Abstract
When and why did cell polarization arise? Recent work in bacteria and yeast suggests that polarization may have evolved to restrict senescence to one daughter during division, by enabling the differential segregation of damaged material. In more complex organisms, polarity functions have diversified to permit the differential inheritance of centrosomes, RNAs, proteins, and membranes, which is essential for the generation of diverse cell types from stem cells and for morphogenesis.
Aging and the origins of cell polarity
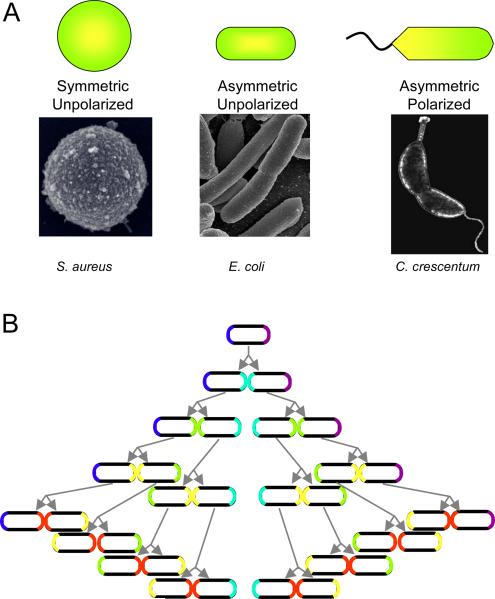
What do we mean by cell polarity? And how and why did it evolve? In broad terms, polarization implies the existence in any object not just of asymmetry but also of directionality. Actin microfilaments, for example, are polarized, while intermediate filaments and septin filaments are not, although all of these structures are asymmetric in the sense that they have a long and short axis. Applied to cells, the idea of directionality distinguishes morphologically unpolarized organisms from those that possess a clear polarity. This is most easily seen in unicellular organisms. For example, while S. aureus is spherical, the bacterium E. coli and the fission yeast S. pombe are asymmetric in the sense that their cell shapes are cylindrical, but the two poles of the cylinder appear to be identical. Morphologically, therefore, they are unpolarized. On the other hand, Vibrio cholerae or Caulobacter crescentum provide instances of prokaryotes that are highly polarized: each has a flagellum at only one pole (Figure 1A).
Figure 1.
Types of cell polarity. (A) Degrees of cell polarization, with examples. (Electron micrograph of S. aureus by Taeok Bae, Univ. of Chicago) (B) Inheritance of old poles by a symmetrically dividing cell, such as E. coli. The oldest pole is purple. One cell will always retain this pole, while the other pole will always be new. If proteins or other cellular components can recognize old from new poles, they can be segregated asymmetrically during each cell division.
Appearances can be deceiving, however, because E. coli and S. pombe exhibit functional polarity at a molecular level. Both organisms divide by extension of the long axis of the cylinder, which is then bisected by the formation of a septum. Nonetheless, the poles of each cylindrical cell are intrinsically different, since one is created de novo in each cell cycle, while one is retained from the mother (Figure 1B). Over several generations, one cell will inevitably inherit an increasingly old pole. Remarkably, this form of polarity, though subtle, turns out to be of crucial importance, because in E. coli the cell that retains the mother pole through several generations ages – that is, it becomes less fit and has a reduced growth rate (Barker and Walmsley, 1999; Stewart et al., 2005). Therefore, the two poles of these apparently unpolarized cells must be functionally distinct. Yet it is not apparent a priori why this should be so – cellular structures are generally dynamic, and the constituents of the old pole could in principle be continually replaced. Indeed, components of the system that defines the division plane in E. coli oscillate rapidly between the two poles (Lutkenhaus, 2007). Why, then, would a cell retain an old pole, and consequently age? An important clue is the recent observation that in E. coli, protein aggregates accumulate preferentially in the daughter that possesses the older pole (Lindner et al., 2008), suggesting that the bacterium uses polarity to ensure deleterious material is differentially inherited by aging cells (Nystrom, 2007). Although it remains to be proven that oxidized proteins or protein aggregates are causative factors in replicative aging, the correlation is strong (Desnues et al., 2003; Maisonneuve et al., 2008).
Replicative senescence also occurs in S. pombe but has not yet been tied directly to pole inheritance. Instead it correlates with an asymmetry in cell diameter (Barker and Walmsley, 1999). However, the spindle poles of S. pombe are distinct, such that during mitosis only one of them recruits a kinase necessary for cytokinesis (Cerutti and Simanis, 1999). It will be of interest to determine if the fatter (older) daughters accumulate oxidized proteins and correspond to daughters that inherit older cellular poles, or a specific spindle pole.
Similar behaviors have been observed in single-cell organisms with a more obviously polarized morphology, such as Caulobacter, or the budding yeast S. cerevisiae, and on the basis of these studies it has been proposed that the evolution of aging was contingent on the development of polarization (Ackermann et al., 2003). However, the coupling between inheritance of old poles and aging in unicellular organisms such as E. coli and S. pombe supports the opposite possibility: that the accumulation of damaged material is a problem common to all cellular organisms, and forced the evolution of cell polarity. Indeed, modeling studies support the rapid emergence of polarized cell division as a strategy to cope with accumulated damage (Ackermann et al., 2007). Without a differential inheritance mechanism that can actively segregate deleterious material into the “older” of two daughter cells either the cells must remove accumulating damage with 100% efficiency or the entire population would age and eventually die out. Lineage survival would be impossible. Polarized cell division, however, enables the rejuvenation of one daughter cell, at the expense of the other, in the absence of perfect damage repair. Hence, we propose that polarity evolved very early and is a universal and essential attribute of cellular organisms. A key test of this idea will be to determine whether S. aureus undergoes replicative senescence, and if the aging progeny accumulate oxidized protein aggregates. This bacterium is not only spherical (Fig. 1A) but divides successively over three generations along orthogonal planes (Giesbrecht et al., 1998), a remarkable process that ought to distribute cell components equally among the progeny, unless there exists a mechanism to anchor damaged material at a position offset from the three axes of division.
How might old poles recruit damaged proteins or other material in cells? No mechanism has yet been uncovered, but proteins targeted to poles might accumulate over time to higher concentrations in older poles versus the new poles that arise as a consequence of cell division. Transmembrane proteins that are attached to the cell wall and are therefore unable to diffuse away from the pole might anchor protein aggregates or other material, and the asymmetry in abundance of these transmembrane proteins between the two ends of the cell might then be sufficient to ensure an asymmetry in aging. If so, one prediction is that the over-expression or deletion of such anchoring proteins would disrupt the asymmetry and lead to eventual extinction of the population. However, if these proteins happen also to be essential for cell division, screens for aging genes in E. coli and S. pombe would be difficult to design.
Cell polarity, differential inheritance, and aging in budding yeast and multicellular organisms
The asymmetric cell divisions of the budding yeast, S. cerevisiae, have provided an important model system with which to investigate the molecular basis both of polarity and senescence. The asymmetry is of two types. Morphologically, the formation of daughter cells does not occur randomly over the surface of the mother cell, but either adjacent to the previous bud site (axial) or, in diploids, at the opposite end to the previous site (bipolar). Functionally, asymmetry arises because the mother can switch mating type during cell division. Moreover, the bud differentially inherits new material produced in the mother cell – RNAs, proteins, and vesicles – and delivered to the bud along actin cables. Older material is retained in the mother.
The overall process of bud formation in yeast involves four steps that generally apply to polarized cell divisions in many contexts (Park and Bi, 2007). First, a landmark, or spatial cue, must identify a unique zone of the cell cortex that will undergo polarized growth or modification – for example, next to the previous bud site. Second, signals must transmit this spatial information to drive asymmetric re-organization of the cytoskeleton – for example, actin cables that extend from the mother to the bud. Third, polarized transport of new materials to the marked region occurs along the cytoskeleton to the polarizing zone. And fourth, anchors and/or barriers are erected to limit the diffusion of these components away from the polarization zone. In S. cerevisiae, a cortical ring forms at the bud neck, composed of GTP-binding proteins called septins, which acts as a diffusion barrier (Gladfelter et al., 2001). In general, much is known about the signaling and polarized transport steps, while less is understood about the assembly of landmarks and anchors or diffusion barriers.
The position at which the bud forms in S. cerevisiae is easily scored because a scar is left in the cell wall after cytokinesis. Importantly, old bud scars are seldom re-used, so they provide not only a convenient marker of cell polarization but also of the age of the mother cell. Pedigree analysis led to the realization that mothers do not live forever (Mortimer and Johnston, 1959). The lifespan of a yeast cell is generally around 20–30 divisions. Moreover, as cells age, they lose certain aspects of asymmetry. Old mothers express genes for both mating types rather than just one, for example, and tend to produce larger daughters, which are short-lived (Sinclair et al., 1998).
Is aging of the mother cell in S. cerevisiae related to its retention of old, and potentially damaged material and by-products, as seems to be the case for E. coli? Aguilaniu and colleagues (Aguilaniu et al., 2003) showed that oxidatively damaged proteins do accumulate with replicative aging and are differentially inherited by the mother cell, through a mechanism that involves the formation of aggregates with the chaperone, Hsp104p (Erjavec et al., 2007). Strikingly, disruption of normal damage segregation reduces the lifespan of the daughter. Moreover, disruption of sir2, a gene essential for normal longevity, results in a failure to properly segregate oxidized proteins, and this defect can be ameliorated by over-expression of Hsp104p, which re-establishes anchoring of damaged proteins by the mother cell (Erjavec et al., 2007).
Why are damaged proteins not simply removed by the normal protein degradation machinery? Some oxidative modifications to proteins are reversible, but carbonylation, which is produced by metal-catalyzed reaction of amino acid side chains with reactive carbonyl groups, is irreversible and can trigger the formation of insoluble aggregates, which might be difficult to degrade. Nonetheless, multicellular organisms have evolved mechanisms to deal with this problem. For example, in the plant Arabidopsis thaliana, oxidation of proteins is sharply reduced prior to flower development (Johansson et al., 2004); and in mice damaged proteins are eliminated during early embryonic development (Hernebring et al., 2006). It is noteworthy that aggregated proteins segregate asymmetrically in the embryonic neuroblasts of Drosophila – but in this case they accumulate not in the daughter that differentiates but into the one that retains its stemness (Rujano et al., 2006). A similar situation has been observed for mammalian embryonic stem (ES) cells (Hernebring et al., 2006). These cells possess high levels of oxidized protein, which disappears when they differentiate, apparently as a result of upregulation of the 20S proteosome. Thus, in some cases it seems possible to rid cells of damaged proteins by degradation. Why budding yeast prefers segregation rather than degradation is unclear. Possibly the difference is related to unicellularity, for if oxidized proteins contribute to replicative aging, then their removal must be very highly efficient to prevent gradual accumulation over multiple generations, and the ultimate loss of the lineage. Replicative aging of stem cells within a multicellular organism is not necessarily deleterious, as long as it occurs predominantly after the production of “clean” germ cells, and progeny. Indeed, one might predict that germ stem cells would accumulate damaged material in order to protect the integrity of the differentiated gametes; and a related argument might even apply to somatic stem cells. Segregation of damaged proteins into one daughter of a unicellular organism might also be energetically more cost-effective than detection and degradation (Ackermann et al., 2007).
Replicative aging is a complex phenomenon, and is not likely to be caused by a single factor, such as protein oxidation. Indeed, aging in budding yeast has also been correlated with the accumulation of small, extrachromosomal DNA circles that arise from the ribosomal RNA locus (Sinclair and Guarente, 1997). There are 100 – 200 tandem repeats of the genes encoding the ribosomal RNAs, and homologous recombination between adjacent repeats occasionally generates a 3 μm circular rDNA molecule (Sinclair et al., 1998). These extrachromosomal rDNA circles (ERCs) accumulate over time, in part because they can replicate themselves once per S-phase. Their accumulation correlates with disintegration of the nucleolus and with senescence. Manipulations that increase the number of ERCs shorten the replicative lifespan of yeast. Moreover, increased expression of Sir2, which is an NAD-dependent histone deacetylase required for transcriptional silencing, reduces rDNA recombination and ERC formation, and increases lifespan. Deletion of sir2, in contrast, increases ERC formation and substantially reduces longevity (Kaeberlein et al., 1999).
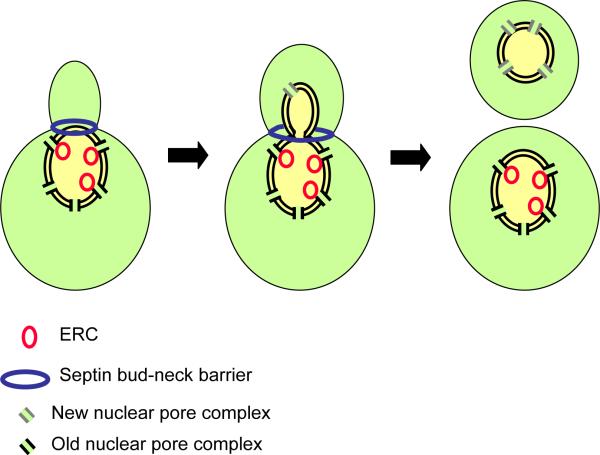
Strikingly, these DNA circles are confined to the mother cell. Thus, as we saw for E. coli, the daughter is protected from premature senescence by cell polarization. Failure to restrict the ERCs to the mother might explain why the daughters of old mothers are short-lived: they are born old. But how is differential inheritance of these ERCs accomplished? The ERCs are nuclear, after all, and the nuclear envelope remains intact during the closed mitosis of budding yeast, so no obvious mechanism could block the DNA circles from entering the daughter, or anchor them in the mother side of the nucleus. Shcheprova and colleagues (Shcheprova et al., 2008) have now provided an answer to this pivotal question. They show that DNA circles are anchored to pore complexes in the nuclear envelope, which are selectively retained by the mother (Figure 2). Diffusion of pre-existing pore complexes from the mother through the bud neck is prevented, perhaps by the septin ring, and new complexes are inserted into the nuclear envelope within the bud. Mutations in bud6, which disrupt the septin barrier, result in buds that fail to rejuvenate, and age prematurely. Thus, aging in yeast is controlled – at least in part – by the compartmentalization of cellular components between mother and daughter (Figure 2).
Figure 2.
A possible mechanism for the differential inheritance of DNA circles by budding yeast. During mitosis the nucleus penetrates into the bud, but a septin-based diffusion barrier at the bud neck prevents the movement of nuclear pore complexes out of the mother. DNA circles, which contribute to aging, are anchored by the nuclear pore complexes and thereby retained by the mother cell (Shcheprova et al. 2008).
Notably, extrachromosomal circles of DNA are not confined to budding yeast, and have been found in other eukaryotic organisms, including flies, frogs, and mammals (Cohen et al., 1999; Cohen et al., 2003; van Loon et al., 1994). Whether they are related to cellular aging will be a key question for future work. For example, during germ cell development in multicellular organisms, are these circles differentially inherited by the stem cells to avoid contamination of the gametes? Are diffusion barriers involved in germ cell development, and might these barriers also be based on septins?
Differential inheritance of centrosomes
The central concept outlined above is that, during an asymmetric cell division, not only is new material differentially inherited, but old components that either cannot be degraded or do not turn over at the same rate as cell division are also segregated in a polarized manner. Although polarity might have arisen originally as a solution to the problem of ensuring rejuvenated progeny, its functions have obviously diversified, particularly in more complex, multicellular organisms. Multiple cellular components are differentially inherited, sometimes to confer differential cell fates, but in other cases for reasons that remain unclear.
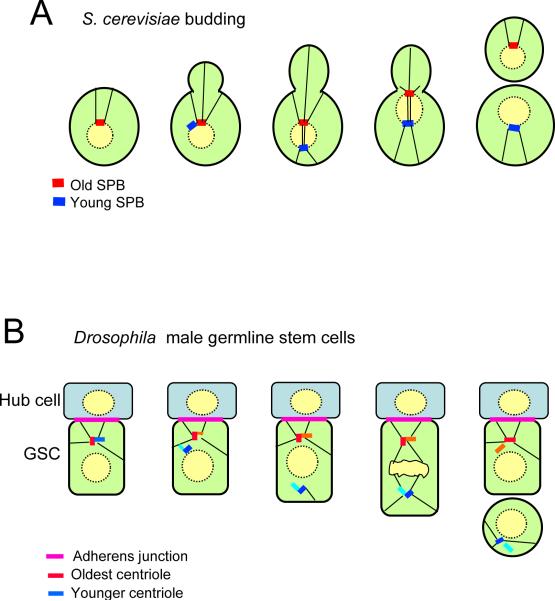
A particularly striking example of differential inheritance of cell components involves the spindle pole bodies (SPBs). These structures are the budding yeast equivalent of centrosomes, and are embedded in the nuclear envelope, which remains intact through mitosis (Figure 3A). They duplicate during interphase by a conservative mechanism, which generates an “old” SPB and a “young” SPB (Bornens and Piel, 2002). The two SPBs then separate to form the opposite poles of the mitotic spindle. Cytoplasmic microtubules orient the SPBs along the mother-bud axis, and begin to pull the nucleus through the bud neck. What is quite remarkable, however, is that it is always the old SPB that aligns with and enters the bud, while the new SPB remains in the mother (Pereira et al., 2001). Therefore, an intrinsic polarity is built into SPB inheritance (Figure 3A). The core components of the SPBs turn over only very slowly, so they may be inherited through multiple generations. Since the mother cell eventually ages and dies, the oldest SPB avoids this fate, since it is always passed on to the new bud, which one might refer to as the “immortal SPB hypothesis”. However, the function of this intriguing asymmetry is still a mystery. Although proteins required for mitotic exit associate specifically with the SPB that migrates into the bud, this asymmetry is unrelated to SPB age, and instead is coupled to proximity to the bud. Possibly the ability of the old and young SPBs to bind microtubules or to sense tension in the microtubules is different, and this might be important to ensure an efficient mitosis. Or perhaps the differential inheritance of other components is somehow dependent on the old SPB entering the bud.
Figure 3.
Asymmetric inheritance of spindle pole bodies (SPB) and centrosomes. (A) In budding yeast the old SPB is inherited by the daughter, while the young SPB is retained by the mother. (B) In the germline stem cells of Drosophila, the old centrosome remains in the stem cell, probably by attachment through microtubules to the adherens junction between the stem cell and the hub cell. The differentiating daughter inherits the young centrosome.
A distinction between the spindle poles is also seen in the fission yeast S. pombe. As mentioned above, in this organism a protein kinase is recruited to only one of the two spindle poles during anaphase. It is recruited by a small GTPase, which is switched off at the opposite pole by the specific recruitment of a GTPase activating protein (GAP) (Cerutti and Simanis, 1999). The kinase is required for septum formation. However, whether the young or old spindle pole recruits the GAP remains to be determined.
In higher organisms the structure equivalent to the SPB is the centrosome (Figure 3B). Each centrosome contains two centrioles, which, like the SPB, undergo conservative duplication (Bornens and Piel, 2002). Thus, the centrioles in any centrosome are non-equivalent: one will always have been assembled at least one generation before the other. Moreover, just as for yeast SPBs, the centrosomes behave differently, and can be differentially inherited. This polarity has perhaps been best described in the germ stem cells of Drosophila (Yamashita and Fuller, 2008). The male germline stem cells are located within a niche that determines their fate (Figure 3B). Cells that remain physically attached to the somatic hub cells in the niche remain as stem cells, while daughters that lose contact with the niche undergo differentiation. This asymmetry is assured by aligning the spindles, such that mitosis occurs in an orientation perpendicular to the hub cells, so one daughter cell can remain attached while the other cannot. How is this orientation arranged? It is probably determined by an adherens junction, which forms between the stem cell and the hub cell, and to which astral microtubules are anchored through the adenomatous polyposis coli (APC2) protein (Yamashita et al., 2003). Loss of adhesion allows the stem cells to drift away from the hub cells and lose their stem cell identify (Song et al., 2002; Wang et al., 2006). Strikingly, the daughter cell that adheres to the hub cells also inherits the old centrosome (Yamashita et al., 2007). Since centriole components turn over only very slowly, the germline stem cells will possess “immortal” centrioles that were assembled many generations earlier – just as happens with the SPB in budding yeast.
This asymmetry arises because young and old centrosomes are, for part of the cell cycle, functionally distinct. In interphase the old centrosome remains attached via microtubules to the adherens junction while the other does not associate with microtubules efficiently and migrates away. This difference in behavior may arise from differences in mature and immature centrioles. Mature centrioles – at least in mammalian cells - possess appendages that can capture microtubules, while immature centrioles lack appendages and can nucleate microtubules but not capture them. Since maturation takes 1. 5 – 2 generations, the two centrosomes differ from one another even though each contains two centrioles. Drosophila centrioles do not appear to contain similar appendages, but young centrioles are consistently shorter than old ones, which might confer a difference in function.
Does a similar inheritance pattern occur in other types of stem cells? During the asymmetric cell divisions of Drosophila larval neuroblasts the centrosomes also show distinct behaviors, in that one remains near the apical cortex, retains pericentriolar material, and maintains a robust attachment to the astral microtubules. The other wanders around the cell until moving just before mitosis to the basal side, where it acquires microtubule-organizing activity (Rebollo et al., 2007; Rusan and Peifer, 2007). However, whether the stationary centrosome always contains the oldest centriole remains to be determined. The development of new activatable fluorescent proteins such as Dronpa should make studies of centrosomal inheritance more tractable.
The intrinsic difference between centrosomes might permit the asymmetric inheritance of fate determinants and other cellular components. For example, as will be discussed below, during mollusc embryogenesis the centrosomes in certain cells differentially associate with specific mRNAs, which are then segregated asymmetrically during division (Kingsley et al., 2007; Lambert and Nagy, 2002). Centrosomes have recently been implicated as a major locus for protein degradation and, unexpectedly, show an asymmetric inheritance of associated polyubiquitinated proteins even during the symmetric cell divisions of somatic cell lines grown in culture (Fuentealba et al., 2008). What the function of such unequal distribution might be remains to be determined, as does the question of whether the young or old centrosome preferentially is associated with proteins marked for degradation. Intriguingly, other forms of unequal distribution have been described during apparently symmetric cell divisions. For example, during cytokinesis the midbody structure that accumulates prior to abcission is often asymmetrically inherited by one daughter cell (Goss and Toomre, 2008; Gromley et al., 2005), although in neural stem cells it is discarded into the extracellular space (Dubreuil et al., 2007).
Asymmetric cell division and differential inheritance of DNA
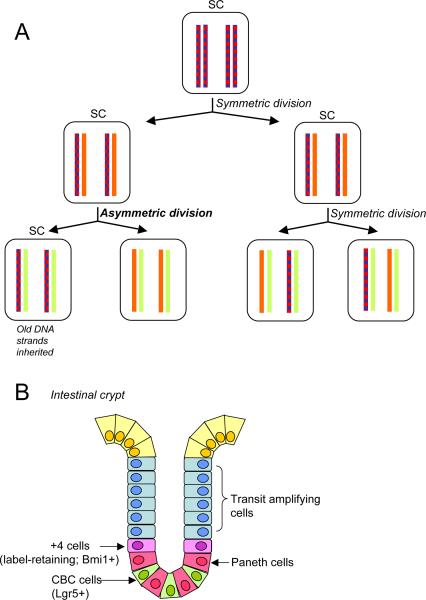
Many years ago, the accumulation of 3H-thymidine into murine embryonic fibroblasts was found to segregate in a nonrandom fashion, which was interpreted as denoting a differential inheritance of sister chromatids (Lark et al., 1966). With the development of the stem cell concept, the idea arose that the integrity of the stem cell genome might be maintained if, during a polarized cell division, the oldest template strand were to be always segregated into the daughter cell that retains stemness (Figure 4A). Errors occurring during DNA replication would be passed on to the differentiating daughter cell (Cairns, 1975, 2006). Epigenetic information would also be differentially inherited by this mechanism.
Figure 4.
The immortal strand hypothesis. (A) A schematic of differential retention of parental DNA strands by the stem cell. The first division shown is symmetric, leading to amplification of the stem cell population. The second shows one asymmetric division in which the stem cell retains the parental strands; while the other is a classical symmetric division. Blue represents BRdU labeling. (B) Schematic of a small intestinal crypt showing the organization of stem cells and other cell types. In this tissue both the +4 cell that shows BrDU label retention (an attribute of the immortal strand hypothesis) and crypt base columnar cells (CBC), which do not retain label, function as intestinal stem cells.
To test the “immortal strand hypothesis”, investigators have used long-term pulse-chase labeling of replicating DNA with BrdU, which identifies cells in which the label is retained over multiple divisions. In a number of cases, notably murine neural, muscle and intestinal stem cells, non-random partitioning of DNA has been reported. For the neural stem cells grown in vitro, the BrdU-retaining cells were actively dividing, and expressed markers for neural progenitors (Karpowicz et al., 2005). In other cases, however, such studies have often failed to detect asymmetric segregation of the DNA – for instance in C. elegans, and mouse embryos (Kiel et al., 2007; Kimble and Crittenden, 2007).
Moreover, recent lineage-tracing experiments have shown that – at least in the small intestinal epithelium – true stem cells do not necessarily retain BrdU label (Barker et al., 2007) (Figure 4B). The absorptive epithelium of the small intestine is folded into villi and crypts, and the label-retaining cells are located at “position +4” immediately above the Paneth cells near the base of the crypts (Potten et al., 2002). However, using lineage tracing from cells that express the G-protein coupled receptor Lgr5, the stem cells were identified as distinct, crypt base columnar cells that are localized between the Paneth cells, and beneath the +4 cells (Barker et al., 2007). Nonetheless, the +4 cells are self-renewing, essential for crypt maintenance, and can give rise - though much more slowly than the Lgr5 cells - to various differentiated cell types (Sangiorgi and Capecchi, 2008). They may function as quiescent, pluripotent progeny of the Lgr5 stem cells, or perhaps as a distinct stem cell population. Whatever the case, label retention can produce false positives, and whether the “immortal strand” is a common or important feature of stem cells remains an open question.
Differential inheritance of cell fate determinants
The differential inheritance of fate determinants is used to generate diverse cell types during development or, in yeast, to switch mating types. The fate determinants can be RNAs or proteins. These determinants have diverse functions, which ultimately control programs of gene expression that either suppress or drive cell differentiation along a particular lineage. For example, in asymmetric division of the fly neural precursors, the formation of differentiated progeny depends on a transcription factor called Prospero, but also on an adapter protein called Numb, which negatively regulates signaling through the Notch pathway (Knoblich, 2008). The mechanisms that ensure the differential inheritance of these factors during neuroblast division are known in considerable detail, although several issues remain unresolved.
Neuroblasts begin life as apico-basally polarized cells of the neuroectoderm (Zhong and Chia, 2008). A complex of conserved polarity proteins– Par-3, Par-6, and atypical protein kinase C (aPKC) – is localized at the apical surface of the ectoderm. Neurogenesis involves the extrusion of the neuroblasts from the epithelial sheet in a basal direction, through a process that is not well understood, but which retains the pre-established polarity. The apical surface of the neuroblast retains contact with the overlying epithelium, which is important for orientation during cell division (Siegrist and Doe, 2006). During neuroblast mitosis, the Par proteins act both to restrict fate determinants to the opposite pole of the cell, away from the epithelium, and to orient the spindle poles along the same axis, such that when the cell divides the Par proteins stay in the apical daughter, which retains stemness, while the fate determinants are segregated into the basal daughter, which differentiates (Knoblich, 2008). The retention of the Par proteins, and in particular aPKC, is important for self-renewal of the neuroblast (Lee et al., 2006).
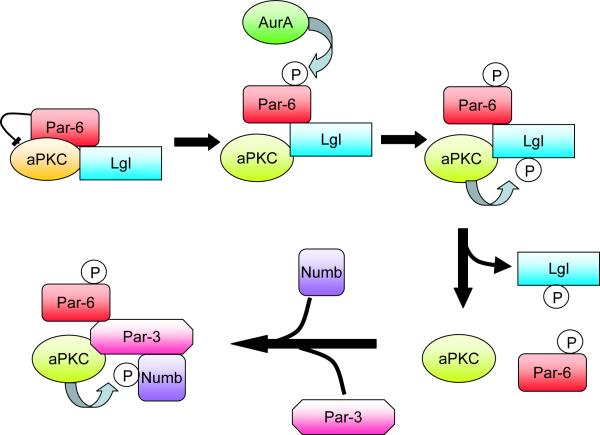
Despite the clear separation of the Par complex and fate determinants to opposite sides of the neural precursors, both sets of proteins are highly dynamic, so there must be an ongoing sorting process that occurs throughout asymmetric cell divisions. Exactly how this sorting occurs has been unclear, but a plausible mechanism has recently been suggested by Knoblich and coworkers (Wirtz-Peitz et al., 2008). They found that Aurora-A, a mitotic kinase required for Numb asymmetry, can phosphorylate Par-6 and release it from aPKC. Since Par-6 inhibits aPKC, the release stimulates aPKC kinase activity, which enables it to phosphorylate and inactivate Numb, releasing it from the cell cortex (Betschinger et al., 2003). Thus, Numb can only associate with the cortex in regions where the Par complex is absent (the basal region in neuroblasts). The underlying mechanism for Numb phosphorylation involves a switch in the specificity of the aPKC, driven by a change in binding partners (Figure 5). Initially, Par-6 and aPKC are associated with a different polarity protein, called Lgl. Phosphorylation of Lgl by active aPKC triggers its disassociation and replacement by Par-3. Par-3 recruits Numb, which is then phosphorylated. Thus, the initial phosphorylation of Par-6 triggers a change in binding partners that alters the substrate specificity of the aPKC (Wirtz-Peitz et al., 2008).
Figure 5.
A model for the regulation of Numb phosphorylation by aPKC during Drosophila asymmetric cell divisions. Phosphorylation of the Par-6 polarity protein releases its inhibitory effect on aPKC, which enables the kinase to phosphorylate Lgl. Lgl is then replaced by Par-3 in the complex, which recruits Numb to be phosphorylated. Phosphorylated Numb is released from the cell cortex. Since Par-3 is restricted to the cortex on one side of the cell, Numb becomes restricted to a cortical crescent on the opposite side (Wirtz-Peitz et al., 2008).
These phosphorylations can explain how fate determinants can be excluded from the cortical region where aPKC is localized, but they do not explain how they are anchored at the opposite pole. Two other fate determinants, Prospero and Brat, bind to an adapter that is essential for their basal localization in neuroblasts, but it remains unclear how the adapter is anchored. Moreover, Prospero needs to enter the nucleus after cell division in order to switch on the differentiation program (Shen et al., 1997), but what triggers this switch in localization remains enigmatic. Finally, we need to understand how the apical cap of Par proteins is maintained in the neuroblast after delamination. What prevents them from dispersing over the entire cell cortex? A likely possibility is that in fact the Par proteins do diffuse away from the apical region, but are excluded from the basal cortex. There is evidence for this type of mechanism in Drosophila epithelial cells, where a basolateral polarity protein called Par-1 phosphorylates Par-3, triggering its disassociation from the cortex. In mutants lacking functional Par-1, the Par-3 becomes dispersed down the lateral membrane (Benton and Johnston, 2003). Another possible mechanism to restrict dispersal is oligomerization, which could trap proteins in large complexes that would have low diffusion rates. There is evidence that Par-3 can form such oligomers (Feng et al., 2007). In addition, Par-3 has been reported to bind to phosphoinositides, which might contribute to its cortical localization (Wu et al., 2007). Possibly other polarity proteins and cell fate determinants use similar mechanisms for membrane attachment.
Oriented mitosis, coupled with the polarization of fate determinants, is a fundamental mechanism for generating diverse cell types that is used throughout the Eukaryotic domain. In many cases the molecular machinery required for orientation and polarization has been conserved, though not universally. For instance, the Par-3/Par-6/aPKC group of polarity proteins operates in the first asymmetric cell divisions of C. elegans, in Drosophila, and in vertebrates; and another set of proteins, including Pins, NuMA, and the Gαi subunit of heterotrimeric G-proteins, plays a key role in orienting the mitotic spindle in this same range of organisms; but none of these proteins are found in fungi or plants (Goldstein and Macara, 2007; Gonczy, 2008). Moreover, multiple mechanisms operate within different cell populations in the same organism. For example, the asymmetric divisions of Drosophila neuroblasts are cell autonomous, but those of the male germline stem cells are determined non-cell autonomously by adhesion to, and signaling from, adjacent hub cells (Figure 2B). Other signaling pathways also impact on spindle orientation. In the neuroblast, Par-3 at the apical crescent recruits an adapter protein called Inscuteable which in turn recruits Pins (Partner of Inscuteable) and Mud (the fly version of NuMA), which somehow – perhaps through dynein – attaches astral microtubules to the apical cell cortex, to ensure correct spindle orientation (Knoblich, 2008). However, a separate, redundant system is present that can rescue orientation defects later in the cell cycle, through the attachment of astral microtubules to Discs large, a polarity protein that can also bind to Pins (Siegrist and Doe, 2005). This same system operates in a different stem cell model – the sensory organ precursor cells of Drosophila - but in this case the localization of Discs large and Pins depends on a distinct signaling pathway, which controls planar cell polarity (PCP) (Bellaiche et al., 2004; Lu et al., 1999). This pathway might be of widespread importance in differential inheritance, and has been implicated in the orientation of cell division during gastrulation of zebrafish (Gong et al., 2004), and in asymmetric cell divisions of C. elegans (Kidd et al., 2005; Korswagen, 2002). In general, it seems likely that the core polarity proteins have evolved multiple functions, coupling to different signaling pathways in a context-dependent fashion. It will be important to dissect out these contextual functions since some might be independent of polarity, or link polarity to apoptosis or cell proliferation.
Differential inheritance of RNAs
Partitioning of RNA molecules into only one daughter cell at mitosis is another mechanism contributing to differential cell fate specification during asymmetric cell division (Figure 6). The budding yeast, S. cerevisiae, offers one of the best-characterized examples of this phenomenon. The ASH1 mRNA concentrates in the newly formed bud and is inherited by the daughter cell (Gonsalvez et al., 2005). The resulting Ash1p protein prevents mating type switching of the daughter cell. Mother cells do not express Ash1p and can switch mating type, thus allowing mating and formation of diploid cells. Formation of such diploid cells can contribute to survival of a population, because diploid cells are more efficient in repairing DNA damage and can meiotically divide and give rise to spores upon nutritional starvation.
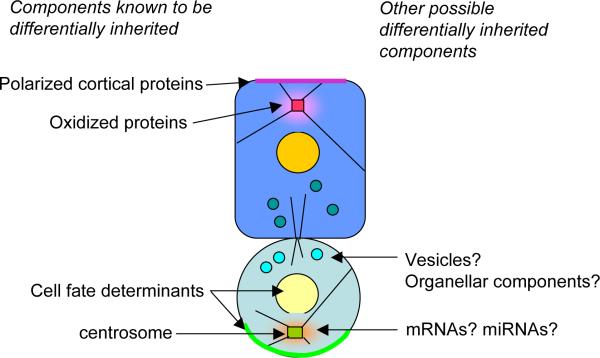
Figure 6.
Summary of known and potential cellular components that are differentially inherited during asymmetric stem cell divisions.
Asymmetrically distributed RNAs are also widespread in oocytes and early embryos, where they function as localized determinants that control axis formation and cell fate specification (King et al., 2005; Palacios and St Johnston, 2001). For example, P-granules in the C. elegans zygote, which are differentially inherited by the P1 cell during the first asymmetric cell division, contain a number of RNAs that are essential for gametogenesis. In Drosophila, similar granules that localize to the posterior pole of the oocyte (from which the germline will develop) contain a RNA responsible for transcriptional repression of somatic genes. A recent global analysis has revealed that during early Drosophila embryogenesis, 71% of mRNAs exhibit a variety of distinct localization patterns, pointing to a remarkable complexity of localization mechanisms and functions (Lecuyer et al., 2007).
The mechanisms underlying RNA localization have been intensely studied and have led to a general model according to which, localization involves active transport of the RNA on microtubules or actin filaments followed by anchoring at the final destination (Gonsalvez et al., 2005; St Johnston, 2005). Targeting is directed by specific sequence elements within the RNA. These elements are recognized by RNA-binding proteins, which in turn couple the RNA, directly or indirectly, to molecular motors (kinesin, dynein or myosin) that transport the RNA to the site of anchoring. However, this is likely a simplified version of the in vivo situation.
For instance, in several different systems, there is a little understood connection between RNA transport and endoplasmic reticulum (ER) trafficking. Yeast mRNAs are co-transported with cortical ER into the incipient bud; in Xenopus oocytes vegetal cortex-localized RNAs associate with ER membranes; gurken mRNA in Drosophila oocytes localizes to ER exit sites and proper secretion of Gurken protein requires a ribonucleoprotein complex that is found at ER exit sites and mediates their formation (Gerst, 2008). Interestingly, a connection with ER trafficking is not only exhibited by mRNAs encoding secreted or trans-membrane factors but also by mRNAs encoding soluble cytosolic or nuclear proteins. For example, the mRNAs encoding Ash1, a transcriptional repressor, and Sro7, a SNARE regulator and member of the lethal giant larvae (Lgl) tumor suppressor family, associate with and are co-transported with the cortical ER into the growing yeast bud (Aronov et al., 2007; Schmid et al., 2006). Therefore, mRNA-ER co-trafficking does not appear to be simply a means of increasing the efficiency of targeting of nascent proteins to the secretory pathway. Similar observations of mRNAs for soluble cytosolic proteins partitioning with the ER have also been reported. However, the functional significance of this partitioning remains unclear (Gerst, 2008; Nicchitta et al., 2005).
An additional point of complexity concerns the association and transport of RNAs on cytoskeletal tracks. High resolution, live-cell imaging experiments in mammalian cells have suggested that the presence of a localization element/zipcode within an mRNA does not simply mediate association of the RNA with cytoskeletal motor complexes (Bullock et al., 2006; Fusco et al., 2003). On the contrary, even non-localized, uniformly distributed RNAs associate with and randomly move along cytoskeletal tracks, and this appears to be important for the ability of these RNAs to uniformly diffuse in the viscous cytoplasmic environment. Polarized RNA distribution, when a localization element is present, results from alterations in the frequency, velocity and duration of movement towards a specific direction.
Once at their final destination, RNAs are anchored through various mechanisms. In some cases, a role for actin or actin-associated proteins has been suggested but without knowledge of the exact molecular details. A distinct mechanism operates in the case of gurken and of pair rule mRNAs in Drosophila. Anchoring depends on the microtubule motor dynein,which is converted to a static anchor at the final destination (Delanoue and Davis, 2005; Delanoue et al., 2007). Another microtubule-based mechanism involves anchoring of a large group of RNAs at microtubule plus ends through the APC tumor suppressor (Mili et al., 2008). Finally, in Xenopus oocytes, coding and non-coding RNAs have been suggested to exert structural roles in maintaining the integrity of the cytokeratin cytoskeleton at the vegetal cortex. Destruction of these transcripts leads to disruption of the cytokeratin network and, in turn, prevents anchoring of other RNAs at the vegetal cortex (Kloc et al., 2005).
Apart from targeted transport of RNAs or their anchoring at sites that will be inherited by one daughter cell, an additional mechanism leading to asymmetric inheritance of RNAs has been described during early embryonic cleavages of mollusk embryos. It involves association of different mRNAs with centrosomes which are subsequently asymmetrically inherited upon cell division (Lambert and Nagy, 2002). Centrosomal RNA has been detected also in other organisms (Lecuyer et al., 2007; Pederson, 2006). It will be of interest to determine whether centrosomal RNA inheritance correlates with differential inheritance of young and old centrosomes.
The functional significance of asymmetrically inherited RNAs is clear only in certain cases. For example, localization of ash1 mRNA in the yeast bud is required for expression of the Ash1 protein specifically in the daughter cell. Failure to localize the RNA leads to distribution of Ash1p throughout the mother and daughter cells and prevents mating type switching in both of them. In other cases, however, RNA localization is not required for asymmetric protein distribution. Mutants that cannot properly localize prospero mRNA in Drosophila neuroblasts, can still segregate the Prospero protein through the use of protein targeting signals, and develop normal ganglion mother cells. In such cases, RNA localization may serve as a back-up mechanism that ensures efficient protein segregation. Other roles are likely carried out by RNAs that are localized in the yeast bud but give rise to proteins that are not asymmetrically distributed (Shepard et al., 2003). Mislocalization of these RNAs does not appear to significantly affect the fitness of the progeny. It is possible that the role of such RNAs is not directly related to localized protein translation but rather to translation-independent functions. Alternatively, the increased RNA concentration or the “new” proteins produced from them might provide the daughter cell with a fitness advantage that has not been detected with the assays used thus far.
Are messenger RNAs the only types of RNAs involved in cell fate specifications? Likely not; piRNAs, a class of 21-nucleotide small RNAs, enriched in the germline, are additional candidates. In C. elegans, they interact with the Piwi family member PRG-1, which localizes to P granules, cytoplasmic structures that are asymmetrically transferred to daughter cells and specify the germ cell lineage (Batista et al., 2008).
Differential inheritance and vesicular traffic
A priori, one might imagine that polarized vesicle traffic would play a major role in asymmetric cell divisions, but with the exception of budding yeast there has been remarkably little work to investigate this possibility (Figure 6). For example, before the Drosophila embryonic neuroblast delaminates from the epithelium it possesses an apical/basal polarity generated by polarized membrane sorting (Knoblich, 2008). Therefore, different transmembrane proteins will populate the apical cortex of the neuroblast as compared to the basolateral cortex, and the neuroblast daughter retains these apical proteins during each asymmetric division. However, whether specific membrane proteins function in these divisions, for instance to retain the Par complex in the apical crescent, remains to be determined. A similar membrane polarity is necessary for the asymmetric cell divisions of the male germline stem cells of Drosophila. As discussed above, these cells form an adherens junction with the hub cells adjacent to them. During cell division, the daughter that retains the junction remains as a stem cell, while the other daughter differentiates. Polarized vesicular delivery of E-cadherin to the region of the cortex in contact with the hub cell is necessary to assemble and maintain the adherens junction (Song et al., 2002). However, it is not known whether other types of vesicles are delivered specifically to the differentiating daughter cells in these examples.
In S. cerevisiae, bud formation depends absolutely on the polarized delivery of new vesicles to the nascent bud (Casamayor and Snyder, 2002). The vesicles, which carry enzymes for cell wall synthesis as well as landmark factors and other transmembrane proteins, are transported by a myosin motor along actin cables to the bud, where they are anchored prior to fusion with the plasma membrane. Delivery is initially focused on the apical tip of the bud, but later spreads over the entire bud surface, until just prior to cytokinesis, when it repolarizes in the opposite direction, to deliver vesicles to the bud neck between mother and daughter. Since the septin ring forms a diffusion barrier at the neck, newly synthesized proteins delivered to the bud plasma membrane may be trapped in the bud, undiluted by old proteins from the mother.
Anchoring of the vesicles involves an interaction between a large protein complex on the plasma membrane, called the exocyst, and a Rab GTPase associated with the vesicle. Exocyst localization to the bud tip depends on the localized activation of Rho family GTPases (Brennwald and Rossi, 2007). Fusion of the vesicles to the plasma membrane is driven by the formation of a complex of SNARE proteins derived from the two membrane compartments. Interestingly, a yeast homologue of the Lgl polarity protein is believed to inhibit exocytosis by sequestering the SNARE on the plasma membrane (Hattendorf et al., 2007) – a mechanism perhaps related to the role of Lgl in inhibiting Par-6/aPKC function in neural precursors of Drosophila.
There are hints that polarized vesicle transport is an important component of differential inheritance during asymmetric divisions in both Drosophila and C. elegans. For example, retrograde traffic from the endosome to the Golgi has recently been implicated in the terminal asymmetric division of epithelial stem cells in C. elegans (Kanamori et al., 2008). Differential inheritance of β-catenin between daughters of these cells determines their subsequent fate. Although β-catenin is a soluble protein, other components of the Wnt polarity pathway are transmembrane proteins that are delivered to the plasma membrane in vesicles. The retrograde transport pathway might reduce the sorting of these polarity proteins into lysosomes, where they would otherwise be degraded. A phospholipase A1, which could regulate membrane traffic, appears to have the opposite effect, either inhibiting retrograde transport or stimulating lysosomal targeting (Kanamori et al., 2008). Strikingly, in addition to controlling the polarized distribution of β-catenin, the phospholipase and the retrograde transport system are also required for correct spindle orientation in these stem cells. Possibly there is a differential delivery of vesicles containing a cortical factor that interacts with the astral microtubules.
During the first asymmetric division of sensory organ precursor cells in Drosophila, recycling endosomes accumulate around the centrosome of one daughter cell (pIIb) but not the other, and this is a key event in controlling Notch/Delta signaling (Emery et al., 2005). Two regulators of the Notch signaling pathway are segregated into the pIIb cells. These proteins both act on the Notch pathway by promoting endocytosis either of Notch itself, or of the Notch ligand, Delta. The differential signaling between the daughter cells then determines their fate in subsequent divisions. Interestingly, Jafar-Nejad et al. found that a mutation in a subunit of the exocyst causes a defect in fate determination during these divisions that is consistent with a loss of Notch signaling (Jafar-Nejad et al., 2005). The implication is that differential vesicle trafficking in the two daugher cells ensures that only one of them (pIIb) becomes a signal-sending cell, while the other receives the signal, and this directionality determines subsequent cell fates.
These two examples hint at a broad involvement of polarized vesicle traffic in asymmetric cell division and differential inheritance of cellular components – an involvement that has barely begun to be investigated.
Conclusions
We have discussed several ideas in this review that we feel are worth exploring experimentally. First, we suggest that polarity is a universal attribute of cellular organisms, that arose initially not to control morphogenesis but as a solution to the pivotal problem of lineage senescence. A prediction of this idea is that even S. aureus, an apparently symmetric and unpolarized organism, will exhibit replicative senescence. Polarized cell divisions enable the segregation of damaged materials into one daughter cell, enabling the other daughter to survive through multiple future divisions, but how is this segregation accomplished? Are nuclear pore complexes involved in the segregation of cellular components in organisms other than budding yeast? Does the septin ring at the yeast bud neck function to trap DNA circles and oxidized protein/chaperone complexes in the mother cell? Are there membrane-bound receptors for oxidized proteins? In eukaryotic cells, are centrosomes involved in sorting damaged proteins between daughters? Second, we suggest that differential inheritance of cellular components might be much more widespread than is generally appreciated. How many RNAs are differentially inherited during stem cell divisions? Are RNAs segregated between the old and young centrosomes in stem cell divisions? Are micro-RNAs segregated into different daughter cells? How important is polarized vesicle transport in such divisions? Do transmembrane proteins function as receptors to recruit polarity proteins or attach astral microtubules during asymmetric cell divisions? Do such receptors define the segregation of fate determinants between daughters? Are other cellular components such as mitochondria or endoplasmic reticulum, or Golgi, ever differentially inherited, for instance such that newly synthesized organelles are preferentially segregated into one daughter (Figure 6)? The advent of photo-activatable and switchable fluorescent proteins should enable some of these questions to be answered fairly easily. The answers to other questions will require sophisticated screens that will be more difficult to implement. Finally, given the central importance of stem cells in all aspects of the development and morphogenesis of higher organisms, and in many types of cancer, the most pressing need for studies in mammals is the development of a robust, experimentally tractable, in vitro model for asymmetric cell division.
Acknowledgements
We thank Deborah Lannigan, Anne Spang, the reviewers, and members of the Macara lab for critical reading of the manuscript and many helpful suggestions. Supported by grant GM 070902 (to IM) and Leukemia and Lymphoma Society fellowship 5028-06 (to SM).
REFERENCES
- Ackermann M, Chao L, Bergstrom CT, Doebeli M. On the evolutionary origin of aging. Aging Cell. 2007;6:235–244. doi: 10.1111/j.1474-9726.2007.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M, Stearns SC, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3441–3455. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MG, Walmsley RM. Replicative ageing in the fission yeast Schizosaccharomyces pombe. Yeast. 1999;15:1511–1518. doi: 10.1002/(sici)1097-0061(199910)15:14<1511::aid-yea482>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Mol Cell. 2008 doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaiche Y, Beaudoin-Massiani O, Stuttem I, Schweisguth F. The planar cell polarity protein Strabismus promotes Pins anterior localization during asymmetric division of sensory organ precursor cells in Drosophila. Development. 2004;131:469–478. doi: 10.1242/dev.00928. [DOI] [PubMed] [Google Scholar]
- Benton R, Johnston DS. Drosophila PAR-1 and 14-3-3 Inhibit Bazooka/PAR-3 to Establish Complementary Cortical Domains in Polarized Cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Bornens M, Piel M. Centrosome inheritance: birthright or the privilege of maturity? Curr Biol. 2002;12:R71–73. doi: 10.1016/s0960-9822(01)00678-9. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Rossi G. Spatial regulation of exocytosis and cell polarity: yeast as a model for animal cells. FEBS Lett. 2007;581:2119–2124. doi: 10.1016/j.febslet.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–1072. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Simanis V. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J Cell Sci. 1999;112(Pt 14):2313–2321. doi: 10.1242/jcs.112.14.2313. [DOI] [PubMed] [Google Scholar]
- Cohen S, Menut S, Mechali M. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol Cell Biol. 1999;19:6682–6689. doi: 10.1128/mcb.19.10.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 2003;13:1133–1145. doi: 10.1101/gr.907603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13:523–538. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Desnues B, Cuny C, Gregori G, Dukan S, Aguilaniu H, Nystrom T. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 2003;4:400–404. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Brauninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu H, Chan LN, Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. Embo J. 2007;26:2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci U S A. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst JE. Message on the web: mRNA and ER co-trafficking. Trends Cell Biol. 2008;18:68–76. doi: 10.1016/j.tcb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gong Y, Mo C, Fraser SE. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430:689–693. doi: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Urbinati CR, Long RM. RNA localization in yeast: moving towards a mechanism. Biol Cell. 2005;97:75–86. doi: 10.1042/BC20040066. [DOI] [PubMed] [Google Scholar]
- Goss JW, Toomre DK. Both daughter cells traffic and exocytose membrane at the cleavage furrow during mammalian cytokinesis. J Cell Biol. 2008;181:1047–1054. doi: 10.1083/jcb.200712137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattendorf DA, Andreeva A, Gangar A, Brennwald PJ, Weis WI. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature. 2007;446:567–571. doi: 10.1038/nature05635. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Johansson E, Olsson O, Nystrom T. Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem. 2004;279:22204–22208. doi: 10.1074/jbc.M402652200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T, Inoue T, Sakamoto T, Gengyo-Ando K, Tsujimoto M, Mitani S, Sawa H, Aoki J, Arai H. Beta-catenin asymmetry is regulated by PLA1 and retrograde traffic in C. elegans stem cell divisions. Embo J. 2008;27:1647–1657. doi: 10.1038/emboj.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P, Morshead C, Kam A, Jervis E, Ramunas J, Cheng V, van der Kooy D. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd AR, 3rd, Miskowski JA, Siegfried KR, Sawa H, Kimble J. A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell. 2005;121:761–772. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, Morrison SJ. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- Kingsley EP, Chan XY, Duan Y, Lambert JD. Widespread RNA segregation in a spiralian embryo. Evol Dev. 2007;9:527–539. doi: 10.1111/j.1525-142X.2007.00194.x. [DOI] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Korswagen HC. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. Bioessays. 2002;24:801–810. doi: 10.1002/bies.10145. [DOI] [PubMed] [Google Scholar]
- Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420:682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- Lark KG, Consigli RA, Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154:1202–1205. doi: 10.1126/science.154.3753.1202. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci U S A. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Usui T, Uemura T, Jan L, Jan YN. Flamingo controls the planar polarity of sensory bristles and asymmetric division of sensory organ precursors in Drosophila. Curr Biol. 1999;9:1247–1250. doi: 10.1016/s0960-9822(99)80505-3. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Ezraty B, Dukan S. Protein aggregates: an aging factor involved in cell death. J Bacteriol. 2008;190:6070–6075. doi: 10.1128/JB.00736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Lerner RS, Stephens SB, Dodd RD, Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- Nystrom T. A bacterial kind of aging. PLoS Genet. 2007;3:e224. doi: 10.1371/journal.pgen.0030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Johnston D. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The centrosome: built on an mRNA? Nat Cell Biol. 2006;8:652–654. doi: 10.1038/ncb0706-652. [DOI] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. Embo J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jaedicke A, Du TG, Jansen RP. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- Sinclair D, Mills K, Guarente L. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Song X, Zhu CH, Doan C, Xie T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science. 2002;296:1855–1857. doi: 10.1126/science.1069871. [DOI] [PubMed] [Google Scholar]
- Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon N, Miller D, Murnane JP. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 1994;22:2447–2452. doi: 10.1093/nar/22.13.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Singh SR, Zheng Z, Oh SW, Chen X, Edwards K, Hou SX. Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell. 2006;10:117–126. doi: 10.1016/j.devcel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]