and inhibit the anti-apoptotic proteins Bcl-2, Bcl-XL, and Mcl-1. Furthermore, JNK can also phosphorylate and activate several BH-3 only proteins, including Bid and Bim, to promote apoptosis (Weston and Davis, 2007).
There are several additional parallel pathways that might contribute to ER-stress-induced apoptosis that will not be reviewed in depth here. They include ER Ca2+ release regulated by ER-resident Bcl-2 family members (Kim et al., 2008), as well as interactions between Bcl-2 family members and IRE1 (Hetz and Glimcher, 2009). The vast numbers of factors that can transmit signals from the ER to mitochondria suggests that tight regulation of these signals is crucial for ensuring that only irremediably ER-stressed cells undergo apoptosis.
Speculative model for the UPR-mediated homeostatic-apoptotic switch
The UPR simultaneously transmits survival and apoptotic signals. Understanding the interplay between these competing signals is necessary to elucidate the mechanism by which cells decide whether to continue to attempt adaptation or to initiate apoptosis. This decision could ultimately depend on how Bcl-2 proteins interpret the mix of survival and apoptotic signals transmitted by the UPR: such interpretation results in cell survival under conditions of remedial stress and cell death when homeostasis cannot be restored following catastrophic ER protein misfolding. To make this decision, cells might incorporate a time factor in which sustained UPR signaling (as could occur during chronic ER stress) increases the likelihood of apoptosis. In support of such a model, the mRNA and protein half-lives of proapoptoic CHOP were found to be short lived compared with pro-survival UPR outputs such as the ER chaperone BiP (Rutkowski et al., 2006). Sustained PERK activity (which is primarily responsible for CHOP upregulation) might thus be necessary to build CHOP levels to a required threshold to stimulate Bcl-2 proteins to commit to apoptosis. In addition, sustained PERK activity should result in protracted translation attenuation, which should be incompatible with survival. Similarly, sustained mRNA degradation mediated by IRE1 might deplete ER cargo and protein folding activities (Han et al., 2009). In support of this notion, overexpression of PERK or IRE1, which leads to their spontaneous oligomerization and activation, is typically sufficient to cause apoptosis. This is reminiscent of apoptosis that occurs during the sustained activation of other protein kinases such as JNK (Ventura et al., 2006).
In addition, the severity of ER stress might alter the relative activation levels of certain UPR output pathways to influence cell-fate decisions. For example, IRE1 has at least three established outputs: XBP1 mRNA splicing, non-specific mRNA cleavage and JNK activation (Han et al., 2009; Hollien et al., 2009; Hollien and Weissman, 2006; Urano et al., 2000). It is possible that different degrees of protein misfolding differentially affect which of these IRE1 outputs are realized by differentially altering its oligomerization state.
Perspectives
Over the past 20 years since the UPR was first described, many of its molecular components have been identified and characterized. To move forward, it will be necessary to investigate how these individual components function as a signaling network to direct cell-fate decisions. To this end, we will need to develop quantitative tools to study various UPR components dynamically in individual living cells as they experience ER stress. In addition, it will be important to challenge cells with physiologically relevant stressors to understand how the UPR contributes to cellular physiology and pathogenesis of protein-misfolding diseases. It is likely that the elucidation of key components of the UPR's homeostatic-apoptotic signaling network will reveal pharmacological targets for drug discovery and potential therapeutics for ER-stress-related diseases.
Acknowledgments
We apologize to those authors whose work was not directly cited due to space limitations. We thank Scott Oakes and Dan Han for critically reading the manuscript. This work was funded through the National Institutes of Health: Director's New Innovator Award DP2 OD001925 (F.R.P), RO1 DK080955 (F.R.P); the Burroughs Wellcome Foundation (F.R.P.); the Partnership for Cures (F.R.P.); and the National Science Foundation Graduate Research Fellowship (P.I.M). Deposited in PMC for release after 12 months.
References
- Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326-332 [DOI] [PubMed] [Google Scholar]
- Brunelle J. K., Letai A. (2009). Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 122, 437-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92-96 [DOI] [PubMed] [Google Scholar]
- Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. (2005). On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 102, 18773-18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Lerner A. G., Vande Walle L., Upton J. P., Xu W., Hagen A., Backes B. J., Oakes S. A., Papa F. R. (2009). IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Ron D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271-274 [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619-633 [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787-3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Glimcher L. H. (2009). Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell 35, 551-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J., Weissman J. S. (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104-107 [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J. H., Li H., Stevens N., Walter P., Weissman J. S. (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186, 323-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Xu W., Reed J. C. (2008). Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 7, 1013-1030 [DOI] [PubMed] [Google Scholar]
- Kohno K. (2007). How transmembrane proteins sense endoplasmic reticulum stress. Antioxid. Redox Signal. 9, 2295-2303 [DOI] [PubMed] [Google Scholar]
- Li J., Lee B., Lee A. S. (2006). Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J. Biol. Chem. 281, 7260-7270 [DOI] [PubMed] [Google Scholar]
- Liu C. Y., Schroder M., Kaufman R. J. (2000). Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275, 24881-24885 [DOI] [PubMed] [Google Scholar]
- Merksamer P. I., Trusina A., Papa F. R. (2008). Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell 135, 933-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaka S., Okada T., Yoshida H., Mori K. (2006). A role of disulfide bridges formed in the lumenal domain of ATF6 in sensing endoplasmic reticulum stress. Mol. Cell. Biol. 27, 1027-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. (2002). ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16, 1345-1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S., Mori M. (2004). Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11, 381-389 [DOI] [PubMed] [Google Scholar]
- Puthalakath H., O'Reilly L. A., Gunn P., Lee L., Kelly P. N., Huntington N. D., Hughes P. D., Michalak E. M., McKimm-Breschkin J., Motoyama N., et al. (2007). ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 129, 1337-1349 [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell. Biol. 8, 519-529 [DOI] [PubMed] [Google Scholar]
- Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006). Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4, e374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Snapp E. L., Lippincott-Schwartz J., Prywes R. (2005). Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol. Cell. Biol. 25, 921-932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W., Welihinda A. A., Kaufman R. J. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusina A., Papa F. R., Tang C. (2008). Rationalizing translation attenuation in the network architecture of the unfolded protein response. Proc. Natl. Acad. Sci. USA 105, 20280-20285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton J. P., Austgen K., Nishino M., Coakley K. M., Hagen A., Han D., Papa F. R., Oakes S. A. (2008). Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol. Cell. Biol. 28, 3943-3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664-666 [DOI] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944-957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura J. J., Hubner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006). Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell 21, 701-710 [DOI] [PubMed] [Google Scholar]
- Wang X. Z., Harding H. P., Zhang Y., Jolicoeur E. M., Kuroda M., Ron D. (1998). Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17, 5708-5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001). Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C. R., Davis R. J. (2007). The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19, 142-149 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007). Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 13, 365-376 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891 [DOI] [PubMed] [Google Scholar]
- Youle R. J., Strasser A. (2008). The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell. Biol. 9, 47-59 [DOI] [PubMed] [Google Scholar]
- Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. (2006). Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587-599 [DOI] [PubMed] [Google Scholar]