Abstract
Background
The first step in invasive disease caused by the normally commensal bacteria Streptococcus pneumoniae, Staphylococcus aureus and Haemophilus influenzae is their colonization of the nasal passages. For any population to colonize a new habitat it is necessary for it to be able to compete with the existing organisms and evade predation. In the case of colonization of these species the competition is between strains of the same and different species of bacteria and the predation is mediated by the host's immune response. Here, we use a neonatal rat model to explore these elements of the ecology of nasal colonization by these occasionally invasive bacteria.
Results
When neonatal rats are colonized by any one of these species the density of bacteria in the nasal passage rapidly reaches a steady-state density that is species-specific but independent of inoculum size. When novel populations of H. influenzae and S. pneumoniae are introduced into the nasal passages of neonatal rats with established populations of the same species, residents and invaders coexisted. However, this was not the case for S. aureus - the established population inhibited invasion of new S. aureus populations. In mixed-species introductions, S. aureus or S. pneumoniae facilitated the invasion of another H. influenzae population; for other pairs the interaction was antagonistic and immune-mediated. For example, under some conditions H. influenzae promoted an immune response which limited the invasion of S. pneumoniae.
Conclusions
Nasal colonization is a dynamic process with turnover of new strains and new species. These results suggest that multiple strains of either H. influenzae or S. pneumoniae can coexist; in contrast, S. aureus strains require a host to have no other S. aureus present to colonize. Levels of colonization (and hence the possible risk of invasive disease) by H. influenzae are increased in hosts pre-colonized with either S. aureus or S. pneumoniae.
Background
The first step in a bacterial disease is the successful establishment of a bacterial population in a host: colonization. The conditions that determine whether a bacterial population can colonize a particular site and the density achieved are fundamental to determining the likelihood of invasive disease, transmission to other hosts and the presence of mutants resistant to antibiotics. How these conditions are affected by prior colonization by bacteria of the same or different species has wide spread consequences for determining the sequelae of the wide-scale use of vaccines directed at specific strains or species (as the vaccine strain/species can potentially be replaced by other potentially invasive strains and species [1]) as well as for evaluating probiotics [2] and understanding epidemiological changes in invasive bacterial diseases [3,4].
Whether bacteria can colonize or not is determined by many ecological factors including the availability of resources (i.e. nutrients, space, attachment space), host immune responses and the presence of toxins or harmful substances. As the presence of established bacteria populations can influence all of these factors, it seems reasonable to assume that co-inhabitants often determine whether colonization can occur. In fact co-inhabitants that are ecologically similar, should limit the colonization as the one that is better at exploiting the habitat should exclude the others through resource limitation [5]. However, as a consequence of even subtle differences in resource (ie nutrients, space or metabolic byproducts) utilization or availability, multiple strains and species of bacteria can co-exist [6-12]. The ability to colonize can also be influenced by interference, which includes residents populations producing harmful substances (like bacterocins [13,14]) or inducing an immune response [15,16]. In the case of three bacterial species which colonize the human nasopharynx (Streptococcus pneumoniae, Staphylococcus aureus and Haemophilus influenzae), epidemiological studies show that co-colonization is rarer than expected [17-21]. These co-inhabitation patterns suggest that there may be interference or competition occurring.
In this report we apply an ecological framework to elucidate the factors contributing to the nasal colonization of neonatal rats of three bacterial species that typically colonize humans: S. pneumoniae, H. influenzae and S. aureus. First we consider the population dynamics of each strain separately. We provide evidence that all three species colonize the nasal passages of neonatal rats and reach an apparent steady-state density and that this level is independent of inoculum density. To explore the effects of co-inhabitants on colonization, 48 hours after colonizing neonatal rats with one species we pulsed with a second inoculum of a marked strain of the same species. The results of these pulse experiments suggest that resident S. aureus prevents co-colonization of the same strain; while for both H. influenzae and S. pneumoniae the total density is increased to allow for the co-existence of pulsed and established populations. We repeated these experiments with the resident and invading populations being of different species and found that H. influenzae colonizes at a higher density when either S. aureus or S. pneumoniae are present and that immune-mediated competition between S. pneumoniae and H. influenzae is both site and strain specific.
Results and Discussion
Population Dynamics
All three species readily colonize the nasal passages of neonatal rats. Within 48 hours after one of the three species is inoculated, H. influenzae, S. aureus and S. pneumoniae reach and maintain for at least three days a constant population (between 100-10,000 cfu depending on the species) in the nasal epithelium (Figure 1). The population dynamics of nasal colonization did not differ in the nasal wash sample with the nasal epithelium. The nasal epithelium, which represent the persisting colonizing bacteria [22], data are shown.
Figure 1.
Population dynamics of nasal colonization. Population dynamics of nasal colonization. Five-day-old neonatal rats were inoculated with 107 (black circles) or 104 cfu (diamonds) of either S. pneumoniae, H. influenzae or S. aureus. The geometric mean bacteria density in the nasal epithelium of 4-16 rats at each time-point is plotted. Dashed line represents limit of detection. Error bars represent SE.
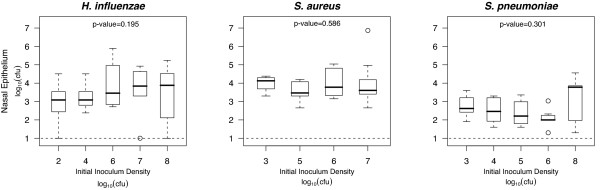
The bacterial load for each of the species was not significantly different from 48 to 96 hours (p-values for each species determined by Kruskal-Wallis rank sum were < 0.05). While the dynamics for both a low and high inoculum density appear to be similar, we ascertained whether bacterial load is inoculum-independent at 48 hours after inoculation. For all three species the bacterial load is invariant over a wide range of inocula (102-108 cfu) (Figure 2), suggesting that nasal colonization rapidly reaches a steady-state that is not limited by how many bacteria are inoculated.
Figure 2.
Bacterial load is independent of inoculum density. Groups of 7-16 five-day-old neonatal rats were inoculated with 102-108 cfu of either S. pneumoniae, H. influenzae or S. aureus. The 25th to 75th percentiles of nasal wash and epithelium samples taken 48 hours after bacterial challenge are represented by the box plots, with the bold horizontal bar indicating the median value, circles outlying values and dotted error bars SE. P values were determined by Kruskal-Wallis rank sum which tested the null hypothesis that the bacterial load are distributed the same in all of the inoculum groups. Dashed line represents limit of detection.
Invasion of Same Species in a Colonized Host
To test whether nasal colonization can occur in the presence of the same species, new populations of bacteria were pulsed (104 cfu inoculated) into rats that were already colonized by bacteria of that species. Antibiotic markers that conferred no in vitro or in vivo fitness costs were used to distinguish the resident and pulsed populations and each experiment was repeated reversing the strains as pulsed or resident to control for any fitness differences. As the population dynamics suggest that the bacterial load for each of these species is tightly controlled, we expected that the total density (resident+pulsed) would return to the bacterial load observed in rats without pulses. Because resident and pulsed strains of the same species utilize the same resource (and attract the same immune responses), co-existence of both strains is expected unless a limiting factor is available only on a first come first serve basis.
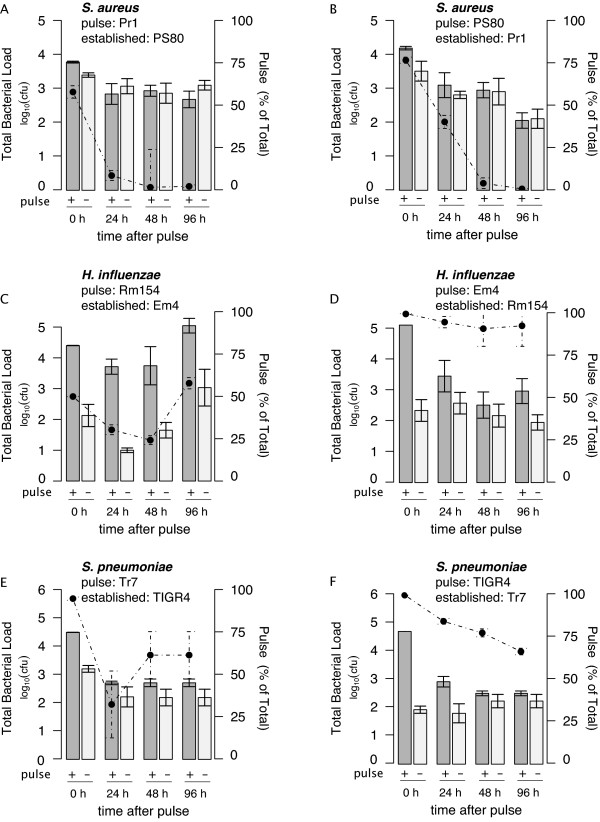
In the case of S. aureus, regardless of whether the marked strain is resident or pulsed, we find that the pulsed strain declines in density (faster relative to the established) over the course of 96 hours (as shown in representative experiments in Figure 3A-B). As the pulsed strain declines (decrease in percent shown in dotted line) the total bacterial load of S. aureus in the rats with the pulsed and established strain (+ pulse) doesn't differ from the total density of S. aureus in the rats with only the established strain (- pulse). For S. aureus the bacterial density does not exceed that observed in rats without a pulse and the resident strain has a competitive advantage.
Figure 3.
Pulse on established populations of same species. Established populations were inoculated into 3-day-old neonatal rats 48 hours prior to pulsing 104 cfu of a marked strain of the same species or PBS. The total bacterial density in nasal epithelium of 6-8 rats with the established and pulsed population (dark grey) and just the established population (light grey) were tracked over 96 hours after the pulse and expressed as the geometric mean with error bars indicating SE. In addition, the percent of the bacterial density that is pulsed is marked with points with dotted error bars indicating SE. Antibiotic marked strains were switched to be either pulsed or established for H. influenzae (in A and B), S. aureus (in C and D) and S. pneumoniae (in E and F).
For both S. pneumoniae and H. influenzae there is an increase in the total density in the rats with the pulse (+ pulse) compared to rats with only the established strain (as shown in representative experiments in Figure 3C-F). We saw the bacterial load increase to varying degrees, more so for H. influenzae than for S. pneumoniae, in each of four replicate experiments (data available upon request). In both of these species, we observe that the pulsed and resident strains co-exist with the pulse strain becoming 25-90% of the population.
For all the species, similar pulse results were obtained in reciprocal experiments (switching pulse and resident strains) confirming that the results were not due to fitness differences in the antibiotic marked strains.
Invasion of Different Species in a Colonized Host
Competition between different strains or species can be defined simply as a reduction in the density of one or both strains when both are present. Competition within the same species and particularly in the case of the same strain (as in the above pulse experiment) is usually mediated through a limiting shared resource. Competition between species, in addition to partitioning of a shared resource, can be mediated through inhibitory agents/toxins (allelopathy) or predators (in this case components of the immune system [23]). Previous studies suggest that production of hydrogen peroxide by S. pneumoniae may affect the densities of other species [24,25] and that immune-mediated competition reduces S. pneumoniae density in the presence of H. influenzae [26]. To evaluate the contributions of these different competitive mechanisms we performed invasion experiments (with one strain of each species: Eagan, TIGR4 and PS80) in which one species was resident and a second was introduced (an invader).
Evidence for synergistic interactions between H. influenzae and S. pneumoniae or S. aureus
Rather than an antagonistic interaction, we found that H. influenzae reached a higher density when invading resident populations of either S. aureus or S. pneumoniae than in the absence of these residents (Figure 4). A similar increase in the bacterial density of H. influenzae was observed in vitro; when mixtures of these strains were grown in broth for 6 hours, H. influenzae density was 20%(± 14) greater with S. pneumoniae and 19%(± 3) greater with S. aureus present than when grown alone (data not shown).
Figure 4.
Invasion of a host colonized with another species. Established populations were inoculated into groups of 10-22 three-day-old neonatal rats 48 hours prior to pulsing 105 cfu of a different species or PBS. The 25th to 75th percentiles of nasal wash and epithelium samples taken 48 hours after bacterial challenge are represented by the box plots, with the bold horizontal bar indicating the median value, circles outlying values and dotted error bars. T-test P values < 0.005 are represented by **. Resident bacterial density was not significantly different from un-invaded rats in any combination of species.
Strain-specific, innate immune-mediated interactions between H. influenzae and S. pneumoniae
We had expected to detect immune-mediated competition between H. influenzae and S. pneumoniae, as observed in a mouse model of colonization by Lysenko and colleagues [26]. However, we saw no evidence of competition between H. influenzae and S. pneumoniae with the strains we initially used: TIGR4 and Eagan (Figure 4).
To investigate further, we tested one additional strain of S. pneumoniae, Poland(6b)-20. We found that this particular strain of S. pneumoniae had a reduced density in the nasal wash, but not the nasal epithelium, when invading in a neonatal rat with an established H. influenzae population (Figure 5). This reduction in Poland-20's population did not occur in neonatal rats which had been depleted of complement or neutrophils.
Figure 5.
Neutrophil- and Complement- Mediated Competition. Three-day-old neonatal rats were treated with either anti-neutrophil serum (-neutrophil) or cobra venom factor (-complement) or PBS and inoculated with either 106cfu of H. influenzae or PBS (alone). Forty-eight hours later, 104 cfu of Poland(6b)-20 S. pneumoniae was inoculated. The 25th to 75th percentiles of nasal wash samples taken 48 hours after S. pneumoniae inoculation are represented by the box plots, with the horizontal bar indicating the median value and circles outlying values. P-value from Mann Whitney U test comparing the bacterial density of previously uninfected rats and those with established populations of H. influenzae. Dashed line represents limit of detection.
To explain why we could only observe this in one of the two strains tested and only then in the nasal wash, we hypothesized that either induction of or susceptibility to the immune response must differ in these strains and locations. We quantified the neutrophil infiltration in the nasal epithelium by measuring the Myeloperoxidase (MPO) activity at 48 hours after inoculation with each strain/species alone or when Poland(6b)-20 was inoculated on an established H. influenzae population (Figure 6A).
Figure 6.
Neutrophil infiltration: comparison of strains and species at 48 hours and dynamics over 96 hours. A) Neutrophils in the nasal epithelium from rats inoculated 48 hours earlier with 104 cfu of bacteria from a single species (Rm154, TIGR4 and Poland(6b)-20) or from rats inoculated 96 hours earlier with 106 cfu of H. influenzae and 48 hours earlier with 104 cfu of Poland(6b)-20 were quantified using the MPO assay. Lines indicate median MPO values. P-value is calculated by the Wilcoxon rank sum test. B) Dynamics of neutrophil infiltration in response to nasal colonization by S. pneumoniae (TIGR4) or H. influenzae. Following inoculation groups of 5-8 rats were sacrificed and neutrophil infiltration was measured by MPO assay. Median MPO Units are plotted. Error bars represent SE. Dashed line represents median MPO of uninoculated rats.
No difference in neutrophil infiltration is observed between rats colonized by the two different S. pneumoniae strains (TIGR4 and Poland(6b)-20). The neutrophil infiltration observed 48 hours after Poland(6b)-20 invaded on an established H. influenzae population (when immune-mediated competition was observed in the nasal wash) was significantly higher than rats with just Poland(6b)-20 colonizing alone. However, neutrophil infiltration was not significantly higher than in rats with only H. influenzae. While these results suggest that H. influenzae is primarily responsible for the neutrophil infiltration that reduces the nasal lumen populations of some strains of S. pneumoniae, S. pneumoniae may still have a role in eliciting the immune response (perhaps with slower dynamics than H. influenzae). We observed that the neutrophil infiltration in response to S. pneumoniae colonizing alone increases from 48-96 hours after inoculation, compared to the constant neutrophil presence with H. influenzae (Figure 6B).
Discussion
Population Dynamics
All three species that we studied (S. aureus, S. pneumoniae and H. influenzae) can colonize the nasal passages of neonatal rats and each reaches a bacterial load that is independent of the initial inoculum size; they increase in density when initially below this level and decline when initially above it. This indicates that the steady-state density is tightly controlled - perhaps by a limiting resource or the host's immune response. The total density of each of these colonizing species is relatively low and there is wide-spread variation in the densities of individuals, similar to what has been observed in colonized humans [27].
Invasion of Same Species in a Colonized Host
In accord with classical ecological theory of competition [5] we would anticipate that established populations of one strain of a species would either prevent colonization of another strain of that species or be eliminated. The outcome would depend on which of the strains was more fit. In the case of genetically marked strains of the same species with equal fitness we would expect to observe co-existence. In a series of pulse experiments with genetically marked colonizing (pulsed) and resident (established) strains we found the situation to be more complicated than this simple interpretation.
For both H. influenzae and S. pneumoniae, the resident and the pulsed strains co-existed. Surprisingly in all replicates of these experiments, the invasion of a second population of the same species was followed by an increase in the total bacterial density. Given that the steady-state bacterial densities were independent of the initial inoculum density following a single inoculation, we had expected that the bacterial density after a second inoculation would decline to the original bacterial load. These results might be attributable to a second inoculation leading to an expansion in the colonization area, increased immune suppression or the release of new resources - perhaps associated with an inflammatory response.
On first consideration it would seem that the results of the S. aureus pulse experiments are consistent with classical ecological theory; the established population of this species inhibited the colonization of a new strain. Moreover, as expected following the pulse by a second strain the total density of S. aureus returned to a level similar to that observed in the single inoculation experiments. However the resident strain had the advantage no matter which marker it carried (the competitive exclusion observed was not due to difference in fitness between the marked strains). We interpret these results as suggesting that S. aureus is limited by a localized resource available on a 'first-come, first-serve' basis - perhaps attachment sites [28,29]. This ecological hypothesis would account for the observations that competing strains of S. aureus were excluded from burn wounds [30] and from nasal colonization in persistent human carriers [31]. While these results do not exclude the possibility that variation within S. aureus strains may allow for coexistence as occasionally has been observed in humans [32], they do suggest that prior nasal colonization with S. aureus can exclude similar S. aureus strains from colonizing.
Invasion of Different Species in a Colonized Host
Ecologically, strains of different species would be anticipated to be more divergent than those of the same species and therefore they would be expected to occupy different niches. The results of our experiments are consistent with this interpretation as any pairwise combination of the three species can co-exist. While we had expected some sharing of resources by these different species, we found no evidence that the presence of one species reduced the density colonizing the nasal epithelium of another species.
The only evidence for inter-specific interactions between these species observed at the nasal epithelium was in fact synergistic rather than antagonistic; H. influenzae reaches a higher density when invading resident populations of either S. aureus or S. pneumoniae than it achieves in rats not colonized by these bacteria. Since this result is also observed in vitro it seems likely due to some host-independent mechanism such as S. aureus and S. pneumoniae providing nutrients that would otherwise limit H. influenzae. Indeed, in the past H. influenzae has been identified and cultured due to the fact that it grew as satellites off of S. aureus colonies [33]. To our knowledge this is the first evidence for S. aureus and S. pneumoniae increasing the density of H. influenzae during nasal colonization.
We had expected to see inter-specific antagonism not only due to resource sharing but also because of interference by toxins and harmful substances. In fact, it has been proposed that the production of hydrogen peroxide by S. pneumoniae may affect the densities of S. aureus and H. influenzae as both are susceptible to hydrogen peroxide killing [24,25,34,35]. However in this and a previous work that specifically addressed this issue [36] we found no evidence that hydrogen peroxide produced by S. pneumoniae limits the colonizing populations of either of the two species. This may be because the density of S. pneumoniae is too low for sufficient hydrogen peroxide production or the nasal epithelium inactivates the hydrogen peroxide produced. Taken at large, we found no ecological interaction between S. aureus and S. pneumoniae colonization that would account for the epidemiological observation that S. aureus-S. pneumoniae co-colonization is rarer than expected [4,18,20,37,38]. We postulate that this epidemiological observation may be due to the bacteria preferring different hosts rather than competitive interactions within hosts [39], or that competitive exclusion may only occur in immunologically mature individuals. Others have suggested that there may still be an ecological interaction based on the pneumococcal pilus [35] or by induction of phage release [40].
Neutrophil-mediated Competition
Previous experiments by Lysenko and colleagues in a mouse model have shown that when H. influenzae and S. pneumoniae co-colonize, S. pneumoniae's density in the nasal wash is lower than when inoculated alone due to immune-mediated competition [26]. At one level, the results of our rat model experiments with H. influenzae and S. pneumoniae are consistent with their results [26]. However, our results also suggest that this immune-mediated competitive interaction may only affect the colonizing S. pneumoniae population in the nasal wash (not the population adhering to nasal epithelium) and is strain-specific. We observed immune-mediated competition with the clinical strain of S. pneumoniae Poland(6b)-20 but not with TIGR4. We hypothesize that pneumococcal strains vary either in their ability to elicit an immune response or in their suscebtibility to the immune response induced by H. influenzae. Although we can't exclude the possibility that the two strains we tested elicited different immune responses, our results suggest that there is no difference in the extent of neutrophil infiltration of the epithelium in response to colonization by either of these strains or any synergism [41] between the two species. Together our results suggest that the immune response primarily elicited by H. influenzae is responsible for reducing the density of S. pneumoniae in the nasal wash and that S. pneumoniae strains may vary in their susceptibility to this innate immune response. While we found limited evidence for immune-mediated competition, since the nasal epithelium bacterial populations of S. pneumoniae are un-altered by this innate immune response this competition may not effect the long-term carriage of S. pneumoniae in the nasal passage.
Limitations
Perhaps the most significant limitation and caveat associated with this study is that the neonatal rat immune system is changing during the course of these experiments, thereby restricting our ability to draw inferences about the role of the immune response and long-term colonization dynamics. While arguably a decent model for young infants, the neonatal rats are unlikely to be an accurate model of the nasal passages of older children or adults. Another limitation of this study is that the results obtained may be strain-specific and only one or two strains for each species was tested. The limited number of strains does not likely reflect the within species diversity in colonization strategies and this diversity should be investigated in further studies. Finally, our ability to draw inferences about the factors influencing the ecology of colonization in these neonatal rats was limited by the substantial amount of variation in densities observed in individual rats.
Conclusion
Caveats and limitations aside, we believe that the application of an ecological framework to the colonization of neonatal rat model with S. aureus, S. pneumoniae and H. influenzae contributes to our understanding of the epidemiology of carriage, disease processes and the impact of vaccination on these bacteria species. These results begin to address the mechanisms responsible for the dynamic process of nasal colonization with turnover and replacement of species, serotypes and strains in the complex community (Figure 7). For example the pulse experiments results suggest that for S. pneumoniae and H. influenzae the presence (and turnover) of multiple strains and serotypes would be expected in carriers as has been observed in humans [42]. Further, our results suggest that that H. influenzae colonization will be more successful (and hence possibly more likely to cause disease) when preceded by either S. aureus or S. pneumoniae. Ultimately the ecology of nasal colonization informs whether vaccination (or antibiotic treatment) directed at one particular species will lead to the unintended consequences of increased colonization by competing (and possibly more pathogenic) species, serotypes or strains.
Figure 7.
Dynamic Process of Nasal Colonization. Graphical interpretation of Pulse and Invasion Experiments.
Methods
Bacterial strains, media and inoculum preparation
A laboratory bacterial strain of each species was selected based on capsular type and invasive potential. S. pneumoniae TIGR4 (serotype 4) [43] and Poland(6b)-20(serotype 6b) [44] were provided by Lesley McGee. Tr7 was selected as a spontaneous rifampicin resistant mutant of TIGR4. S. aureus PS80 (serotype 8 ATTC 27700) was obtained from American Type Culture and Pr1 was selected as a spontaneous mutant of PS80 exhibiting resistance to rifampin. H. influenzae type b Eagan and its streptomycin resistant mutant Rm154 were provided by Richard Moxon. Em4 was selected as a spontaneous mutant of Eagan exhibiting resistance to nalidixic acid.
S. pneumoniae strains were grown in Todd-Hewitt broth (Becton Dickinson) supplemented with 0.5% w/v of yeast extract (THY) and plates were supplemented with 4% v/v of sheep blood (BBL). Broth cultures and agar plates of S. pneumoniae were incubated at 37°C with 5% CO2 H. influenzae strains were grown in brain heart infusion broth (Becton Dickinson) supplemented with 10 μg of hemin (sigma) and 2 μg of βNAD (sigma) per ml (sBHI). S. aureus strains were cultivated in Luria-Bertani (LB; Becton Dickinson,) broth cultures.
Equal fitness of antibiotic marked strains was confirmed by mixing equal densities of cultures in exponential phase and sampling the initial densities and the densities 6 hours later in broth or 48 hours later in nasal passages of neonatal rats. For all combinations (i.e. TIGR4/Tr7, PS80/Pr1, Rm154/Em4), there was no significant fitness difference in vitro or in vivo (data not shown). The spontaneous antibiotic resistant mutant strains were repeatedly grown alone in broth and consistently showed 100% plating efficiencies when plated on media with antibiotics versus media alone.
To determine if synergistic interaction between H. influenzae occurred in vitro when co-cultured with either S. pneumoniae or S. aureus, H. influenzae was grown in sBHI with or without another species and the intial densities and the densities 6 hours later were compared.
Inoculum for all the infant rat experiments were prepared by initially growing strains to late logarithmic phase (OD620:0.35-0.8). These were stored at -80°C and then thawed before suspending in 2 ml of either LB, THY or sBHI. Mid-exponential phase cultures were centrifuged (5,000 g × 3 min) and resuspended in phosphate-buffered saline with 0.1% gelatin (PBSG). Note the addition of gelatin did not lead to an increase in the inoculation density for any of these bacteria. Bacterial densities were estimated by plating dilutions of S. aureus on LB Agar plates or LB plates supplemented with rifampicin (40 mg/L); S. pneumoniae on THY blood plates supplemented with either streptomycin (40 mg/L) or rifampicin (50 mg/L) or H. influenzae on sBHI plates supplemented with bacitracin (0.3 g/L) and either streptomycin (4 mg/L) or nalidixic acid (5 mg/L).
Infant Rat Model
Although neonatal rats do not naturally carry S. aureus, S. pneumoniae and H. influenzae, they can be reproducibly colonized with these species. All animal experiments were performed under the guidelines approved by the Emory Institutional Animal Care and Use Committee. Three-day-old pups, born of timed-pregnant Sprague-Dawley rats (Charles River Laboratories), were randomly reassigned to dams. At 3 or 5 days of age, rats were intranasally inoculated by touching a drop of 102 - 108 bacteria of either S. aureus, S. pneumoniae or H. influenzae (that had been spun down and re-suspended in 5 μl PBS supplemented with 0.1% gelatin (PBS-G)) to the right and then another 5 μl to the left external nares [45]. The nasal flora of un-inoculated neonatal rats, determined by colony morphology on blood plates, appeared to consist primarily of non-hemolytic streptococci and coagulase-negative staphylococci. No S. aureus, S. pneumoniae and H. influenzae colonies were isolated from un-inoculated neonatal rats and all of these strains colonized in spite of the presence of this nasal flora.
Two days after the innoculation, nasal wash was collected from 200 μl of PBS-G instilled into a 5 cm intramedic polyetylene tubing (PE50, intramedic, Clay Adams) placed into the trachea, and nasal epithelium was scraped from the nasal passages after a second wash of 200 μl of PBSG and removal of the frontal bones. 3 sequential nasal washes of 200 μl of PBS-G contained no significant decrease in the bacteria density compared to the first wash. The nasal epithelium was homogenized in 1 ml of PBS-G.
In all experiments, 100 μl of the nasal wash and nasal epithelium samples were plated directly and serially diluted onto selective plates. The limit for detection was 10 cfu/ml. Nasal wash densities were converted to cfu in rat by multiplying cfu/ml by 5 (200 uL total vol.) and nasal epithelium by multiplying by 1 (1 ml total vol.). With the exception of the H. influenzae -S. pneumoniae interaction, data from the nasal wash and nasal epithelium data are in agreement and only the nasal epithelium data are presented; as nasal epithelium likely represents the persistent colonizing population [22].
Experimental Design
For the population dynamics of nasal colonization, groups of 4-16 5-day-old rats were intranasally inoculated with either 104 or 107 cfu bacteria of S. aureus, S. pneumoniae or H. influenzae and sampled 12-144 hours after inoculation. Inoculum independence was confirmed by inoculating groups of 7-16 5-day-old rats with 102- 108 cfu bacteria of S. aureus, S. pneumoniae or H. influenzae and sampling at 48 hours.
For intra-species invasion, one marked variant of a particular strain was intranasally inoculated into two groups of 24-36 3-day-old rats. Fourty-eight hours later one group was intranasally inoculated with the same species with the alternative antibiotic marker while the other inoculated with PBS. At 0, 24, 48 and 96 hours after pulsing with the same species 6-8 rats were sacrificed and sampled. For each pairing between antibiotic marked strains of the same species (i.e. TIGR4/Tr7, PS80/Pr1, Rm154/Em4), this experiment was repeated with the reverse strain being established and pulsed.
For the inter-species invasion, experiments testing, groups of 8-12 3-day-old rats were inoculated in both nostrils with either one species (S. aureus, S. pneumoniae or H. influenzae) or with PBS. All of these rats were then inoculated 48 hours later with 106- 107 of another species (S. aureus, S. pneumoniae or H. influenzae), and then sacrificed 48 hours after the inoculation of second species.
Immune Depletion
For systemic complement reduction, cobra venom factor (CVF; Advanced Research Technologies, San Diego, CA) was administered to 4-day-old neonatal rat by intraperitoneal injection of 500 μg/kg of weight (dissolved in 0.1 M PBS) [46]. Systemic complement reduction was confirmed by the EZ Complement CH50 Test kit (Diamedix, Miami, FL) [47]. Serum from age matched un-inoculated control rats had CH50 of 40.94 ± 6.6, while CVF treated rats had a CH50 of 21.6 ± 3.9 until 5 days after CVF treatment.
For systemic neutrophil depletion, anti-neutrophil serum (ANS, absorbed rabbit anti-rat PMN; Accurate Chemical, Westbury, NY) was administered to 4-day-old neonatal rat by subcutaneous injection of 6 μL/g of weight (diluted 1:1 in PBS) [48]. Systemic neutrophil depletion was confirmed by FACS analysis of blood and local depletion confirmed in the nasal passages using a myeloperoxidase (MPO) assay of nasal epithelium [49]. In ANS treated un-inoculated rats nasal epithelium MPO was 0.002 ± 0.01 U, compared to control rats 0.072 ± 0.02 U.
Statistical Analysis
The bacterial densities (and the log10 transformed densities) during colonization were not normally distributed. To determine whether inoculum size altered the median bacterial density or whether the density varied from 48 to 96 hours post-inoculation, a Kruskal-Wallis rank sum test was used to compare the ranks for each inoculum size or time point. A Wilcoxon rank-sum test was used to evaluate the statistical significance in inter-species competitions or the myeloperoxidase results for different strains.
Authors' contributions
EM conceived of, undertook and analyzed all of the experiments. AY assisted in the conception and analysis of the pulse experiments. BRL was a supportive kibitzer and advised the conception and interpretation of all the experiments. All three authors contributed to the writing of this manuscript.
Contributor Information
Elisa Margolis, Email: emargol@emory.edu.
Andrew Yates, Email: andrew.yates@einstein.yu.edu.
Bruce R Levin, Email: blevin@emory.edu.
Acknowledgements
We would like to thank Richard Moxon for his continued support, ideas and encouragement in this endeavor. We are particularly grateful to Lesley McGee and Bill Shafer for generously providing strains. Thanks to Lynn Huynh and William Margolis for critically reading an earlier version of the manuscript and to Winston Lee for providing invaluable advice on the MPO assay. This research was supported by NIH AI40662 (Bruce Levin) and NIH T32GM08169 (Emory Medical Scientist Training Program; Elisa Margolis).
References
- Lipsitch M. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg Infect Dis. 1999;5(3):336–345. doi: 10.3201/eid0503.990304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Fattah ASA, Maruchi N. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 2000;44(2):127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- Schuchat A, Robinson K, Wenger JD, Harrison LH, Farley M, Reingold AL, Lefkowitz L, Perkins BA. Bacterial meningitis in the United States in 1995. Active Surveillance Team. N Engl J Med. 1997;337(14):970–976. doi: 10.1056/NEJM199710023371404. [DOI] [PubMed] [Google Scholar]
- Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rumke HC, Verbrugh HA, Hermans PWM. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363(9424):1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- Hardin G. The competitive exclusion principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- Paine R. Food web complexity and species diversity. American Naturalist. 1966;100:65–75. doi: 10.1086/282400. [DOI] [Google Scholar]
- Tilman D. Competition and biodiversity in spatially structured habitats. Ecology. 1994;75:2–16. doi: 10.2307/1939377. [DOI] [Google Scholar]
- Levin. Coexistence of two asexual strains on a single resource. Science. 1972;175:1272–1274. doi: 10.1126/science.175.4027.1272. [DOI] [PubMed] [Google Scholar]
- Helling RB, Vargas CN, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116(3):349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S, Lenski. Tests of ecological mechanisms promoting the stable coexistence of two bacterial genotypes. Ecology. 1996;77:2119–2129. doi: 10.2307/2265706. [DOI] [Google Scholar]
- Bohannan, Lenski. The Relative Importance of Competition and Predation Varies with Productivity in a Model Community. American Naturalist. 2000;156:329–340. doi: 10.1086/303393. [DOI] [PubMed] [Google Scholar]
- Chesson P. General theory of competitive coexistence in spatially-varying environments. Theor Popul Biol. 2000;58(3):211–237. doi: 10.1006/tpbi.2000.1486. [DOI] [PubMed] [Google Scholar]
- Chao L, Levin BR. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA. 1981;78(10):6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Gordon DM. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 1999;7(3):129–133. doi: 10.1016/S0966-842X(99)01459-6. [DOI] [PubMed] [Google Scholar]
- Graham AL. Ecological rules governing helminth-microparasite coinfection. Proc Natl Acad Sci USA. 2008;105(2):566–570. doi: 10.1073/pnas.0707221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AB, Fenton A. Emphasizing the ecology in parasite community ecology. Trends Ecol Evol. 2007;22(3):133–139. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4(10):e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi SA, Adrian P, Kuwanda L, Cutland C, Albrich WC, Klugman KP. Long-term effect of pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae-and associated interactions with Staphylococcus aureus and Haemophilus influenzae colonization-in HIV-Infected and HIV-uninfected children. J Infect Dis. 2007;196(11):1662–1666. doi: 10.1086/522164. [DOI] [PubMed] [Google Scholar]
- Jacoby P, Watson K, Bowman J, Taylor A, Riley TV, Smith DW, Lehmann D, Team KOMRP. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007;25(13):2458–2464. doi: 10.1016/j.vaccine.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, Rahav G, Rubinstein E. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA. 2004;292(6):716–720. doi: 10.1001/jama.292.6.716. [DOI] [PubMed] [Google Scholar]
- Melles DC, Bogaert D, Gorkink RFJ, Peeters JK, Moorhouse MJ, Ott A, van Leeuwen WB, Simons G, Verbrugh HA, Hermans PWM, van Belkum A. Nasopharyngeal co-colonization with Staphylococcus aureus and Streptococcus pneumoniae in children is bacterial genotype independent. Microbiology. 2007;153(Pt 3):686–692. doi: 10.1099/mic.0.2006/002279-0. [DOI] [PubMed] [Google Scholar]
- Briles DE, Novak L, Hotomi M, van Ginkel FW, King J. Nasal colonization with Streptococcus pneumoniae includes subpopulations of surface and invasive pneumococci. Infect Immun. 2005;73(10):6945–6951. doi: 10.1128/IAI.73.10.6945-6951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilyugin S, Antia R. Modeling immune responses with handling time. Bull Math Biol. 2000;62(5):869–90. doi: 10.1006/bulm.2000.0181. [DOI] [PubMed] [Google Scholar]
- Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun. 2000;68(7):3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. Interference between Streptococcus pneumoniae and Staphylococcus aureus: In vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol. 2006;188(13):4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1:e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg CO. A study of carriers of Staphylococcus aureus with special regard to quantitative bacterial estimations. Acta Med Scand Suppl. 1965;436:1–96. [PubMed] [Google Scholar]
- Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10(3):243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- Shuter J, Hatcher VB, Lowy FD. Staphylococcus aureus binding to human nasal mucin. Infect Immun. 1996;64:310–318. doi: 10.1128/iai.64.1.310-318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickman K. Studies of bacterial interference in experimentally produced burns in guinea pigs. Acta Pathol Microbiol Scand [B] Microbiol Immunol. 1970;78:15–28. doi: 10.1111/j.1699-0463.1970.tb04266.x. [DOI] [PubMed] [Google Scholar]
- Nouwen J, Boelens H, van Belkum A, Verbrugh H. Human factor in Staphylococcus aureus nasal carriage. Infect Immun. 2004;72(11):6685–6688. doi: 10.1128/IAI.72.11.6685-6688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cespedes C, Said-Salim B, Miller M, Lo SH, Kreiswirth BN, Gordon RJ, Vavagiakis P, Klein RS, Lowy FD. The clonality of Staphylococcus aureus nasal carriage. J Infect Dis. 2005;191(3):444–452. doi: 10.1086/427240. [DOI] [PubMed] [Google Scholar]
- Davis D. The accessory factors in bacterial growth. V. The value of the satellite (or symbiosis) phenomenon for the classification of hemophilic bacteria. Journal of Infectious Disease. 1921;29:187–191. [Google Scholar]
- Tano K, Hakansson EG, Holm SE, Hellstrom S. Bacterial interference between pathogens in otitis media and alpha-haemolytic Streptococci analysed in an in vitro model. Acta Otolaryngol. 2002;122:78–85. doi: 10.1080/00016480260349827. [DOI] [PubMed] [Google Scholar]
- Regev-Yochay G, Lipsitch M, Basset A, Rubinstein E, Dagan R, Raz M, Malley R. The pneumococcal pilus predicts the absence of Staphylococcus aureus co-colonization in pneumococcal carriers. Clin Infect Dis. 2009;48(6):760–763. doi: 10.1086/597040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis E. Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J Bacteriol. 2009;191(2):571–575. doi: 10.1128/JB.00950-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally LM, Jeena PM, Gajee K, Sturm AW, Tomkins AM, Coovadia HM, Goldblatt D. Lack of association between the nasopharyngeal carriage of Streptococcus pneumoniae and Staphylococcus aureus in HIV-1-infected South African children. J Infect Dis. 2006;194(3):385–390. doi: 10.1086/505076. [DOI] [PubMed] [Google Scholar]
- Watson K, Carville K, Bowman J, Jacoby P, Riley TV, Leach AJ, Lehmann D, Team KOMRP. Upper respiratory tract bacterial carriage in Aboriginal and non-Aboriginal children in a semi-arid area of Western Australia. Pediatr Infect Dis J. 2006;25(9):782–790. doi: 10.1097/01.inf.0000232705.49634.68. [DOI] [PubMed] [Google Scholar]
- Lee GM, Huang SS, Rifas-Shiman SL, Hinrichsen VL, Pelton SI, Kleinman K, Hanage WP, Lipsitch M, McAdam AJ, Finkelstein JA. Epidemiology and risk factors for Staphylococcus aureus colonization in children in the post-PCV7 era. BMC Infect Dis. 2009;9:110. doi: 10.1186/1471-2334-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa I, Novick RP, Penades JR. Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci USA. 2009;106(4):1234–1238. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner AJ, Lysenko ES, Paul MN, Weiser JN. Synergistic proinflammatory responses induced by polymicrobial colonization of epithelial surfaces. Proc Natl Acad Sci USA. 2005;102(9):3429–3434. doi: 10.1073/pnas.0500599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauver JS, Marrs CF, Foxman B, Somsel P, Madera R, Gilsdorf JR. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg Infect Dis. 2000;6(6):622–630. doi: 10.3201/eid0606.000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293(5529):498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Overweg K, Hermans PW, Trzcinski K, Sluijter M, de Groot R, Hryniewicz W. Multidrug-resistant Streptococcus pneumoniae in Poland: identification of emerging clones. J Clin Microbiol. 1999;37(6):1739–1745. doi: 10.1128/jcm.37.6.1739-1745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis E, Levin BR. Within-host evolution for the invasiveness of commensal bacteria: an experimental study of bacteremias resulting from Haemophilus influenzae nasal carriage. J Infect Dis. 2007;196(7):1068–1075. doi: 10.1086/520934. [DOI] [PubMed] [Google Scholar]
- Cowell RM, Plane JM, Silverstein FS. Complement activation contributes to hypoxic-ischemic brain injury in neonatal rats. J Neurosci. 2003;23(28):9459–9468. doi: 10.1523/JNEUROSCI.23-28-09459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassiter HA, Walz BM, Wilson JL, Jung E, Calisi CR, Goldsmith LJ, Wilson RA, Morgan BP, Feldhoff RC. The administration of complement component C9 enhances the survival of neonatal rats with Escherichia coli sepsis. Pediatr Res. 1997;42:128–136. doi: 10.1203/00006450-199707000-00020. [DOI] [PubMed] [Google Scholar]
- Hudome S, Palmer C, Roberts RL, Mauger D, Housman C, Towfighi J. The role of neutrophils in the production of hypoxic-ischemic brain injury in the neonatal rat. Pediatr Res. 1997;41(5):607–616. doi: 10.1203/00006450-199705000-00002. [DOI] [PubMed] [Google Scholar]
- Zen K, Liu Y, McCall IC, Wu T, Lee W, Babbin BA, Nusrat A, Parkos CA. Neutrophil migration across tight junctions is mediated by adhesive interactions between epithelial coxsackie and adenovirus receptor and a junctional adhesion molecule-like protein on neutrophils. Mol Biol Cell. 2005;16(6):2694–2703. doi: 10.1091/mbc.E05-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]