Abstract
Septin complexes at the bud neck in Saccharomyces cerevisiae serve as a scaffold for proteins involved in signaling, cell cycle control, and cell wall synthesis. Many of these bind asymmetrically, associating with either the mother- or daughter-side of the neck. Septin structures are inherently apolar so the basis for the asymmetric binding remains unknown. Bni4, a regulatory subunit of yeast protein phosphatase type 1, Glc7, binds to the outside of the septin ring prior to bud formation and remains restricted to the mother-side of the bud neck after bud emergence. Bni4 is responsible for targeting Glc7 to the mother-side of the bud neck for proper deposition of the chitin ring. We show here that Bni4 localizes symmetrically, as two distinct rings on both sides of the bud neck following energy depletion or activation of cell cycle checkpoints. Our data indicate that loss of Bni4 asymmetry can occur via at least two different mechanisms. Furthermore, we show that Bni4 has a Swe1-dependent role in regulating the cell morphogenesis checkpoint in response to hydroxyurea, which suggests that the change in localization of Bni4 following checkpoint activation may help stabilize the cell cycle regulator Swe1 during cell cycle arrest.
Keywords: Bni4, Glc7, Cell stress, Septins
Introduction
Septins are a conserved family of cytoskeletal GTP-binding proteins that form cortical filaments and ring-like structures in animals and fungi (Kinoshita, 2006; Versele and Thorner, 2005; Weirich et al., 2008). In metazoans, septins have been implicated in diverse physiological processes. They are required for cytokinesis in some Drosophila (Fares et al., 1995; Neufeld and Rubin, 1994) and Caenorhabditis elegans cells (Nguyen et al., 2000), and for morphogenesis of neurons (Tada et al., 2007; Xie et al., 2007) and sperm (Ihara et al., 2005). Septin mutations have roles in the etiology of cancer (Hall and Russell, 2004) and in neurodegenerative diseases (Kuhlenbaumer et al., 2005). In the budding yeast Saccharomyces cerevisiae, where septins were first identified and have been studied in the most detail, septins form filaments at the bud neck that are required for cytokinesis, for proper cell morphogenesis, and for the regulation of several cell cycle checkpoints (reviewed by Longtine and Bi, 2003). In other fungi, septins have roles in cell morphogenesis and cytokinesis (reviewed by Douglas et al., 2005).
What are the roles of septins in these diverse processes? In yeast, cortical septin filaments at the bud neck form a diffusion barrier that serves to compartmentalize the mother cell from the bud (Dobbelaere and Barral, 2004; Takizawa et al., 2000). This barrier is necessary for normal polarized growth and division and prevents newly formed daughter cells from acquiring the age of their mother cells (Shcheprova et al., 2008). A similar function has been proposed to account for the role of septins in the morphogenesis of mammalian neurons and sperm (Caudron and Barral, 2009). Septins also provide a docking site for protein complexes involved in processes ranging from cell wall synthesis to cell cycle control. In yeast, septins tether the chitin synthase III complex, formin proteins, components of the morphogenesis checkpoint, and components of the actomyosin ring during cytokinesis (reviewed by Gladfelter et al., 2001; Longtine and Bi, 2003). Importantly, septins act as a scaffold for proteins required for the DNA-damage checkpoint in yeast and metazoans (Kremer et al., 2007; Smolka et al., 2006).
Details of the basic structure of septin filaments are rapidly emerging. X-ray crystallographic and electron microscopy studies of septin filaments have led to an understanding of the basic septin architecture (Bertin et al., 2008; John et al., 2007; Sirajuddin et al., 2007). Filaments are polymerized from a core oligomeric structure that contains two, three or four different septins, depending on the species. Importantly, the core structures are non-polar. In yeast, five septins, Shs1, Cdc3, Cdc10, Cdc11 and Cdc12, are expressed during vegetative growth and first form a ring prior to bud emergence at the presumptive bud site. This ring rearranges into an hourglass-shaped collar at bud emergence and remains in this arrangement at the bud neck until cytokinesis, when the septin complex splits into two separate rings, occupying opposite sides of the neck (Longtine and Bi, 2003). The use of orientationally constrained green fluorescent protein (GFP)-septin fusion proteins with polarized fluorescence microscopy has revealed that septin filaments are aligned parallel to the mother-daughter axis in the septin collar and may reorient 90° when the collar splits at cytokinesis (Vrabioiu and Mitchison, 2006). Further analysis of a collection of orientationally constrained fusions reveals that the filaments have no net polarity (Vrabioiu and Mitchison, 2007), consistent with the apolarity of septin filaments polymerized in vitro.
In remarkable contrast to the apolar nature of the septin filaments are the observations that many septin-binding proteins associate asymmetrically with the septin collar. Some are located exclusively on the daughter-side of the collar, whereas others are located on the mother-side (for a review, see Gladfelter et al., 2001). An apolar septin cytoskeleton excludes the most obvious mechanism to explain the asymmetric localization of binding proteins. To investigate the mechanism of asymmetric protein binding to septin filaments, we have focused on the yeast protein Bni4. This protein localizes to the outside of the septin ring prior to budding and remains restricted to the mother-side of the neck as the septin ring develops into a collar during bud emergence (DeMarini et al., 1997; Kozubowski et al., 2005; Kozubowski et al., 2003; Sanz et al., 2004). This asymmetry is maintained in mutants that have defects in septin architecture, including cla4, gin4 and elm1 (Kozubowski et al., 2005). Localization of Bni4 to septins requires phosphorylation by the G1-cyclin-dependent kinase Pcl1,2-Pho85 (Zou et al., 2009). In addition, Bni4 associates with the type 1 protein phosphatase Glc7 and, together, this complex recruits chitin synthase III to the cell cortex on the mother-side of the septin collar (Kozubowski et al., 2003; Larson et al., 2008). The asymmetric localization of Bni4 and Glc7 is essential for proper morphological development, as expression of a septin Cdc10-Bni4 chimera targets Glc7 and chitin synthase III to both sides of the bud neck, resulting in slow growth and elongated buds (Larson et al., 2008). In this study we show that cell cycle arrest and energy deprivation lead to dramatic loss of Bni4 asymmetry. Our data provide insight into how Bni4 is asymmetrically restricted to the mother-side of the bud neck.
Results
Bni4 loses asymmetry during cell cycle arrest
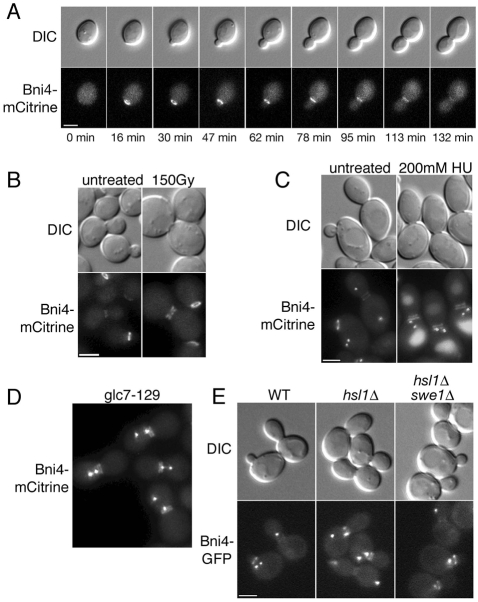
Bni4 forms a ring on the cell cortex prior to bud emergence and remains on the mother-side of the neck after the bud has formed. Levels of Bni4 at the neck gradually fall as the bud grows in size, as shown in time-lapse images of Bni4-mCitrine in living cells (Fig. 1A). During our studies, we noted that Bni4 localizes to the bud neck as two bright, symmetric rings in a minor population of cells. Time-lapse imaging revealed that these cells are uniformly large-budded, growth arrested, and often have short spindles, as determined by the localization of Spc42-GFP, a spindle pole body protein (data not shown). To define the conditions that lead to this altered localization, we examined the location of Bni4-mCitrine in cells arrested at several points in the cell cycle. In many cases, cell cycle arrest induced nearly symmetric localization of Bni4. This was true for activation of the DNA-damage and DNA-replication checkpoints following γ-irradiation and treatment by hydroxyurea (HU), respectively (Fig. 1B,C). Activation of the spindle assembly checkpoint in glc7-129 mutant cells (Bloecher and Tatchell, 1999) also induced symmetric localization of Bni4-mCitrine (Fig. 1D). We assayed the morphogenesis checkpoint by testing localization of Bni4-GFP in hsl1Δ mutants. Activation of this checkpoint by stabilizing the Swe1 kinase causes a cell cycle delay in G2. Swe1 degradation is inhibited in cells deleted for the Hsl1 kinase, whose binding to septins leads to the phosphorylation and subsequent degradation of Swe1 (Lew, 2003). We examined the localization of Bni4 in hsl1Δ mutant cells and found that Bni4 is present at higher levels at the bud necks, as previously reported (Kozubowski et al., 2003), but remains on the mother-side of the bud neck in cycling cells. However, in arrested cells containing elongated buds, Bni4 is present as two bright rings (Fig. 1E). Deletion of SWE1 suppresses the elongated phenotype of hsl1Δ mutant cells and loss of Bni4 asymmetry was no longer detected (Fig. 1E). Interestingly, Bni4-GFP levels remain elevated in hsl1Δ swe1Δ mutant cells (data not shown), indicating that the influence of Hsl1 on Bni4 protein levels at the neck is independent of Swe1.
Fig. 1.
Bni4 is targeted to mother- and daughter-sides of the bud neck following cell cycle arrest. (A) DIC and fluorescence microscopy showing Bni4-mCitrine localization throughout the cell cycle. Strain JRL265 (BNI4-mCitrine) was grown to mid-log phase in synthetic complete (SC) medium then examined at the indicated times. (B) DIC and fluorescence images of Bni4-mCitrine in strain JRL264. Cells were grown to mid-log phase and either left untreated or exposed to 150 Gy γ-radiation. Cells were examined after a 3 hour incubation at 30°C following irradiation. (C) DIC and fluorescence images of cells from diploid strain JRL1000 (BNI4-mCitrine/BNI4-mCitrine SPC42-GFP(3X)/SPC42). Cells were grown to mid-log phase in SC medium and incubated for 2 hours in the presence of 200 mM hydroxyurea (HU). (D) DIC and fluorescence images of cells from strain KT2592 (BNI4-mCitrine SPC42-GFP(3X) glc7-129) grown to log phase at 24°C in synthetic medium. (E) DIC and fluorescence images of cells from strains JRL240 (BNI4-GFP SPC42-GFP(3X)), JRL236 (hsl1Δ BNI4-GFP SPC42-GFP(3X)), and JRL238 (hsl1Δ swe1Δ BNI4-GFP SPC42-GFP(3X)). Cells were grown to mid-log phase in SC medium. Scale bars: 3 μm.
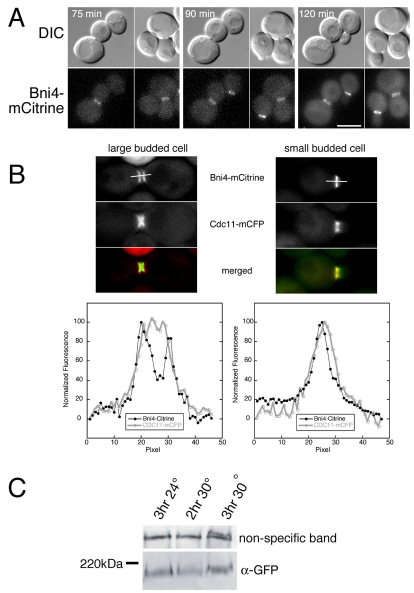
Activation of the DNA-damage checkpoint by incubating temperature-sensitive cdc13-1 cells above the permissive temperature also induces symmetric binding of Bni4 to the septin collar (Fig. 2A). To determine whether the loss of Bni4 asymmetry occurs in response to checkpoint activation or as a consequence of cell cycle arrest, we imaged cdc13-1 cells that had been grown at the permissive temperature and then shifted the cells on the microscope slide to the non-permissive temperature using a heated objective. Bni4-GFP lost asymmetry only as cells arrested with large buds, irrespective of the time at the restrictive temperature (Fig. 2A). This result indicates that the movement of Bni4 to the daughter-side of the neck occurs as a consequence of cell cycle arrest rather than as a consequence of activation of some upstream component in the checkpoint pathway.
Fig. 2.
Bni4-mCitrine localization in large budded cells. (A) Time-lapse imaging of DIC and fluorescence images of cells from strain KT2575 (BNI4-mCitrine cdc13-1) after a shift from 24°C to 32°C. The time indicated in each panel is the time after the temperature shift. Note the small budded cells at the 90 minute and 120 minute time points still retain Bni4-mCitrine on the daughter-side of the bud neck. (B) Upper panels: fluorescence images of septin (Cdc11-mCFP) and Bni4-mCitrine in diploid strain JRL265/JRL574 (BNI4-mCitrine homozygous CDC11-mCFP heterozygous). Cells were grown to mid-log phase in synthetic medium and then incubated for 1.5 hours in 150 mM hydroxyurea. Lower panels: the relative fluorescence of the CFP and mCitrine was measured through the middle of the bud neck along the mother-bud axis for 3 μm as illustrated by the bars in the upper panels. (C) Immunoblot analysis of Bni4-mCitrine using α-GFP. Strain KT2575 (BNI4-mCitrine cdc13-1) was grown to mid-log phase in YPD at 24°C and then incubated for the indicated times at either 24°C or 30°C. A non-specific band was used as a load control.
Cell cycle arrest at G2/M results in the accumulation of neck-spanning septin collars rather than split rings (Cid et al., 2001), and so the two bright rings of Bni4 in arrested cells should not be due to splitting of the underlying septin scaffold. We confirmed this in our background by imaging Bni4-mCitrine and Cdc10-mCFP in cdc13-1 and HU-arrested cells (data not shown). Here, Bni4 appears to be located on the outside edges of the septin collar, rather than directly over the septins. We imaged Cdc11-mCFP and Bni4-mCitrine after HU treatment and measured the fluorescence intensity through the neck along the mother-bud axis in small and large budded cells (Fig. 2B). In large budded, arrested cells, Bni4 is located in two rings at the outer edges of the septin collar whereas, in small budded cells, Bni4 remains on the mother-side of the septin collar. The implications of this result are discussed below (see Discussion).
If the septin collar is not physically altered following cell cycle arrest, is it possible that Bni4 is modified following cell cycle arrest, allowing symmetric binding to the septin collar? Bni4 is phosphorylated in a cell-cycle-dependent manner (Kozubowski et al., 2003), in part by the Pcl1,2-Pho85 cyclin-dependent kinase (Zou et al., 2009). Bni4 phosphorylation can be monitored by a reduction in its electrophoretic mobility. However, immunoblot analysis showed that the mobility of Bni4-mCitrine does not change following activation of the DNA-damage checkpoint in cdc13-1 mutant cells (Fig. 2C).
bni4Δ partially suppresses the hypersensitivity to HU caused by deletion of HSL1
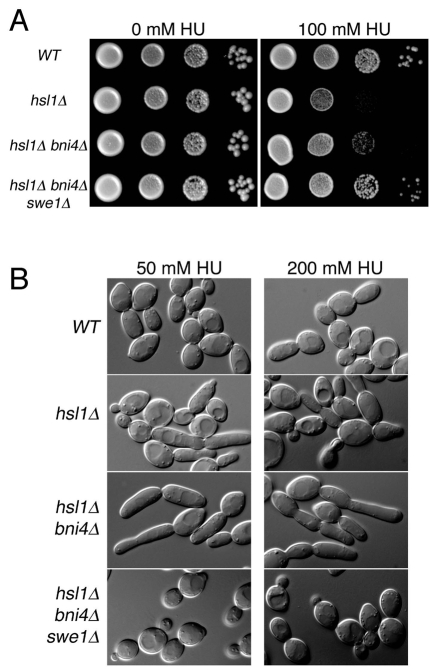
What is the physiological reason for loss of Bni4 asymmetry during cell cycle arrest? Septins have a central role in the cell morphogenesis checkpoint (Lew, 2003) and have been more recently implicated in the cellular response to DNA replication stress (Enserink et al., 2006; Liu and Wang, 2006; Smolka et al., 2006). Rad53, an essential DNA-damage checkpoint kinase, was recently found to associate with the septin complex in the presence of genotoxic stresses such as methyl methanesulphonate (MMS) or HU (Smolka et al., 2006). In addition, deletion of RAD53 results in elongated buds due to an accumulation of Swe1 (Enserink et al., 2006). Deletion of the Hsl1 kinase, which is responsible for targeting Swe1 to the bud neck, results in HU hypersensitivity (Enserink et al., 2006; Liu and Wang, 2006). To test the possibility that Bni4 has a role in this response, we assayed bni4Δ mutants for altered sensitivity to HU. bni4Δ mutants exhibit a wild-type response to HU (data not shown) but the bni4Δ mutation partially suppresses the hypersensitivity of hsl1Δ strains to HU (Fig. 3A). In liquid medium, the buds of hsl1Δ bni4Δ mutant cells are also more uniform in size and shape (Fig. 3B). As with the plate assay, the elongated bud phenotype is completely dependent on SWE1 (Fig. 3B). Together, these results suggest that Bni4 has a role in modulating Swe1 activity in response to replication stress.
Fig. 3.
Genetic interactions between BNI4 and HSL1. (A) Cultures of diploid strains KT1112xKT1113 (WT), JRL1002xJRL1005 (hsl1Δ), JRL1003xJRL1004 (hsl1Δ bni4Δ), KT2801xKT2803 (hsl1Δ bni4Δ swe1Δ) were serially diluted onto SC medium or SC medium containing 100 mM hydroxyurea (HU) and incubated at 33°C for 72 hours before imaging. (B) DIC images of cells from strains listed in A. Cells were grown to mid-log phase in SC medium and then incubated for 5 hours in SC medium containing the designated concentrations of HU.
Bni4 loses asymmetry when cells are starved for carbohydrates or depleted for energy
We monitored the location of Bni4-mCitrine under conditions that induce nutritional stress. Cells were grown in SC medium containing glucose, washed in water, and resuspended in a variety of media lacking either glucose, yeast nitrogen base, amino acids or uracil, for which the strain is auxotrophic. We also arrested cells with rapamycin, which is thought to mimic starvation conditions (Crespo and Hall, 2002).
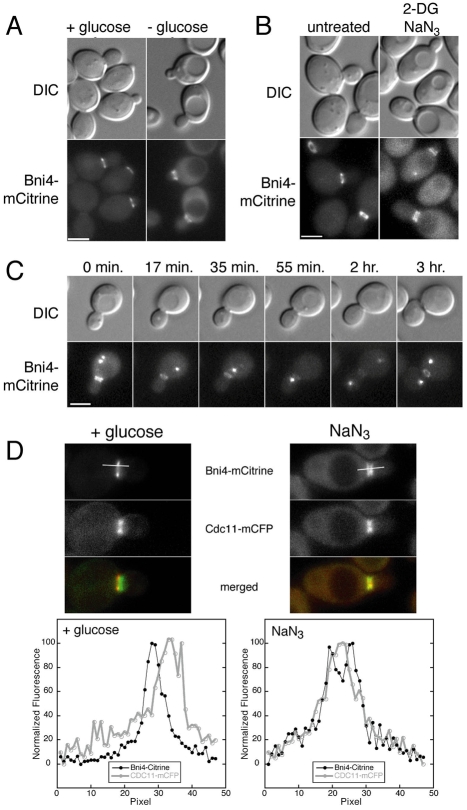
Glucose starvation was the only condition that led to symmetric localization of Bni4-mCitrine. Within 30 minutes of transfer to glucose-deficient medium, we observed cells with Bni4-mCitrine located on both sides of the bud neck. After 1.5 hours in glucose-deficient medium, Bni4-mCitrine localized to both sides of the bud neck in most budded cells (89%, n=148) (Fig. 4A). This loss of asymmetry occurred in cells containing buds of all sizes, indicating that the loss of asymmetry after glucose starvation occurs independently of the cell cycle. Close examination of larger diploid cells revealed that Bni4-mCitrine is present as two rings, one on each side of the bud neck, rather than as an hourglass-shaped collar, as in the septin complex (data not shown). Loss of Bni4 asymmetry also occurs in cells in YP media lacking glucose (data not shown). In cells treated with the metabolic inhibitors 2-deoxyglucose (2-DG) and sodium azide (NaN3), Bni4 asymmetry is lost in nearly all budded cells (93%, n=263) after 30 minutes of exposure (Fig. 4B). To determine whether loss of Bni4 asymmetry is a reversible process, we incubated cells in media lacking glucose for 1.5 hours, then placed them on an agarose pad of medium containing glucose and followed the cells by time-lapse microscopy. Bni4-mCitrine asymmetry is restored approximately 15 minutes after cells are exposed to glucose, prior to the time at which they resume growth and cell division (Fig. 4C). The effect of metabolic inhibitors is also rapidly reversible, with restoration of Bni4 asymmetry within minutes after washing cells with fresh media containing glucose (data not shown). In conclusion, Bni4 relocates to the mother- and daughter-side of the septin collar when cells are subjected to energy depletion, but upon energy restoration, asymmetry is rapidly re-established.
Fig. 4.
Localization of Bni4 following energy depletion. (A) DIC and fluorescence microscopy of cells from strains JRL264 (BNI4-mCitrine). Cells were grown to mid-log phase in synthetic medium then placed in synthetic medium lacking glucose for 1.5 hours. Scale bar: 3 μm. (B) DIC and fluorescence microscopy analysis of homozygous diploid strain JRL876 (BNI4-mCitrine). Cells were grown to mid-log phase in synthetic medium then incubated in synthetic medium lacking glucose and containing 100 mM 2-deoxyglucose/10 mM NaN3 for 30 minutes. Scale bar: 3 μm. (C) Time-lapse microscopy of strain JRL326 [BNI4-mCitrine SPC42-GFP(3X)]. Cells were grown to mid-log phase in synthetic medium then placed in synthetic medium lacking glucose for 1.5 hours. Cells were then followed by time-lapse on synthetic medium containing 2% glucose. The Spc42-GFP protein marks the spindle pole body, which duplicates and separates during S phase. In many images only one spindle pole body is visible in the focal plane. Scale bar: 3 μm. (D) Upper panels: fluorescence images of Cdc11-mCFP and Bni4-mCitrine in diploid strain JRL265/JRL574 growing in synthetic medium (left panels) or incubated for 30 minutes in 10 mM sodium azide (right panels). Lower panels: the relative fluorescence of the mCFP and mCitrine for 3 μm through the middle of the bud neck along the mother-bud axis as illustrated by the bars in the upper panels.
As during cell cycle arrest, during glucose starvation Bni4 appears to be located on the outside edges of the septin collar, rather than directly over the septins. We imaged Cdc11-mCFP and Bni4-mCitrine in growing and sodium-azide-arrested cells and measured the fluorescence intensity through the neck along the mother-bud axis. In growing cells, Bni4 is located at the outer edge of the septin collar in the mother cell whereas, in the energy-depleted cells, Bni4 is located at the outer edges of the septin collar in both mother and daughter cells (Fig. 4D).
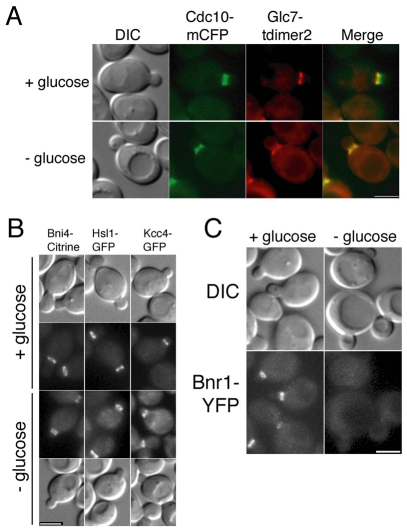
Many proteins bind asymmetrically to the septin scaffold at the bud neck (Gladfelter et al., 2001). We examined the locations of other bud neck proteins following energy deprivation. The Bni4-binding protein Glc7 remains associated with Bni4 at the neck following arrest (Fig. 5A) but Chs4, an activator of chitin synthase III, does not (data not shown). Hsl1 and Kcc4 are Nim1-related kinases normally restricted to the daughter-side of the bud neck (Barral et al., 1999; Gladfelter et al., 2001). Following glucose starvation, Hsl1 and Kcc4 remain localized to the daughter-side of the bud neck in the majority of cells (Fig. 5B). Bnr1, a formin protein responsible for nucleating actin filaments at the bud neck (Evangelista et al., 2002; Pruyne et al., 2004; Sagot et al., 2002), is restricted to the mother-side of the neck but dissociates from the bud neck following glucose starvation (Fig. 5C). By contrast, Bnr1 remains associated with the bud neck following cell cycle arrest (data not shown). Thus, energy depletion does not affect the location of all proteins associated with the septin scaffold in the same manner.
Fig. 5.
Energy deprivation changes the location of some, but not all, bud neck-associated proteins. (A) DIC and fluorescence images of septin (Cdc10-mCFP) and Glc7-tdimer2 in diploid strain JRL884/KT2450. Cells were grown to mid-log phase in synthetic medium then incubated for 1.5 hours in synthetic medium with or without 2% glucose. (B) DIC and fluorescence images of cells from strains JRL264 (BNI4-mCitrine), YLK173 (HSL1-GFP) and YLK179 (KCC4-GFP). Cells were treated as in A. (C) DIC and fluorescence images of cells from strain JRL443 (BNR1-YFP). Cells were treated as in A. Scale bars: 3 μm.
The septin architecture is not altered by energy depletion
The observed changes in Bni4 localization following energy depletion could be caused by a structural change in the septin collar. Alternatively, the change could be due to an alteration or modification of Bni4 or an undefined regulatory protein. To test the former possibility, we investigated the structure of the septin collar in more detail. At the end of the cell cycle, the septin collar splits into two rings with a concomitant change in the conformation of septins or the orientation of septin filaments (Vrabioiu and Mitchison, 2006). One explanation for loss of Bni4 asymmetry is that energy depletion might lead to premature splitting of septin rings. However, septin collars, as observed using fluorescence microscopy, appeared normal following energy depletion (data not shown).
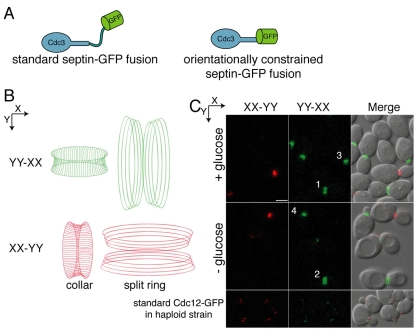
To examine changes in septin structure or orientation, we used a conformationally constrained Cdc3-GFP fusion protein expressed from the native CDC3 promoter (Vrabioiu and Mitchison, 2006). The α-helix at the C-terminus of Cdc3 was attached directly to the N-terminal α-helix of GFP (Fig. 6A), creating a GFP moiety constrained in a direction dictated by the orientation of the septin filaments. Vrabioiu and Mitchison used this septin fusion, together with polarized fluorescence microscopy, to confirm the filamentous nature of the septins in vivo (Vrabioiu and Mitchison, 2006). In the septin collar of budded cells, filaments are oriented parallel to the mother-bud axis. After the collar splits into rings, septins are oriented perpendicular to the long axis. As described previously (Vrabioiu and Mitchison, 2006), we assayed polarized fluorescence in diploid cells heterozygous for the orientationally constrained Cdc3-GFP fusion. Using polarized filters for fluorescence excitation and emission that are oriented orthogonally in the X- and Y-axes, the signal from one axis was subtracted from that from the other (XX-YY or YY-XX) to determine the polarized fluorescence signal, and hence the direction of Cdc3-GFP orientation. As shown in Fig. 6B, septin collars in cells whose mother-bud axis is orientated along the Y-axis emit fluorescence in the YY-XX filter configuration (pseudocolored green in Fig. 6C), whereas septin collars orientated along the X-axis are fluorescent in only the XX-YY configuration (pseudocolored red). By contrast, the split septin rings have the orthogonal orientation (Fig. 6B). As shown in Fig. 6C, septins in budded cells with collars are oriented along the mother-bud axis, whereas septins in large budded cells with split rings are oriented perpendicular to the mother-bud axis. After 1.5 hours of glucose starvation, when Bni4 should be present as a double ring, the septin orientation appears unchanged compared with that in control cells growing in the presence of glucose (Fig. 6C). For example, cells 3 and 4 (Fig. 6C) are oriented along the Y-axis and their septin filaments, in the collar form, are also oriented along the Y-axis. By contrast, cells 1 and 2 in Fig. 6C are oriented along the X-axis, but their septin collars have already split and, therefore, the filaments are oriented along the Y-axis. These results demonstrate that the septins do not reorient upon glucose starvation. Thus, an underlying change in septin filament orientation cannot explain the loss of Bni4 asymmetry.
Fig. 6.
Loss of Bni4 asymmetry is not due to changes in septin architecture. (A) The orientationally constrained septin-GFP fusion protein. The standard septin-GFP fusions contain a flexible linker connecting the septin and GFP portions of the fusion protein (left). In the orientationally constrained septin-GFP fusion, the last α-helix of the coiled-coil domain of Cdc3 was fused to the first α-helix of GFP, creating a single α-helix. Consequently, the orientation of Cdc3 dictates the orientation of GFP (right). (B) Diagram of septin filaments and their orientation as visualized by polarized fluorescence microscopy. Filaments oriented in the direction of the Y-axis are colored green and filaments oriented in the X-axis are colored red. (C) Polarized fluorescence images of strain JRL794 (CDC3-GFP heterozygous diploid). Strain YLK68 (CDC12-GFP), which does not contain an orientationally constrained GFP-tag, was used as a negative control. Cells were treated as in Fig. 4A and then imaged by polarized fluorescence microscopy. The X-axis signal subtracted from the Y-axis signal (YY-XX) and the Y-axis signal subtracted from the X-axis signal (XX-YY) are pseudocolored green and red, respectively, and correspond to the orientations shown in B. Scale bar, 3 μm.
Bni4 phosphorylation correlates with its change in location following energy depletion
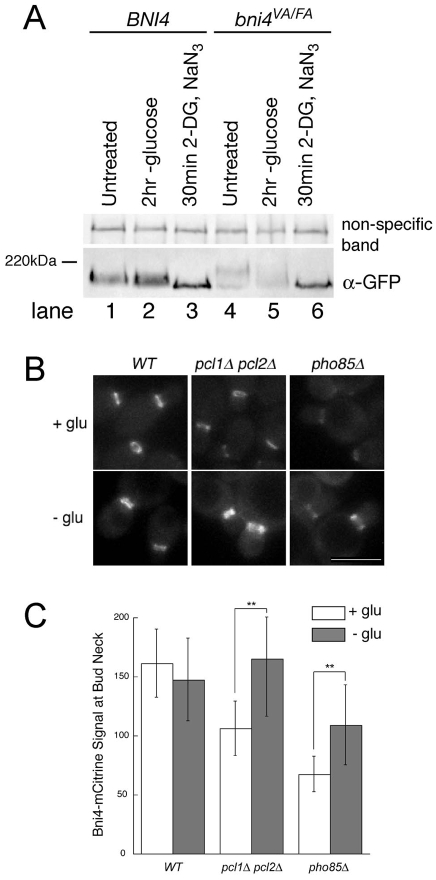
We next assayed the phosphorylation state of Bni4 following energy depletion by monitoring the electrophoretic mobility of Bni4-GFP. Interestingly, immunoblot analysis of Bni4-GFP revealed that its electrophoretic mobility is increased following glucose starvation (Fig. 7A, lane 2) or energy depletion (lane 3). We also tested the phosphorylation state of Bni4V831A/F833A, a Bni4 variant that is normally hyperphosphorylated (Kozubowski et al., 2003). This variant also exhibits an increased mobility on glucose-depleted medium (Fig. 7A, lane 5) and a more dramatic increase after energy depletion (lane 6).
Fig. 7.
Bni4 and Bni4V831A/F833A become hypophosphorylated following energy depletion. (A) Immunoblot of Bni4-GFP and Bni4V831A/F833A-GFP using α-GFP. Strains JRL369 (BNI4-GFP) (lanes 1-3) and YLK85 (bni4V831A/F833A-GFP) (lanes 4-6) were grown to mid-log phase in synthetic medium containing glucose. Cells were then left untreated (lanes 1 and 4) or incubated in synthetic media lacking glucose (lanes 2 and 5) with the addition of 100 mM 2-deoxyglucose and 10 mM NaN3 (lanes 3 and 6) for the indicated times. A non-specific band was used as a load control. (B) Fluorescence microscopy of Bni4-mCitrine at the neck in wild-type (JRL265), pcl1Δ pcl2Δ (JRL685) and pho85Δ (JRL653) cells. Cells were imaged at mid-log phase in synthetic medium (+ glu) or after incubation for 1.5 hours in synthetic medium without glucose (− glu). Scale bar: 5 μm. (C) Quantitative analysis of relative fluorescent signal at the bud neck of the cells in B (n>50). Double asterisks denote significantly different levels. **P<0.0001, according to the unpaired Student's t-test.
The change in Bni4 electrophoretic mobility following energy depletion suggests that dephosphorylation of Bni4 is responsible for the loss of asymmetry. However, pcl1Δ pcl2Δ and pho85Δ mutants, which substantially reduce the level of Bni4 phosphoryation, reduce the levels of Bni4 at the neck (Zou et al., 2009) but do not alter its location on the mother-side of the neck. However, Bni4 retains some phosphorylated residues in pcl1Δ pcl2Δ and pho85Δ mutants, consistent with the abundance of non-Pho85-dependent phosphorylation sites in Bni4 (Bodenmiller et al., 2008). If these remaining phosphorylated residues are responsible for the asymmetric localization of Bni4 and energy depletion results in global dephosphorylation of Bni4, then we would expect that pcl1Δ pcl2Δ and pho85Δ mutants would behave similarly to the wildtype when energy depleted. As predicted, glucose depletion completely restores Bni4 to the bud neck in the pcl1Δ pcl2Δ mutant (Fig. 7B,C). Bni4 levels were only partially restored in the pho85Δ strain, but the pho85 mutation is pleiotropic, and so it is possible that the failure of glucose depletion to ameliorate the defect of pho85 is due to off-target effects.
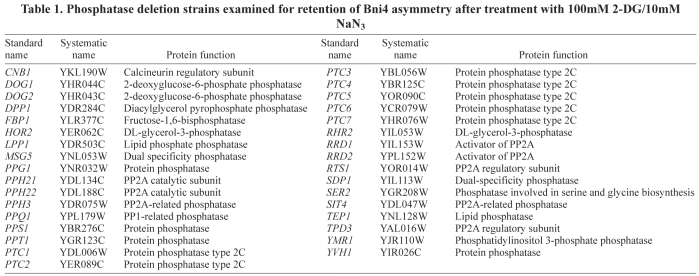
The dephosphorylation of Bni4 following energy depletion suggests that activities of Bni4 protein kinases and/or Bni4 phosphatases are regulated in response to energy depletion. Glc7 has been implicated as a Bni4 phosphatase because a Bni4 variant that cannot bind to Glc7 (Bni4V831A/F833A) is hyperphosphorylated (Kozubowski et al., 2003). However, following energy depletion, Bni4V831A/F833A-GFP is hypophosphorylated (Fig. 7A) and still localizes to both sides of the neck (data not shown), which suggests that a rapid increase in Glc7 activity is probably not responsible for the decrease in Bni4 phosphorylation following energy depletion. We also examined the localization of Bni4-GFP in a collection of deletion mutants of phosphatase-related genes in the presence of 2-DG and NaN3 (Table 1). However, Bni4 was asymmetrically distributed under normal growth conditions and rapidly lost asymmetry in the presence of the metabolic inhibitors in all strains tested. These results do not eliminate the hypothesis that one or more phosphatases regulate Bni4 localization due to the functional redundancy of the phosphatases and their regulators.
Table 1.
Phosphatase deletion strains examined for retention of Bni4 asymmetry after treatment with 100mM 2-DG/10mM NaN3
Discussion
In budding yeast, septins act as a scaffold to tether a diverse array of proteins to the bud neck. These proteins carry out many important and, in some cases essential, functions but it is unknown how they bind to septins at the molecular level. Furthermore, the temporal and spatial association of these proteins with septin filaments is complex, exhibiting cell-cycle-specific association with septins or localization to one side of the neck. This asymmetry could be accounted for if the septin filaments were inherently polar. However, recent studies indicate that septin filaments are fundamentally apolar structures, thus eliminating the most convenient model to explain this asymmetry. Our observations that the asymmetric localization of Bni4 can be disrupted by either energy deprivation or cell cycle arrest shed new light on this complex process.
The loss of Bni4 asymmetry during energy deprivation is independent of the cell cycle and is rapidly reversible. This alteration appears to be due largely to Bni4 dephosphorylation rather than to any gross change in the underlying septin architecture. The observation that decreased levels of Bni4 at the bud neck in mutants with reduced Pcl1,2-Pho85 kinase activity can be reversed by glucose depletion leads us to propose that Bni4-septin association is regulated both positively and negatively by phosphorylation. Phosphorylation by Pcl1,2-Pho85 allows Bni4 to associate with the neck, whereas phosphorylation at additional sites is required to prevent Bni4 from localizing to the daughter-side of the neck. In this model, energy deprivation results in nearly complete dephosphorylation of Bni4, allowing it to localize to both sides of the neck in a cell-cycle-independent manner. A Bni4 variant lacking ten putative Pcl1,2-Pho85 sites (Bni4-10A) binds at very low levels to the neck (Zou et al., 2009) but association with the neck is not recovered by glucose deprivation (J.R.L. and K.T., unpublished). This result is not strictly compatible with our model, but it is possible that the ten amino acid substitutions disrupt the activity of Bni4 for reasons other than preventing Pcl1,2-Pho85 phosphorylation. Identification and alteration of the in vivo phosphorylation sites in Bni4 will be necessary to test this model. The model also predicts that one or more protein phosphatases are involved in dephosphorylating Bni4 upon energy deprivation but the nature of these activities is unknown at present.
An interesting feature of the association of Bni4 with the bud neck is that Bni4 does not strictly co-localize with septins. Bni4 is located at the inner edge of the septin collar during vegetative growth and at the inner and outer edges of the collar following cell cycle arrest or energy deprivation (Fig. 2A,B). Although an accurate assessment of Bni4 localization will require spatial resolution beyond that of the light microscope, our results suggest that Bni4 is binding to the interface of the septin filaments and the membrane cortex. In this regard, we have not been able to demonstrate a direct interaction between Bni4 and septins in immune complexes and no Bni4–septin interactions have been reported, other than a two-hybrid interaction between Bni4 and Cdc10 (DeMarini et al., 1997). An association of Bni4 with the cortical edge of the septin collar would fit with its biological role in targeting chitin synthase III. To effectively target this transmembrane complex from a cytoplasmic endosomal compartment to the plasma membrane, Bni4 would need to be located at the extreme edge of the septin collar. We do not know the physiological role of targeting Bni4 to the mother- and daughter-side of the neck following energy deprivation. Wild-type and bni4Δ cells recover from arrest following energy depletion with similar kinetics and bni4Δ mutants do not lose viability more rapidly following energy depletion (K.T., unpublished). Furthermore, Chs4, which is targeted to Bni4 during vegetative growth, dissociates from the neck following energy deprivation.
We believe that the loss of Bni4 asymmetry by depletion of cellular energy and cell cycle arrest occurs via two different mechanisms. Energy depletion induces a loss of asymmetry throughout the cell cycle, whereas cell cycle arrest leads only to the symmetric targeting of Bni4 to the neck in large budded cells. In addition, Bnr1, another protein restricted to the mother-side of the bud neck, dissociates from the neck upon energy depletion but remains unchanged in large-budded cells during cell cycle arrest. Energy depletion also results in an increased electrophoretic mobility of Bni4, which suggests a decrease in its phosphorylation state. By contrast, no obvious change in Bni4 electrophoretic mobility occurs upon cell cycle arrest.
Previous studies indicate that one physiological role of Bni4 is to target the Glc7 phosphatase to the bud neck and activate it towards substrates involved in chitin synthesis (Kozubowski et al., 2003; Larson et al., 2008). Genetic studies also support a role for Bni4 in the regulation of cell wall synthesis (Lesage et al., 2004). In addition, Bni4 was identified as a regulator of cortical septin organization through the use of a sensitized assay (Gladfelter et al., 2005). The results presented here support a further role for Bni4 in the morphogenesis or DNA replication checkpoints. Swe1 is stabilized in response to DNA replication stress and alterations in components of the morphogenesis checkpoint that negatively regulate Swe1, such as Hsl1, Hsl7 and Cla4, lead to hypersensitivity to HU and an elongated bud phenotype (Enserink et al., 2006; Liu and Wang, 2006). We show here that bni4Δ mutations partially suppress the HU sensitivity of hsl1Δ and influence bud morphology.
Since Glc7 is targeted to the bud neck by Bni4, a logical inference is that Glc7 activity is important for this checkpoint process. It is worth noting that both Hsl1 and Hsl7 are located on the daughter-side of the bud neck. During vegetative growth, Bni4 is located on the opposite side of the septin collar but relocates to the daughter-side of the neck following checkpoint activation, bringing it into proximity with components of the morphogenesis checkpoint. The implication here is that Bni4-Glc7 could now dephosphorylate one or more components of the checkpoint pathway. Glc7 could also dephosphorylate septins. Septins were found to associate with Rad53, a key component of DNA-damage and replication checkpoints, and null mutants in the septin Shs1 are sensitive to HU (Smolka et al., 2006). Consistent with the possibility that Glc7 activity targets septins, Gladfelter et al. identified Bni4 in a screen for proteins that regulate septin assembly (Gladfelter et al., 2005). We have also found that expression of the Glc7-binding domain of Bni4 (Bni4-823-892) fused to the septin Cdc10 causes slow growth and accumulation of cells with elongated buds (Larson et al., 2008). This phenotype is dependent upon Glc7 binding, as a Cdc10 fusion containing a Bni4 fragment with the V831A/F833A amino acid substitutions, which prevents Glc7 binding, fails to induce the elongated bud phenotype. Furthermore, directly targeting Glc7 to the bud neck via a Cdc10 fusion causes the redirection of septins to the bud tip and accumulation of cells with elongated buds (J.R.L. and K.T., unpublished).
In conclusion, we have shown that the temporal and spatial restriction of Bni4 on the septin collar can be altered in response to energy deprivation and cell cycle arrest. Elucidation of the mechanisms responsible for these dynamic changes will most likely require a detailed analysis of the phosphorylated residues in Bni4 and septins. However, given our current lack of understanding, in general, of how proteins associate with septins, such studies could have broader implications for understanding the roles of septins in animals and fungi.
Materials and Methods
Yeast strains and media
The yeast strains used in this work are listed in Table 2 and are congenic to KT1112 (Stuart et al., 1994) or KT1358 (MATα leu2 his3 ura3 trp1) (Bloecher and Tatchell, 2000), except the deletion panel strains (Winzeler et al., 1999), which were obtained from Research Genetics (Open Biosystems, Huntsville, AL). Primers are listed in Table 3. Yeast strains were grown on YPD medium (2% Bacto peptone, 1% yeast extract, 2% glucose) at 30°C, except where noted. Strains were sporulated at 24°C on medium containing 2% Bacto peptone, 1% yeast extract, and 2% potassium acetate. Synthetic complete medium and media lacking supplements were prepared as described previously (Sherman et al., 1986). Yeast transformation, manipulation of Escherichia coli, and the preparation of bacterial growth media were performed as described previously (Kaiser et al., 1994; Maniatis et al., 1989). PCR was used to generate gene deletion and fluorescent protein fusion strains. Each amplified cassette was introduced into a diploid strain; drug-resistant or amino acid-prototrophic transformants were sporulated; and haploid meiotic segregants were isolated by tetrad analysis. Deletion of HSL1 was described previously (Kozubowski et al., 2003). To generate hsl1Δ::NatMX6, YLK52 (MATα leu2 his3 ura3 trp1 hsl1Δ::KanMX6) was transformed with EcoRI-digested pAG25 (Goldstein and McCusker, 1999). To generate swe1Δ::KanMX4, a deletion cassette was amplified using DNA from the swe1Δ Research Genetics panel strain and primers SP27-F and SP28-R. The cassette was then used to transform YLK239 (MATa/MATα leu2/leu2 his3/his3 ura3/ura3 trp1/trp1) and haploids were recovered after tetrad dissection. BNI4-yEmCitrine was generated with primers SP63-F and SP66-R, using pKT211 (Sheff and Thorn, 2004) as the template. Integrated GFP-CHS4 and CDC10-mCFP were described previously (Larson et al., 2008). Construction of GLC7-tdimer2 was described previously (Bharucha et al., 2008). The SPC42-GFP(3X) allele (a gift from Mark Winey, University of Colorado, Boulder, CO), the cdc16-1 allele, cdc27-1 allele and cdc13-1 allele (a gift from Ted Weinert, University of Arizona) were introduced into the JC482 background by at least seven serial backcrosses. Ectopically integrated BNI4-GFP and bni4V831A/F833A-GFP were described previously (Kozubowski et al., 2003; Larson et al., 2008). BNI4-CFP was previously described (Kozubowski et al., 2003). The polarized CDC3-GFP fusion was constructed and characterized by Vrabioiu and Mitchison (Vrabioiu and Mitchison, 2006). Plasmid pLK10 (pRS315 CEN LEU2 BNI4-GFP) and construction of KCC4-GFP and CDC12-GFP were described previously (Kozubowski et al., 2003). BNR1-YFP was generated with primers BNRGFP1 and BNRGFP2 with pDH5 (Yeast Resource Center, University of Washington, Seattle, WA) as the template. HSL1-GFP was made using pLK3 (Kozubowski et al., 2003) as the template and primers HSL1F and HSL1R.
Table 2.
Yeast strains used in this study
Table 3.
Primers used in this study
Microscopic analysis and biochemical procedures
Cells were placed onto a pad of 2% agarose in synthetic medium containing 2% glucose or 100 mM 2-deoxyglucose, 10 mM NaN3, as noted, and imaged for GFP, CFP, YFP and RFP, as previously described (Kozubowski et al., 2003). Image analysis, filter wheels, shutters, and Z-axis stepping motor were controlled by Slidebook software (Olympus Imaging Systems, Melville, NY). Time lapse imaging was performed as previously described (Bloecher and Tatchell, 2000).
To assess Bni4-GFP phosphorylation state, total protein was prepared from cultures by lysis in TCA (Davis et al., 1993; Stuart et al., 1994). Protein extracts were electrophoresed on 6% polyacrylamide-SDS gels. Immunoblot analysis was performed as described (Stuart et al., 1994) using the Living Colors A.v. monoclonal anti-GFP (JL-8) antibody (Clontech, Mountain View, CA), with subsequent detection using the enhanced chemiluminescence system (Amersham ECL Plus, GE Health Care, UK).
Acknowledgments
We thank Mark Winey and Ted Weinert for reagents and Lucy Robinson for critically reading the manuscript. A portion of this work was supported by National Institutes of Health grant GM-47789. Deposited in PMC for release after 12 months.
References
- Barral Y., Parra M., Bidlingmaier S., Snyder M. (1999). Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Gene. Dev. 13, 176-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A., McMurray M. A., Grob P., Park S. S., Garcia G., 3rd, Patanwala I., Ng H. L., Alber T., Thorner J., Nogales E. (2008). Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA 105, 8274-8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha J. P., Larson J. R., Gao L., Daves L. K., Tatchell K. (2008). Ypi1, a Positive Regulator of Nuclear Protein Phosphatase Type 1 Activity in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 1032-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A., Tatchell K. (1999). Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Gene. Dev. 13, 517-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A., Tatchell K. (2000). Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 149, 125-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P., Schlapbach R., Aebersold R. (2008). PhosphoPep-a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 26, 1339-1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F., Barral Y. (2009). Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell 16, 493-506 [DOI] [PubMed] [Google Scholar]
- Cid V. J., Adamikova L., Sanchez M., Molina M., Nombela C. (2001). Cell cycle control of septin ring dynamics in the budding yeast. Microbiology 147, 1437-1450 [DOI] [PubMed] [Google Scholar]
- Crespo J. L., Hall M. N. (2002). Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66, 579-591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N. G., Horecka J. L., Sprague G. F., Jr (1993). Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 122, 53-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini D. J., Adams A. E. M., Fares H., De Virgilio C., Valle G., Chuang J. S., Pringle J. R. (1997). A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139, 75-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. (2004). Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305, 393-396 [DOI] [PubMed] [Google Scholar]
- Douglas L. M., Alvarez F. J., McCreary C., Konopka J. B. (2005). Septin function in yeast model systems and pathogenic fungi. Eukaryotic Cell 4, 1503-1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J. M., Smolka M. B., Zhou H., Kolodner R. D. (2006). Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J. Cell Biol. 175, 729-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 32-41 [DOI] [PubMed] [Google Scholar]
- Fares H., Peifer M., Pringle J. R. (1995). Localization and possible functions of Drosophila septins. Mol. Biol. Cell 6, 1843-1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick D. L., Tatchell K. (1996). The REG2 gene of Saccharomyces cerevisiae encodes a type1 protein phosphatase-binding protein that functions with Reg1p and the Snf1p protein kinase to regulate growth. Mol. Cell. Biol. 16, 2922-2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681-689 [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Kozubowski L., Zyla T. R., Lew D. J. (2005). Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 118, 1617-1628 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553 [DOI] [PubMed] [Google Scholar]
- Hall P. A., Russell S. E. (2004). The pathobiology of the septin gene family. J. Pathol. 204, 489-505 [DOI] [PubMed] [Google Scholar]
- Ihara M., Kinoshita A., Yamada S., Tanaka H., Tanigaki A., Kitano A., Goto M., Okubo K., Nishiyama H., Ogawa O., et al. (2005). Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell 8, 343-352 [DOI] [PubMed] [Google Scholar]
- John C. M., Hite R. K., Weirich C. S., Fitzgerald D. J., Jawhari H., Faty M., Schlapfer D., Kroschewski R., Winkler F. K., Walz T., et al. (2007). The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 26, 3296-3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. (1994). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Kinoshita M. (2006). Diversity of septin scaffolds. Curr. Opin. Cell Biol. 18, 54-60 [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Panek H., Rosenthal A., Bloecher A., DeMarini D. J., Tatchell K. (2003). A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 14, 26-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L., Larson J. R., Tatchell K. (2005). Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 3455-3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer B. E., Adang L. A., Macara I. G. (2007). Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell 130, 837-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlenbaumer G., Hannibal M. C., Nelis E., Schirmacher A., Verpoorten N., Meuleman J., Watts G. D., De Vriendt E., Young P., Stogbauer F., et al. (2005). Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat. Genet. 37, 1044-1046 [DOI] [PubMed] [Google Scholar]
- Larson J. R., Bharucha J. P., Ceaser S., Salamon J., Richardson C. J., Rivera S. M., Tatchell K. (2008). Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell 29, 3040-3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Sdicu A. M., Menard P., Shapiro J., Hussein S., Bussey H. (2004). Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167, 35-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J. (2003). The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15, 648-653 [DOI] [PubMed] [Google Scholar]
- Liu H., Wang Y. (2006). The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis. Mol. Biol. Cell 17, 2746-2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Bi E. (2003). Regulation of septin organization and function in yeast. Trends Cell Biol. 13, 403-409 [DOI] [PubMed] [Google Scholar]
- Maniatis Sambrook T. J., Fritsch E. F. (1989). Molecular Cloning: A Laboratory Manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Neufeld T. P., Rubin G. M. (1994). The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77, 371-379 [DOI] [PubMed] [Google Scholar]
- Nguyen T. Q., Sawa H., Okano H., White J. G. (2000). The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J. Cell Sci. 113, 3825-3837 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. (2004). Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15, 4971-4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Klee S. K., Pellman D. (2002). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42-50 [DOI] [PubMed] [Google Scholar]
- Sanz M., Castrejon F., Duran A., Roncero C. (2004). Saccharomyces cerevisiae Bni4p directs the formation of the chitin ring and also participates in the correct assembly of the septum structure. Microbiology 150, 3229-3241 [DOI] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S. B., Gonnet G., Barral Y. (2008). A mechanism for asymmetric segregation of age during yeast budding. Nature 454, 728-734 [DOI] [PubMed] [Google Scholar]
- Sheff M. A., Thorn K. S. (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661-670 [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. (1986). Methods in Yeast Genetics Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kuhlmann D., Macara I. G., Weyand M., Stark H., Wittinghofer A. (2007). Structural insight into filament formation by mammalian septins. Nature 449, 311-315 [DOI] [PubMed] [Google Scholar]
- Smolka M. B., Chen S. H., Maddox P. S., Enserink J. M., Albuquerque C. P., Wei X. X., Desai A., Kolodner R. D., Zhou H. (2006). An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J. Cell Biol. 175, 743-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart J. S., Frederick D. L., Varner C. M., Tatchell K. (1994). The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 14, 896-905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Simonetta A., Batterton M., Kinoshita M., Edbauer D., Sheng M. (2007). Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. 17, 1752-1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P. A., DeRisi J. L., Wilhelm J. E., Vale R. D. (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341-344 [DOI] [PubMed] [Google Scholar]
- Versele M., Thorner J. (2005). Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15, 414-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu A. M., Mitchison T. J. (2006). Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature 443, 466-469 [DOI] [PubMed] [Google Scholar]
- Vrabioiu A. M., Mitchison T. J. (2007). Symmetry of septin hourglass and ring structures. J. Mol. Biol. 372, 37-49 [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Barral Y. (2008). The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell. Biol. 9, 478-489 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906 [DOI] [PubMed] [Google Scholar]
- Xie Y., Vessey J. P., Konecna A., Dahm R., Macchi P., Kiebler M. A. (2007). The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 17, 1746-1751 [DOI] [PubMed] [Google Scholar]
- Zou J., Friesen H., Larson J., Huang D., Cox M., Tatchell K., Andrews B. (2009). Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 3239-3250 [DOI] [PMC free article] [PubMed] [Google Scholar]