Abstract
Phospholipases are a large and diverse set of enzymes that metabolize the phospholipid components of cell membranes and function in key lipid signaling pathways. The molecular characterization of novel phospholipases would benefit from chemical probes that selectively target these enzymes based on their distinct substrate specificity and catalytic properties. Here, we present the synthesis and characterization of a set of activity-based protein profiling (ABPP) probes that contain key recognition and reactivity elements for targeting phospholipases of the serine hydrolase superfamily. We show that these probes accurately report on the sn-1 and sn-2 substrate specificities of phospholipases in cell and tissue proteomes, including the sn-1 selective phoshpolipase DDHD1 and a calcium-dependent transacylase activity implicated in endocannabinoid biosynthesis. We anticipate that these phospholipase-directed ABPP probes will facilitate the discovery of new lipid-metabolizing enzymes and provide valuable insights into their substrate preferences.
Phospholipases are a large and diverse set of enzymes that metabolize the phospholipid components of cell membranes1,2 and regulate key lipid signaling pathways, such as those that generate eicosanoids3 and endocannabinoids4. Although many phospholipases have been molecularly characterized, others remain poorly defined as unenriched activities in crude cell or tissue extracts. Phospholipases also share considerable sequence homology with other hydrolytic enzymes that accept distinct substrates, such as neutral lipids2. The identification of novel phospholipases would thus benefit from chemical probes that selectively target these enzymes based on their distinct substrate specificity and catalytic properties. Here, we present the synthesis and characterization of a set of activity-based protein profiling (ABPP5) probes that contain key recognition and reactivity elements for targeting functional subclasses of phospholipases. We show that the labeling profiles of these probes accurately report on the substrate specificity of both characterized and uncharacterized phospholipases in diverse proteomic backgrounds.
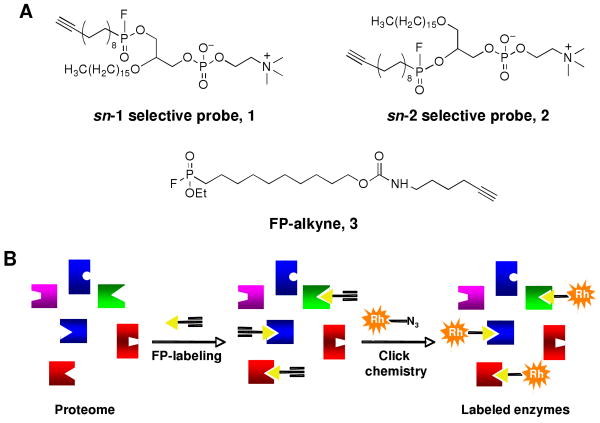
A substantial number of phospholipases belong to the serine hydrolase (SH) superfamily of enzymes, imparting upon them susceptibility to proteomic profiling using the fluorophosphonate (FP) class of ABPP probes. Original FP probes6,7 and variants thereof8 are rather generic in structure, which grants them broad reactivity across the SH class, but does not inform on the possible substrate preferences of individual SH enzymes. To target serine phospholipases, we designed and synthesized phosphatidylcholine (PC) probes containing reactive FP elements at either the sn-1 or sn-2 positions of a PC scaffold (1 and 2, Figure 1A and Supporting Figure 1) connected to an alkyne for click chemistry conjugation to azide-reporter tags9,10 (Figure 1B). We hypothesized that these probes would prove more selective for reacting with phospholipases (over other SHs) than a general FP-alkyne probe (3, Figure 1A and Supporting Figure 2), as they resemble the natural substrates of these enzymes. Probes 1 and 2 might further distinguish between phospholipase A1 (PLA1) and A2 (PLA2) activities, thereby providing additional information on the substrate specificity of targeted enzymes.
Figure 1.
ABPP strategy for serine phospholipases. (A) FP probes that target functional subclasses of phospholipases. (B) Detection of probe-labeled enzymes by click chemistry. Rh, rhodamine.
For initial characterization of probes 1 and 2, we assessed their reactivity with the mammalian serine phospholipase DDHD1, which shows selective PLA1 (but not PLA2) activity11. We also assessed the probe reactivity of DDHD2, a homologous enzyme to DDHD1 with less well-characterized phospholipase activity. HEK293T cells were transfected with cDNAs encoding FLAG-tagged DDHD1 and DDHD2 enzymes. DDHD1 and DDHD2 were then affinity purified with an anti-FLAG antibody to afford enriched enzyme samples (Supporting Figure 3), and these enzymes were then treated with probes 1, 2 or 3 (10 μM) at 37 °C for 1 h. Labeled enzymes were subjected to click chemistry with rhodamine–N3 (Rh–N3) and analyzed by SDS-PAGE and in-gel fluorescence scanning (Figure 2). Both enzymes readily reacted with FP-alkyne 3, as expected for serine phospholipases. Notably, DDHD1 showed much greater reactivity with probe 1 compared to 2, consistent with the reported PLA1-selective catalytic activity of this enzyme11. In contrast, DDHD2 displayed similar reactivity with both probes 1 and 2. To quantify the relative reactivity of 1 and 2 with DDHD lipases, we treated DDHD1 and DDHD2 with probes at concentrations ranging from 3 nM to 50 μM and quantified the labeling signals. Probe 1 potently reacted with DDHD1, showing half-maximal labeling at ∼300 nM, while probe 2 exhibited much weaker labeling (partial labeling at 20-50 μM, Supporting Figure 4). Both probes reacted similarly with DDHD2, showing half-maximal labeling at low μM concentrations (1-3 μM). These data demonstrate that probes 1 and 2 accurately report on the PLA1-selective catalytic activity of DDHD1, and suggest further that DDHD2, which displayed similar reactivity with both probes, might exhibit both PLA1 and PLA2 activity12.
Figure 2.
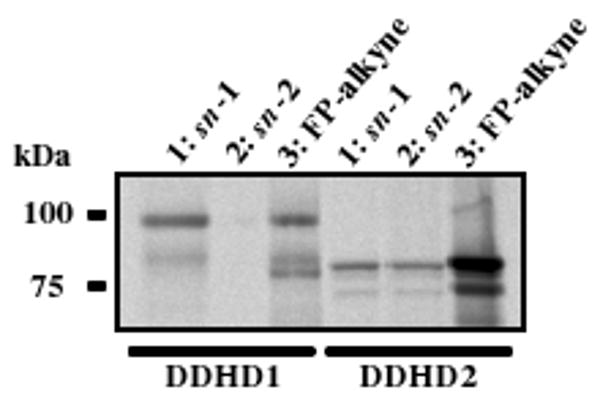
DDHD1 and DDHD2 labeling with FP probes. 1-3 (10 μM) were incubated with DDHD1 and DDHD2 for 1 h at 37 °C, reacted by click chemistry with Rh–N3, and reactions visualized by SDS-PAGE and in-gel fluorescence scanning (fluorescent gel shown in grayscale).
We next compared the labeling profiles of probes 1-3 in mouse tissue proteomes (Figure 3). Several proteins were detected that showed selective reactivity with either probe 1 (red asterisks) or 2 (blue asterisks). Interestingly, a subset of 2-labeled proteins detected in the soluble testis proteome did not react with either probes 1 or 3, suggesting that they may be PLA2-type enzymes that preferentially react with substrate-like ABPP probes.
Figure 3.
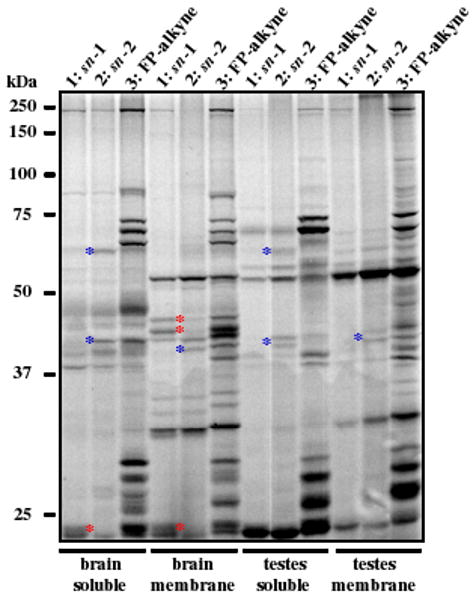
ABPP of mouse tissue proteomes with probes 1-3. Soluble and membrane proteomes (2 μg/mL) were incubated with 1 and 2 (10 μM) or 3 (2 μM) for 1 h at 37 °C. Probe-labeled enzymes were reacted with Rh–N3 by click chemistry, resolved by SDS-PAGE, and detected by in-gel fluorescence scanning. Red and blue stars mark proteins selectively labeled with the sn-1 or sn-2 probes, respectively.
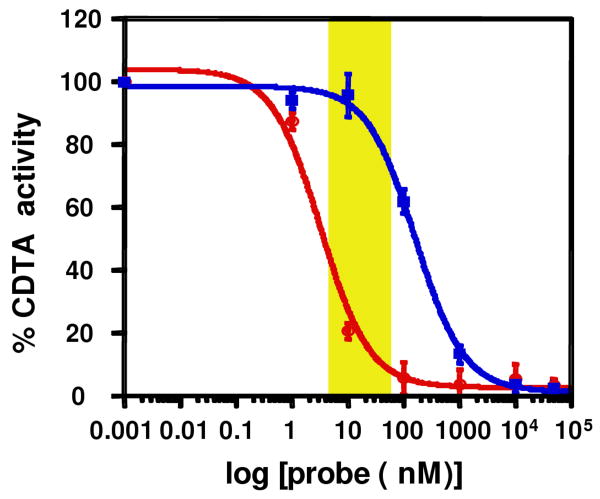
Some important phospholipase activities have to date only been characterized in tissue/cell extracts, and the enzyme(s) responsible for these activities remain unidentified. One such phospholipase activity is the calcium-dependent transacylase (CDTA) enzyme implicated in the biosynthesis of N-acyl ethanolamines (NAEs), including the endocannabinoid anandamide4. CDTA activity has been detected in specific rodent tissues, such as brain13, where it transfers the sn-1 acyl chain of PC to phosphatidylethanolamine (PE) to form N-acyl PE (NAPE), which is then converted to NAEs by one of multiple pathways4. Despite extensive research efforts13,14, the molecular identification of CDTA has not yet been achieved, likely indicating that it is a very low abundance enzyme. Affinity probes that selectively react with CDTA could prove of value for enriching and identifying this enzyme from tissue proteomes. Toward this end, we found that CDTA is inhibited by the sn-1 probe 1 with much greater potency than the sn-2 probe 2 (IC50 values of 3 nM and 165 nM, respectively; Figure 4). Future comparative ABPP studies that characterize the protein targets of probes 1 and 2 over a concentration range where they show differential reactivity with CDTA (yellow box, Figure 4) may facilitate the identification of this enzyme.
Figure 4.
CDTA inhibition with probes 1 and 2. Rat brain cortical membrane proteome (50 μg) was incubated with 1 or 2 (0.001 nM-50 μM) in the presence of 2 mM DTT, 6 mM CaCl2 and 0.5% NP40 for 1 h at 37 °C and assayed for CDTA activity using 1,2-[14C]-PC. Formation of NAPE was monitored by thin layer chromatography and visualized by autoradiography. Curves represent average values ± standard error (SE) for three experiments (see Supporting Figure 5 for primary autorad data). The yellow box marks a concentration range over which probe 1 selectively inhibited CDTA activity.
In conclusion, we have synthesized and characterized ABPP probes that target functional subclasses of serine phospholipases. Placement of an alkyne-conjugated FP reactive group in the sn-1 or sn-2 position of PC provided probes 1 and 2, whose reactivity profiles accurately reported on the sn-1 specificity of both the DDHD1 and CDTA enzymes. Considering that several additional proteins that selectively reacted with 1 or 2 were identified in tissue proteomes, we anticipate that these probes, and variants thereof15, will facilitate the discovery of new serine phospholipases with sn-1 and/or sn-2 specificity.
Supplementary Material
Acknowledgments
This work was supported by the Damon Runyon Cancer Foundation (S.E.T., DRG: 1978-08), the NIH (CA087660), and the Skaggs Institute for Chemical Biology.
Footnotes
Supporting Information Available: Synthesis, experimental protocols and figures showing overexpression of DDHD1 and DDHD2 and substrate assays with CDTA, DDHD1 and DDHD2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Schaloske RH, Dennis EA. Biochim Biophys Acta. 2006;1761:1246–59. doi: 10.1016/j.bbalip.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Kienesberger PC, Oberer M, Lass A, Zechner R. J Lipid Res. 2009;50(Suppl):S63–8. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith WL, Marnett LJ, DeWitt DL. Pharmacol Ther. 1991;49:153–79. doi: 10.1016/0163-7258(91)90054-p. [DOI] [PubMed] [Google Scholar]
- 4.Ahn K, McKinney MK, Cravatt BF. Chem Rev. 2008;108:1687–707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Patricelli MP, Cravatt BF. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Schicher M, Jesse I, Birner-Gruenberger R. Methods Mol Biol. 2009;580:251–66. doi: 10.1007/978-1-60761-325-1_14. [DOI] [PubMed] [Google Scholar]
- 9.Speers AE, Adam GC, Cravatt BF. J Amer Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 10.Speers AE, Cravatt BF. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Higgs HN, Han MH, Johnson GE, Glomset JA. J Biol Chem. 1998;273:5468–77. doi: 10.1074/jbc.273.10.5468. [DOI] [PubMed] [Google Scholar]
- 12.We attempted to characterize DDHD2's activity with PC substrates containing sn-1 ester/sn-2 -ether or sn-1 ether/sn-2 ester linkages (Supporting Figure 6), but, unlike DDHD1, DDHD2 did not accept substrates where ether linkages were included in the sn-1/sn-2 position.
- 13.Cadas H, di Tomaso E, Piomelli D. J Neurosci. 1997;17:1226–42. doi: 10.1523/JNEUROSCI.17-04-01226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin XH, Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N. J Biol Chem. 2007;282:3614–23. doi: 10.1074/jbc.M606369200. [DOI] [PubMed] [Google Scholar]
- 15.Phospholipases can also show head-group selectivity, and we therefore envision that variants of probes 1 and 2, where the choline group is replaced with serine, ethanolamine, or inositol, may also prove useful for characterizing additional types of phospholipases.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.