Abstract
Significant hurdles remain to large-scale implementation of medical interventions in the developing world due to the lack of a modern diagnostic infrastructure. This is especially pertinent to the international roll-out of antiretroviral drugs to treat HIV, which ideally includes a CD4 t-cell to determine eligibility. We designed a novel technique to estimate mature t-cell numbers by calculating the amount of rearranged t-cell receptor β genes from dried blood spots of HIV-infected individuals in the United States and Uganda. It was observed that the rearranged t-cell receptor β count correlated well with total lymphocyte counts from both study populations (Baltimore R=0.602, Uganda R=0.497; p<0.001) and the ability for this measurement to determine antiretroviral initiation was similar to total lymphocyte counts, which can be used to determine eligibility in HIV+ children. This technique as well as other dried blood spot based technologies could increase the diagnostic and monitoring capabilities in resource-limited settings.
1. Introduction
The recent expansion of anti-retroviral treatment (ART) for the human immunodeficiency virus (HIV) in the developing world has highlighted a growing chasm between resource-limited and resource-rich countries in the area of diagnostic medicine(Barnett et al., 2008; Redd and Quinn, 2008; Stevens et al., 2008). Many modern diagnostic techniques are only accessible to patients in areas that are in close proximity to fully equipped state-of-the-art medical facilities, which are either non-existent or only found in major urban centers in most resource-limited countries. This lack of diagnostic laboratory infrastructure has proved to be a major hurdle for the scale-up of modern medical treatment in the developing world. This is especially true in sub-Saharan Africa where a significant proportion of the population lives in rural environments(Parker and Cubitt, 1999; UNFPA, 2007; Redd and Quinn, 2008). In the case of ART, initiation is guided by combining clinical staging with a CD4+ T-cell count, which requires fresh whole blood, a reliable cold-transport chain, and a laboratory based flow cytometer within approximately a half day’s travel from the patient(Spacek et al., 2006; Kagaayi et al., 2007; Hallett et al., 2008). The World Health Organization (WHO) has recommended that in areas where CD4 counts are unavailable that total lymphocyte count (TLC) can be used to help determine ART eligibility in children(WHO, 2006 accessed Dec 10, 2009). However, TLC assays suffer from many of the same constraints as CD4 tests, in particular, the requirements of a fresh blood sample and a reliable cold-transport chain. Due to these requirements, expanding the diagnostic and clinical monitoring capability in resource-limited settings to reach the nearly 70% of ART-eligible individuals currently without access to treatment would require a massive expansion of the medical and transportation infrastructure in these areas, which currently is not a feasible option(UNAIDS, 2007). Consequently, new approaches are needed to increase the diagnostic capacity given the laboratory and logistical limitations in resource-limited settings(Redd and Quinn, 2008).
One possible strategy is to use dried blood spots (DBS) as a sample collection method since these are stable at room temperature, can be obtained from a finger prick, and cellular and viral proteins and nucleic acids can be easily recovered(Parker and Cubitt, 1999; Mwaba et al., 2003; Ziemniak et al., 2006; McDade et al., 2007). Additionally, DBS would eliminate the necessity for a cold-transport chain and allow samples to be easily shipped to a centrally located laboratory for testing.
The TCR-β gene undergoes a gene rearrangement process during T-cell maturation to obtain a functional TCR protein(Siu et al., 1984). Due to junctional flexibility and the somatic cell mutations that occur during rearrangement, the DNA and protein sequence of the rearranged TCR-β genes (rTCR-β) are highly diverse making direct quantification difficult. However, it has been demonstrated that accurate amounts of unrearranged TCR-β genes can be determined from cellular DNA(Chain et al., 2005). Thus, we hypothesized that DNA derived from DBS could be used to accurately determine an individual’s total lymphocyte count (TLC) by calculating the amounts of total cell DNA using a housekeeping gene and subtracting the number of TCR-β genes remaining in the unrearranged germline configuration, thereby obtaining a rTCR-β count.
2. Materials and Methods
2.1. Study Populations
Venous blood samples were collected in EDTA purple top vacutainer tubes (BD Bioscience) from HIV+ subjects attending the Moore Clinic at Johns Hopkins Hospital in Baltimore (n=64) or participating in either the ART service program or the Rakai Community Cohort Study (RCCS) in Rakai, Uganda (n=245) (Table 1). Institutional Review Board approvals were obtained from Uganda Virus Research Institute’s Scientific and Ethics Committee, the Uganda National Council for Science and Technology, and from the Institutional Review Boards of collaborating US institutions (National Institutes of Health and Johns Hopkins University). These patients have previously provided consent for collection of samples for the use in HIV research. Eight 50μL blood aliquots from the tube used for the FACS derived CD4 count were blotted on two Whatman 903 filter paper cards for each subject and dried overnight at room temperature.
Table 1.
Population dynamics of the study sites with median values and interquartile range shown.
| Study site | Percent female | Median Age | Total Lymphocyte Count/μL | CD4 count/μL |
|---|---|---|---|---|
| Baltimore, MD (n=64) | 36.5% | 48 (44–52) | 1960 (1585–2615) | 370.5 (240–511) |
| Rakai, Uganda (n=245) | 73.3% | 32 (28–39) | 1910 (1520–2432.5) | 580.5 (408–803) |
2.2. RTCR-β quantification assay
To obtain the rearranged rTCR-β count six 3mm holes were punched out of ~3/4 of a blood spot and used for the DNA isolation. Genomic DNA (gDNA) was isolated according to the manufacturer’s recommended protocol included in the Purelink Genomic DNA Mini kit (Invitrogen), and eluted in 100μL of nuclease-free water (Promega). The resulting DNA samples were then stored at −20°C.
Two primer-probe pairs were used for the subsequent quantitative real-time PCR (QPCR). The germline configuration of the VD junction region (VD-J) of the TCR-β was calculated using previously reported primers and probes(Chain et al., 2005). Albumin (Alb) was used as a housekeeping gene to determine total cell number, and calculated with primers Alb-2 forward 5′-CACTTGTTGAGCTCGTGAAAC-3′, Alb-2 reverse 5′-CAGCCTTGCAGCACTTCTC-3′, and Alb-2 Probe 5′ FAM-CAAGCCCAAGGCAACAAAAGAGCA-TAMRA-3′ (Sigma). Final primer and probe concentrations for both QPCR reactions are 1μM, and 0.5μM respectively. HeLa cell genomic DNA, which has 100% TCR-β genes in germline configuration, was isolated from cell culture using the same extraction kit as above and eluded in nuclease-free water. This HeLa cell DNA was used as a standard for both target genes at a concentration of 6 ng/μL for an equivalent cellular DNA count of 2000 genomes/μL(Chain et al., 2005). A four step 1:10 serial dilution of the standard was performed and 5μL of each used in a 50μL reaction for a range of detection from 10,000-10 genomic copies. The samples and standards were run in a 50μL reaction on an ABI 7900 real-time PCR machine for 40 cycles with and annealing temperature of 57°C for 60 seconds. In addition, a 25% mixture of gDNA derived from the T-cell clone Een 217, which has 98.3% of its TCR-β genes rearranged according to preliminary real-time PCR analysis, and 75% HeLa cell DNA was created based on spectrophotometer derived gDNA levels, and was used as a control for QPCR assay variability and to examine the assay accuracy.
The QPCR reaction were run in a 96-well format with each of the four HeLa controls, ddH2O, and a control sample of high and low HeLa cell DNA were run in duplicate for each target gene. Each plate set (18 samples) was run in triplicate. Standard curves were created for each run according to the HeLa cell dilutions, and values determined for each sample replicate. The resulting duplicate counts for VD-J were averaged and subtracted from the Alb average to obtain the final rearranged TCR-β count. The three replicates are then averaged and normalized for that plate set [(Sample rTCR count – mean of set rTCR counts)/standard deviation of set rTCR counts] to minimize inter-assay variability.
Aliquots of the 25% T-cell clone mixture were diluted 1:5 with water, and the resulting mixture was serially diluted 5 times at a 1:2 ratio. The resulting samples were estimated to contain 1400, 700, 350, 175, and 87.5 counts of rTCR-β. These dilutions were run using the protocol described above to determine the range and accuracy of the assay. The assay was found to be highly variable when the dilutions were below an estimated 22 rTCR-β counts.
2.3. Statistical analysis
For both populations normalized rTCR-β counts were compared to flow cytometer derived TLC using a Spearman rank order correlation analysis. Receiver operating curve (ROC) analysis was performed for normalized rTCR-β counts and TLC with a cut-off of a CD4 count < 250 cells/μL for determining ART eligibility. Significance for both tests was a p<0.05.
3. Results
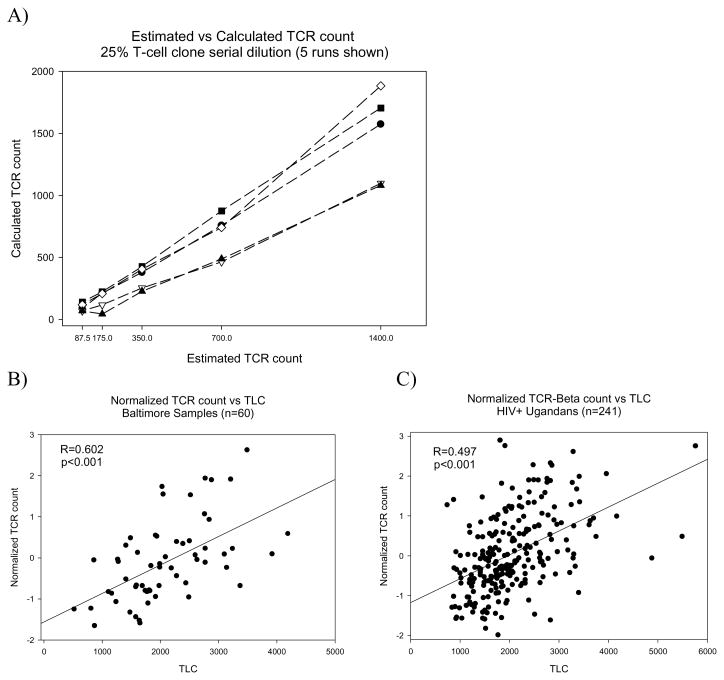
The 25% mixture of gDNA derived from the T-cell clone Een 217, which has 98.3% of its TCR-β genes rearranged, were used to examine assay variability and accuracy. These dilutions were run using the protocol described above to determine the range and accuracy of the assay (Figure 1A). It was observed that the assay varies significantly at the higher end of the range examined, but the linearity of the results of each run were consistent suggesting that this was due to inter-assay variation. To account for this, the rTCR-β counts of the subsequent samples were normalized for each real-time PCR plate of 18 samples.
Figure 1.
The calculated rTCR-β raw count for a 1:2 serial dilution of a 25% rearranged T-cell mixture is shown compared to the estimated starting amount (A). The normalized rTCR-β count is shown for the subjects from Baltimore (B) and Uganda (C) compared to the flow cytometer derived total lymphocyte count. Coefficients for correlation (R) with significance indicated (Spearman rank order correlation).
The normalized rTCR-β counts were compared to the TLC for the HIV+ patients from Baltimore and Uganda (Figure 1B&C respectively). The normalized rTCR count correlated significantly for both groups, with the Ugandan population having a slightly lower coefficient of correlation (R) than the Baltimore samples (R=0.497 & 0.602 respectively, p<0.001; Spearman rank test). The normalized rTCR count also correlated significantly with CD4 counts from these same subjects (Uganda R=0.288, Baltimore R=0.387; p<0.01).
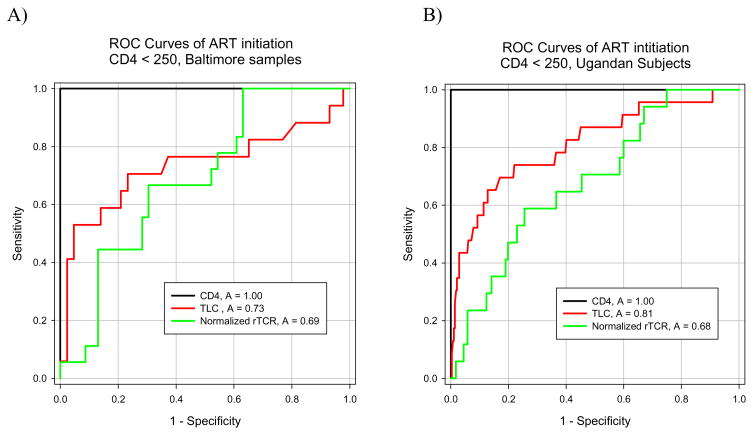
The World Health Organization (WHO) recommends that TLC can be used as a surrogate for CD4 counts in children in resource limited settings. Therefore, ROC analyses were performed to examine if the normalized rTCR-β count could be used as an indicator for ART initiation, and how well this and TLC compared to the gold standard of CD4 counts. In both populations, a CD4 count of <250 was used as the cut-off for ART eligibility, which is consistent with the standard of care at the Rakai Health Science Program. It was found that the normalized rTCR-β counts from both populations were approximately equal in their predictive ability in determining ART initiation in subjects with CD4<250 cells/uL (Figure 2). Additionally, it was found that the rTCR-β count predicted ART initiation at a similar accuracy as TLC in the Baltimore subjects (p=0.581), however, in the Ugandan population the TLC was significantly more accurate (p<0.05) (Figure 2). Using a cutoff value of −0.5 for the normalized rTCR-β count it was observed that the sensitivity and specificity were similar between study populations, but the positive predictive value of the assay was markedly higher in the Baltimore population (Table 2).
Figure 2.
ROC analyses of TLC and rTCR-β counts for ART initiation using a cut-off of a CD4 count < 250 cells/μL for the Baltimore (A) and Ugandan (B) populations are shown. The TLC and normalized rTCR-β count were significantly different in the Ugandan population (p<0.05).
Table 2.
Sensitivity, specificity, positive and negative predictive values for Baltimore and Rakai populations using a normalized rTCR count of −0.5 and a CD4 count of 250 as the cut-off for ART eligibility.
| Study Site | Sensitivity | Specificity | Positive predictive value | Negative predictive value |
|---|---|---|---|---|
| Baltimore, MD (n=64) | 61.1 | 69.6 | 44.0 | 82.1 |
| Rakai, Uganda (n=245) | 58.8 | 71.5 | 13.3 | 95.9 |
4. Discussion
DBS are an ideal sample collection method for large scale monitoring in the developing world due to simple collection procedures and the high stability of the sample in the absence of refrigeration(Parker and Cubitt, 1999; McDade et al., 2007). Additionally, DBS have been used for decades to diagnose adult and neonatal HIV-1 infection using either antibody assays or PCR, and can be used to detect a range of other infectious diseases and genetic abnormalities(Parker and Cubitt, 1999). DBS-based tests could allow the limited number of existing laboratories and trained staff in the developing world to greatly increase their coverage with little additional capital investment. This would have the added benefit of streamlining and centralizing quality control which is an essential component in the expansion of diagnostic monitoring in resource-limited settings(Redd and Quinn, 2008; Stevens et al., 2008). This technology could also possibly be adapted to a point of care test for children, which is the ideal solution for ART rollout in resource poor settings.
While examining multiple extractions from the same subject’s blood it was observed that the end rTCR-β count was varied between different extraction protocols, as well as within individual protocols performed at different times. This problem could possibly be eliminated by automating the extraction process. This assay also does not take into account the effects of residual T-cell receptor excision circles or TCR allelic exclusion on the rTCR-β counts and it is possible that due to the high rate of lymphocyte turnover in HIV disease these two factors could be artificially increasing or decreasing the rTCR count respectively(Hazenberg et al., 2000; McCune, 2001). Additionally, only mature T-cells that have undergone an extensive selection process in the thymus are found in the bloodstream with a rearranged TCR-β gene. Thus, the TCR-β assay is most likely detecting only fully mature or close to fully mature T-cells, and not CD4−/CD8− dual negative T-cells, γ/δ TCR T-cells, or lymphocyte progenitor cells, all of which can be counted as lymphocytes in a TLC(Mathiot et al., 2001).
CD4 counts are highly predictive of HIV disease progression, and significant benefit to the long-term survival is seen in patients who begin ART earlier or before their CD4 count drops below 200(Palella et al., 2003; Hallett et al., 2008; Kitahata et al., 2009). However, the CD4 test requires a FACS based reader, fresh whole blood, and a reliable cold-transport chain to obtain an accurate measurement. These constraints impede large-scale roll-out of ART in developing nations(Stevens et al., 2008). The ROC and sensitivity analysis of the rTCR-β counts, as well as the cost and logistical requirements of real-time PCR, suggest that it may be possible to use this measurement as a tool for epidemiological studies, but further optimization of the assay is necessary before the utility of this technique as a diagnostic alternative for ART initiation is possible.
5. Conclusion
Despite these limitations, the results presented here describe the first technique designed to assess total mature T-cell numbers from gDNA alone, and provide a proof-of-concept that this technique can accurately estimate T-cell number from a DBS. This and other DBS based technologies could increase the diagnostic and monitoring capabilities in resource-limited settings.
Acknowledgments
The authors would like to thank all the participants of the study in Rakai Uganda and at Johns Hopkins Hospital, as well as the staff of the RHSP and the Moore clinic. The following reagent was obtained through the AIDS Research and Reference Program, Division of AIDS, NIAID, NIH: Een 217 from Dr Robert F. Siliciano. AD Redd designed and performed the research, analyzed the data and wrote the paper; EJ Ciccone performed the research and analyzed the data; G Nakigozi designed the research; JC Keruly performed the research; A Ndyanabo and B Iga analyzed the data; RH Gray, D Serwadda, and TC Quinn designed the research. Funding for this study was provided by Johns Hopkins Center for Global Health and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnett D, Walker B, Landay A, Denny TN. CD4 immunophenotyping in HIV infection. Nature Reviews Microbiology. 2008;6:S7. doi: 10.1038/nrmicro1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain JL, Joachims ML, Hooker SW, Laurent AB, Knott-Craig CK, Thompson LF. Real-time PCR method for the quantitative analysis of human T-cell receptor gamma and beta gene rearrangements. J Immunol Methods. 2005;300:12–23. doi: 10.1016/j.jim.2005.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett TB, Gregson S, Dube S, Garnett GP. The impact of monitoring HIV patients prior to treatment in resource-poor settings: insights from mathematical modelling. PLoS Med. 2008;5:e53. doi: 10.1371/journal.pmed.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazenberg MD, Hamann D, Schuitemaker H, Miedema F. T cell depletion in HIV-1 infection: how CD4+ T cells go out of stock. Nat Immunol. 2000;1:285–9. doi: 10.1038/79724. [DOI] [PubMed] [Google Scholar]
- Kagaayi J, Makumbi F, Nakigozi G, Wawer MJ, Gray RH, Serwadda D, Reynolds SJ. WHO HIV clinical staging or CD4 cell counts for antiretroviral therapy eligibility assessment? An evaluation in rural Rakai district, Uganda. Aids. 2007;21:1208–10. doi: 10.1097/QAD.0b013e32810c8dce. [DOI] [PubMed] [Google Scholar]
- Kitahata MM, Gange SJ, Abraham AG, Merriman B, Saag MS, Justice AC, Hogg RS, Deeks SG, Eron JJ, Brooks JT, Rourke SB, Gill MJ, Bosch RJ, Martin JN, Klein MB, Jacobson LP, Rodriguez B, Sterling TR, Kirk GD, Napravnik S, Rachlis AR, Calzavara LM, Horberg MA, Silverberg MJ, Gebo KA, Goedert JJ, Benson CA, Collier AC, Van Rompaey SE, Crane HM, McKaig RG, Lau B, Freeman AM, Moore RD. Effect of early versus deferred antiretroviral therapy for HIV on survival. N Engl J Med. 2009;360:1815–26. doi: 10.1056/NEJMoa0807252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiot ND, Krueger R, French MA, Price P. Percentage of CD3+CD4−CD8−gammadeltaTCR- T cells is increased HIV disease. AIDS Res Hum Retroviruses. 2001;17:977–80. doi: 10.1089/088922201750290096. [DOI] [PubMed] [Google Scholar]
- McCune JM. The dynamics of CD4+ T-cell depletion in HIV disease. Nature. 2001;410:974–9. doi: 10.1038/35073648. [DOI] [PubMed] [Google Scholar]
- McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography. 2007;44:899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- Mwaba P, Cassol S, Pilon R, Chintu C, Janes M, Nunn A, Zumla A. Use of dried whole blood spots to measure CD4+ lymphocyte counts in HIV-1-infected patients. Lancet. 2003;362:1459–60. doi: 10.1016/S0140-6736(03)14693-4. [DOI] [PubMed] [Google Scholar]
- Palella FJ, Jr, Deloria-Knoll M, Chmiel JS, Moorman AC, Wood KC, Greenberg AE, Holmberg SD. Survival benefit of initiating antiretroviral therapy in HIV-infected persons in different CD4+ cell strata. Ann Intern Med. 2003;138:620–6. doi: 10.7326/0003-4819-138-8-200304150-00007. [DOI] [PubMed] [Google Scholar]
- Parker SP, Cubitt WD. The use of the dried blood spot sample in epidemiological studies. J Clin Pathol. 1999;52:633–9. doi: 10.1136/jcp.52.9.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AD, Quinn T. Foreword: CD4 immunodiagnostics for HIV in the developing world. Nature Reviews: Microbiology. 2008:S1. doi: 10.1038/nrmicro1996. [DOI] [PubMed] [Google Scholar]
- Siu G, Kronenberg M, Strauss E, Haars R, Mak TW, Hood L. The structure, rearrangement and expression of D beta gene segments of the murine T-cell antigen receptor. Nature. 1984;311:344–50. doi: 10.1038/311344a0. [DOI] [PubMed] [Google Scholar]
- Spacek LA, Gray RH, Wawer MJ, Sewankambo NK, Serwadda D, Wabwire-Mangen F, Kiwanuka N, Kigozi G, Nalugoda F, Quinn TC. Clinical illness as a marker for initiation of HIV antiretroviral therapy in a rural setting, Rakai, Uganda. Int J STD AIDS. 2006;17:116–20. doi: 10.1258/095646206775455766. [DOI] [PubMed] [Google Scholar]
- Stevens WS, Gelman R, Glencross DK, Scott LE, Crowe SM, TS Evaluating new CD4 enumeration technologies for resource-constrained countries. Nature Reviews Microbiology. 2008;6:s29. doi: 10.1038/nrmicro2000. [DOI] [PubMed] [Google Scholar]
- UNAIDS. AIDS Epidemic Update. 2007. [Google Scholar]
- UNFPA. Unleashing the potential of urban growth. UNFPA; 2007. UNFPA: State of World Population 2007. [Google Scholar]
- WHO. Recommendations for a public health approach. World Health Organization; 2006. [accessed Dec 10, 2009]. Antiretroviral therapy of HIV infection in infants and children in resource-limited settings: towards universal access. [PubMed] [Google Scholar]
- Ziemniak C, George-Agwu A, Moss WJ, Ray SC, Persaud D. A sensitive genotyping assay for detection of drug resistance mutations in reverse transcriptase of HIV-1 subtypes B and C in samples stored as dried blood spots or frozen RNA extracts. J Virol Methods. 2006;136:238–47. doi: 10.1016/j.jviromet.2006.05.030. [DOI] [PubMed] [Google Scholar]