Abstract
In vertebrates, overexpression of facioscapulohumeral muscular dystrophy (FSHD) region gene 1 (FRG1) recapitulates the pathophysiology exhibited by FSHD patients, although the role of FRG1 in FSHD remains controversial and no precise function for FRG1 has been described in any organism. To gain insight into the function and potential role of FRG1 in FSHD, we analyzed the highly conserved Caenorhabditis elegans ortholog, frg-1. C. elegans body-wall muscles contain two distinct subcellular pools of FRG-1: nuclear FRG-1, concentrated in the nucleoli; and cytoplasmic FRG-1, associated with the Z-disk and costamere-like structures known as dense bodies. Functionally, we demonstrate that FRG-1 is an F-actin-bundling protein, consistent with its localization to dense bodies; this activity is conserved in human FRG1. This is particularly intriguing because it places FRG-1 along side the list of dense-body components whose vertebrate orthologs are involved in the myriad myopathies associated with disrupted costameres and Z-disks. Interestingly, overexpressed FRG-1 preferentially accumulates in the nucleus and, when overexpressed specifically from the frg-1 promoter, disrupts the adult ventral muscle structure and organization. Together, these data further support a role for FRG1 overexpression in FSHD pathophysiology and reveal the previously unsuspected direct involvement of FRG-1 in muscle structure and integrity.
Keywords: FSHD, Dense body, Muscle, FRG1, Muscular dystrophy, Actin
Introduction
FSHD is an autosomal dominant myopathy typically characterized by progressive atrophy of muscles in the face, upper arms and shoulder girdle, with progression to the abdominal, pelvic girdle and foot extensor muscles in more severe cases (Pandya et al., 2008; van der Maarel and Frants, 2005; van der Maarel et al., 2007). The genetic lesions responsible for more than 95% of FSHD patients are deletions within DNA repeats on chromosome 4q35, resulting in epigenetic changes and the disrupted regulation of gene expression (Lunt et al., 1995; Tawil, 2008; van Deutekom et al., 1993; Wijmenga et al., 1992; Zeng et al., 2009). FRG1 is a candidate gene for causing the FSHD pathophysiology, based initially on its proximity to the FSHD genetic lesion (van Deutekom et al., 1996). However, multiple expression analyses have produced inconclusive, and often contradictory, results with respect to altered mRNA expression levels in FSHD-affected muscle (Arashiro et al., 2009; Gabellini et al., 2006; Jiang et al., 2003; Osborne et al., 2007; Winokur et al., 2003). Circumventing the inconsistencies of the RNA expression data, both mice and Xenopus transgenically overexpressing FRG1 in skeletal muscle exhibit FSHD-like phenotypes (Gabellini et al., 2006; Hanel et al., 2009; Wuebbles et al., 2009). In addition, ubiquitous FRG1 overexpression not only disrupts normal vertebrate muscle development, but also leads to a tortuous vasculature similar to the retinal vasculopathy exhibited by more than 50% of FSHD patients (Gieron et al., 1985; Wuebbles et al., 2009). Thus, overexpression of FRG1 alone has the capacity to recapitulate the major clinical pathophysiology seen in FSHD; the molecular mechanism underlying how increased FRG1 expression levels could promote this is not known.
FRG1 is expressed in and required for normal development of the vertebrate musculature and vasculature (Hanel et al., 2009; Wuebbles et al., 2009). Although the precise function of FRG1 is not known, overexpression studies in human cell culture have characterized it as a nuclear protein implicated in RNA biogenesis (Rappsilber et al., 2002; van Koningsbruggen et al., 2004; van Koningsbruggen et al., 2007). Supporting a role in RNA processing, FRG1-overexpressing mice and cell lines exhibit mis-splicing of some muscle-related genes, similar to that seen in some muscle cells derived from FSHD patients (Davidovic et al., 2008; Gabellini et al., 2006; van Koningsbruggen et al., 2007). However, analysis of the inferred FRG1 protein sequence indicates that, in addition to two nuclear localization signals (NLSs), FRG1 also contains a lipocalin motif, suggesting potential involvement in the transport of hydrophobic particles (Flower, 1996), and a predicted fascin domain, which is found in various proteins that bundle and organize actin filaments into structures that stabilize cellular processes such as cell mobility and cell adhesion (Edwards and Bryan, 1995; Kureishy et al., 2002). This suggests that FRG1 might have functions other than or in addition to RNA biogenesis.
To gain insight into how FRG1 overexpression could lead to FSHD, we have sought to first understand the normal function of FRG1 during development. Because FRG1 is very highly conserved from invertebrates to vertebrates (Fig. 1) (Grewal et al., 1998) and invertebrate striated muscle, including C. elegans body-wall muscle, resembles vertebrate skeletal muscle (Lecroisey et al., 2007; Moerman and Williams, 2006), investigating the function and potential role of FRG1 in mediating muscular dystrophy is highly amenable to analysis in invertebrate model organisms. Here, we analyzed the C. elegans ortholog of FRG1, frg-1 (ZK1010.3), with respect to biological activity, expression and subcellular distribution in body-wall muscle and the effects of FRG-1 overexpression on the adult body-wall musculature.
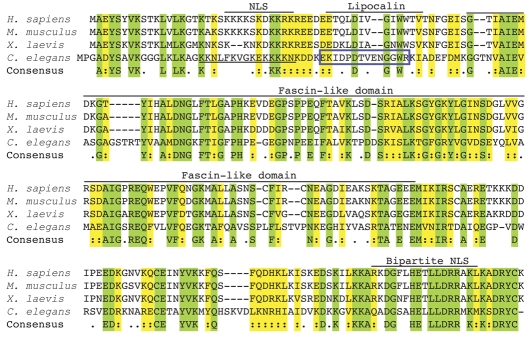
Fig. 1.
FRG1 is highly conserved evolutionarily. The deduced amino acid sequence alignment of FRG1 homologs using ClustalW shows that human FRG1 is 97% identical to the mouse protein, 81% identical to Xenopus and 42% identical to C. elegans. Green residues are conserved in all four species; yellow residues are similar in all four species; unshaded residues are not conserved. Putative domains characterized for human FRG1 are shown above. The predicted C. elegans NLS is underlined. The peptide used to generate the FRG-1 antibody is in the blue box.
Results
FRG-1 is a conserved actin-bundling protein
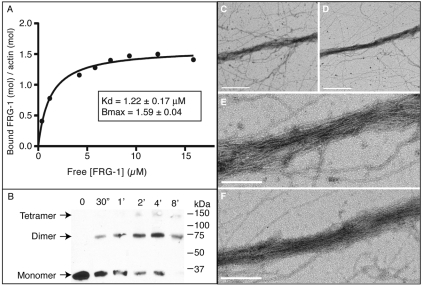
FRG1 is highly evolutionarily conserved among vertebrates and invertebrates (Grewal et al., 1998), with human FRG1 sharing 42% identity (58% similarity) with C. elegans FRG-1 over the full length of the protein (Fig. 1); however, its function has remained elusive. Analysis of the predicted amino acid sequence for the presence of conserved domains (Pfam 23.0, http://pfam.sanger.ac.uk/) revealed that FRG-1, as with all other FRG1 orthologs, contains a single fascin-like domain (PF06220), indicative of an actin-binding protein (Adams, 2004; Edwards and Bryan, 1995). To determine whether FRG-1 could, in fact, bind F-actin, high-speed actin co-sedimentation assays were performed (Fig. 2A). Recombinant FRG-1 (supplementary material Fig. S1A) was incubated with F-actin for 2 hours and then centrifuged at 100,000×g to pellet all F-actin and F-actin-bound FRG-1. The amount of FRG-1 in the pellets (F-actin–FRG-1) and supernatants (free FRG-1) was determined by gel electrophoresis. Saturable F-actin binding was reproducibly achieved by the co-sedimentation of increasing concentrations of recombinant FRG-1 with a fixed amount of F-actin. Nonlinear regression analysis of the binding data from four independent experiments yielded an average apparent dissociation constant (Kd) of 1.22±0.17 μM, with saturation (Bmax) at 1.59±0.04 mol/mol (corresponding to two FRG-1 per actin monomer). The Hill slope is near h=1 (0.87±0.16), indicating FRG-1 does not bind with positive cooperativity. Overall, we conclude that FRG-1 is an actin-binding protein.
Fig. 2.
FRG-1 and FRG1 proteins bind and bundle F-actin. (A) High-speed F-actin cosedimentation assays were performed at varying concentrations of FRG-1 with a constant concentration of F-actin. (B) Western blot analysis of a representative glutaraldehyde crosslinking time course using recombinant FRG-1. Monomer (30 kDa), dimer (60 kDa) and tetramer (120 kDa) species were present. (C-F) Electron micrographs of actin bundles formed with recombinant (C,E) C. elegans FRG-1 or (D,F) human FRG1. (C,D) Scale bars: 500 nm. (E,F) Scale bars: 200 nm.
FRG-1 contains a single fascin-like domain. Fascins are a conserved family of actin-bundling proteins that stabilize actin filaments for cellular processes. Each vertebrate fascin protein contains four fascin domains, two of which contain single actin-binding sites (Edwards and Bryan, 1995; Kureishy et al., 2002). In order to bundle F-actin, proteins must either possess multiple actin-binding domains or multimerize to generate the necessary multiple actin-binding sites. To determine whether FRG-1 functions similarly to fascin as an actin-bundling protein, we first performed a glutaraldehyde crosslinking experiment on purified recombinant FRG-1 and found that FRG-1 forms homodimers and homotetramers in solution (Fig. 2B), thus providing the multiple actin-binding sites necessary for F-actin bundling. To establish whether FRG-1 is an actin-bundling protein, in vitro F-actin-bundling assays were performed. It has been previously shown that F-actin does not sediment under low-speed centrifugation (<20,000 g) conditions unless complexed into supermolecular aggregates by a crosslinking protein (Meyer and Aebi, 1990). Recombinant FRG-1 was incubated with polymerized F-actin for 1 hour, then centrifuged at 12,000 g and the amount of FRG-1 in the pellets and supernatants was similarly quantified by gel electrophoresis (supplementary material Fig. S1B). Saturable F-actin bundling was achieved by increasing concentrations of recombinant FRG-1 with a fixed amount of F-actin. To confirm that FRG-1 was bundling F-actin and not merely forming disorganized aggregates, actin-bundling reactions were visualized by negative staining through electron microscopy. The micrographs clearly show the formation of F-actin bundles stimulated by C. elegans FRG-1 (Fig. 2C,E), as well as human FRG1 (Fig. 2D,F). We conclude that C. elegans FRG-1 has bona fide F-actin-bundling activity and that this is evolutionarily conserved in human FRG1.
FRG-1 is both a nuclear protein and a cytoplasmic protein associated with the dense body
The vertebrate FRG1 protein has been characterized as a nuclear protein in cell culture (van Koningsbruggen et al., 2004). An analysis of the predicted C. elegans FRG-1 amino acid sequence (Prosite release 20.49, http://ca.expasy.org/prosite/) identified a bipartite NLS in the N terminus (Fig. 1), in a similar location to one of the vertebrate FRG1 NLSs. To determine the distribution and subcellular localization of endogenous FRG-1 during muscle development, an antibody against FRG-1 (Fig. 1; supplementary material Fig. S2) was generated and used for immunostaining. During C. elegans embryogenesis, FRG-1 was in fact primarily nuclear; however, diffuse cytoplasmic staining was also evident (Fig. 3A-C; supplementary material Fig. S2A-C). In body-wall muscle of larval-stage animals, cytoplasmic FRG-1 began to form foci and showed a surprisingly striated and punctate immunostaining (Fig. 3D-F) suggestive of the dense body. Similarly, in adult animals, cytoplasmic FRG-1 immunostaining retained its striking dense-body-like pattern (Fig. 3M,N, arrowheads). C. elegans dense bodies are homologous to vertebrate focal adhesions. They physically anchor the actin cytoskeleton of the muscle cell to the basal lamina and hypodermis, and orient in long rows along the muscle, appearing at regular intervals alternating with the M-lines (Moerman and Williams, 2006). Because a cytoplasmic pool of FRG1 had never previously been described in any organism, much less at the muscle-attachment sites, the C. elegans genome was analyzed for FRG-1-related proteins that could potentially be misleading. There are no other FRG-1-like proteins known or predicted in the C. elegans genome. All FRG1 proteins contain a fascin-like domain, which places FRG1 orthologs in the fascin family of cytoplasmic actin-bundling proteins. Interestingly, analysis of the C. elegans genome for fascin-domain-containing proteins revealed that FRG-1 is the only such protein in C. elegans (supplementary material Fig. S3) and is most closely related to the fascin-like domain of vertebrate FRG-1, suggesting that FRG-1 might be the ancestral fascin domain family protein. Together with our antibody characterization (supplementary material Fig. S2), we conclude that this cytoplasmic immunostaining represents FRG-1.
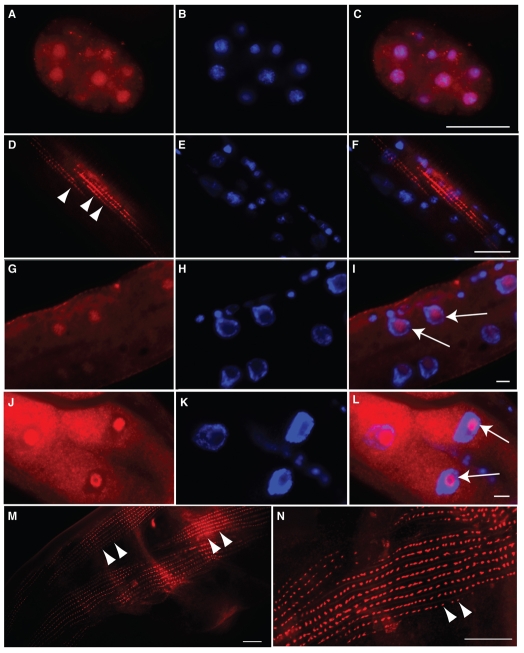
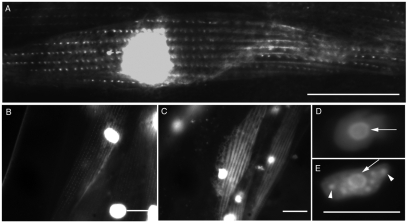
Fig. 3.
Immunostaining identifies endogenous FRG-1 as both a nuclear and a cytoplasmic protein displaying a pattern indicative of body-wall muscle dense bodies. N2 worms immunostained for FRG-1 (A,D,G,J,M,N) in red and co-stained with DAPI (B,E,H,K) in blue, with images merged (C,F,I,L). In embryo (A-C), larval (D-I) and adult (J-N) stages, FRG-1 was localized in the DAPI-poor regions of the nuclei (I and L, arrows). Cytoplasmic staining for FRG-1 resembled that of body-wall muscle cell dense bodies (D,M,N, arrowheads). Scale bars: 10 μm.
Determining whether larvae and adults retained a nuclear FRG-1 pool in body-wall muscle required the immunostaining procedure to be altered as described for other nuclear proteins at these stages. Unfortunately, this procedure destroyed the cytoplasmic structures, but successfully revealed the presence of FRG-1 in body-wall muscle nuclei in larva (Fig. 3G-I, arrows) and adults (Fig. 3J-L, arrows; supplementary material Fig. S2D-F), localized primarily to the DAPI-poor regions indicative of nucleoli (Shaw and Doonan, 2005). Thus, endogenous FRG-1 in C. elegans is both a nuclear protein and a cytoplasmic protein throughout development, appearing to concentrate in the nucleoli and dense bodies, respectively, of body-wall muscle cells.
To confirm the association of FRG-1 with dense bodies, coimmunostaining for PAT-3 (β-integrin), a transmembrane extracellular matrix (ECM) receptor that is concentrated in all dense bodies and M-lines (Gettner et al., 1995), was carried out (Fig. 4). Both FRG-1 and PAT-3 immunostaining produced identical but only partially overlapping and generally adjacent punctate patterns (Fig. 4A-C), indicating that FRG-1 is a component of all of the body-wall muscle dense bodies. Using confocal microscopy and focusing on only the dense bodies positioned at the edge of the organism allowed visualization of proteins positioned along the dense-body structure as it protruded into the muscle cytoplasm (Fig. 4D-F). Clearly, FRG-1 does not overlap with PAT-3 at the base of the dense body; instead FRG-1 immunostaining appeared adjacent to PAT-3 along the cytoplasmic finger-like projection of the dense body, where actin thin filaments attach (Fig. 4M). Considering that FRG-1 is an F-actin-bundling protein, it was not surprising that FRG-1 was absent from M-lines, structures that anchor myosin thick filaments (Fig. 4B,C, arrows) (Moerman and Williams, 2006).
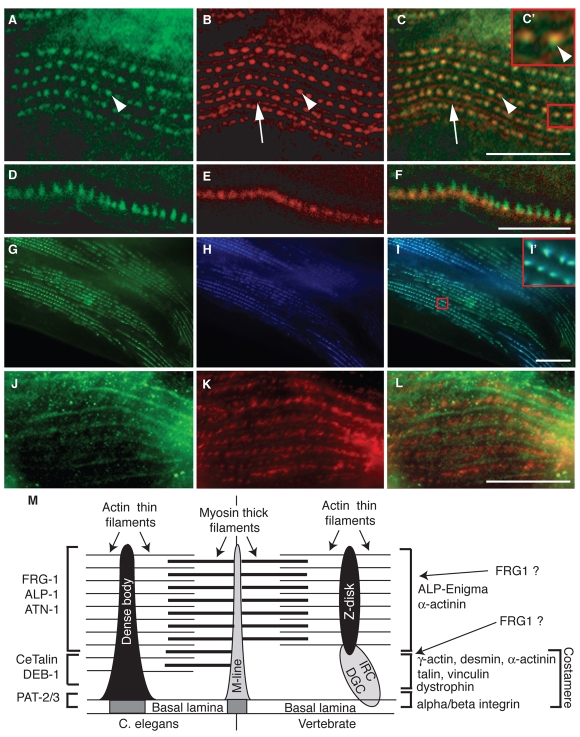
Fig. 4.
FRG-1 associates with body-wall muscle cell dense bodies, adjacent to PAT-3, and colocalizes with ALP-1. (A-F) Adult animals co-immunostained for FRG-1 (A,D, green) and PAT-3 (B,E, red) show FRG-1 present at all body-wall muscle dense bodies (C, arrowheads), whereas it is absent from M-lines (C, arrows). FRG-1 and PAT-3 show adjacent, partially overlapping signals at dense bodies when merged (C,C′, arrowheads). (D-F) Confocal images of dense body immunostaining. (G-I) Double transgenic animals for Pmyo-3::frg-1::yfp and Pmyo-3::alp-1α::cfp show that expression patterns for FRG-1 (G) and ALP-1 (H) are precisely colocalized in the image merge (I and I′). (J-L) Mutant C. elegans strain containing a homozygous partial deletion for atn-1 was co-immunostained for FRG-1 (J, green) and PAT-3 (K, red), showing disrupted FRG-1 localization (L). (M) Model depicting FRG-1 localization in dense bodies with the analogous vertebrate components (based on Moerman and Williams, 2006; Fluck et al., 2002). Scale bars: 20 μm. DGC, dystrophin-glycoprotein complex; IRC, integrin receptor complex.
To further confirm the immunological determination that endogenous FRG-1 is associated with the dense body and further characterize its localization, transgenic strains coexpressing fluorescently tagged FRG-1 (Pmyo-3::frg-1::yfp) and α-actinin-associated LIM protein 1α (ALP-1α) (Pmyo-3::alp-1α::cfp) from the strong muscle-specific myosin heavy chain (myo-3) promoter were generated (Fig. 4G-I). ALP-1α is a conserved dense-body protein that interacts with α-actinin (ATN-1), a conserved actin-bundling protein similarly localized to the dense bodies and distal to the sarcolemma (Barstead et al., 1991; Han and Beckerle, 2009; Xia et al., 1997). Pmyo-3::frg-1::yfp adult animals showed a cytoplasmic dense-body-like pattern (Fig. 4G, green), confirming the immunocharacterization of endogenous FRG-1 (Fig. 3M,N). However, unlike the case with PAT-3 (Fig. 4C,F), the FRG-1::YFP fusion protein precisely colocalized with ALP-1α::CFP in body-wall muscle dense bodies (Fig. 4G-I,I′), supporting the localization of FRG-1 proteins to the distal part of the dense bodies (Fig. 4M). Finally, we evaluated FRG-1 localization in the atn-1(ok84) mutant (Fig. 4J-L), which contains a 1.1 kb deletion within atn-1 and is predicted to be a null mutant (Han and Beckerle, 2009). In atn-1(ok84) mutants, the dense bodies are less well defined, but retain a periodic punctate appearance following PAT-3 staining (Fig. 4K). In contrast, the FRG-1 immunostaining pattern was no longer restricted to the dense bodies and appeared filamentous on M-lines (Fig. 4L compared with Fig. 4C), indicating that ATN-1 is required for proper localization of FRG-1 to the dense bodies and that integrins do not directly anchor FRG-1 at dense bodies. These data also place FRG-1 distal to PAT-3 and the basal lamina, where ALP-1α and ATN-1 accumulate; actin thin filaments attach to these dense-body projections (Fig. 4M) (Han and Beckerle, 2009), further supporting FRG-1 as a component of the actin-associated machinery of the dense body.
Overexpressed FRG-1 shows an altered subcellular distribution in body-wall muscle
In Pmyo-3::frg-1::gfp adult worms, FRG-1::GFP strongly localized to the nucleus and, with a much lower intensity, the dense bodies (Fig. 5A), suggesting that overexpressed FRG-1 preferentially accumulates in the nucleus. To investigate a range of FRG-1 overexpression (Fig. 5B-E) in all FRG-1-expressing cell types, including body-wall muscle, transgenic lines expressing FRG-1::GFP under the control of the C. elegans frg-1 promoter (Pfrg-1::frg-1::gfp) were generated. Consistently, all animals expressing FRG-1::GFP showed very strong nuclear localization regardless of overall expression levels (Fig. 5A-C). However, different expression levels of FRG-1 affected the cytoplasmic and nuclear distributions. When the expression level of FRG-1 was visibly low, cytoplasmic FRG-1 localized strictly to the dense bodies (Fig. 5B); however, when the expression level of FRG-1 was notably higher, FRG-1 appeared to spread along the actin filaments, well beyond its normal distribution pattern (Fig. 5C compared with Fig. 5A,B, Fig. 4A,G and Fig. 3J). This was in contrast to the Pmyo-3::frg-1::gfp lines, which did not show the aberrant coating of the F-actin cytoskeleton, indicating that FRG-1 expression from the more ubiquitously expressed frg-1 promoter was somehow different from FRG-1 expression from the muscle-specific myo-3 promoter. Similarly, expression levels affected the distribution of nuclear FRG-1. Cells overexpressing low levels of FRG-1 contained the majority of nuclear FRG-1 in the granular component of nucleoli (Fig. 5D). Cells overexpressing high levels of FRG-1 contained additional non-nucleolar aggregates of FRG-1 (Fig. 5E) that were not detected in endogenous nuclear FRG-1 staining. Therefore, the normal cytoplasmic and nuclear distributions are both affected by FRG-1 expression levels from the frg-1 promoter.
Fig. 5.
Overexpressed FRG-1 preferentially localizes to the nucleus and shows neomorphic localization. (A) Pmyo-3::frg-1::gfp transgenic animals express FRG-1::GFP strongly in body-wall muscle nuclei and weakly in the dense bodies. Animals transgenic for Pfrg-1::frg-1::gfp show visually low (B,D) and high (C,E) levels of FRG-1::GFP expression in body-wall muscle cells (B and C) and nuclei (D and E). High levels of FRG-1 expression lead to coating of the F-actin filaments (compare C, FRG-1-stained striations with B, punctate dense-body staining) and nuclear aggregate formation (E, arrowheads, compared with D) in addition to normal nucleolar staining (arrows). Scale bars: 10 μm.
Overexpression of FRG-1 from the frg-1 promoter disrupts the muscular organization of ventral body-wall muscle
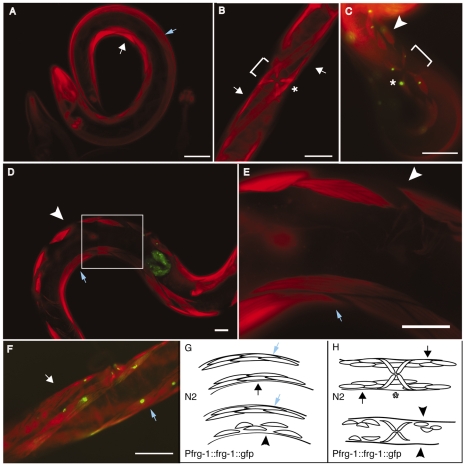
FSHD pathophysiology might result from increased FRG1 gene expression; however, transgenic C. elegans overexpressing FRG-1 from the muscle-specific myo-3 promoter showed no obvious movement or muscle phenotype (Fig. 6F). It should be noted that, in addition to skeletal muscle, FRG1 expression has been detected in all human tissues tested (Bodega et al., 2009; van Deutekom et al., 1996), raising the possibility that FRG1 expression in non-muscle cells might be important for generating FSHD pathology. Similarly, C. elegans Serial Analysis of Gene Expression (SAGE) data (http://elegans.bcgsc.ca/home/sage.html) confirmed that frg-1 is expressed not only in muscle cells, but also in hypodermal cells, pharynx cells, pharyngeal marginal cells, pharyngeal gland cells and neurons. In addition, in situ hybridization data (Yuji Kohara: The nematode expression pattern database http://nematode.lab.nig.ac.jp/db2/ShowGeneInfo.php?celk=CELK04952) indicates that expression of frg-1 starts as early as the two-cell stage and decreases during larval development, but increases in adults. Thus, because FRG-1 expressed in non-muscle cells and from early points in development might impact the integrity of the adult musculature, the Pfrg-1::frg-1::gfp transgenic adult animals were assayed for muscle phenotypes (Fig. 6). During muscle development, myoblasts migrate to four quadrants, two dorsal and two ventral, and flatten both basally against the hypodermis and laterally against neighboring muscle cells, forming four continuous double rows of muscle cells along much of the length of the animal (Hresko et al., 1994; Moerman and Williams, 2006). Phalloidin staining revealed that ~25% of Pfrg-1::frg-1::gfp adult transgenic animals showed disruption of the body-wall musculature, manifested as smaller, misaligned, missing and disconnected muscle cells primarily at the muscle-muscle junctions running down the center of the ventral muscle quadrants (Fig. 6D,E compared with 6A,B). At the vulva, phalloidin staining of transgenic adult animals (Fig. 6C) shows that ventral body-wall muscle cells are discontinuous underneath the vulval muscles (bracket) and body-wall muscle cells are smaller compared with control animals (Fig. 6B). Interestingly, the muscle defects were exclusively on the ventral side of the animal and never on the dorsal side, despite FRG-1 expression being confirmed throughout the dorsal and ventral musculature by GFP visualization. Overall, FRG-1 overexpression, specifically in the spatiotemporal pattern dictated by the frg-1 promoter and not in muscle cells alone, disrupts the integrity of the adult ventral body-wall musculature in C. elegans.
Fig. 6.
Overexpression of FRG-1 specifically from the frg-1 promoter disrupts the ventral muscle-muscle lateral junctions. Phalloidin staining of (A) wild-type N2 adults and (B) pRF4(rol-6) transgenic animals shows the normal organization and structure of the ventral (white arrows) and dorsal (blue arrows) body-wall and vulval (asterisk) muscles. Transgenic animals overexpressing FRG-1::GFP from the frg-1 promoter visualized by phalloidin and GFP merge (C-E) show specific disruption of ventral muscle-muscle lateral junctions and absence of some muscle cells (white arrowheads, bracket at vulva), whereas the dorsal musculature (blue arrows) remains intact. The boxed area in D is shown magnified 3× in E. (F) Transgenic animals overexpressing FRG-1::GFP from the myo-3 promoter showed normal muscle structure, as visualized by phalloidin staining. Diagrams depicting a lateral view (G) and a ventral view (H) of the dorsal (blue arrows) and ventral (black arrows, normal; black arrowheads, disrupted) musculature of normal (top) and transgenic (bottom) animals. Scale bars: 50 μm.
Discussion
A top priority in FSHD research is determining which gene (or genes) is misexpressed as a result of the 4q35 lesion and how its altered expression results in FSHD pathogenesis. Among the candidate genes, the role of FRG1 in FSHD has been controversial and understanding impeded by a lack of knowledge of the biological function of FRG1. Here, we have identified a conserved activity for FRG1, F-actin bundling. However, FRG1 in vertebrates had been characterized as a nuclear protein involved in some aspect of RNA biogenesis (based on studies in cell culture), whereas overexpressed FRG1 strongly localized to the nucleus of these cells (Rappsilber et al., 2002; van Koningsbruggen et al., 2004; van Koningsbruggen et al., 2007). Thus, the biological implications of identifying F-actin-bundling activity for a nuclear protein such as FRG1 were initially not clear because of the controversial nature of both the form and function of nuclear actin (Pederson and Aebi, 2005). Characterizing endogenously expressed C. elegans FRG-1 as both a nuclear protein accumulating in the nucleolus, consistent with a role in RNA biogenesis, and associated with the dense body, a plasma-membrane-bound muscle adhesion structure that attaches to the actin cytoskeleton (Fig. 3M), suggests an explanation; FRG-1 is a multifunctional protein residing in distinct subcellular niches, primarily the nucleolus and the dense body. During C. elegans larval stages, there is an increase in the number of body-wall muscle sarcomeres and filaments and the dense bodies begin making projections into the cytoplasm, accumulating proteins such as the F-actin-anchoring proteins ATN-1 and CeTalin (Francis and Waterston, 1985; Moerman and Williams, 2006). The appearance of dense-body FRG-1 immunostaining in the cytoplasm similarly coincides with this dense-body proliferation, maturation and actin-associated protein accumulation, and supports a possible role for FRG-1 in stabilizing F-actin at the dense body.
C. elegans dense bodies primarily function in muscles to transduce mechanical force from the muscle by attaching the sarcomeres to the muscle membrane and underlying ECM through PAT-2 (α-integrin) and PAT-3 heterodimers (Gettner et al., 1995; Williams and Waterston, 1994), essentially forming the mechanical anchor between sarcomeric actin and the ECM. Therefore, the dense body is a structure that performs functions analogous to the vertebrate Z-disk and costameres together (Lecroisey et al., 2007; Moerman and Williams, 2006; Pardo et al., 1983; Street, 1983). Identifying the association of FRG-1 with dense bodies is a compelling result, because FSHD pathophysiology includes reorganization of the sarcolemma (Reed et al., 2006). In addition, numerous other muscular dystrophies, including Duchenne (Hoffman et al., 1987; Monaco et al., 1986), Becker (Kunkel et al., 1986), certain limb-girdle dystrophies (LGMD2B, 2G, 2J) (Guglieri et al., 2008) and some rare congenital muscular dystrophies (Hayashi et al., 1998), as well as multiple other Z-disk diseases (Selcen and Carpen, 2008), result from mutations affecting vertebrate orthologs of dense-body components. Thus, the localization of FRG-1 to muscle-attachment sites raises new possibilities regarding the role of FRG-1 in muscle development and disease beyond RNA biogenesis. FRG-1 might perform two independent functions: stabilization of F-actin through actin bundling at the dense body and RNA processing in the nucleus. Alternatively, the two pools of FRG-1 might be functionally linked. For example, dense bodies and focal adhesions have an additional role, serving as platforms for transducing biochemical and mechanical signals from the ECM and surrounding tissues to the nucleus (Lecroisey et al., 2007; Romer et al., 2006); FRG-1 would be particularly well suited to functioning in this capacity. Interestingly, when FRG-1 was overexpressed in the Pmyo-3::frg-1::gfp and Pfrg-1::frg-1::gfp transgenic lines, nuclear FRG-1 was extremely intense when compared with dense-body FRG-1. It might be that overexpressed FRG-1 preferentially accumulates in the nucleus, because it was unable to be efficiently retained in the cytoplasm. Even small increases in the levels of FRG-1 could be detrimental, strictly by altering its subcellular distribution, thus affecting its nuclear protein levels and in turn its nuclear functions, such as the regulation of the alternative splicing of genes required for muscle development.
Translational impact relating to FSHD pathophysiology
Here, we have found conserved activity for FRG1 (actin bundling) and identified FRG-1 as associated with the C. elegans body-wall muscle dense body, a highly conserved structure required for muscle function. Components of the dense body have vertebrate orthologs that are intimately involved in numerous myopathies, rendering FRG1 the only FSHD candidate gene with a direct link to muscle structure. Recently, human primary myoblasts were found to have increased levels of FRG1 during myogenic differentiation; however, FSHD-patient-derived primary myoblasts showed aberrantly higher levels of FRG1 expression during differentiation (Bodega et al., 2009). In C. elegans, increased levels of FRG-1 during development affect its subcellular distribution, becoming neomorphic (spread along actin filaments), and disrupt the lateral muscle-muscle attachments of the ventral muscle quadrants. Considering the high degree of conservation between dense-body components and their vertebrate counterparts (Moerman and Williams, 2006), the evolutionarily conserved muscle phenotype of overexpressed FRG1 orthologs in Xenopus, M. musculus and now C. elegans (Gabellini et al., 2006; Hanel et al., 2009), and the conserved actin-bundling activity of the human FRG1 protein, we propose that FRG1 is likely to localize and function similarly in human muscle (Fig. 4M). These data support a model whereby increases in FRG1 protein levels within the muscle and associated tissues contribute to FSHD pathology, potentially mediated by changes in the subcellular localization of FRG1 that result in altered increased or aberrant FRG1 activity.
Materials and Methods
C. elegans strains and genetics
Standard methods were used for culturing C. elegans (Brenner, 1974). Bristol N2 was used as the wild type. Strain RB1812:atn-1(ok84) was provided by the C. elegans Genetics Center, funded by the NIH National Center for Research Resources (NCRR), at the University of Minnesota. Transgenic lines were generated by standard microinjection techniques, using pRF4 (rol-6) as a marker when indicated, and the plasmids described below.
Molecular biology
Vectors were from the Fire Lab kit (Addgene, Cambridge, MA). Reverse transcriptase (RT)-PCR products were amplified with SuperScript III-Platinum Taq one-step RT-PCR kit (Invitrogen Corp., Carlsbad, CA) from N2 total RNA isolated by the Trizol (Invitrogen Corp., Carlsbad, CA) method as per the manufacturer's instructions, cloned into pGEM-T Easy vector (Promega Corp., Madison, WI) and sequenced. Primers are listed in supplementary material Table S1. To generate Pmyo-3::frg-1::gfp, the frg-1 cDNA fragment was amplified by RT-PCR (primers #1 and #2) and subcloned into vector pPD118.20 by NotI and KpnI digestion. To generate Pfrg-1::frg-1::gfp, 600 bp upstream of the frg-1 transcription start site was PCR amplified (primers #3 and #4) from N2 genomic DNA, and cloned into plasmid Pmyo-3::frg-1::gfp by PstI and NotI digestion. To generate Pmyo-3::alp-1α::cfp, the alp-1α cDNA was amplified by RT-PCR (primers #10 and #11), and subcloned into pPD136.61 digested with BamHI and KpnI. The plasmid Pmyo-3::frg-1::yfp was similarly generated using vector pPD136.64 and the frg-1 cDNA.
Antibodies
Rabbit polyclonal CeFRG-1 antibody (GenScript USA Inc., Piscataway, NJ) was raised against the peptide EKIDPDTVENGGWRC conjugated to keyhole limpet hemocyanin (KLH), affinity purified against a GST-EKIDPDTVENGGWR protein generated in Escherichia coli, and then negatively absorbed against recombinant sepharose-bound GST. The MH25 monoclonal antibody against PAT-3 (Francis and Waterston, 1985), developed by R. H. Waterston, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained at the University of Iowa Department of Biological Sciences, Iowa City, IA, USA.
Phalloidin staining
Worms were fixed in 3.7% formaldehyde solution in PBS at room temperature (RT) for 10 minutes, washed three times with PBS, soaked in 100% acetone at −20°C for 5 minutes and then washed three times with PBS. Rhodamine phalloidin (Invitrogen Corp., Carlsbad, CA) was added (5 Units/ml) for 30 minutes at RT, then washed with PBS and mounted for microscopy.
Immunostaining
For embryo staining, embryos were collected by bleaching, washed four times with M9 buffer, fixed (3.7% formaldehyde, 0.1 M NaPhosphate buffer pH 7.0, 0.1 mM EDTA) for 10 minutes at RT, washed two times with PBS pH 7.0, transferred to PBST (PBS pH 7.0 plus 0.5% Tween-20), incubated for 5 minutes at RT and pelleted. FRG-1 antibody (1:500 in 70% PBST and 30% normal goat serum) was added and incubated overnight at 4°C with rocking. After four PBST washes of 5 minutes each at RT, secondary antibody (AlexaFluor488 goat anti-rabbit IgG; Invitrogen Corp., Carlsbad, CA) was diluted 1:1000 and incubated with embryos for at least 1 hour at RT. Immunostained embryos were washed three times with PBST and mounted for microscopy.
For immunostaining of adult animals (Finney and Ruvkun, 1990), worms were suspended in a 4% paraformaldehyde, quick frozen on dry ice, thawed and incubated on ice for 1 hour. Worms were washed three times in 1% Triton X-100, 100 mM Tris (pH 7.5) and incubated in 1% Triton X-100, 100 mM Tris (pH 7.5) and 1% β-mercaptoethanol at 37°C for 2 hours for cytoplasmic or 4 hours for nuclear staining. After three washes in 10 mM NaBO3 (pH 9.2), the worms were incubated in 10 mM NaBO3 and 0.3% H2O2 for 1 hour for cytoplasmic or 3 hours for nuclear staining, washed three times with 10 mM NaBO3 (pH 9.2) and stored for further processing in Ab-A buffer (1× PBS, 0.1% Triton X-100, 1% BSA, 0.05% NaN3). FRG-1 antibody (1:500) or MH25 monoclonal antibody (1:10) in Ab-A buffer was added and incubated at 4°C overnight, then washed three times in Ab-B buffer (1× PBS, 0.1% Triton X-100, 0.1% BSA, 0.05% NaN3) prior to incubation with secondary antibody (AlexaFluor488 goat anti-rabbit IgG, AlexaFluor594 goat anti-mouse IgG; Invitrogen Corp., Carlsbad, CA), as above. Immunostained animals were washed three times with Ab-B buffer, pelleted as above and mounted for microscopy.
Fluorescence microscopy
C. elegans were examined by fluorescence microscopy using an Olympus BX60 microscope equipped with a 100× NA 1.35 oil immersion lens. Pictures were acquired with a SpotRT monochrome model 2.1.1 camera and Spot Advanced software (Diagnostic Instruments, Sterling Heights, MI). Images were processed using Adobe Photoshop CS2, ImageJ to adjust brightness, contrast, size and merge or split channel.
Actin-binding and -bundling assays
To generate recombinant protein, cDNAs for human FRG1 and C. elegans FRG-1 were PCR amplified (primers #8 and #9, and #6 and #7, respectively, supplementary material Table S1), subcloned between the NdeI and XhoI restriction sites of pET-23b vector (Novagen, Gibbstown, NJ), transformed into E. coli BL21(DE3) and induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG). The protein was purified using TALON resin (Clontech, Mountain View, CA) as per the manufacturer's instructions. Proteins were bound to MonoS resin using an AKTA-FPLC (GE Healthcare, Piscataway, NJ) for chromatography, and step eluted between 400 and 600 mM NaCl in 20 mM Tris pH 8.0 and 10% glycerol. Actin was purified from rabbit skeletal muscle as described (Pardee and Spudich, 1982).
Actin cosedimentation assays were carried out as described with minor modifications (Ono et al., 1997). For high-speed sedimentation assays, increasing amounts of recombinant FRG-1 (3-30 μM as monomer) were incubated with G-actin (final concentration 6 μM) in a total volume of 50 μl at RT for 2 hours. The reactions were subjected to centrifugation at 100,000×g for 20 minutes at RT. The amounts of FRG-1 and F-actin in the supernatants and resuspended pellets were determined by densitometry from Coomassie-Blue-stained 10% SDS-PAGE. The intensities of the scanned bands were quantified by volume integration after local background subtraction using Bio-Rad Quantity One software (Bio-Rad Laboratories). Subtracting the percentage of FRG-1 pelleted without actin normalized the pelleted FRG-1. The binding data from four independent experiments were analyzed by fitting to the Michaelis-Menton equation, Y=Bmax*X/(Kd+X), using the nonlinear regression function of Prism 5 (GraphPad Software), where Y is pelleted FRG-1 per F-actin (mol/mol) and X is the supernatant FRG-1 concentration (micromolar) of the samples. For low-speed cosedimentation assays, increasing amounts of recombinant FRG-1 (1 to 9 μM as monomer) were incubated with G-actin (final concentration 11.4 μM) in a total volume of 60 μl at RT for 1 hour. The reactions were subjected to centrifugation at 12,000×g for 10 minutes at RT and quantified as described for the high-speed assays. The binding data from three independent experiments were analyzed by fitting to the ‘one site-specific binding with Hill slope’ equation, Y=Bmax*X^h/(K ^dh+X^h), using the nonlinear regression function of Prism 5.
For electron microscopy, G-actin (1 μM final) was incubated with 1 μM of recombinant human FRG1 or C. elegans FRG-1 in 10 mM Hepes pH 7.8, 50 mM KCl, 2 mM MgCl2 and 2 mM EGTA for 30 minutes at 4°C. The reactions were spotted onto carbon-coated grids, glow discharged, washed and stained in 2% uranyl acetate for electron microscopy using a JOEL 2100 transmission electron microscope at 80 KV. Images were captured with a Gatan CCD (2000×2000).
Glutaraldehyde crosslinking
Recombinant FRG-1 protein (0.2 mg/ml) was incubated with 10 mM HEPES pH 7.6, 2 mM MgCl2 and 0.01% glutaraldehyde in 1 ml at RT. Samples (50 μl) were removed at the indicated time intervals and the reactions were stopped by the addition of Laemmli SDS-PAGE buffer. Reactions were then boiled and electrophoresed on a 10% SDS-PAGE. Protein was detected by western blotting using the CeFRG-1 antibody described above.
Supplementary Material
Acknowledgments
We thank Michel Bellini for help with microscopy, and Meredith Hanel, Ryan Wuebbles, Jessica Sun and Steven Long for helpful discussions and ideas. The electron microscopy was carried out in part at the Frederick Seitz Materials Research Laboratory, Central Facilities, University of Illinois, which is partially supported by the US Department of Energy under grants DE-FG02-07ER46453 and DE-FG02-07ER46471. P.L.J. is funded by grant #1RO1AR055877 from NIAMS-NIH. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/7/1116/DC1
References
- Adams J. C. (2004). Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16, 590-596 [DOI] [PubMed] [Google Scholar]
- Arashiro P., Eisenberg I., Kho A. T., Cerqueira A. M., Canovas M., Silva H. C., Pavanello R. C., Verjovski-Almeida S., Kunkel L. M., Zatz M. (2009). Transcriptional regulation differs in affected facioscapulohumeral muscular dystrophy patients compared to asymptomatic related carriers. Proc. Natl. Acad. Sci. USA 106, 6220-6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead R. J., Kleiman L., Waterston R. H. (1991). Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil. Cytoskeleton 20, 69-78 [DOI] [PubMed] [Google Scholar]
- Bodega B., Ramirez G. D., Grasser F., Cheli S., Brunelli S., Mora M., Meneveri R., Marozzi A., Mueller S., Battaglioli E., et al. (2009). Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 7, 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovic L., Sacconi S., Bechara E. G., Delplace S., Allegra M., Desnuelle C., Bardoni B. (2008). Alteration of expression of muscle specific isoforms of the fragile X related protein 1 (FXR1P) in facioscapulohumeral muscular dystrophy patients. J. Med. Genet. 45, 679-685 [DOI] [PubMed] [Google Scholar]
- Edwards R. A., Bryan J. (1995). Fascins, a family of actin bundling proteins. Cell Motil. Cytoskeleton 32, 1-9 [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895-905 [DOI] [PubMed] [Google Scholar]
- Flower D. R. (1996). The lipocalin protein family: structure and function. Biochem. J. 318, 1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M., Ziemiecki A., Billeter R., Muntener M. (2002). Fibre-type specific concentration of focal adhesion kinase at the sarcolemma: influence of fibre innervation and regeneration. J. Exp. Biol. 205, 2337-2348 [DOI] [PubMed] [Google Scholar]
- Francis G. R., Waterston R. H. (1985). Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101, 1532-1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D., D'Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M. A., Bottinelli R., et al. (2006). Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 439, 973-977 [DOI] [PubMed] [Google Scholar]
- Gettner S. N., Kenyon C., Reichardt L. F. (1995). Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol. 129, 1127-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieron M. A., Korthals J. K., Kousseff B. G. (1985). Facioscapulohumeral dystrophy with cochlear hearing loss and tortuosity of retinal vessels. Am. J. Med. Genet. 22, 143-147 [DOI] [PubMed] [Google Scholar]
- Grewal P. K., Todd L. C., van der Maarel S., Frants R. R., Hewitt J. E. (1998). FRG1, a gene in the FSH muscular dystrophy region on human chromosome 4q35, is highly conserved in vertebrates and invertebrates. Gene 216, 13-19 [DOI] [PubMed] [Google Scholar]
- Guglieri M., Straub V., Bushby K., Lochmuller H. (2008). Limb-girdle muscular dystrophies. Curr. Opin. Neurol. 21, 576-584 [DOI] [PubMed] [Google Scholar]
- Han H. F., Beckerle M. C. (2009). The ALP-Enigma protein ALP-1 functions in actin filament organization to promote muscle structural integrity in Caenorhabditis elegans. Mol. Biol. Cell 20, 2361-2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel M. L., Wuebbles R. D., Jones P. L. (2009). Muscular dystrophy candidate gene FRG1 is critical for muscle development. Dev. Dyn. 238, 1502-1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y. K., Chou F. L., Engvall E., Ogawa M., Matsuda C., Hirabayashi S., Yokochi K., Ziober B. L., Kramer R. H., Kaufman S. J., et al. (1998). Mutations in the integrin alpha7 gene cause congenital myopathy. Nat. Genet. 19, 94-97 [DOI] [PubMed] [Google Scholar]
- Hoffman E. P., Brown R. H., Jr, Kunkel L. M. (1987). Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919-928 [DOI] [PubMed] [Google Scholar]
- Hresko M. C., Williams B. D., Waterston R. H. (1994). Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 124, 491-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Yang F., van Overveld P. G., Vedanarayanan V., van der Maarel S., Ehrlich M. (2003). Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 12, 2909-2921 [DOI] [PubMed] [Google Scholar]
- Kunkel L. M., Hejtmancik J. F., Caskey C. T., Speer A., Monaco A. P., Middlesworth W., Colletti C. A., Bertelson C., Muller U., Bresnan M., et al. (1986). Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature 322, 73-77 [DOI] [PubMed] [Google Scholar]
- Kureishy N., Sapountzi V., Prag S., Anilkumar N., Adams J. C. (2002). Fascins, and their roles in cell structure and function. BioEssays 24, 350-361 [DOI] [PubMed] [Google Scholar]
- Lecroisey C., Segalat L., Gieseler K. (2007). The C. elegans dense body: anchoring and signaling structure of the muscle. J. Muscle Res. Cell Motil. 28, 79-87 [DOI] [PubMed] [Google Scholar]
- Lunt P. W., Jardine P. E., Koch M. C., Maynard J., Osborn M., Williams M., Harper P. S., Upadhyaya M. (1995). Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum. Mol. Genet. 4, 951-958 [DOI] [PubMed] [Google Scholar]
- Meyer R. K., Aebi U. (1990). Bundling of actin filaments by alpha-actinin depends on its molecular length. J. Cell Biol. 110, 2013-2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D. G., Williams B. D. (2006). Sarcomere assembly in C. elegans muscle. WormBook 16, 1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A. P., Neve R. L., Colletti-Feener C., Bertelson C. J., Kurnit D. M., Kunkel L. M. (1986). Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature 323, 646-650 [DOI] [PubMed] [Google Scholar]
- Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., Matsumura F. (1997). Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272, 2527-2533 [DOI] [PubMed] [Google Scholar]
- Osborne R. J., Welle S., Venance S. L., Thornton C. A., Tawil R. (2007). Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology 68, 569-577 [DOI] [PubMed] [Google Scholar]
- Pandya S., King W. M., Tawil R. (2008). Facioscapulohumeral dystrophy. Phys. Ther. 88, 105-113 [DOI] [PubMed] [Google Scholar]
- Pardee J. D., Spudich J. A. (1982). Purification of muscle actin. Methods Enzymol. 85, 164-181 [DOI] [PubMed] [Google Scholar]
- Pardo J. V., Siliciano J. D., Craig S. W. (1983). A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc. Natl. Acad. Sci. USA 80, 1008-1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T., Aebi U. (2005). Nuclear actin extends, with no contraction in sight. Mol. Biol. Cell 16, 5055-5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231-1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed P., Porter N. C., Strong J., Pumplin D. W., Corse A. M., Luther P. W., Flanigan K. M., Bloch R. J. (2006). Sarcolemmal reorganization in facioscapulohumeral muscular dystrophy. Ann. Neurol. 59, 289-297 [DOI] [PubMed] [Google Scholar]
- Romer L. H., Birukov K. G., Garcia J. G. (2006). Focal adhesions: paradigm for a signaling nexus. Circ. Res. 98, 606-616 [DOI] [PubMed] [Google Scholar]
- Selcen D., Carpen O. (2008). The Z-disk diseases. Adv. Exp. Med. Biol. 642, 116-130 [DOI] [PubMed] [Google Scholar]
- Shaw P., Doonan J. (2005). The nucleolus. Playing by different rules? Cell Cycle 4, 102-105 [DOI] [PubMed] [Google Scholar]
- Street S. F. (1983). Lateral transmission of tension in frog myofibers: a myofibrillar network and transverse cytoskeletal connections are possible transmitters. J. Cell Physiol. 114, 346-364 [DOI] [PubMed] [Google Scholar]
- Tawil R. (2008). Facioscapulohumeral muscular dystrophy. Neurotherapeutics 5, 601-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel S. M., Frants R. R. (2005). The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 76, 375-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel S. M., Frants R. R., Padberg G. W. (2007). Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta 1772, 186-194 [DOI] [PubMed] [Google Scholar]
- van Deutekom J. C., Wijmenga C., van Tienhoven E. A., Gruter A. M., Hewitt J. E., Padberg G. W., van Ommen G. J., Hofker M. H., Frants R. R. (1993). FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 2, 2037-2042 [DOI] [PubMed] [Google Scholar]
- van Deutekom J. C., Lemmers R. J., Grewal P. K., van Geel M., Romberg S., Dauwerse H. G., Wright T. J., Padberg G. W., Hofker M. H., Hewitt J. E., et al. (1996). Identification of the first gene (FRG1) from the FSHD region on human chromosome 4q35. Hum. Mol. Genet. 5, 581-590 [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S., Dirks R. W., Mommaas A. M., Onderwater J. J., Deidda G., Padberg G. W., Frants R. R., van der Maarel S. M. (2004). FRG1P is localised in the nucleolus, Cajal bodies, and speckles. J. Med. Genet. 41, e46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S., Straasheijm K. R., Sterrenburg E., de Graaf N., Dauwerse H. G., Frants R. R., van der Maarel S. M. (2007). FRG1P-mediated aggregation of proteins involved in pre-mRNA processing. Chromosoma 116, 53-64 [DOI] [PubMed] [Google Scholar]
- Wijmenga C., Hewitt J. E., Sandkuijl L. A., Clark L. N., Wright T. J., Dauwerse H. G., Gruter A. M., Hofker M. H., Moerer P., Williamson R., et al. (1992). Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 2, 26-30 [DOI] [PubMed] [Google Scholar]
- Williams B. D., Waterston R. H. (1994). Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124, 475-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winokur S. T., Chen Y. W., Masny P. S., Martin J. H., Ehmsen J. T., Tapscott S. J., van der Maarel S. M., Hayashi Y., Flanigan K. M. (2003). Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 12, 2895-2907 [DOI] [PubMed] [Google Scholar]
- Wuebbles R. D., Hanel M. L., Jones P. L. (2009). FSHD region gene 1 (FRG1) is crucial for angiogenesis linking FRG1 to facioscapulohumeral muscular dystrophy-associated vasculopathy. Dis. Model Mech. 2, 267-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Winokur S. T., Kuo W. L., Altherr M. R., Bredt D. S. (1997). Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 139, 507-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., de Greef J. C., Chen Y. Y., Chien R., Kong X., Gregson H. C., Winokur S. T., Pyle A., Robertson K. D., Schmiesing J. A., et al. (2009). Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 5, e1000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.