Abstract
PALB2 physically and functionally connects the proteins encoded by the BRCA1 and BRCA2 breast and ovarian cancer genes into a DNA-damage-response network. However, it remains unclear how these proteins associate with chromatin that contains damaged DNA. We show here that PALB2 binds directly to a conserved chromodomain protein, MRG15, which is a component of histone acetyltransferase-deacetylase complexes. This interaction was identified by analysis of purified MRG15- and PALB2-containing protein complexes. Furthermore, MRG15 interacts with the entire BRCA complex, which contains BRCA1, PALB2, BRCA2 and RAD51. Interestingly, MRG15-deficient cells, similarly to cells deficient in PALB2 or BRCA2, showed reduced efficiency for homology-directed DNA repair and hypersensitivity to DNA interstrand crosslinking agents. Additionally, knockdown of MRG15 diminished the recruitment of PALB2, BRCA2 and RAD51 to sites of DNA damage and reduced chromatin loading of PALB2 and BRCA2. These results suggest that MRG15 mediates DNA-damage-response functions of the BRCA complex in chromatin.
Keywords: MRG15, PALB2, BRCA1, BRCA2, DNA repair
Introduction
Increased genetic instability is linked to cancer and a poorer prognosis (Hartwell and Kastan, 1994). DNA double-strand breaks (DSBs) are among the most deleterious lesions to cells and homologous recombination (HR) is crucial for error-free repair of DSBs (Venkitaraman, 2001). BRCA1 and BRCA2, the major breast and ovarian cancer susceptibility genes, are essential for HR and for resistance to DNA interstrand crosslinkers, such as mitomycin C (MMC) (Venkitaraman, 2001). It has recently been demonstrated that the product of another breast cancer susceptibility gene, PALB2, physically and functionally links BRCA1 and BRCA2 into the BRCA complex, which also includes RAD51 and functions in DNA-damage responses (Sy et al., 2009b; Zhang et al., 2009a; Zhang et al., 2009b). BRCA1 acts upstream in the network to organize PALB2 (Zhang et al., 2009a; Zhang et al., 2009b). PALB2, which was identified as a BRCA2-interacting protein, in turn recruits BRCA2 to sites of DNA damage in chromatin (Xia et al., 2006). BRCA2 then regulates the assembly of RAD51 into nucleoprotein filaments with single-strand DNA to initiate HR (Venkitaraman, 2001).
MRG15 is a conserved chromodomain protein that specifically binds to Lys36-methylated histone H3 (Zhang et al., 2006), and has an established role in transcriptional regulation (Carrozza et al., 2005). In particular, MRG15, and the closely related protein MRGX, are integral components of the NuA4/Tip60 histone acetyltransferase (HAT) and also histone deacetylase (HDAC) complexes (Doyon et al., 2004; Hayakawa et al., 2007). Although several lines of evidence suggest that MRG15 and its related proteins are also involved in DNA-repair processes (Garcia et al., 2007; Nakayama et al., 2003), the mechanism is unknown.
Here, we demonstrate that MRG15 binds directly to a key component of the machinery for DSB repair, PALB2. Furthermore, MRG15 is required for HR and resistance to MMC. Additionally, we show that the recruitment of PALB2 and PALB2-associated proteins to sites of DNA damage and chromatin depends on MRG15. These results suggest that MRG15 participates in the response to DSBs by recruiting the BRCA complex to sites of damaged DNA.
Results
Co-immunopurification of MRG15 and PALB2 in a protein complex
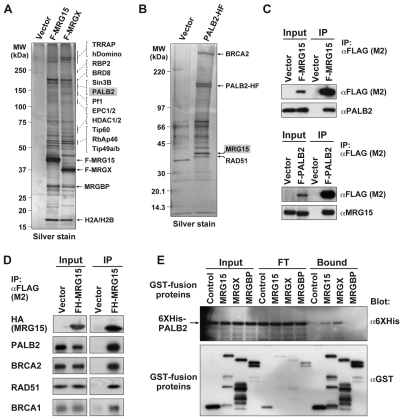
Previously, using a HeLa cell line expressing FLAG-tagged MRG15, we found that MRG15 associates with HAT- and HDAC-containing multiprotein complexes (Hayakawa et al., 2007). Interestingly, additional mass spectrometric analysis identified PALB2 as another stable component of the FLAG-MRG15 complex (Fig. 1A; supplementary material Fig. S1A). PALB2 was also detected in an immunopurified complex with FLAG-MRGX, a closely related MRG family protein.
Fig. 1.
MRG15 associates with PALB2 in a protein complex. (A) Immunopurified FLAG-MRG15- or FLAG-MRGX-containing complexes resolved by SDS-PAGE and silver stained. A mock purification from cells containing the empty expression vector, with the FLAG epitope tag, is shown as a control. Proteins identified by LC-MS/MS analysis are indicated on the right. (B) An immunopurified FLAG-HA-PALB2-containing complex resolved and analyzed as in A. (C) Whole-cell lysates (Input) of HeLa cells expressing FLAG-MRG15 (top) or FLAG-PALB2 (bottom) and fractions immunoprecipitated (IP) using anti-FLAG M2 beads subjected to western blotting using the antibodies indicated. (D) Whole-cell lysates (Input) of HEK293T cells expressing FLAG-MRG15 and fractions immunoprecipitated (IP) using anti-FLAG M2 beads subjected to immunoblotting using the antibodies indicated. (E) Each purified GST fusion protein was examined for its ability to bind 6×His-PALB2. Input, non-bound (FT), and bound fractions were analyzed by western blotting with anti-6×His or anti-GST antibodies.
To confirm a possible functional link between MRG15 and PALB2, we expressed FLAG-HA-tagged PALB2 in HeLa cells and immunopurified the PALB2-containing complex using anti-FLAG and then anti-HA antibodies. As previously reported, BRCA2 and RAD51 were identified in the PALB2-containing complex (Fig. 1B) (Xia et al., 2006). Importantly, mass spectrometric analysis also revealed that MRG15 was specifically immunoprecipitated with FLAG-HA-PALB2, but not with the FLAG-HA epitope tag alone (Fig. 1B; supplementary material Fig. S1B). This result suggests that MRG15 associates with the BRCA complex.
The association of MRG15 and PALB2 with FLAG-PALB2 and FLAG-MRG15, respectively, was confirmed by reciprocal immunoprecipitation and western blotting analysis using specific antibodies (Fig. 1C). Incubation of extracts with ethidium bromide can be used to test for DNA-independent protein associations (Lai and Herr, 1992). Using this approach, we found that the interaction of MRG15 with PALB2 was not disrupted by ethidium bromide and is therefore not mediated by DNA (supplementary material Fig. S2).
Interaction of MRG15 with PALB2 and the BRCA complex
Although BRCA1, BRCA2 and RAD51 were not identified by mass spectrometric analysis of FLAG-MRG15-containing complexes (Fig. 1A), we found that these BRCA proteins were clearly detected in the isolated FLAG-HA-MRG15 complex by immunoblotting (Fig. 1D). This result further suggests that MRG15 associates with the entire BRCA complex.
To determine whether MRG proteins interact directly with PALB2, we performed an in vitro binding assay using recombinant proteins. We found that 6×His-PALB2 interacted with GST-MRG15 and GST-MRGX, but not with GST alone or GST-MRGBP (Fig. 1E). MRGBP is a partner protein of MRG proteins (Cai et al., 2003; Hayakawa et al., 2007). Thus, PALB2 might act as a scaffold that links MRG15 to the entire BRCA complex.
MRG15 has a crucial role in HR and resistance to MMC
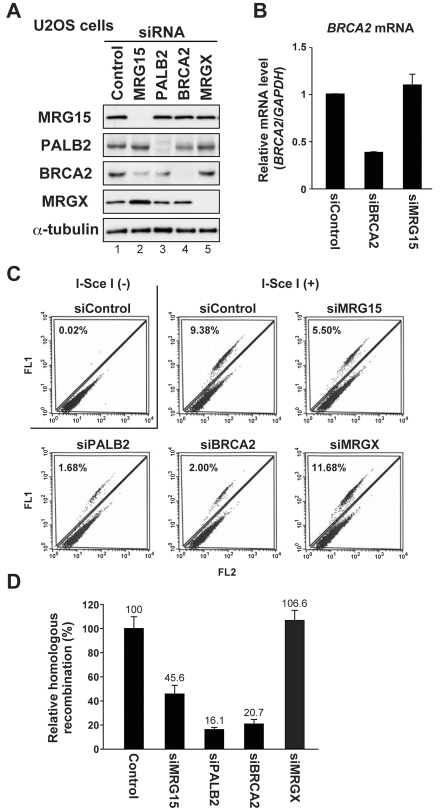
BRCA1, PALB2, BRCA2 and RAD51 are all required for HR (Stark et al., 2002; Sy et al., 2009b; Xia et al., 2006). To determine whether MRG15 also has a role in HR-based DSB repair, we used specific siRNAs and depleted endogenous MRG15, MRGX, PALB2 or BRCA2. As previously observed, PALB2 depletion led to decreased levels of BRCA2 protein (Xia et al., 2006) (Fig. 2A, lane 3; Fig. 3B, lane 3). Interestingly, depletion of MRG15, but not depletion of MRGX, resulted in a similar reduction of BRCA2 levels (Fig. 2A lane 2; Fig. 3B, lane 2). This reduction was not attributed to the off-target effect of siRNAs for MRG15, because abundance of BRCA2 mRNA was not affected by transfection with a siRNA directed against MRG15 (Fig. 2B). In addition, the BRCA2 protein level was partially recovered by transient expression of siRNA-resistant MRG15 (supplementary material Fig. S3B). This result supports the possibility that MRG15, along with PALB2, is required for the proper function of BRCA2.
Fig. 2.
MRG15 stimulates homologous recombination. (A) U2OS cells were treated with control or indicated siRNAs and the levels of endogenous MRG15, PALB2, BRCA2, MRGX or α-tubulin (loading control) were analyzed by western blotting. (B) Relative levels of BRCA2 mRNA were analyzed in U2OS cells following a control transfection or transfection with cocktails of siRNAs directed against BRCA2 or MRG15. (C) U2OS-DR cells were treated with control or indicated cocktails of siRNAs and assayed for HR. Representative data obtained by FACS analysis are shown with average levels of GFP-positive cells indicated. (D) Relative HR ratios (% ± s.d.) calculated from three independent experiments. Cocktails of siRNAs directed against MRG15, MRGX, PALB2 or BRCA2 were used.
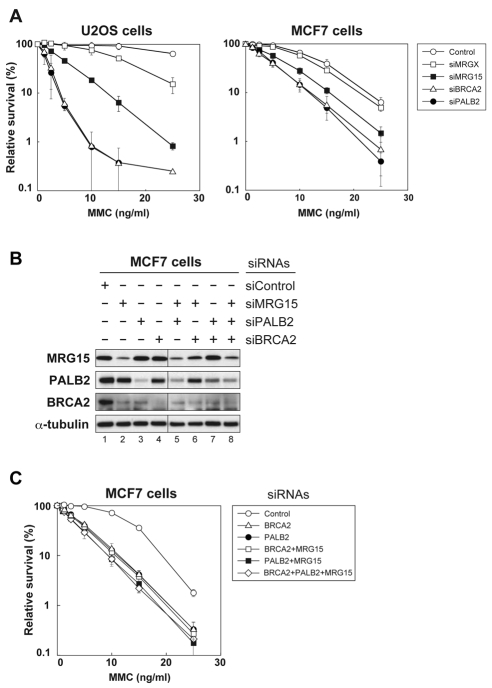
Fig. 3.
MRG15 has a crucial role in resistance to MMC. (A) Survival of U2OS or MCF7 cells pretreated with control or indicated siRNAs determined using a clonogenic assay following treatment with a range of concentrations of MMC. Results are means (± s.d.) of at least three independent experiments. (B) MCF7 cells were treated with control or indicated siRNAs and the levels of endogenous MRG15, PALB2, BRCA2 or α-tubulin (loading control) were analyzed by western blotting. Individual proteins, or a combination of two or three proteins together, were depleted. (C) Survival of MCF7 cells pretreated with control or indicated siRNAs determined using a clonogenic assay following treatment with a range of concentrations of MMC. Cocktails of siRNAs directed against MRG15, MRGX, PALB2 or BRCA2 were used (A-C). Results are means ± s.d. of at least three independent experiments.
Depletion of either PALB2 or BRCA2 in U2OS cells carrying a stably integrated copy of the DR-GFP HR reporter severely reduced the repair efficiency (Fig. 2C,D). Of interest, although the effect was milder than depletion of either PALB2 or BRCA2, depletion of MRG15 also reduced the repair efficiency by more than 50%. By contrast, knockdown of MRGX had no effect. During preparation of our paper, a report appeared suggesting that depletion of MRG15 instead increases the gene conversion rate (Sy et al., 2009a). Although further research will be required to understand this discrepancy, our results are supported by a parallel HR-reporter assay using two additional independent siRNAs (supplementary material Fig. S4A,B), and by a report that MRG15-deficient MEFs display a repair defect measurable using a comet assay (Garcia et al., 2007). The requirement of MRG15 in HR-based DSB repair is consistent with, and is strengthened by, our novel findings of a direct interaction between MRG15 and PALB2 (Fig. 1E), and our findings that MRG15 has roles in resistance to MMC and recruitment of the BRCA complex to sites of DNA damage (see below).
HR is required for sister chromatid exchange (SCE) (Sonoda et al., 1999). Since our results indicate that MRG15 stimulates HR, we also tested for the involvement of MRG15 in SCE (supplementary material Fig. S5). We found that depletion of MRG15 decreased the level of SCE in U2OS cells by nearly 40%, further supporting a role for MRG15 in HR.
Because components of the BRCA complex are required for resistance to MMC (Sy et al., 2009c; Zhang et al., 2009a), we tested whether MRG15 and MRGX also have a role in this process. Depletion of MRG15 caused hypersensitivity of two different cell lines, U2OS and MCF7 cells, to MMC (Fig. 3A). The effect of MRG15 knockdown was milder than that seen upon knockdown of BRCA2 or PALB2. The direct involvement of MRG15 in MMC resistance was confirmed by using a different siRNA (supplementary material Fig. S4C). In addition, this MMC hypersensitivity was partially inhibited by transient expression of siRNA-resistant MRG15 (supplementary material Fig. S3D). Consistent with the HR assay, knockdown of MRGX only weakly affected sensitivity to MMC (Fig. 3A). Thus, MRG15, but not the closely related protein MRGX, is required for HR and resistance to MMC.
To corroborate the functional link between MRG15 and BRCA proteins, we simultaneously depleted these proteins and analyzed MMC sensitivity. Double knockdown of MRG15 and either PALB2 or BRCA2, or of all three, showed, at most, limited additivity (Fig. 3B,C). Since knockdown efficiencies were comparable in single-, double- and triple-knockdown experiments, we consider it unlikely that incomplete knockdown limited the amount of additivity. Although MRG15 could have a role in this process that is independent of PALB2 and BRCA2, our results suggest that MRG15 cooperates with these proteins to mediate resistance to MMC.
Disruption of MRG15 impairs targeting of PALB2 to sites of damage in chromatin
The recruitment of BRCA2 to nuclear foci and chromatin requires PALB2 (Xia et al., 2006). To further investigate the role of MRG15 in DSB repair, and to determine whether MRG15 regulates this PALB2-dependent process, we performed a chromatin fractionation assay. Briefly, cells were lysed, subjected to fractionation and chromatin-bound proteins were analyzed by western blotting. In this assay, a portion of the pools of MRG15, PALB2 and BRCA2 were detected in the chromatin-bound fraction (ppt) to different degrees (Fig. 4A, lane 3). U2OS cells transfected with various siRNAs were then subjected to cellular fractionation to detect chromatin-bound proteins. As previously observed (Xia et al., 2006), knockdown of PALB2 caused a loss of chromatin-bound BRCA2 (Fig. 4A, compare lanes 3 and 9). Interestingly, MRG15 knockdown diminished the chromatin association of both PALB2 and BRCA2 (Fig. 4A, compare lanes 3 and 6).
Fig. 4.
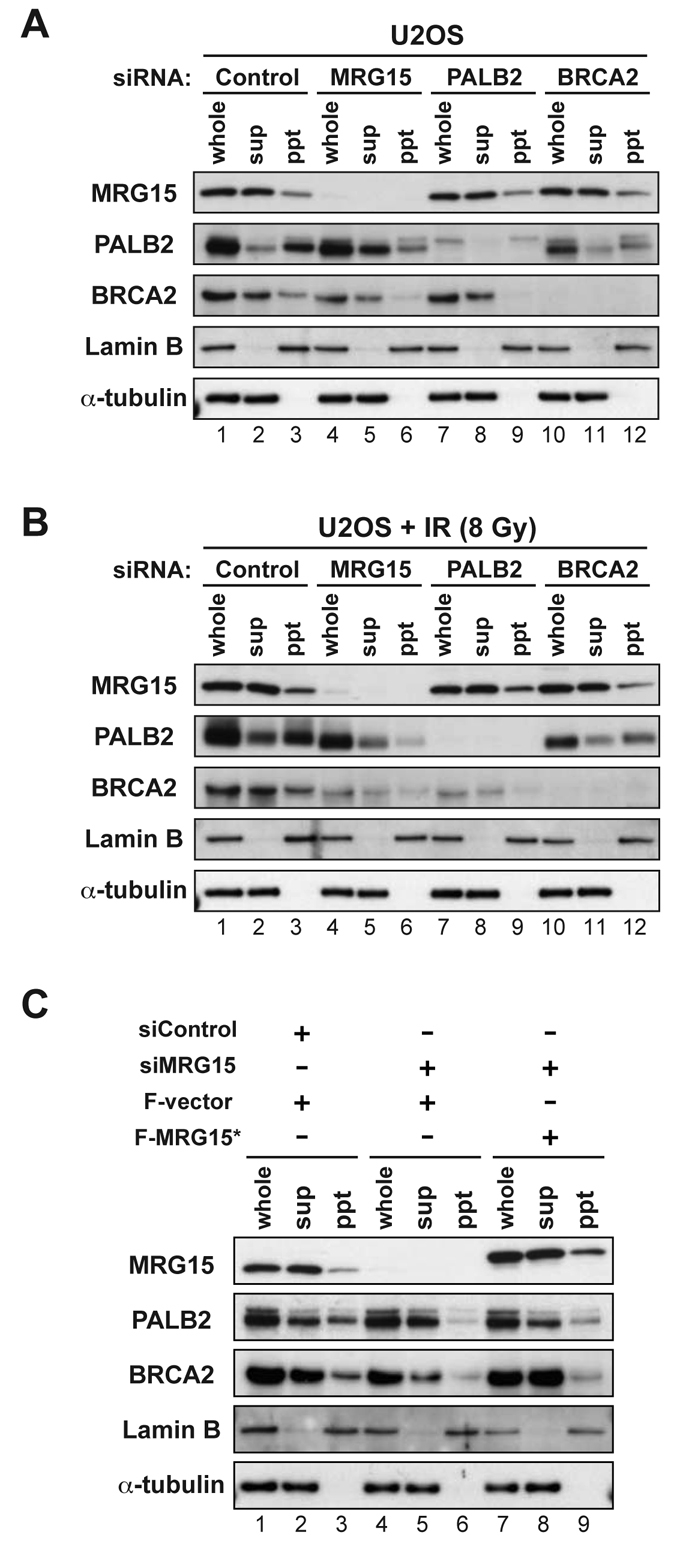
MRG15 is involved in the localization of PALB2 to chromatin. (A,B) U2OS cells transfected with control or the indicated siRNA cocktails were extracted to obtain whole cell lysates (whole), soluble supernatant (sup) and chromatin-enriched pellet (ppt) fractions. Proteins in each fraction were resolved by SDS-PAGE and analyzed by immunoblotting using indicated antibodies. α-tubulin and lamin B are markers for detergent-soluble and insoluble fractions, respectively. Cells were untreated (A) or were harvested 6 hours after exposure to 8 Gy IR (B). (C) Expression of siRNA-resistant MRG15 (F-MRG15*) in U2OS cells to rescue depletion of MRG15 mediated by a siMRG15 cocktail. Cells were separated into whole-cell lysates, soluble supernatants and insoluble fractions and analyzed by immunoblotting for levels of MRG15, PALB2 and BRCA2. α-tubulin and lamin B are markers for detergent-soluble and -insoluble fractions, respectively.
The levels of PALB2 and BRCA2 associated with chromatin were somewhat increased after exposure to IR (Fig. 4B, lane 3). Depletion of MRG15 also decreased the levels of PALB2 and BRCA2 that were associated with chromatin following exposure to IR (Fig. 4B, compare lanes 3 and 6). Additionally, depletion of MRG15 in a different cell line, MCF7, also decreased the association of BRCA2 with chromatin in untreated cells (supplementary material Fig. S6A, compare lanes 3 and 6). Furthermore, depletion of MRG15 using a distinct siRNA, directed against the 3′-UTR of MRG15, decreased the levels of PALB2 and BRCA2 that were associated with chromatin in a manner similar to results obtained with the siMRG15 cocktail (supplementary material Fig. S6B, compare lanes 3 and 6). Importantly, the reduction of chromatin-bound PALB2 and BRCA2 was largely impeded by transient expression of siRNA-resistant MRG15 (Fig. 4C, compare lanes 6 and 9; supplementary material Fig. S6B, compare lanes 6 and 9). Together, these results, obtained in two different cell lines and using two different sets of siRNAs, suggest that MRG15 acts to promote the association of PALB2 and BRCA2 with chromatin. We propose that PALB2, once positioned in an MRG15-dependent manner, recruits BRCA2 to chromatin.
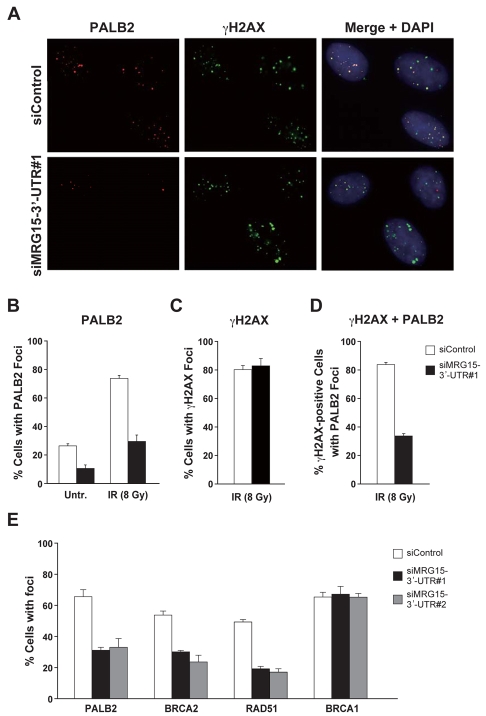
Since PALB2 assembles into DNA-damage foci in chromatin (Xia et al., 2006), we tested the role of MRG15 in the recruitment of PALB2 to nuclear foci. Depletion of MRG15 decreased the localization of PALB2 to foci, both in untreated cells and in cells exposed to IR (Fig. 5A,B). By contrast, depletion of MRG15 had no effect on the assembly of a marker for DSBs, γH2AX, into nuclear foci after exposure of cells to IR (Fig. 5C). However, MRG15 was required for the efficient recruitment of PALB2 to DSBs, as indicated by colocalization of PALB2 foci with γH2AX foci (Fig. 5D). This result is of interest given that incorrect localization of PALB2 is associated with defects in HR and sensitivity to MMC (Zhang et al., 2009a; Zhang et al., 2009b).
Fig. 5.
MRG15 is involved in the localization of PALB2, BRCA2 and RAD51, but not BRCA1, to DNA-damage foci. (A-D) U2OS cells transfected with siGFP or siMRG15-3′-UTR#1 were fixed 16 hours after exposure to 8 Gy IR or were left untreated. Cells were labeled with anti-PALB2 or γ-H2AX antibodies and counterstained with DAPI to visualize nuclei. Examples of colocalization in merged images are shown (A), along with quantification of the percentage of cells (mean ± s.d. of three counts) with five or more PALB2 foci (B), the percentage of cells with five or more γ-H2AX foci (C), or in which PALB2 foci were detected in γ-H2AX-positive cells (D). (E) The percentage of cells transfected with siGFP, siMRG15-3′-UTR#1, or siMRG15-3′-UTR#2 with five or more PALB2, BRCA2, RAD51 or BRCA1 foci 16 hours after exposure to 8 Gy IR.
PALB2 is required for the assembly of BRCA2 and RAD51 nuclear foci (Xia et al., 2007; Xia et al., 2006), which ultimately culminates in DSB-repair by HR. Since MRG15 regulates the assembly of PALB2 foci (Fig. 5A,B), we also examined whether it is required for the assembly of BRCA2, RAD51 and BRCA1 foci. Indeed, depletion of MRG15 using a siRNA (siMRG15-3′-UTR#1) strongly decreased the assembly of BRCA2 and RAD51 foci in U2OS cells exposed to IR (Fig. 5E). Direct involvement of MRG15 in the recruitment of these proteins was further corroborated by using a different siRNA (siMRG15-3′-UTR#2). Furthermore, the ability of ectopically expressed MRG15 to largely correct the siRNA-mediated decrease in PALB2, BRCA2 and RAD51 foci strengthens our conclusion that MRG15 mediates the recruitment of these proteins (supplementary material Fig. S7). However, depletion of MRG15 did not affect the assembly of BRCA1 foci. Since it has recently been reported that BRCA1 is required for the recruitment of PALB2 to sites of DNA damage (Zhang et al., 2009a; Zhang et al., 2009b), our results suggest that MRG15 and BRCA1 independently regulate the recruitment of PALB2, BRCA2 and RAD51 to DNA-damage foci.
Discussion
In this report, we have shown that a conserved chromodomain protein, MRG15, directly binds to PALB2 and is required for HR, MMC resistance and recruitment of PALB2, BRCA2 and RAD51 to sites of DNA damage. MRG15 forms complexes with the Tip60 HAT that has been implicated in DNA-damage responses (Murr et al., 2006) and also with HDACs. It is important to note that we have not been able to detect Tip60 or TRRAP, another component of the NuA4 HAT, in PALB2 immunoprecipitates (data not shown). Since it binds directly, perhaps the association of MRG15 with PALB2 is more readily detectable than the interaction of Tip60 and TRRAP with PALB2. Considering that Tip60 and related proteins are implicated in the HR-directed DNA repair pathway (Murr et al., 2006), we propose that the interaction of MRG15 with PALB2 regulates recruitment of the BRCA complex to sites of DNA damage where chromatin is relaxed following histone acetylation by the NuA4 HAT (Fig. 6). BRCA1 is required to position PALB2 (Zhang et al., 2009a; Zhang et al., 2009b). As such, MRG15 might determine the accessibility or affinity of PALB2 for chromatin, rather than identifying the sites of damage where PALB2 is recruited. Another possibility is that MRG15 is stably bound to chromatin containing specific histone marks and might therefore recruit various chromatin remodeling complexes necessary to manage distinct steps in DNA repair.
Fig. 6.
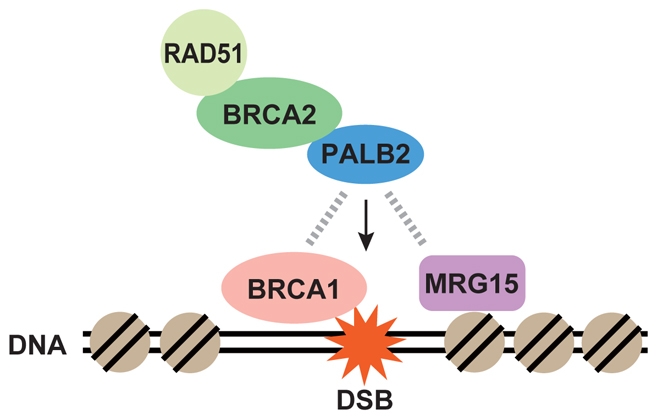
Proposed model for the role of PALB2 in linking MRG15 and the BRCA complex into a pathway of HR. We propose that by directly binding PALB2, BRCA1 and MRG15 independently regulate site selection and chromatin accessibility, respectively, in PALB2 recruitment to sites of DNA damage. Once it is localized, PALB2 is proposed to be responsible for the recruitment of BRCA2 and RAD51.
During preparation of our paper, a report appeared from another group demonstrating a functional link between MRG15 and PALB2 (Sy et al., 2009a). Although we agree that MRG15 physically interacts with PALB2, Sy and co-workers suggested that a mutant PALB2 lacking the MRG15-binding domain is associated with hyper-recombination and resistance to MMC. This is in contrast to our findings that depletion of MRG15 using siRNAs decreases the efficiency of gene conversion and sensitizes cells to MMC. Depletion of MRG15 might relax chromatin and could have effects on the recruitment of BRCA2 and RAD51 that are independent from the interaction of MRG15 with PALB2. Since MRG15 is a component of three types of protein complexes, including the Tip60 HAT complex, the Sin3-containing HDAC complex and the BRCA complex, it is possible that depletion of MRG15 disturbs the function and/or stability of these HAT and HDAC complexes, in addition to affecting the recruitment of the BRCA complex. At the same time, deletion of a sizable portion of a protein, as was done in the experiments reported by Sy and colleagues, could juxtapose domains of the protein that do not normally interact and could disrupt protein folding. Such differences in approach might, in part, explain why our findings contrast with their results.
Importantly, Sy and co-workers also reported that depletion of MRG15 does not affect the level of BRCA2 protein and increases the gene conversion rate using an HR assay that was similar to the one we used. This is seemingly contradictory to our findings and further research will be required to understand this discrepancy. It should be noted, though, that we reproducibly observed that depletion of MRG15 leads to decreased levels of BRCA2 protein. Xia and colleagues (Xia et al., 2006) showed that siRNA-mediated depletion of PALB2 disrupts both the association of BRCA2 with chromatin and its stability. This is similar to our findings that depletion of MRG15 disrupts both BRCA2 stability and the association of BRCA2 with chromatin. Perhaps by regulating the access of BRCA2 to chromatin, MRG15 also regulates its stability. It is important to note, however, that because BRCA2 regulates the RAD51 recombinase, instability of BRCA2 will diminish the efficiency of HR and might not reflect a direct role for MRG15 in this process.
In this study, we demonstrate the direct involvement of MRG15 in DSB-mediated homologous recombination (Fig. 2D), resistance to MMC (Fig. 3A), the association of PALB2 and BRCA2 with chromatin using a nuclear fractionation assay (Fig. 4) and the assembly of BRCA proteins into nuclear foci (Fig. 5). Our conclusion is supported also by a report that MRG15-deficient MEFs display a repair defect measurable using a comet assay (Garcia et al., 2007). Further studies are necessary to better delineate the mechanisms by which MRG15 contributes to the DNA-damage response network.
Materials and Methods
Cells and antibodies
Cells were grown in Dulbecco's modified Eagle's medium or Eagle's minimum essential medium, each supplemented with 10% fetal calf serum. Antibodies used were: anti-FLAG M2 (Sigma), anti-6×His (Qiagen), anti-HA (HA.11; Covance), anti-BRCA2 (Ab-1 and Ab2; Calbiochem), anti-RAD51 (H-92; Santa Cruz), anti-BRCA1 (D9; Santa Cruz) and anti-γH2AX (UBI). Antibodies against MRG15 and MRGX were previously described (Hayakawa et al., 2007). Anti-PALB2 rabbit polyclonal antibodies were prepared using GST fused with residues 1-200 or 601-880 of PALB2 (Zhang et al., 2009a).
Purification of PALB2- or MRG15-interacting proteins
Affinity purification of PALB2- and MRG15-containing complexes was performed as previously described (Hayakawa et al., 2007; Zhang et al., 2009a). To examine potential involvement of DNA in PALB2-MRG15 interaction, extracts of HEK293T cells expressing FLAG-MRG15 or FLAG-PALB2 were treated with or without 50 μg/ml ethidium bromide for 30 min on ice before immunoprecipitation with anti-FLAG M2 beads.
In vitro binding assay
Recombinant 6×His-tagged proteins and GST-fusion proteins were produced in E. coli and purified according to the manufacturer's instructions (Clontech and GE Healthcare). The eluted materials were dialyzed against phosphate-buffered saline (PBS) with 10% glycerol, divided into aliquots, and stored at −80°C before use. Each purified GST-fusion protein was incubated with 6×His-PALB2 in pull-down buffer (25 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% Nonidet P-40) for 4 hours at 4°C. GST-fusion proteins and bound proteins were pulled down using glutathione beads (GE Healthcare) and analyzed by western blotting.
RNA interference
siRNAs synthesized by B-Bridge International or Dharmacon were introduced into MCF-7 cells using electroporation by Gene Pulsar II (Bio-Rad) or into U2OS cells using Lipofectamine reagents (Invitrogen), as described previously (Zhang et al., 2009a). After 24-72 hours, siRNA-treated cells were used for western blotting. Genomic DNA in cell lysates was digested using 0.05 U/μl Benzonase (Novagen) for 30 minutes on ice. For assays of DSB-induced HR, SCE and immunofluorescence microscopy, cells were collected for analysis 4-5 days after transfection. siRNAs used in this studies were: siMRG15 (cocktail), 5′-gcagaaacagcgagaacuuTT-3′ (ORF: 398-416); 5′-caagaaaucucguggaaacTT-3′ (ORF: 788-806); 5′-ggcucaaauuguugaagaaTT-3′ (3′-UTR: 1658-1676); siMRG15-3′-UTR#1, 5′-gggauaugcuguagaguguTT-3′ (3′-UTR: 1393-1411); siMRG15-3′-UTR#2, 5′-gguucaauucaguguauauTT-3′ (3′-UTR: 1532-1550); siPALB2 (cocktail), 5′-ccaaagagcugaaaagauuTT-3′ (ORF: 314-322); 5′-ggaaaagacuaaaggaacaTT-3′ (ORF: 718-736); 5′-ggaaagagccgguuguaaaTT-3′ (ORF: 2831-2849); siBRCA2 (cocktail), 5′-gcaaagaccacauuggaaaTT-3′ (ORF: 1063-1081); 5′-cagacaagcucaaagguaaTT-3′ (ORF: 2611-2629); 5′-ggaaagagauacagaauuuTT-3′ (ORF: 9234-9252); siMRGX (cocktail), 5′-gcagagaaguaaaaugagaTT-3′ (ORF: 540-560); 5′-ccagaaagaacaagcagaaTT-3′ (ORF: 679-697); 5′-gaggaggcguuuaagaauaTT-3′ (ORF: 789-807).
Quantification of BRCA2 mRNA levels
Total RNA of U2OS cells transfected each siRNA was extracted by TRIZOL reagent (Invitrogen). One Step SYBR PrimeScript RT-PCR Kit II (TaKaRa) was used for amplification of cDNA fragments according to the manufacturer's instructions. Real-time RT-PCR was carried out using an ABI PRISM 7300 Realtime PCR System (Applied Biosystems). The primers used in these analyses were: BRCA2-RT-Fw, 5′-ACTTGCCCCTTTCGTCTATTTG-3′ and BRCA2-RT-Rv, 5′-TGCAGCAATTAACATATGAGGCTTA-3′ for BRCA2 cDNA, and GAPDH-RT-Fw, 5′-CATGGCCTCCAAGGAGTAAG-3′ and GAPDH-RT-Rv, 5′-TTCCTCTTGTGCTCTTGCTG-3′ for GAPDH cDNA. All PCR reactions were performed at least three times.
Clonogenic survival assay
MMC sensitivity was assayed as previously described (Schoenfeld et al., 2004) with minor modification. Briefly, MCF-7 or U2OS cells were transfected with siRNAs for each factor and incubated for 24 hours. 2000-10,000 siRNA-treated cells were plated in 90 mm dishes, incubated with 0-25 ng/ml MMC for 24 hours, and allowed to grow for 10-14 days in fresh medium before colonies were counted. Cells in colonies were fixed with 10% methanol and 10% acetic acid, stained with 1% crystal violet in methanol, and counted using ImageJ 1.38X software (NIH).
Assay of DSB-induced HR
HR assays in U2OS-DR cells were performed as previously described (Zhang et al., 2009a) with minor modification. Briefly, cells were transfected with siRNAs for each factor and incubated for 48-72 hours. pCBASce was then introduced using lipofectamine reagent or by electroporation. Cells were collected after 72 hours and analyzed using a FACSCalibur instrument (Becton-Dickinson, San Jose, CA). Doublets were gated out and the percentage of GFP-positive cells was calculated from three independent experiments as a measure of the efficiency of HR.
Chromatin fractionation assay
siRNA-treated cells were collected and extracted in lysis buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM sodium orthovanadate, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with protease inhibitor cocktail (Complete, Roche). After 5 minutes on ice, supernatant and chromatin-enriched pellet fractions were separated by centrifugation at 8000 r.p.m. for 2 minutes. Whole and pellet fractions were treated with 0.05 U/μl benzonase (Novagen) for 30 minutes on ice.
Immunofluorescence microscopy
Analysis using U2OS cells transfected with siRNAs was as described previously (Zhang et al., 2009a), except that cells were incubated with extraction buffer described above for chromatin fractionation for 2 minutes on ice before fixation for 20 minutes at room temperature in PBS containing 2% paraformaldehyde. Cells were washed for 5 minutes with PBS, then incubated with primary antibodies diluted in PBS with 3% bovine serum albumin, 0.05% Tween 20, 0.04% sodium azide for 60 minutes at 37°C. After three washes with PBS, cells were incubated for 30 minutes at 37°C with secondary antibodies diluted in the buffer described above. Cells were washed twice with PBS and mounted with Vectashield containing DAPI. Cells were examined using a Zeiss Axiovert 200M microscope.
To analyze rescue of siRNA-mediated defects in the assembly of foci, U2OS cells were stably transduced with pOZ or pOZ-FLAG-HA-MRG15. The pOZ vector was obtained from Y. Nakatani (Dana-Farber Cancer Institute, Boston, MA). Selection of cells was as described previously (Zhang et al., 2009a).
Sister chromatid exchange assay
48 hours following transfection with siRNAs, cells were incubated with 10 μM BrdU for 24 hours, washed and incubated for 20 hours in BrdU-free medium. Cells were then incubated with 1 μg/ml nocodazole for 4 hours and mitotic cells were selectively detached. Cells were swollen in 75 mM KCl, fixed with 75% methanol, 25% acetic acid, and spread on slides as described previously (Fan et al., 2009). DNA was denatured, cells were washed, and BrdU was detected using an anti-BrdU antibody (Becton-Dickinson), as described previously (Pinkel et al., 1985). Cells were processed for immunofluorescence microscopy as described above.
Complementation assay using siRNA-resistant MRG15
Mutated MRG15 cDNA that escapes from the siMRG15 cocktail was constructed by ligation-mediated site-directed mutagenesis (Sawano and Miyawaki, 2000) using two mutagenesis primers (398-GCAGAAACAGCGAGAACTT-416, and 788-CAAGAAATCTCGTGGAAAC-806). The mutagenized MRG15 (designated MRG15*) was inserted into the pFLAG-C1 vector (pFLAG-MRG15*). Cells were seeded onto six-well plates and transfected with siRNAs using RNAiMAX (Invitrogen). Cells were incubated for 1 day after siRNA transfection, and pFLAG-MRG15* or pFLAG-C1 plasmid was then transfected using Lipofectamine LTX (Invitrogen). Cells were subcultured into 60 mm dishes 6 hours after the transfection of the plasmids described above and cultured for 1 day before harvest and further analyses.
Supplementary Material
Acknowledgments
We thank K. Komatsu and M. Jasin for the DR-GFP system, Y. Nakatani for the pOZ vector, and J. Kobayashi for critical experimental advice. Grant support: NIH R01 HL085587 (P. R. Andreassen). Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (J. Nakayama). Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/7/1124/DC1
References
- Cai Y., Jin J., Tomomori-Sato C., Sato S., Sorokina I., Parmely T. J., Conaway R. C., Conaway J. W. (2003). Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278, 42733-42736 [DOI] [PubMed] [Google Scholar]
- Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., et al. (2005). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581-592 [DOI] [PubMed] [Google Scholar]
- Doyon Y., Selleck W., Lane W. S., Tan S., Cote J. (2004). Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24, 1884-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Zhang F., Barrett B., Ren K., Andreassen P. R. (2009). A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 37, 1740-1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S. N., Kirtane B. M., Podlutsky A. J., Pereira-Smith O. M., Tominaga K. (2007). Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to gamma ionizing radiation. FEBS Lett. 581, 5275-5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Kastan M. B. (1994). Cell cycle control and cancer. Science 266, 1821-1828 [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Ohtani Y., Hayakawa N., Shinmyozu K., Saito M., Ishikawa F., Nakayama J. (2007). RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells 12, 811-826 [DOI] [PubMed] [Google Scholar]
- Lai J. S., Herr W. (1992). Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89, 6958-6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R., Loizou J. I., Yang Y. G., Cuenin C., Li H., Wang Z. Q., Herceg Z. (2006). Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 8, 91-99 [DOI] [PubMed] [Google Scholar]
- Nakayama J., Xiao G., Noma K., Malikzay A., Bjerling P., Ekwall K., Kobayashi R., Grewal S. I. (2003). Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 22, 2776-2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Thompson L. H., Gray J. W., Vanderlaan M. (1985). Measurement of sister chromatid exchanges at very low bromodeoxyuridine substitution levels using a monoclonal antibody in Chinese hamster ovary cells. Cancer Res. 45, 5795-5798 [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. (2000). Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28, E78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld A. R., Apgar S., Dolios G., Wang R., Aaronson S. A. (2004). BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444-7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki M. S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S. (1999). Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19, 5166-5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark J. M., Hu P., Pierce A. J., Moynahan M. E., Ellis N., Jasin M. (2002). ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277, 20185-20194 [DOI] [PubMed] [Google Scholar]
- Sy S. M., Huen M. S., Chen J. (2009a). MRG15 is a novel PALB2 interacting factor involved in homologous recombination. J. Biol. Chem. 284, 21127-21131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy S. M., Huen M. S., Chen J. (2009b). PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA 106, 7155-7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy S. M., Huen M. S., Zhu Y., Chen J. (2009c). PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol. Chem. 284, 18302-18310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman A. R. (2001). Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J. Cell Sci. 114, 3591-3598 [DOI] [PubMed] [Google Scholar]
- Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006). Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719-729 [DOI] [PubMed] [Google Scholar]
- Xia B., Dorsman J. C., Ameziane N., de Vries Y., Rooimans M. A., Sheng Q., Pals G., Errami A., Gluckman E., Llera J., et al. (2007). Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39, 159-161 [DOI] [PubMed] [Google Scholar]
- Zhang F., Fan Q., Ren K., Andreassen P. R. (2009a). PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 7, 1110-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X. (2009b). PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 19, 524-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., Huang Q., Liu Q., Hao Q., Ding J. (2006). Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 34, 6621-6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.