Abstract
Progesterone Receptor Membrane Component-1 (PGRMC1) is present in both the cytoplasm and nucleus of spontaneously immortalized granulosa cells (SIGCs). PGRMC1 is detected as a monomer in the cytoplasm and a DTT-resistant PGRMC1 dimer in the nucleus. Transfected PGRMC1-GFP localizes mainly to the cytoplasm and does not form a DTT-resistant dimer. Moreover, forced expression of PGRMC1-GFP increases the sensitivity of the SIGCs to progesterone (P4) 's anti-apoptotic action, indicating that the PGRMC1 monomer is functional. However, when endogenous PGRMC1 is depleted by siRNA treatment and replaced with PGRMC1-GFP, P4 responsiveness is not enhanced, although overall levels of PGRMC1 are increased. P4's anti-apoptotic action is also attenuated by actinomycin D, an inhibitor of RNA synthesis, and P4 activation of PGRMC1 suppresses Bad and increases Bcl2a1d expression. Taken together, the present studies suggest a genomic component to PGRMC1's anti-apoptotic mechanism of action, which requires the presence of the PGRMC1 dimer.
Keywords: membrane progesterone receptor, apoptosis, progesterone, granulosa cell
1. Introduction
Progesterone (P4) is synthesized and secreted from the ovary and influences the function of numerous tissues that regulate reproductive function (Conneely and Lydon 2000; Stouffer 2003; Gellersen et al. 2009). P4's actions on the uterus (Conneely and Lydon 2000) and mammary gland (Mote et al. 2007; Kariagina et al. 2008) as well as the pituitary and hypothalamus (Scott et al. 2002) have been well characterized. Interestingly, P4 is not only synthesized within the ovary but also plays essential roles in regulating its function (Stouffer 2003). Specifically, P4 acting through the classic nuclear P4 receptor (PGR) plays an essential role in ovulation (Conneely and Lydon 2000). P4 also stimulates cholesterol metabolism (Rung et al. 2005), regulates steroidogenesis (Rothchild 1996) and promotes the survival of granulosa cells isolated from developing (Peluso et al. 2005; Peluso et al. 2006) and preovulatory follicles (Shao et al. 2003; Rung et al. 2005; Friberg et al. 2009) and luteal cells (Peluso et al. 2005).
The anti-apoptotic action of P4 on granulosa cells isolated from developing rat follicles and rat luteal cells is not mediated through PGR, since these cells do not express this P4 receptor (Park and Mayo 1991; Natraj and Richards 1993; Park-Sarge and Mayo 1994; Shao et al. 2003). Similarly, spontaneously immortalized granulosa cells (SIGCs) do not express PGR (Peluso et al. 2002). Importantly, rat granulosa cells of developing follicles, rat luteal cells and SIGCs all express another P4 receptor, progesterone receptor membrane component-1 (PGRMC1) (Peluso et al. 2006). Like granulosa cells isolated from developing rat follicles (Peluso and Pappalardo 1998), spontaneously immortalized granulosa cells (SIGCs) undergo apoptosis within 5 h of being placed in serum-free culture media and P4's anti-apoptotic effect on these cells is mediated by PGRMC1 (Peluso et al. 2006; Peluso et al. 2008). This conclusion is based on studies involving the depletion of PGRMC1 using siRNA approaches (Peluso et al. 2008; Peluso et al. 2008; Peluso et al. 2009) and over expression of PGRMC1-GFP fusion protein (Mansouri et al. 2008; Peluso et al. 2008).
Although cloned from porcine liver in 1998 (Gerdes et al. 1998), relatively little is known about PGRMC1. PGRMC1 is a small 22–26 kDa protein that has a short extracellular domain, a single transmembrane domain and a cytoplasmic domain that consists largely of a cytochrome P450 b5/heme binding domain (Cahill 2007; Peluso 2007; Losel et al. 2008). Preliminary studies have shown that depletion on any of these components significantly reduces the capacity of PGRMC1-GFP fusion protein to bind P4 and to transduce P4's anti-apoptotic action (Peluso et al. 2008). PGRMC1 also forms dimers (Falkenstein et al. 2001) but the physiological significance of PGRMC1 dimerization is not known.
Immunocytochemical studies have localized PGRMC1 to the cytoplasm and nucleus of rat granulosa cells (Peluso et al. 2006), SIGCs (Peluso et al. 2008), human granulosa cells (Engmann et al. 2006; Peluso et al. 2009) and human ovarian cancer cells (Peluso et al. 2008). However, little attention has been directed toward understanding PGRMC1's role in the nucleus. Interestingly, immunocytochemical studies of ovarian cancer cells revealed that the amount of nuclear PGRMC1 is variable, ranging from non-detectable to abundant levels (Peluso 2007; Peluso et al. 2008). This suggests that the presence of PGRMC1 within the nucleus is regulated and may be an essential part of PGRMC1's mechanism of action.
To begin to elucidate PGRMC1's putative nuclear or genomic action, the levels of endogenous PGRMC1 and transfected PGRMC1-GFP fusion protein within the cytoplasmic and nuclear cell fractions were assessed by Western analysis. Since PGRMC1-GFP fusion protein did not significantly localize to the nucleus, the role of nuclear PGRMC1 was assessed by depleting endogenous PGRMC1 from both the cytoplasm and nucleus and increasing cytoplasmic PGRMC1 by over-expressing PGRMC1-GFP fusion protein. We then monitored the ability of P4 to prevent apoptosis and to influence gene expression.
2. Materials and Methods
2.1. Spontaneously Immortalized Granulosa Cell Culture
SIGCs, which were derived from rat granulosa cells isolated from preovulatory follicles (Stein et al. 1991), were used in these studies. Unless otherwise indicated, SIGCs were plated at 2 × 105 cells/ml in 35 mm culture dishes for studies involving RNA or protein analysis or 24 well culture plates for studies of apoptosis. The cultures were maintained for 24 h in DMEM-F12 supplemented with 5% FBS. After 24 h, cells were treated according to the protocols outlined in each experiment.
To assess apoptosis SIGCs were placed in serum-free medium supplemented with different doses of P4 (0–1000 nM) depending on the experimental design. After 5 h, DAPI was added directly to the culture media and the cells incubated for 10 min at 37°C in the dark as previously described (Peluso et al. 2008). After staining, the cells were observed under epi-fluorescent optics. Cells were considered apoptotic if DAPI staining revealed nuclei with condensed and/or fragmented (Peluso et al. 2008).
2.2. Immunological Detection of Progesterone Receptor Membrane Component-1
Endogenous PGRMC1 was detected by immunocytochemistry and Western blot analysis. Immunocytochemistical localizations were performed as previously described (Peluso et al. 2008) using an antibody to PGRMC1 at a dilution of 1: 100 (HPA002877, Sigma Chemical Co., St. Louis, MO). The secondary antibody was a goat anti-rabbit IgG labeled with either Alexa 488 or 546. Western blots were conducted using the same PGRMC1 antibody at a dilution of 1:250. PGRMC1 was revealed using either an anti-rabbit HRP-labeled antibody (1:2000) and ECL detection (GE Health, Buckinghamshire, UK) or a goat anti-rabbit IRdye 800-labeled antibody (1:20,000) and the fluorescent-based Li-Cor Odessey detection system. As negative controls, immunocytochemical preparations and Western blots were conducted without the primary antibody.
Whole cell lysates were prepared by lysing cells in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA and NP-40) as previously described (Peluso et al. 2008). The RIPA buffer was supplemented with protease and phosphatase inhibitors (Peluso et al. 2008). All procedures were conducted on ice. Total protein in the lysate was determined using the BCA assay kit from Pierce Biotechnology (Cat Number 23225, Rockford, IL).
Western blot analysis was also used to assess the cellular localization of PGRMC1. This was accomplished by isolating cytoplasmic and nuclear proteins using the Qproteome nuclear protein kit and protocol provided from Qiagen (Valencia, CA). Briefly, SIGCs were lysed in hypotonic buffer containing 0.1 M DTT. The lysate was then centrifuged and the cytoplasmic proteins collected from the supernatant and the nuclear proteins from the cell pellet. The nuclear proteins were then extracted by treating the cell pellet with high-salt buffer. This resulted in the separation of the nucleic-acid proteins from the nucleic acids. After centrifugation, the nuclear proteins were collected from the supernatant. This nuclear protein fraction contained nucleic-acid binding proteins, including those proteins that function as transcription factors. To assess the purity of the cytoplasmic and nuclear fraction, the presence of heterogeneous nuclear ribonucleoprotein A1 (HnRNP A1), a protein that localizes almost exclusively to the nucleus, was monitored by Western blot analysis using an antibody (sc-10029) provided by Santa Cruz Biotechnology (Santa Cruz, CA). This antibody was used at a 1:200 dilution and detected by a donkey anti-goat secondary antibody (sc-2020, 1:2000). Similarly, the presence of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a cytoplasmic protein, was detected by Western blot using an antibody (AM4300; 1:4000) provided by Ambion, Inc (Austin, TX).
2.3. Generation and Expression of PGRMC1-GFP Fusion Protein
The expression vector that encodes PGRMC1-GFP fusion protein has been described (Peluso et al. 2008). Briefly, total mRNA was isolated from SIGCs and cDNA was generated. The entire coding region of PGRMC1 was then amplified using the following primer pair: forward: TTCTCGAGATGGCTGCCGAGGATGTG (with XhoI site) and reverse: AGAAGCTTGTCACTCTTCCGAGC (with HindIII site). PGRMC1 was then cloned into pEGFP-N1 vector (Clontech, Mountain View, CA) at the XhoI and HindIII restriction sites. The resulting construct, referred to as PGRMC1-GFP, was sequenced to ensure that it correctly encoded PGRMC1.
GFP or the PGRMC1-GFP fusion protein was detected in situ by observing the cells under fluorescent optics and a GFP filter set. In some studies involving the localization of PGRMC1-GFP, cells were fixed in 4% paraformaldehyde for 5 min at 4°C and then for 10 min at room temperature (≈ 23° C). The cells were then washed three times in Hanks Balanced Salt solution at −20°C, permeabilized with 0.1% Triton x for 7 min at room temperature, and counter stained with DAPI (1 μg/ml) and examined under epi-fluorescence using the GFP and DAPI filter sets. For Western blot analysis, a GFP antibody (Cell Signaling, Beverly, MA Cat # 2555) was used at a dilution of 1:1000.
2.4. Depletion of Endogenous PGRMC1 using PGRMC1 siRNA
PGRMC1 was depleted from SIGCs as follows. SIGCs (4 × 105 cells) were plated in 35 mm culture dishes. After culture for 24 h, the medium was removed and half of the culture dishes were transfected with PGRMC1 siRNA (Cat. No. AM16708 siRNA ID# 253165, Ambion, Austin, TX). This PGRMC1 siRNA targeted the sequence from position 1329 to 1348 (cctggtaattggcagttgg), which is within the non-coding region of PGRMC1. The remaining dishes were transfected with scramble siRNA (Cat. No. AM4611, Ambion, Austin, TX). All transfections were done using the Lipofectamine 2000 transfection kit (Invitrogen,Carlsbad, CA) according to manufacturer's instructions. PGRMC1 and scramble siRNAs were used at a final concentration of 0.6 pM. For some experiments, SIGCs were also transfected with either pEGFP empty vector or pEGFP-PGRMC1 (1.2 μg/ml of culture media). After transfection the cells were incubated for 24 h and then either assessed for PGRMC1 expression as described above or monitored for P4-regulated apoptosis.
To assess the effect of PGRMC1 on apoptosis, SIGCs were transfected as outlined above. After 24 h, the serum-supplemented medium was removed and the cells were placed in serum-free medium supplemented with a suboptimal dose of P4 (0.1 nM P4). After 5 h, the cells were stained with DAPI. To determine the effect of PGRMC1-GFP on the ability of SIGCs to respond to P4, random areas within each cell culture were sequentially observed under a GFP filter set and a DAPI filter set. Images of each area were captured and stored in a computer. By comparing the images from the same area the transfection status (GFP-green fluorescence) and viability (apoptosis; DAPI-blue fluorescence) of each cell could be determined. Approximately 100 transfected cells per culture well were evaluated for apoptosis. The percentage of transfected apoptotic cells/treatment dish was calculated (Peluso et al. 2008).
2.5. PCR Analysis of Endogenous PGRMC1 and PGRMC1-GFP Fusion Protein
Quantitative measurements of endogenous Pgrmc1 mRNA were done as follows. Briefly, primers to rat Pgrmc1 were designed using NCBI Primer blast and products were designed to detect across the coding /non-coding sequence. Primers used were: forward 5'- TGCTCGGAAGAGTGACTGAA-3' and reverse 5'- TTCACTTTCTGGATTTGTGACG-3'. The forward primer was within the PGRMC1 coding sequence (672–691 bp) and the reverse primer (871–892 bp) was outside the coding sequence. The PCR product from these primers was 221 bp and could only be generated by amplification of endogenous Pgrmc1 mRNA, which possesses a 3' non-coding sequence. Real-time PCR was performed on total RNA with the appropriate primers using the Script one-site RT-PCR kit with SYBR green (BioRad Laboratories, Hercules, CA). Relative level of Pgrmc1 mRNA was evaluated with BioRad iCycler software using the ΔCT method. Values were expressed as a percentage of the scramble control treatment.
To detect Pgrmc1-Gfp mRNA, the forward primer was 5'- TCGGATGATGAAGAACCAAA-3' (Pgrmc1 coding sequence 643–663 bp) and the reverse primer was 5'-TCCAGCTCGACCAGGATG-3', which targeted the GFP coding sequence. The PCR product from these primers was 161 bp and could only be generated in the presence of Pgrmc1-Gfp mRNA, which has a GFP coding sequence.
2.6. Effect of P4 and PGRMC1 on the Expression of Genes Involved in Apoptosis
Some of the apoptosis-related genes whose expression is regulated by P4 and PGRMC1 were identified using the rat specific apoptosis RT2 Profiler PCR Array (PARN-012A) following the manufacturer's instructions (SABiosciences, Frederick, MD). Briefly, SIGCs were transfected with either PGRMC1 or scramble siRNA as previously described and 24 h later treated with or without 1 μM P4 in serum-free medium. Five h later total RNA was isolated using the RNeasy Plus mini kit (Qiagen, Valencia, CA). For each of the four treatments, 25 ng of total RNA was added to each well of the 96 well assay plate. This entire experiment was repeated 3 times. The expression level of each gene was normalized to the house keeping genes using PCR array data analysis software available on line from SAbiosciences. The each gene expression was presented as a percentage of the scramble siRNA control.
2.7. Statistical Analysis
All experiments were repeated two to three times with each experiment yielding essentially identical results. When appropriate, data from each replicate was pooled and analyzed by a one-way ANOVA followed by a Student-Newman-Keuls test.
3. Results
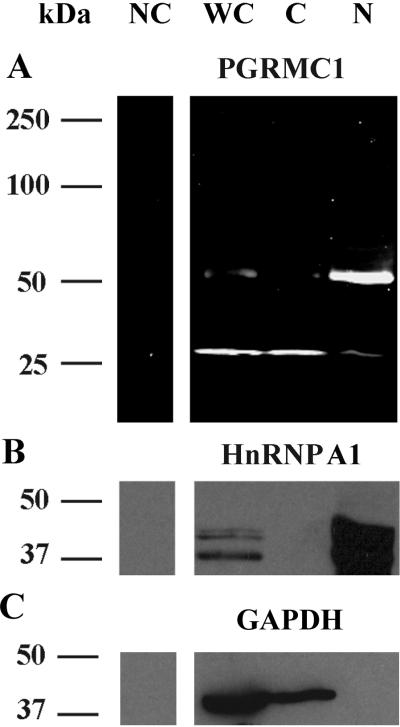
Western blot analysis of whole cell lysate from SIGCs detected PGRMC1 as ≈ 26 kDa protein with a minor band at ≈ 52 kDa (Figure 1). The ≈ 52 kDa band was not always detected in whole cell lysates. To assess the cellular distribution of PGRMC1, SIGCs were lysed in hypotonic buffer containing DTT (0.1 M) and separated into cytoplasmic and nuclear fractions by centrifugation. The total amount of protein isolated from the cytoplasmic fraction averaged 650 ± 84 μg/107cells while 83 ± 20 μg of protein/107cells was isolated from the nuclear fraction (p < 0.05). When equal amounts of proteins were loaded onto gels, the ≈ 26 kDa PGRMC1 form (i.e. monomer) was predominately found in the cytoplasmic fraction, while the ≈ 52 kDa PGRMC1 band was the major form present in the nuclear fraction (Figure 1A). The ≈ 52 kDa band presumably represents a PGRMC1 dimer. Note that the efficiency of this cellular fractionation protocol was confirmed by Western blot analysis of heterogeneous nuclear ribonucleoprotein (HnRNP A1) (Figure 1B), which is one of the most abundant proteins in the nucleus (Siomi and Dreyfuss 1995). The antibody used in the western blots detected HnRNP A1 (≈ 37 kDa) as well as other HnRNP family members. Similarly, GAPDH was detected in the cytoplasmic fraction and not the nuclear fraction (Figure 1C), which is consist with its role in cytosolic energy production (Mazzola and Sirover 2003).
Figure 1.
Detection of PGRMC1 (A) HnRNP A1 (B) and GAPDH (C) in lysates prepared from whole cells (WC), cytoplasmic (C) or nuclear (N) fractions. WC lysate was prepared in RIPA buffer, while the C and N fractions were prepared in hypotonic buffers supplemented with 0.1M DTT. NC indicates a negative control in which a Western blot was conducted in the absence of the primary antibody.
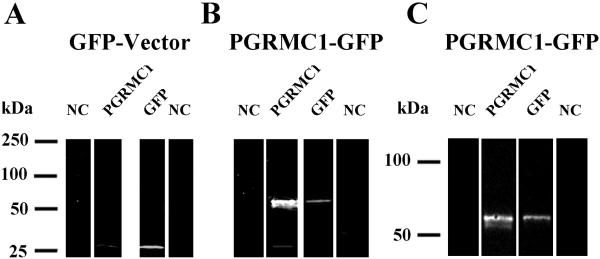
To determine the localization and dimerization status of PGRMC1-GFP, SIGCs were transfected with either empty GFP vector or a vector that encodes the PGRMC1-GFP fusion proteins. After transfection with the empty GFP-vector, GFP was readily detected in Western blots of whole cell lysates as a single band, which is similar in size to the endogenous PGRMC1 monomer (Figure 2A). After transfection with the PGRMC1-GFP vector, the PGRMC1 antibody detected an ≈ 26 kDa, an ≈ 52 kDa and an ≈ 54 kDa band (Figure 2B). In Figure 2B the ≈ 52 kDa was difficult to resolve due to the intense staining of the ≈ 54 kDa band. By reducing the exposure time and running the gel longer, the ≈ 52 and ≈ 54 kDa bands were readily identified (Figure 2C). The ≈ 26 and ≈ 52 kDa bands represent the monomeric and dimeric forms of PGRMC1, while the ≈ 54 kDa band represents the PGRMC1-GFP fusion protein, since it is also detected by the GFP antibody (Figure 2B,C). In PGRMC1-GFP transfected cells, the amount of PGRMC1-GFP fusion protein is several fold greater than endogenous PGRMC1 levels. Note that a PGRMC1-GFP dimer, which would be > 100 kDa, was not detected (Figure 2B,C).
Figure 2.
Detection of endogenous PGRMC1 and PGRMC1-GFP fusion protein by Western blots conducted on whole cell lysates prepared in RIPA buffer from cells either transfected with empty GFP vector (A) or a vector encoding PGRMC1-GFP (B,C). Each sample was probed with an antibody to either PGRMC1 or GFP. The Western blot in panel C is restricted to the 50 to 100 kDa range in order to more clearly reveal the 52 and 54 kDa bands. NC indicates a negative control in which a Western blot was conducted in the absence of the primary antibody.
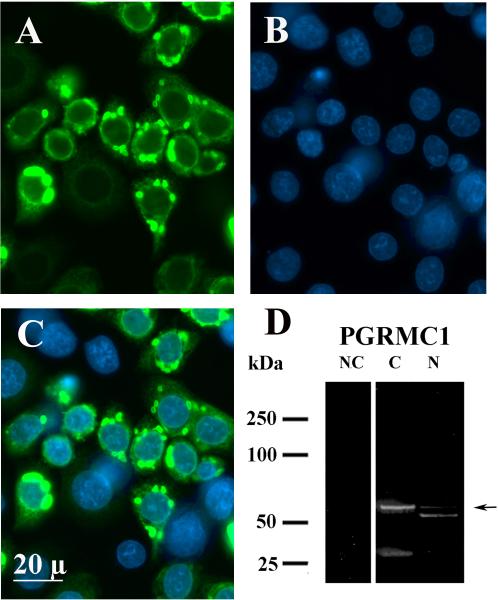
Twenty-four h after transfecting cells with either the GFP empty vector or the PGRMC1-GFP expression vector, about 40 to 50% of the cells were transfected. After transfection with the empty GFP vector, GFP was observed throughout the cell including the nucleus as has been shown previously (Peluso et al. 2008). In contrast the majority of the PGRMC1-GFP fusion protein was localized to the cytoplasm as revealed by GFP fluorescence (Figure 3A–C). Western blots of the cytoplasmic fraction after transfection with the PGRMC1-GFP vector detected an ≈ 26 kDa band (i.e. endogenous PGRMC1 monomer) and an ≈ 54 kDa band, which represents the PGRMC1-GFP fusion protein (Figure 3D). Western blots of the nuclear fraction revealed an ≈ 52 kDa (i.e. endogenous PGRMC1 dimer) and an ≈ 54 kDa band (i.e. PGRMC1-GFP fusion protein). The intensity of the ≈ 54 kDa band in the nuclear fraction was several fold less than the corresponding band in the cytoplasm (Figure 3D), even through equal amounts of cytoplasmic and nuclear proteins were loaded onto the respective lanes. Additional studies failed to show an effect of P4 on the distribution of either PGRMC1 or PGRMC1-GFP fusion protein (data not shown).
Figure 3.
Cellular localization of PGRMC1-GFP fusion protein as assessed by GFP-fluorescence or Western blot analysis. After PGRMC1-GFP transfection, some cells were fixed, counter stained with DAPI and examined under epi-fluorescence using the GFP and DAPI filter sets to reveal the localization of PGRMC1-GFP (A) or the nucleus (B). A merged image is shown in C. The remaining cells were lysed and separated into cytoplasmic (C) or nuclear (N) fractions. The Western blots were probed with an antibody to PGRMC1. The PGRMC1-GFP fusion protein is identified with an arrow. NC indicates a negative control in which a Western blot was conducted in the absence of the primary antibody.
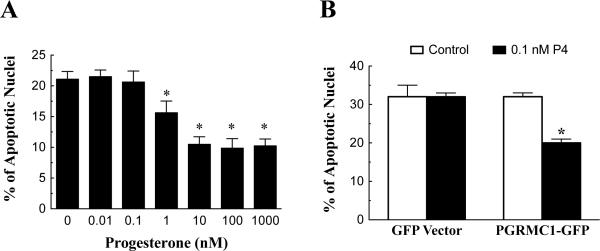
P4 inhibited SIGC apoptosis in a dose-dependent manner with a partially effective dose of 1 nM (p < 0.05 compared to zero P4) and a maximally effect dose of 10 nM (p < 0.05 compared to zero P4; Figure 4A). SIGCs transfected with the empty GFP vector underwent apoptosis in the absence of P4 and as expected treatment with an ineffective dose of P4 (0.1 nM) failed to reduce the percentage of GFP-transfected SIGCs that underwent apoptosis (Figure 4B). In contrast, 0.1 nM P4 was effective in reducing the percentage of PGRMC1-GFP transfected cells that underwent apoptosis compared to control (p < 0.05) (Figure 4B). This confirms previous studies (Peluso et al. 2008) and indicates that the elevated levels of PGRMC1-GFP fusion protein sensitized SIGCs to the anti-apoptotic effects of P4.
Figure 4.
The effect of P4 and PGRMC1-GFP on SIGC apoptosis. In panel A, the effect of increasing concentrations of P4 on the percentage of SIGCs that undergo apoptosis within 5 h in serum-free medium is shown. Panel B shows the effect of either the empty GFP vector or PGRMC1-GFP on the percentage of transfected SIGCs that undergo apoptosis in the presence or absence of 0.1 nM P4. In these graphs, an * indicates a value that is significantly different (p < 0.05) from control (no P4 group).
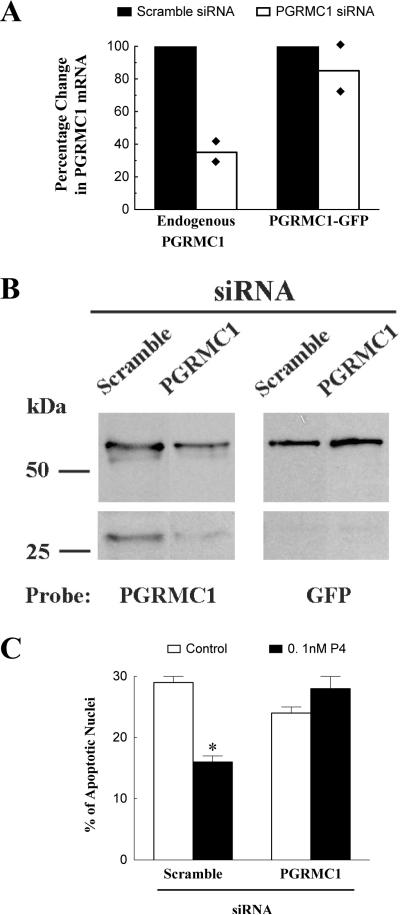
To determine whether over-expression of PGRMC1-GFP could maintain P4 responsiveness in the relative absence of endogenous PGRMC1, a combined PGRMC1 siRNA and PGRMC1-GFP transfection protocol was employed. The effectiveness of this approach was assessed by immunocytochemistry (Figure 5), real-time PCR (Figure 6A) and Western blot analysis (Figure 6B). Specifically, immunocytochemical analysis revealed that after combined PGRMC1 siRNA and PGRMC1-GFP transfection, PGRMC1 could not be detected in either the cytoplasm or nucleus of SIGCs that were not transfected with PGRMC1-GFP. Thus, most if not all of the PGRMC1 within transfected cells is the PGRMC1-GFP fusion protein. Moreover, the design of the PGRMC1 siRNA was such that the mRNA that encodes endogenous PGRMC1 but not PGRMC1-GFP fusion was reduced (i.e. targeting non-coding regions of Pgrmc1 mRNA present only in the endogenous form) (Figure 6A). The specificity of the siRNA was also confirmed by Western blots in which treatment with the PGRMC1 siRNA depleted both the ≈ 26 and 52 kDa bands (i.e. the endogenous monomeric and dimeric forms of PGRMC1) without significantly depleting the PGRMC1-GFP fusion proteins (Figure 6B). Note that a shorter exposure was required to clearly detect both the 52 and 54 kDa bands, while a longer exposure was needed to detect the 26 kDa PGRMC1 band. This is the reason for the break in the Western blot shown in Figure 6B.
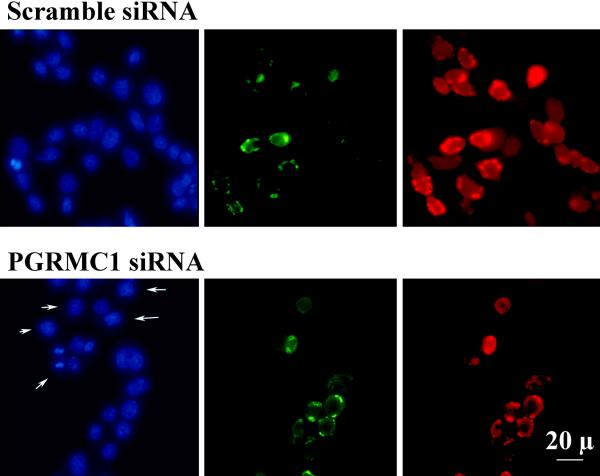
Figure 5.
The effect of treatment with PGRMC1-GFP fusion protein and either scramble or PGRMC1 siRNA on the levels of endogenous PGRMC1 and PGRMC1-GFP fusion protein as assessed by immunocytochemistry. After treatment the cells were fixed, stained for PGRMC1 and counter stained with DAPI (1 μg/ml). In the upper panel, cells were transfected with PGRMC1-GFP and scramble siRNA. The same field of cells was observed for DAPI staining (Blue-total cells), GFP fluorescence (Green-presence of PGRMC1-fusion protein) or PGRMC1 using an Alexa 546-labeled secondary antibody (Red-both endogenous and PGRMC1-fusion protein). The images in the upper panel reveal that a select population of cells expressed PGRMC1-GFP but all cells expressed endogenous PGRMC1. The lower panel shows cells transfected with PGRMC1-GFP and PGRMC1 siRNA. In cells not successfully transfected with the PGRMC1-GFP expression vector (some of these cells are marked by arrows in the DAPI-stained image), endogenous PGRMC1 is not detected in either the cytoplasm or nucleus. PGRMC1 was only detected in PGRMC1-GFP transfected cells with most of the PGRMC1-GFP fusion protein localizing to the cytoplasm. A negative control was conducted for the immunocytochemical localization of PGRMC1 by omitting the primary antibody. The negative control did not show any staining.
Figure 6.
The effect of PGRMC1 siRNA and PGRMC1-GFP expression on endogenous and PGRMC1-GFP mRNA levels as assessed by real-time PCR (A) and Western blot analysis (B). In panel A, values are shown as a mean of two and the diamond (◆) indicates the individual value of each replicate. Panel B shows a Western blot that used aliquots of whole cell lysates prepared from PGRMC1-GFP transected cells treated with either scramble or PGRMC1 siRNA. Panel B is divided into four panels. The two panels on the right were probed with the PGRMC1 antibody and represent the same blot exposed for 30 sec (upper panel) and 60 seconds (lower panel). The shorter exposure was required to clearly distinguish the 52 and 54 kDa PGRMC1 bands. The two panels on the left were probed with a GFP antibody. Panel C shows the effect of PGRMC1-GFP and either scramble or PGRMC1 siRNA on the percentage of transfected SIGCs that undergo apoptosis in the presence or absence of 0.1 nM P4. The * indicates a value that is significantly different (p < 0.05) from control (no P4 group).
PGRMC1-GFP transfected SIGCs treated with scramble siRNA were responsive to the anti-apoptotic action of 0.1 nM P4 (Figure 6C), confirming the data shown in Figure 4B. Under conditions in which endogenous PGRMC1 was depleted by PGRMC1 siRNA treatment and replaced by PGRMC1-GFP fusion protein, 0.1 nM P4 did not inhibit apoptosis of transfected SIGCs that expressed PGRMC1-GFP fusion protein (Figure 6C). This suggests that elevated levels of PGRMC1-GFP fusion protein without endogenous PGRMC1 are not sufficient to enhance the sensitivity of SIGCs to P4's anti-apoptotic action.
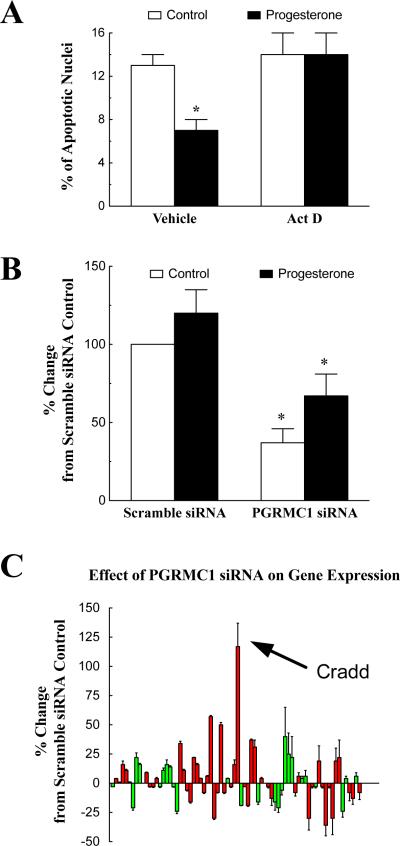
To gain further insight PGRMC1's mechanism of action, SIGCs were cultured with P4 and either actinomycin D (Figure 7A) or cylcoheximide (not shown). Treatment with either of these macromolecular synthesis inhibitors attenuated P4's anti-apoptotic activity. These studies imply that P4's ability to inhibit apoptosis was dependent on de novo RNA and protein synthesis. In order to determine whether PGRMC1 plays a role in P4-regulated mRNA synthesis, endogenous PGRMC1 levels were reduced using a PGRMC1 siRNA approach. This approach reduced endogenous Pgrmc1 mRNA levels by ≈ 50% (p < 0.05) and this suppression was not effected by P4 treatment (Figure 7B).
Figure 7.
The effect of actinomycin D (Act D; 4 μM) and P4 (1 μM) on SIGC apoptosis (A). In panel B, the effect of PGRMC1 siRNA and P4 on endogenous levels of PGRMC1 mRNA is shown. In panels A and B the * indicates a value that is significantly different (p < 0.05) from control (no P4 group). The effects of PGRMC1 siRNA treatment of the expression profile of various apoptosis-related genes is shown in panel C. The mRNA levels of both endogenous PGRMC1 and apoptosis-related genes were assessed by real-time PCR and shown as a percentage change from scramble control values. The values are means (± standard error) of three experiments. Genes associated with the induction of apoptosis are shown in red and the anti-apoptotic genes shown in green.
Real-time PCR revealed that 74 of the 84 apoptosis-related genes in the microarray were reliably detected. Compared to the gene profile of SIGCs treated with scramble siRNA, PGRMC1 siRNA significantly altered the gene profile (p < 0.001). Specifically, PGRMC1 siRNA increased the expression of a number of genes known to stimulate apoptosis as well as a few anti-apoptotic genes (Figure 7C). For example the expression of Cradd mRNA, which encodes a protein that recruits caspase 2 into a cell death-signaling complex (Ahmad et al. 1997) is dramatically elevated.
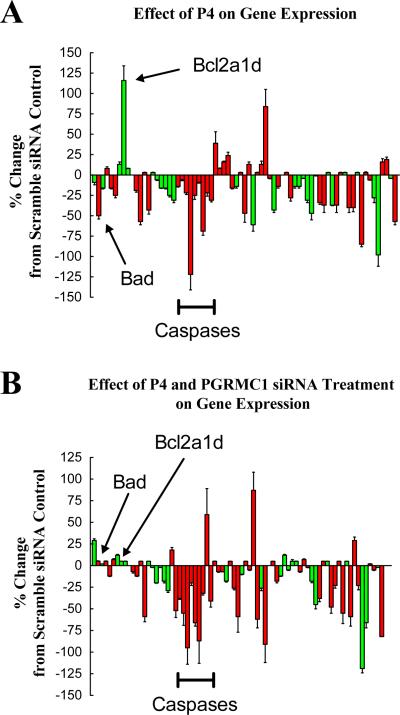
The addition of P4 to scramble siRNA-treated SIGCs also altered the gene profile compared to SIGCs treated only with scramble siRNA (p < 0.001). The overall effect of P4 was to suppress several genes that promote apoptosis (Figure 8A). Most noticeably, the mRNAs that encode Bad and several caspase family members were suppressed, while Bcl2a1d mRNA levels were increased. Importantly, the mRNA levels of several genes were within 5% of scramble control values, which indicates that P4's effects were not indiscriminant.
Figure 8.
The effects of 1μM P4 plus scramble siRNA (A) or 1μM P4 plus PGRMC1 siRNA treatment (B) on the expression profile of various apoptosis-related genes. The mRNA levels of apoptosis-related genes were assessed by real-time PCR and shown as a percentage change from scramble siRNA control values. The values are means (± standard error) of three experiments. Genes associated with the induction of apoptosis are shown in red and the anti-apoptotic genes shown in green.
P4's effect on the gene expression profile of PGRMC1-deplete SIGCs was very similar to that observed in scramble siRNA-treated SIGCs, including the suppression of the mRNAs that encode the caspases (Compare Figure 8A to 8B). However, P4 was not able to increase Bcl2a1d mRNA levels and decrease Bad mRNA levels in PGRMC1-depleted SIGCs (Compare Figure 8A to 8B).
4. Discussion
The present Western blot studies confirm immunocytochemical observations that PGRMC1 localizes to the cytoplasm and nucleus of SIGCs (Peluso et al. 2006). Interestingly, the major band detected in PGRMC1 western blots of whole cell and the cytoplasmic lysates is ≈ 26 kDa, while most of PGRMC1 in the nucleus is detected as a ≈ 52 kDa protein. When equal amounts of protein are loaded onto gels, PGRMC1 is easily detected in both cellular fractions but there is 8 times more protein in the cytoplasm than in the nucleus. Therefore, most PGRMC1 localizes to the cytoplasm. What is somewhat surprising is that the PGRMC1 in the nuclear lysate appears as a dimer, even though the nuclear lysate was prepared in 0.1 M DTT, which is known to disrupt PGRMC1 dimers (Falkenstein et al. 2001). That the ≈ 52 kDa protein is a PGRMC1 dimer is supported by the immunocytochemical analysis which reveals that PGRMC1 siRNA treatment depletes PGRMC1 from both cytoplasm and nucleus. This is confirmed by Western blot analysis, which shows that the PGRMC1 siRNA depletes both the 26 and 52 kDa forms of PGRMC1 [present study; (Peluso et al. 2009). This was also observed when siRNA that targeted different sequences of PGRMC1 was used (Peluso et al. 2008).
The observation that PGRMC1 is present at different cellular localizations allows for the possibility of different cellular sites of action. To begin to explore this, PGRMC1-GFP fusion protein was transfected into SIGCs. This results in a several fold increase in PGRMC1-GFP compared to endogenous PGRMC1. PGRMC1-GFP localizes predominately to the cytoplasm with only a very small amount detected within the nucleus. In addition PGRMC1-GFP is not detected as a dimer (i.e. a band of ≥ 100 kDa). The reason why PGRMC1-GFP does not form a dimer is unknown but could be due to the presence of GFP. Regardless, PGRMC1-GFP is capable of binding P4 through a single high-affinity binding site (Meyer et al. 1998; Mansouri et al. 2008; Peluso et al. 2008; Peluso et al. 2009) and increasing its level enhances the sensitivity of SIGCs to P4 (present study; Peluso et al. 2006; Peluso et al. 2008). These observations imply that P4 binds to PGRMC1-GFP within the cytoplasm and that this interaction is sufficient to enhance cell responsiveness to P4's anti-apoptotic action.
Since transfecting SIGCs increases the levels of PGRMC1-GFP several fold over endogenous PGRMC1 levels and in turn the responsiveness to P4, it was hypothesized that over-expression of the PGRMC1-GFP fusion protein would be sufficient to mediate P4's anti-apoptotic action even when endogenous PGRMC1 levels were depleted. To test this concept, endogenous PGRMC1 was reduced and replaced with PGRMC1-GFP by using a combined PGRMC1 siRNA and PGRMC1-GFP transfection protocol. Although this combined protocol effectively replaced endogenous PGRMC1 with PGRMC1-GFP fusion protein, the cells treated with this combined protocol failed to respond to the anti-apoptotic effect of P4. The failure to respond to P4 was not likely due to a lack of PGRMC1 but rather to the relative absence of PGRMC1 dimers, which would normally be present within the nucleus. This suggests that in addition to the cytoplasmic site of action, PGRMC1 has a nuclear site of action, which probably involves gene transcription. This possibility is supported by the finding that actinomycin D, a well-characterized inhibitor of RNA synthesis, attenuates P4's ability to prevent SIGC apoptosis. Collectively, these studies support a nuclear or genomic component of PGRMC1's mechanism of action.
In an attempt to identify some of the genes whose expression might be part of the putative genomic action of PGRMC1, mRNA levels of numerous genes involved in apoptosis were assessed using real-time PCR microarrays. This approach yielded several interesting findings. First, treatment with PGRMC1 siRNA in the absence of P4 significantly altered that pattern of gene expression compared to treatment with scrambled (control) siRNA. The overall effect was to increase the mRNA levels of many genes like Cradd that are involved in promoting apoptosis. This suggests that PGRMC1, which is not bound to P4, regulates gene expression in a manner that would make the cells more susceptible to undergoing apoptosis. This finding fits well with PGRMC1's cell survival function (Cahill 2007; Losel et al. 2008) and is consistent with the observations that depletion of PGRMC1 makes ovarian (Peluso et al. 2008), endometrial (Pru et al. 2009) and breast cancer (Crudden et al. 2006; Pru et al. 2009) cells more sensitive to the apoptotic effects of various chemotherapeutic agents.
The second finding is that P4 alters the gene expression profile such that the mRNAs that encode many genes involved in promoting apoptosis are suppressed. Some of suppressed genes include caspase 1,2,3 and 14 as well as Bad. The only anti-apoptotic gene showing an increase in expression is Bcl2a1d. BCL2A1D is a BCL2 family member and functions like BCL2 to prevent apoptosis (Rasooly et al. 2005; Yin et al. 2005). Since elevating the ratio of BCL2 to BAD is known to promote granulosa cell survival (Kaipia et al. 1997), P4's ability to differentially regulate the expression of these two proteins could be an important part of it's anti-apoptotic action. Moreover, the overall effect of P4 is to promote the genes known to promote cell survival, which would make the cells more resistant to apoptosis. This fits with P4's well-characterized anti-apoptotic action (Cahill 2007). Interesting, the actinomycin D studies suggest that specific genes are induced as part of P4's anti-apoptotic mechanism. With the exception of Bcl2a1d, P4 treatment generally suppressed gene expression. The most likely mechanism to explain this contradiction is that P4 induces the expression of one or more transcription factors which in turn act to suppress the expression of the genes that are directly involved in stimulating apoptosis.
The third observation from the microarray studies is that the P4-regulated expression of Bcl2a1d and Bad is dependent on PGRMC1. This conclusion is based on observation that P4's ability to induce Bcl21d mRNA and to suppress Bad mRNA is completely attenuated by pretreatment with PGRMC1 siRNA. However other P4-induced changes in the gene profile do not appear to be dependent on PGRMC1. Most noticeably the changes related to the mRNA levels of several caspase family members. The mechanism through which P4 influences the expression of these genes is unknown but does not involve the nuclear P4 receptor (PGR), since SIGCs do not express these receptors (Peluso et al. 2006; Peluso et al. 2008).
In summary, the present studies suggest that the mechanism through which PGRMC1 regulates cell viability has at least two components. The first component involves P4's interaction with PGRMC1 that localizes to the cytoplasm and probably the plasma membrane. This site of action likely accounts for P4's ability to induce protein kinase G activity (Peluso and Pappalardo 2004; Peluso et al. 2007) and to suppress Erk1/2 activity (Peluso et al. 2003) as well as intracellular free calcium levels (Peluso et al. 2002), since these three cytoplasmic events are involved in P4's anti-apoptotic action. The genomic component of PGRMC1's action appears to require the DTT-resistant dimeric form of PGRMC1, which is present in the nucleus and likely functions to change the gene expression profile such that the net effect is to promote cell survival. Whether the PGRMC1 dimer functions as a transcription factor to alter gene expression is presently under investigation. Finally, it is clear that both cytoplasmic and nuclear components of PGRMC1's mechanism of action are required to mediate P4's anti-apoptotic action but the precise molecular signaling events of each component remain to be elucidated.
Supplementary Material
5. Acknowledgment
The authors would like to thank Dr. Robert C. Burghardt of Texas A&M University for providing the spontaneously immortalized granulosa cells and Dr. Jim Pru of Washington State University for his critical reading this manuscript. These studies were supported by a grant from the National Institutes of Health (HD 052740), which was awarded to JJP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad M, Srinivasula SM, Wang L, Talanian RV, Litwack G, Fernandes-Alnemri T, Alnemri ES. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57:615–619. [PubMed] [Google Scholar]
- Cahill MA. Progesterone receptor membrane component 1: an integrative review. J Steroid Biochem Mol Biol. 2007;105:16–36. doi: 10.1016/j.jsbmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Conneely OM, Lydon JP. Progesterone receptors in reproduction: functional impact of the A and B isoforms. Steroids. 2000;65:571–577. doi: 10.1016/s0039-128x(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Crudden G, Chitti RE, Craven RJ. Hpr6 (heme-1 domain protein) regulates the susceptibility of cancer cells to chemotherapeutic drugs. J Pharmacol Exp Ther. 2006;316:448–455. doi: 10.1124/jpet.105.094631. [DOI] [PubMed] [Google Scholar]
- Engmann L, Losel R, Wehling M, Peluso JJ. Progesterone regulation of human granulosa/luteal cell viability by an RU486-independent mechanism. J Clin Endocrinol Metab. 2006;91:4962–4968. doi: 10.1210/jc.2006-1128. [DOI] [PubMed] [Google Scholar]
- Falkenstein E, Eisen C, Schmieding K, Krautkramer M, Stein C, Losel R, Wehling M. Chemical modification and structural analysis of the progesterone membrane binding protein from porcine liver membranes. Mol Cell Biochem. 2001;218:71–79. doi: 10.1023/a:1007269507856. [DOI] [PubMed] [Google Scholar]
- Friberg PA, Larsson DG, Billig H. Dominant role of nuclear progesterone receptor in the control of rat periovulatory granulosa cell apoptosis. Biol Reprod. 2009;80:1160–1167. doi: 10.1095/biolreprod.108.073932. [DOI] [PubMed] [Google Scholar]
- Gellersen B, Fernandes MS, Brosens JJ. Non-genomic progesterone actions in female reproduction. Hum Reprod Update. 2009;15:119–138. doi: 10.1093/humupd/dmn044. [DOI] [PubMed] [Google Scholar]
- Gerdes D, Wehling M, Leube B, Falkenstein E. Cloning and tissue expression of two putative steroid membrane receptors. Biol Chem. 1998;379:907–911. doi: 10.1515/bchm.1998.379.7.907. [DOI] [PubMed] [Google Scholar]
- Kaipia A, Hsu SY, Hsueh AJ. Expression and function of a proapoptotic Bcl-2 family member Bcl-XL/Bcl-2-associated death promoter (BAD) in rat ovary. Endocrinology. 1997;138:5497–5504. doi: 10.1210/endo.138.12.5588. [DOI] [PubMed] [Google Scholar]
- Kariagina A, Aupperlee MD, Haslam SZ. Progesterone receptor isoform functions in normal breast development and breast cancer. Crit Rev Eukaryot Gene Expr. 2008;18:11–33. doi: 10.1615/critreveukargeneexpr.v18.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losel RM, Besong D, Peluso JJ, Wehling M. Progesterone receptor membrane component 1--many tasks for a versatile protein. Steroids. 2008;73:929–934. doi: 10.1016/j.steroids.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Mansouri MR, Schuster J, Badhai J, Stattin EL, Losel R, Wehling M, Carlsson B, Hovatta O, Karlstrom PO, Golovleva I, Toniolo D, Bione S, Peluso J, Dahl N. Alterations in the expression, structure and function of progesterone receptor membrane component-1 (PGRMC1) in premature ovarian failure. Hum Mol Genet. 2008;17:3776–3783. doi: 10.1093/hmg/ddn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola JL, Sirover MA. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim Biophys Acta. 2003;1622:50–56. doi: 10.1016/s0304-4165(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Meyer C, Schmieding K, Falkenstein E, Wehling M. Are high-affinity progesterone binding site(s) from porcine liver microsomes members of the sigma receptor family? Eur J Pharmacol. 1998;347:293–299. doi: 10.1016/s0014-2999(98)00103-4. [DOI] [PubMed] [Google Scholar]
- Mote PA, Graham JD, Clarke CL. Progesterone receptor isoforms in normal and malignant breast. Ernst Schering Found Symp Proc. 2007:77–107. [PubMed] [Google Scholar]
- Natraj U, Richards JS. Hormonal regulation, localization, and functional activity of the progesterone receptor in granulosa cells of rat preovulatory follicles. Endocrinology. 1993;133:761–769. doi: 10.1210/endo.133.2.8344215. [DOI] [PubMed] [Google Scholar]
- Park OK, Mayo KE. Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol Endocrinol. 1991;5:967–978. doi: 10.1210/mend-5-7-967. [DOI] [PubMed] [Google Scholar]
- Park-Sarge OK, Mayo KE. Regulation of the progesterone receptor gene by gonadotropins and cyclic adenosine 3',5'-monophosphate in rat granulosa cells. Endocrinology. 1994;134:709–718. doi: 10.1210/endo.134.2.8299566. [DOI] [PubMed] [Google Scholar]
- Peluso JJ. Non-genomic actions of progesterone in the normal and neoplastic mammalian ovary. Semin Reprod Med. 2007;25:198–207. doi: 10.1055/s-2007-973432. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Bremner T, Fernandez G, Pappalardo A, White BA. Expression pattern and role of a 60-kilodalton progesterone binding protein in regulating granulosa cell apoptosis: involvement of the mitogen-activated protein kinase cascade. Biol Reprod. 2003;68:122–128. doi: 10.1095/biolreprod.102.007542. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Fernandez G, Pappalardo A, White BA. Membrane-initiated events account for progesterone's ability to regulate intracellular free calcium levels and inhibit rat granulosa cell mitosis. Biol Reprod. 2002;67:379–385. doi: 10.1095/biolreprod67.2.379. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Liu X, Gawkowska A, Johnston-MacAnanny E. Progesterone activates a progesterone receptor membrane component 1-dependent mechanism that promotes human granulosa/luteal cell survival but not progesterone secretion. J Clin Endocrinol Metab. 2009;94:2644–2649. doi: 10.1210/jc.2009-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso JJ, Liu X, Romak J. Progesterone maintains basal intracellular adenosine triphosphate levels and viability of spontaneously immortalized granulosa cells by promoting an interaction between 14-3-3sigma and ATP synthase beta/precursor through a protein kinase G-dependent mechanism. Endocrinology. 2007;148:2037–2044. doi: 10.1210/en.2006-1603. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Liu X, Saunders MM, Claffey KP, Phoenix K. Regulation of ovarian cancer cell viability and sensitivity to cisplatin by progesterone receptor membrane component-1. J Clin Endocrinol Metab. 2008;93:1592–1599. doi: 10.1210/jc.2007-2771. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A. Progesterone mediates its anti-mitogenic and anti-apoptotic actions in rat granulosa cells through a progesterone-binding protein with gamma aminobutyric acidA receptor-like features. Biol Reprod. 1998;58:1131–1137. doi: 10.1095/biolreprod58.5.1131. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A. Progesterone regulates granulosa cell viability through a protein kinase G-dependent mechanism that may involve 14-3-3sigma. Biol Reprod. 2004;71:1870–1878. doi: 10.1095/biolreprod.104.031716. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Losel R, Wehling M. Expression and function of PAIRBP1 within gonadotropin-primed immature rat ovaries: PAIRBP1 regulation of granulosa and luteal cell viability. Biol Reprod. 2005;73:261–270. doi: 10.1095/biolreprod.105.041061. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Losel R, Wehling M. Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone's antiapoptotic action. Endocrinology. 2006;147:3133–3140. doi: 10.1210/en.2006-0114. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Romak J, Liu X. Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology. 2008;149:534–543. doi: 10.1210/en.2007-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pru JK, Friel AM, Zhang L, Blok LJ, Shioda T, Peluso J, Rueda BR. Progesterone receptor membrane component 1 promotes endometrial and breast cancer cell viability in response to chemotherapy in vitro and in vivo. Biol Reprod. 2009;81:98. [Google Scholar]
- Rasooly R, Schuster GU, Gregg JP, Xiao JH, Chandraratna RA, Stephensen CB. Retinoid x receptor agonists increase bcl2a1 expression and decrease apoptosis of naive T lymphocytes. J Immunol. 2005;175:7916–7929. doi: 10.4049/jimmunol.175.12.7916. [DOI] [PubMed] [Google Scholar]
- Rothchild I. The corpus luteum revisited: are the paradoxical effects of RU486 a clue to how progesterone stimulates its own secretion? Biol Reprod. 1996;55:1–4. doi: 10.1095/biolreprod55.1.1. [DOI] [PubMed] [Google Scholar]
- Rung E, Friberg PA, Shao R, Larsson DG, Nielsen E, Svensson PA, Carlsson B, Carlsson LM, Billig H. Progesterone-receptor antagonists and statins decrease de novo cholesterol synthesis and increase apoptosis in rat and human periovulatory granulosa cells in vitro. Biol Reprod. 2005;72:538–545. doi: 10.1095/biolreprod.104.033878. [DOI] [PubMed] [Google Scholar]
- Scott RE, Wu-Peng XS, Pfaff DW. Regulation and expression of progesterone receptor mRNA isoforms A and B in the male and female rat hypothalamus and pituitary following oestrogen treatment. J Neuroendocrinol. 2002;14:175–183. doi: 10.1046/j.0007-1331.2001.00750.x. [DOI] [PubMed] [Google Scholar]
- Shao R, Markstrom E, Friberg PA, Johansson M, Billig H. Expression of progesterone receptor (PR) A and B isoforms in mouse granulosa cells: stage-dependent PR-mediated regulation of apoptosis and cell proliferation. Biol Reprod. 2003;68:914–921. doi: 10.1095/biolreprod.102.009035. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LS, Stoica G, Tilley R, Burghardt RC. Rat ovarian granulosa cell culture: a model system for the study of cell-cell communication during multistep transformation. Cancer Res. 1991;51:696–706. [PubMed] [Google Scholar]
- Stouffer RL. Progesterone as a mediator of gonadotrophin action in the corpus luteum: beyond steroidogenesis. Hum Reprod Update. 2003;9:99–117. doi: 10.1093/humupd/dmg016. [DOI] [PubMed] [Google Scholar]
- Yin W, Raffelsberger W, Gronemeyer H. Retinoic acid determines life span of leukemic cells by inducing antagonistic apoptosis-regulatory programs. Int J Biochem Cell Biol. 2005;37:1696–1708. doi: 10.1016/j.biocel.2005.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.