Abstract
Dynamic microtubules are essential for the process of mitosis. Thus, elucidating when, where, and how microtubule dynamics are regulated is key to understanding this process. One important class of proteins that directly regulates microtubule dynamics is the Kinesin-13 family. Kinesin-13 proteins induce depolymerization uniquely from both ends of the microtubule. This activity coincides with their cellular localization and with their ability to regulate microtubule dynamics to control spindle assembly and kinetochore-microtubule attachments. In this review, we highlight recent findings that dissect the important actions of Kinesin-13 family members and summarize important studies on the regulation of their activity by phosphorylation and by protein-protein interactions.
Keywords: Kinesin-13, Microtubule dynamics, Phosphorylation, Protein-protein interaction, Spindle assembly, Kinetochore-microtubule attachments
1. Introduction
Dynamic microtubules are critical to multiple aspects of cellular function. Most notable is the dramatic rearrangement of the microtubule cytoskeleton as the cell breaks down its interphase microtubule array to assemble the mitotic spindle. Dynamic microtubules are also important for the movements of the chromosomes on the spindle and the accurate partitioning of the DNA to the two daughter cells. The emerging picture is that multiple proteins act in space and time to orchestrate this symphony of cellular events. The Kinesin-13 family members provide excellent examples of the diverse mechanisms that are utilized to regulate microtubule dynamics. Most cells contain multiple members of the Kinesin-13 family that control individual facets of spindle microtubule dynamics. Their activities are regulated by distinct targeting to regions of the spindle, by regulatory phosphorylation events, and by interactions with different binding partners. While work is still in its early stages, these studies demonstrate the complexities of the cytoskeletal architecture and highlight the diverse approaches that are needed to tackle the question of how microtubule dynamics are spatially and temporally regulated during mitotic progression.
1.1 Establishment of Microtubule Dynamics within the Spindle
Microtubules are inherently dynamic polymers that exhibit a behavior known as dynamic instability [1], in which microtubules co-exist in states of growth and shrinkage and interconvert randomly between these two states. The proper regulation of dynamic instability is critical for spindle assembly as there is a dramatic increase in microtubule turnover as cells enter mitosis [2–4] that is correlated with an increase in catastrophe frequency and a decrease in rescue frequency in cells [5]. Microtubules are organized within the spindle with their less dynamic minus-ends at the centrosome and their more dynamic plus-ends extending toward the spindle equator. Within the spindle, there are several classes of microtubules that are defined both by their organization and by their dynamic properties. All classes of microtubules within the spindle exhibit dynamic instability, but both K-fibers and spindle microtubules exhibit an additional behavior called poleward microtubule flux in which tubulin subunits are continuously added at the plus-ends of microtubules and translocated toward the spindle pole where they are removed from the minus-ends [6–8]. Poleward flux may be critical for chromosome segregation [9–11]; however, some recent studies suggest that flux may also contribute to the fidelity of chromosome segregation [12,13]. These studies highlight the complexities of regulating microtubule dynamics that must occur to ensure proper spindle function.
1.2 Regulators of Microtubule Dynamics
Many microtubule regulatory proteins, including both microtubule stabilizing and destabilizing proteins, control microtubule dynamics to achieve proper spindle assembly and chromosome segregation. A lot of interest over the past decade has focused on the activities of the Kinesin-13 family of microtubule destabilizing motors, which play diverse roles in spindle assembly, spindle dynamics, proper chromosome attachment, and accurate chromosome segregation. In this review, we will focus on recent studies that provide insight into how Kinesin-13 family members regulate distinct aspects of microtubule dynamics, as well as how their activities may be temporally and spatially controlled within the spindle.
2. The Kinesin-13 Family of Molecular Motors
The kinesin superfamily members are molecular motors that couple ATP hydrolysis to force production. It was originally thought that all kinesins translocated unidirectionally along the microtubule lattice; however, the observation that the Kinesin-13 family members strictly depolymerize microtubules from both microtubule ends challenged this idea [14]. Since then, it has become clear that other kinesin family members also regulate microtubule dynamics, but appear to do so from only one microtubule end [15–18]. Together these studies highlight the fact that small changes in the conserved kinesin catalytic domain can cause dramatic structural changes in the microtubule polymer that lead to alterations in microtubule dynamics.
2.1 Structure and Function of Kinesin-13 Family Members
Kinesin-13 family members have been classified phylogenetically in multiple studies [19–22]. In human and in mouse there are four Kinesin-13 members, including Kif2A, Kif2B, Kif2C (MCAK), and Kif24. In contrast, there are no Kinesin-13 family members in either Saccharoymces cerevisiae or in S. pombe, although these organisms do have members of the Kinesin-8 and Kinesin-14 families of microtubule depolymerases. Invertebrate species have variable numbers of Kinesin-13 members, and the most well studied of these include Klp10A, Klp59C, and Klp59D in Drosophila. It should be noted that there is no functional data regarding the role of Kif24 in any system, so our discussion is generalized to the other members of the Kinesin-13 family and may not reflect eventual studies on Kif24.
Kinesin-13 proteins are microtubule depolymerizing enzymes that bind to the end of the microtubule, induce a conformational change at the microtubule end that leads to a microtubule catastrophe [14]. Kinesin-13s are microtubule-end stimulated ATPases [23] that can use one-dimensional diffusion along the lattice to reach the microtubule end [24]. Structurally, a consistent feature of the Kinesin-13 family members is that they have an N-terminal globular domain, followed by a positively charged neck, a centrally located catalytic core, and a C-terminal dimerization domain [25]. The N-terminal domain affects the subcellular localization of multiple members of this family [26–28]. The positively charged neck is required for microtubule depolymerization activity [29], and contains a major regulatory site for modulating this activity [30–32]. The catalytic domain contains the conserved ATP and microtubule binding domains found in all kinesins, but several unique structural features, particularly in loop two and helix four, may help explain how Kinesin-13s can induce or stabilize a curved conformation of the tubulin protofilaments [33–35]. The C-terminal domain is required for dimerization [36,37], which contributes to the efficiency of microtubule depolymerization.
3. Kinesin-13 Function in Mitosis
While the originally identified member of the Kinesin-13 family was neuronal Kif2 (now called Kif2A) [38,39], which plays an important role in neurite outgrowth and collateral branch expression in mouse neurons [40,41], a large body of research has focused on the diverse functions of Kinesin-13s during mitosis. We will focus most of our discussion on studies involving the vertebrate members of this family; however, we will point out studies in Drosophila that provide significant insight into our understanding of the important roles of these proteins in mitosis.
3.1 The Subcellular Distribution of Kinesin-13s during Mitosis
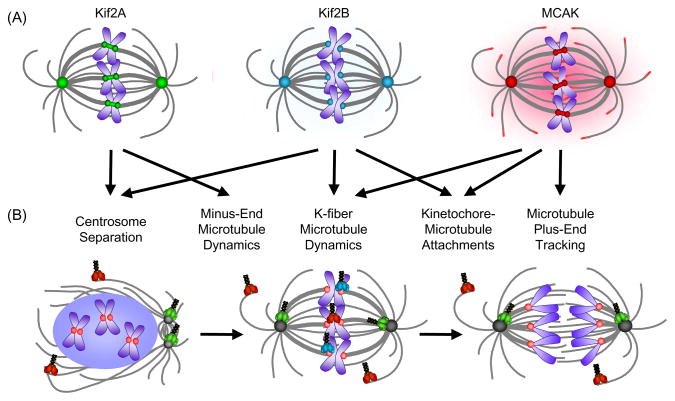
The first mitotic Kinesin-13 described was MCAK (aka Kif2C), which localizes to the spindle poles, centromeres and kinetochores, the plus-tips of microtubules, and the cytoplasm [42,43] (Fig. 1A). Kif2A is found mainly associated with the spindle poles [44–46]; however, it is also associated with centromeres [46,47]. The localization of Kif2B is less clearly established due to its very low expression level in most cell types examined [48]; however over-expressed GFP-Kif2B localizes to kinetochores and spindle poles as well. The diverse localizations highlight the potential for Kinesin-13s to act in multiple processes in spindle assembly. Furthermore, the overlapping localization suggests that they may either functionally substitute for each other at a specific spindle locale or that each Kinesin-13 may actually play distinct roles during each phase of mitosis, and thus their activities are likely precisely regulated.
Fig. 1.
Localization and function of Kinesin-13s in mitosis. (A) The vertebrate Kinesin-13s have overlapping localizations at spindle poles and kinetochores/centromeres. MCAK is also found in the cytoplasm and at the plus-ends of microtubules. Kif2A localizations are shown in green, Kif2B in teal, and MCAK in red. Microtubules are shown in gray and chromosomes are shown in blue. (B) Summary of vertebrate Kinesin-13 function during mitosis. Spindle structures and Kinesin-13 models are colored as in (A) with kinetochores depicted in pink. Models of the Kinesin-13s are shown at their predominant site(s) of function in prophase, metaphase, and anaphase.
3.2 Kinesin-13s are Required for Spindle Assembly
It is well established that Kinesin-13s are required for spindle assembly (Fig. 1B), but the exact processes that they affect vary between systems. Multiple members of the Kinesin-13 family are important in spindle bipolarity [11,45,49–51]. Depletion of MCAK from Xenopus egg extracts results in a high percentage of monopolar spindles [49]; whereas inhibition or RNAi of MCAK in mammalian cells only causes a slight increase in monopolar spindles [52–56]. These findings imply that MCAK may be more critical for bipolarity in embryonic versus somatic cells. In contrast to MCAK inhibition, knockdown of Kif2A in somatic cells causes a dramatic increase in monopolar spindles [45,51], providing strong evidence for this Kinesin-13 in establishing bipolar spindles.
It is well established that microtubule-based sliding plays a major role in centrosome separation and establishing spindle bipolarity. However, the finding that Kif2A inhibition causes monopolar spindles suggests that an imbalance in microtubule dynamics can also lead to defects in centrosome separation. Treatment of Kif2A RNAi cells with low doses of nocodazole that inhibit microtubule polymerization or co-depletion with the microtubule depolymerase MCAK restoring spindle bipolarity [45] is consistent this idea. In addition, kinetochore-based pushing forces contribute to centrosome separation [57]. The observation that co-depletion of Kif2A and Nuf2, a component of the microtubule-kinetochore linker complex, rescues spindle bipolarity [45] supports the idea that the kinetochore-based pushing forces are balanced by minus-end microtubule dynamics. A recent study showing that Kif2B knockdown affects the kinetics of centrosome separation [48] is indicative that multiple Kinesin-13 family members may contribute to spindle bipolarity through different pathways. To elucidate the pathways that contribute to spindle bipolarity and the underlying mechanisms will require quantitative analysis of mitotic progression and careful analysis of microtubule dynamics after manipulation of Kinesin-13 activity.
3.2 Kinesin-13s Regulate Spindle Microtubule Dynamics
It is clear that Kinesin-13s contribute to proper regulation of spindle microtubule dynamics as their inhibition results in alteration of microtubule polymer levels in nearly every model system examined [11,45,49,50,52,53,55,58]; however, the particular subclasses of microtubules that are affected vary depending on the cell type and the particular Kinesin-13 examined (Fig. 1B). For example, RNAi of Klp10A disrupts spindle microtubule turnover specifically near microtubule minus-ends [59], which is likely most critical to regulate poleward microtubule flux. MCAK inhibition on the other hand causes a reduction in K-fiber microtubule turnover, with no effect on spindle microtubules [60,61]. This effect is temporally regulated during mitotic progression as MCAK controls K-fiber dynamics during metaphase, whereas Kif2B contributes to K-fiber turnover during prometaphase [62]. Intriguingly the proper regulation of K-fiber dynamics and chromosome segregation may be critical for genomic stability [62], highlighting the importance of maintaining precise K-fiber microtubule dynamics.
Kinesin-13s may also regulate K-fiber dynamics through their role in controlling poleward microtubule flux. Inhibition of Klp10A suppresses poleward flux at microtubule minus-ends [11,59], which is important in anaphase A-mediated chromosome segregation. Similarly, Kif2A is proposed to regulate microtubule flux in vertebrate cells and extracts [12,44]; although this function remains controversial as others have found no effect on poleward microtubule flux upon Kif2A inhibition [46,47].
The incredibly complex effects of Kinesin-13s on spindle assembly and regulation of microtubule dynamics evokes the idea that perturbing the dynamics of one class of microtubules may indirectly alter the dynamics of another class. The observation that treatment of cells with low levels of paclitaxel does not cause the same alterations in spindle organization and microtubule dynamics as MCAK inhibition [60] suggests that overall suppression of microtubule dynamics is distinct from perturbation of an individual microtubule dynamics regulator. Alternatively, the different Kinesin-13s may simply be functionally redundant. The observation that Kif2A addback does not rescue spindle assembly in MCAK depleted Xenopus egg extracts argues against this idea [46]. Together, these studies illustrate that the activities of each of these Kinesin-13s are likely finely controlled in different areas at different times to insure proper spindle assembly and chromosome segregation.
3.3 Kinesin-13s Regulate Kinetochore-Microtubule Attachments
It has become increasingly clear that Kinesin-13s play a major role in regulating proper kinetochore-microtubule attachments (Fig. 1B). It was initially observed that inhibition of centromeric MCAK leads to lagging chromosomes at anaphase [27], which was later shown to be due to defects in chromosome congression as a result of improperly attached chromosomes [28,63]. However knockdown of MCAK results in only a few lagging chromosomes per cell [27,56,64], whereas knockdown of MCAK and Kif2A together causes a higher percentage of cells with more lagging chromosomes [12]. In contrast, Kif2B knockdown causes a dramatic increase in improperly attached chromosomes leading to defective chromosome segregation [62]. This suggests that Kif2B plays a more critical role in the regulation of kinetochore-microtubule attachments than MCAK. It is not clear whether these defects are due solely to the role of these kinesins in regulating K-fiber turnover dynamics or in poleward microtubule flux, which is postulated to enhance the fidelity of chromosome segregation [12,13]. Another interesting idea is that kinetochore-associated MCAK may regulate the attachment status not solely by releasing the attachment, but rather just by loosening the end states of microtubules embedded in the kinetochore to alter microtubule binding affinity [61]. Together these studies support the idea that Kinesin-13s are indeed important in regulating kinetochore-microtubule attachments and highlight the complexity of mechanisms utilized to monitor these attachments. Important and interesting mechanistic questions for the future are to understand whether the Kinesin-13s actually correct or prevent improper attachments as well as to understand how they sense and respond to attachment defects.
3.4 Kinesin-13s and Chromosome Segregation
Since their initial discovery as proteins that can regulate microtubule dynamics, it has long been suspected that Kinesin-13s may be responsible for the kinetochore-mediated microtubule depolymerization activity during anaphase A chromosome segregation. The observation that inhibition of centromeric MCAK gives rise to anaphase segregation defects provided support for this idea [27]; however, the absence of defects in the rates of anaphase A chromosome to pole movement after MCAK inhibition suggested that the anaphase A defects were more likely a consequence of earlier prometaphase defects [63]. In contrast to vertebrates, work from Drosophila embryos clearly shows that Klp10A and Klp59C account for the majority of chromosome to pole movement during anaphase A [11]. The observation that a Kif2A/MCAK double knockdown increases chromosome segregation defects supports the idea that Kif2A microtubule depolymerization at spindle poles may contribute to anaphase A movements [12]. To date, no kinetochore-associated microtubule depolymerases that contribute directly to anaphase A have been identified in vertebrates, leaving open the question of what molecules actually regulate the kinetochore-mediated microtubule depolymerization that occurs in vertebrate cells.
Very few studies have examined the roles of Kinesin-13 proteins in anaphase B. In Drosophila, suppression of minus-end depolymerization may actually drive spindle elongation in anaphase B [59,65]. In support of this idea, both Klp10A and Kif2A localize to poles and inhibition causes a diminishment of poleward flux, with a concomitant increase in spindle length [11,44,66]. In contrast, spindle length after MCAK inhibition is actually slightly shorter than controls [60,63], which may result from changes in plus-end dynamics at kinetochores [67]. Overall, these results highlight the idea that Kinesin-13s may also be important for anaphase B microtubule dynamics; however, this analysis will require that we find ways to temporally inhibit Kinesin-13 activity so that any defects in anaphase can be dissected from any early defects that occurred during spindle assembly.
4. Regulation of Kinesin-13 Activity
The complexity of actions that are carried out by members of the Kinesin-13 family and their overlapping localizations suggests that their activities must be precisely regulated. In addition, their spatial localization needs to be precisely controlled so that they are at the right place at the right time during mitosis. Kinesin-13 activities and localizations are regulated via phosphorylation by a number of important mitotic kinases and through interaction with a number of important regulatory proteins.
4.1 Phosphorylation Controls Kinesin-13 Activity and Localization
A major advance in our understanding of Kinesin-13 activity came from the observation that Aurora B phosphorylation inhibits the microtubule depolymerization activity of MCAK [30–32]. Aurora B phosphorylates MCAK at multiple sites throughout the N-terminal domain and the neck [30–32,68], but it was shown that S196 is the critical site for regulating this activity [31] (Fig. 2A). S196 is within the neck domain of MCAK, so it is interesting to speculate that phosphorylation at this site will introduce a negative charge into the normally positively charged neck that is important for controlling microtubule depolymerization activity [29]. It is important to note that while phosphorylation at this site is inhibitory; it does not completely abolish activity. One idea is that phosphorylation at this site may act more like a rheostat to fine-tune microtubule depolymerization activity. It is also possible that the ability of MCAK to be phosphorylated at this site is regulated by its accessibility to the kinase, which may be controlled by interactions between the N- and C-terminal domains [69] or by the binding of other regulatory molecules to MCAK [70,71].
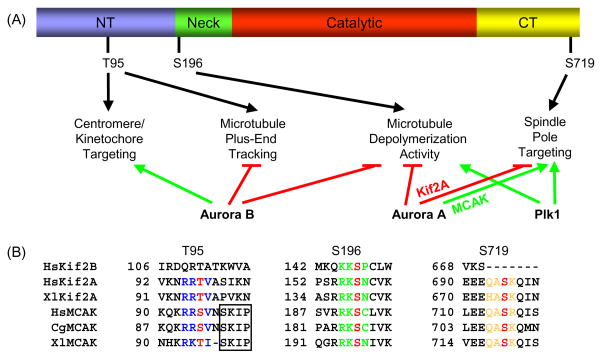
Fig. 2.
Kinesin-13 localization and function is regulated by conserved phosphorylation sites. (A) Schematic of secondary amino acid structure of Kinesin-13s and summary of Aurora B, Aurora A, and Plk1 phosphorylation effects. NT, N-terminal domain; and CT, C-terminal domain. Only the Xenopus MCAK phosphorylation sites are depicted for clarity. Green arrows depict promotion and red lines depict inhibition of localization or activity. (B) The T95, S196, and S719 MCAK phosphorylation sites are highly conserved in Kif2A, whereas only the S196 site is conserved in Kif2B. For HsKif2A, S157 is equivalent to S132 as reported in Knowlton, et al. [70]. Phosphorylation sites are colored red, and the surrounding conserved residues are colored blue for the T95 site, green for the S196 site, and orange for the S719 site. The SxIP microtubule tip localization signal is boxed, and is only conserved in MCAK. Hs, human; Xl, frog (Xenopus laevis); and Cg, Chinese hamster. Genbank accession numbers used for alignment: HsKif2B, NM032559; HsKif2A, NM004520; XlKif2A, BC057698; HsMCAK, NM006845; CgMCAK, U11790; and XlMCAK, BC044976.
S196 and the surrounding residues are highly conserved in most members of the Kinesin-13 family (Fig. 2B), suggesting that other members of the family may also be regulated by phosphorylation at this site. Indeed, Kif2A microtubule depolymerization activity is modulated by Aurora B phosphorylation at the S196 equivalent site [70]. However, this site is not conserved in vertebrate Kif24, which may not be important in mitosis, nor is it conserved in Drosophila Klp59C. Interestingly, in Klp59C the serine is an aspartate residue, which may mimic the phosphorylated status. Because Klp59C is an active microtubule depolymerase [11], this supports the idea that phosphorylation at this site does not abolish microtubule depolymerization activity, but rather moderates activity.
Phosphorylation of MCAK by distinct kinases does not necessarily result in inhibition of activity. For example, while Aurora A kinase suppresses the microtubule depolymerization activities of both MCAK [72] and Kif2A [73,74], Plk1 actually stimulates Kif2A depolymerization activity [73] (Fig. 2A). These stimulating Plk1 phosphorylation site(s) are unknown, and their identification will be important for us to understand the mechanisms by which distinct kinases coordinate Kinesin-13 activities.
In addition to regulating the microtubule depolymerization activity, a major role of phosphorylation is to control subcellular targeting of these kinesin family members (Fig. 2A). Inhibition of Aurora B kinase prevents MCAK targeting to centromeres [30,31,68,75] at least in part by altering MCAK turnover at centromeres [30]. MCAK displays a differential localization between inner centromeres and kinetochores from prometaphase to metaphase depending on the microtubule attachment or congression status of the chromosome [30,61,63]. This spatial and temporal localization is regulated by phosphorylation, suggesting that MCAK may have two distinct binding sites [30,61]. The exact mechanism for this temporal and spatial display is currently unknown, but likely involves phosphorylation promoting inner centromere binding and dephosphorylation promoting kinetochore binding. In Xenopus extracts, MCAK targeting to centromeres/kinetochores is regulated by a combination of sites, wherein phosphorylation at S110 and dephosphorylation at T95 promotes association [68]. This complex regulatory network of interactions may help explain why mutation of all putative Aurora B phosphorylation sites to alanine did not abolish MCAK targeting to centromeres [30,32], and supports the idea that perhaps T95 phosphorylation regulates the differential association of MCAK with kinetochores and centromeres during bi-orientation and congression.
The T95 site is conserved within MCAK proteins from different species, but is also conserved in Kif2A, despite the variable association of Kif2A with centromeres (Fig. 2B). Interestingly, T95 is not conserved in Kif2B, nor is it a predicted Aurora B phosphorylation site [76]. The idea that dephosphorylation of T95 promotes kinetochore association in MCAK is consistent with the idea that the absence of a phosphorylation site in Kif2B could similarly promote kinetochore association. By analogy, the inability of Kif2B to be phosphorylated in this region would also prevent inner centromere localization. Together these studies support the idea that Kif2B functions more distally on the outer kinetochore and MCAK functions more proximally toward the inner centromere, but additional studies are clearly needed to test this hypothesis. In addition, S110 is not highly conserved outside of Xenopus, making it difficult to understand exactly how this could be the primary site that promotes centromere association.
Phosphorylation of MCAK and Kif2A by Aurora A also regulates its association with spindle poles [72,73,77]. MCAK association is promoted at least in part by phosphorylation of S719 [72], a site that is highly conserved in MCAK and Kif2A, but is physically absent in Kif2B (Fig. 2B). However, it is unclear whether this Aurora A regulated mechanism of localization is conserved because Aurora A inhibition actually increases the localization of Kif2A to spindle poles [73]. In contrast, Plk1 acts antagonistically to Aurora A because inhibition of Plk1 decreases the amount of Kif2A associated with spindle poles [73]. The localization of Kif2A to spindle poles is also regulated by DDA3, which increases its targeting efficiency [78]. Whether this interaction is modulated by phosphorylation or if DDA3 is upstream or downstream of Plk1 is unknown.
Together these studies show that there is a complex hierarchy of phosphorylation events that control both Kinesin-13 localization and microtubule depolymerization activity. It will be essential to address when and where each phosphorylation event occurs as well as to elucidate the mechanisms by which individual kinases modulate activity and localization. This analysis is likely going to be complicated, as it will require careful depletion/addback experiments in egg extracts or knockdown/rescue approaches in cells so that protein levels can be precisely controlled. Given the observation that association with additional proteins also modulates Kinesin-13 function, simple reconstitution approaches may not clearly elucidate the full complexities of the in vivo regulatory mechanisms.
4.2 Kinesin-13 Targeting is Regulated through Binding Partners
The complex localization of Kinesin-13 family members suggests that one mechanism to regulate function would be to associate with different binding partners. A large number of binding partners have been identified for Kinesin-13 family members, but with a few notable exceptions, the consequences of these interactions have not been clearly elucidated.
The first regulatory binding partner discovered for a Kinesin-13 was ICIS, which stimulates the microtubule depolymerization activity of MCAK [71]. ICIS localizes to the inner centromere, and its targeting is MCAK-dependent. ICIS may provide a link between MCAK and the Aurora B chromosomal passenger complex because the C-terminus of ICIS interacts with MCAK, whereas the N-terminus of ICIS interacts with parts of the chromosomal passenger complex [70,71]. Surprisingly, ICIS function is not limited to the regulation of MCAK as ICIS and Aurora B also regulate Kif2A [70]. ICIS can stimulate both Kif2A and MCAK microtubule depolymerization activity, but only after they are first phosphorylated at S132 and S196 respectively, which are inhibitory for depolymerization activity. These results suggest that ICIS and Aurora B act antagonistically to modulate microtubule depolymerization activity, and provide yet another mechanism to fine-tune Kinesin-13 activity.
MCAK association with numerous microtubule plus-end binding proteins (+ TIPs) and its ability to track with growing MICROTUBULE ends may also affect its function. MCAK associates with APC in Xenopus extracts [79], wherein APC is a large protein that plays important roles in spindle orientation, microtubule organization, and genomic instability [80,81]. Both MCAK and APC bind kinetochores in Xenopus egg extracts [79,82], and the association between MCAK and APC is mediated by the N-terminal domain of MCAK and the C-terminus of APC [79]. APC and MCAK also interact individually at microtubule plus-ends through an interaction with EB1 [83–86], suggesting a mechanism for APC and MCAK association. The association of Kinesin-13s with EB1 is highly conserved, as both Drosophila Klp10A and Klp59C are both +TIPs [87]. The binding of EB1 to MCAK and Klp10A is direct [83,87], and in MCAK is mediated at least in part through an N-terminal microtubule tip localization signal SxIP [88] (Fig. 2B). Recent studies identified the EB1 binding protein TIP150 as another protein that may modulate the interaction between MCAK, EB1, and microtubules [89]. TIP150 is thought to serve as a linker between EB1 and MCAK because TIP150 enhances MCAK association with EB1 [89], suggesting that the precise interaction between all of these components needs to be carefully examined. One interesting note is that TIP150 shares limited sequence homology to the C-terminal domain of ICIS, raising the question of whether TIP150 also regulates MCAK depolymerization activity.
The ability of Kinesin-13s to plus-tip track on growing microtubules appears to be limited to MCAK, as neither Kif2A or Kif2B possess tip-tracking ability [42,48], which is likely due to the absence of the EB1 microtubule tip localization signal [88] (Fig. 2B). This ability to tip-track by association with EB1 [83,89] is also regulated by phosphorylation [42,88,89] at least in part due to the S92 (T95 equivalent) site [42]. It will therefore be important to address how the phosphorylation status of MCAK regulates its association with EB1 and TIP150, and most crucially to elucidate the importance of MCAK tip-tracking in its many diverse functional roles.
5. Conclusions and Future Directions
It is clear that Kinesin-13 family members play diverse and important roles during mitosis. What is most notable is the diversity of processes that are affected and the complex regulation that occurs. Perhaps this is not surprising given the complex nature of microtubule dynamics within the spindle. While significant progress has been made, it is clear that we have a long way to go before fully understanding this Kinesin family. It will be essential to decipher when and where each Kinesin-13 is found and whether it is active or inactive at that site. It is important to keep in mind that guilt by association may not be accurate here. For example, a particular Kinesin-13 can be localized at a distinct spindle area and yet be phosphorylated at an inhibitory site, implying that its activity is suppressed. Testing this idea will rely on the development of novel biosensors that can detect active and inactive forms of each Kinesin-13. In addition, we require more sophisticated means to measure microtubule dynamics within the spindle so that we can understand how specific subpopulations of microtubules are perturbed upon Kinesin-13 inhibition. Temporal inhibition of Kinesin-13 function would provide a tremendous opportunity to dissect the roles of Kinesin-13s at individual stages of mitosis. Finally, we need to understand how the Kinesin-13 proteins are phosphorylated and how interactions with other regulatory proteins are controlled so that we can elucidate the molecular mechanisms by which these proteins carry out their many actions.
Acknowledgments
The authors would like to thank all members of the lab for thoughtful discussions. The authors are especially grateful to Jim Powers for comments on the manuscript. CEW is supported by the NIH (GM059618).
Abbreviations
- APC
adenomatous polyposis coli
- ATP
adenosine tri-phosphate
- DDA3
identified by differential display and is activated by p53
- EB1
end-binding protein 1
- GFP
green fluorescent protein
- ICIS
inner centromere Kin-I stimulator
- Kif
kinesin family member (2A, 2B, and 2C)
- Klp
kinesin-like protein (10A, 59C, and 59D)
- K-fiber
kinetochore fiber
- MCAK
mitotic centromere-associated kinesin
- Nuf2
Ndc80 kinetochore complex component
- Plk1
polo-like kinase 1
- RNAi
ribonucleic acid interference
- S
serine
- SxIP
microtubule tip localization signal (serine, any amino acid, isoleucine, proline)
- +TIP
microtubule plus-end tracking protein
- TIP150
+TIP of 150 kilodaltons
- T
threonine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mitchison TJ, Kirschner MW. Dynamic instability of microtubule growth. Nature. 1984;312:237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 2.Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–89. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- 3.Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. Tubulin dynamics in cultured mammalian cells. J Cell Biol. 1984;99:2175–86. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: Abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135:201–14. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rusan NM, Fagerstrom CJ, Yvon AM, Wadsworth P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol Biol Cell. 2001;12:971–80. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwok BH, Kapoor TM. Microtubule flux: Drivers wanted. Curr Opin Cell Biol. 2007;19:36–42. doi: 10.1016/j.ceb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Mitchison TJ. Polewards microtubule flux in the mitotic spindle: Evidence from photoactivation of fluorescence. J Cell Biol. 1989;109:637–52. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J Cell Sci. 2005;118:1105–16. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- 9.Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992;119:569–82. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai A, Maddox PS, Mitchison TJ, Salmon ED. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J Cell Biol. 1998;141:703–13. doi: 10.1083/jcb.141.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, et al. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–70. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 12.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–32. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 13.Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J Cell Biol. 2009;186:11–26. doi: 10.1083/jcb.200904153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 15.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase Kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–7. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu HM, Yun M, Anderson DE, Sage H, Park HW, Endow SA. Kar3 interaction with Cik1 alters motor structure and function. EMBO J. 2005;24:3214–23. doi: 10.1038/sj.emboj.7600790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a Kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–23. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 18.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast Kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–62. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005;15:467–76. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Kim AJ, Endow SA. A kinesin family tree. J Cell Sci. 2000;113:3681–2. doi: 10.1242/jcs.113.21.3681. [DOI] [PubMed] [Google Scholar]
- 22.Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–43. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, et al. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–57. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–9. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 25.Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17:82–8. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Wordeman L, Wagenbach M, Maney T. Mutations in the ATP-binding domain affect the subcellular distribution of mitotic centromere-associated kinesin (MCAK) Cell Biol Int. 1999;23:275–86. doi: 10.1006/cbir.1999.0359. [DOI] [PubMed] [Google Scholar]
- 27.Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walczak CE, Gan EC, Desai A, Mitchison TJ, Kline-Smith SL. The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr Biol. 2002;12:1885–9. doi: 10.1016/s0960-9822(02)01227-7. [DOI] [PubMed] [Google Scholar]
- 29.Ovechkina Y, Wagenbach M, Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J Cell Biol. 2002;159:557–62. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–68. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 31.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–86. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 32.Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol Biol Cell. 2004;15:2895–906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niederstasser H, Salehi-Had H, Gan EC, Walczak C, Nogales E. XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J Mol Biol. 2002;316:817–28. doi: 10.1006/jmbi.2001.5360. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers – M type kinesins curl the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- 35.Shipley K, Hekmat-Nejad M, Turner J, Moores C, Anderson R, Milligan R, et al. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–32. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hertzer KM, Ems-McClung SC, Kline-Smith SL, Lipkin TG, Gilbert SP, Walczak CE. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell. 2006;17:700–10. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maney T, Wagenbach M, Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J Biol Chem. 2001;276:34753–8. doi: 10.1074/jbc.M106626200. [DOI] [PubMed] [Google Scholar]
- 38.Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. Kinesin family in murine central nervous system. J Cell Biol. 1992;119:1287–96. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noda Y, Sato-Yoshitake R, Kondo S, Nangaku M, Hirokawa N. KIF2 is a new microtubule-based anterograde motor that transports membranous organelles distinct from those carried by kinesin heavy chain or KIF3A/B. J Cell Biol. 1995;129:157–67. doi: 10.1083/jcb.129.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, et al. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114:229–39. doi: 10.1016/s0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- 41.Morfini G, Quiroga S, Rosa A, Kosik K, Caceres A. Suppression of KIF2 in PC12 cells alters the distribution of a growth cone nonsynaptic membrane receptor and inhibits neurite extension. J Cell Biol. 1997;138:657–69. doi: 10.1083/jcb.138.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, et al. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–7. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–71. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–8. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohi R, Burbank K, Liu Q, Mitchison TJ. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr Biol. 2007;17:953–9. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 47.Cameron LA, Yang G, Cimini D, Canman JC, Kisurina-Evgenieva O, Khodjakov A, et al. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J Cell Biol. 2006;173:173–9. doi: 10.1083/jcb.200601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The Kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–9. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walczak CE, Mitchison TJ, Desai A. XKCM1: A Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 50.Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–16. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–99. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell. 2004;15:1580–90. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmfeldt P, Stenmark S, Gullberg M. Differential functional interplay of TOGp/XMAP215 and the KinI kinesin MCAK during interphase and mitosis. EMBO J. 2004;23:627–37. doi: 10.1038/sj.emboj.7600076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmfeldt P, Zhang X, Stenmark S, Walczak CE, Gullberg M. CaMKIIgamma-mediated inactivation of the Kin I kinesin MCAK is essential for bipolar spindle formation. EMBO J. 2005;24:1256–66. doi: 10.1038/sj.emboj.7600601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–31. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stout JR, Rizk RS, Kline SL, Walczak CE. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biol. 2006;7:26. doi: 10.1186/1471-2121-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184:365–72. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell. 2005;16:3176–86. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rizk RS, Bohannon KP, Wetzel LA, Powers J, Shaw SL, Walczak CE. MCAK and paclitaxel have differential effects on spindle microtubule organization and dynamics. Mol Biol Cell. 2009;20:1639–51. doi: 10.1091/mbc.E08-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–79. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–59. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hedrick DG, Stout JR, Walczak CE. Effects of anti-microtubule agents on microtubule organization in cells lacking the Kinesin-13 MCAK. Cell Cycle. 2008;7:2146–56. doi: 10.4161/cc.7.14.6239. [DOI] [PubMed] [Google Scholar]
- 65.Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci U S A. 2004;101:15938–43. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laycock JE, Savoian MS, Glover DM. Antagonistic activities of Klp10A and Orbit regulate spindle length, bipolarity and function in vivo. J Cell Sci. 2006;119:2354–61. doi: 10.1242/jcs.02957. [DOI] [PubMed] [Google Scholar]
- 67.Waters JC, Mitchison TJ, Rieder CL, Salmon ED. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell. 1996;7:1547–58. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–76. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ems-McClung SC, Hertzer KM, Zhang X, Miller MW, Walczak CE. The interplay of the N-and C-terminal domains of MCAK control microtubule depolymerization activity and spindle assembly. Mol Biol Cell. 2007;18:282–94. doi: 10.1091/mbc.E06-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knowlton AL, Vorozhko VV, Lan W, Gorbsky GJ, Stukenberg PT. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr Biol. 2009 doi: 10.1016/j.cub.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohi R, Coughlin ML, Lane WS, Mitchison TJ. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell. 2003;5:309–21. doi: 10.1016/s1534-5807(03)00229-6. [DOI] [PubMed] [Google Scholar]
- 72.Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control Ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19:2752–65. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang CY, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci. 2009;122:1334–41. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katayama H, Sasai K, Kloc M, Brinkley BR, Sen S. Aurora kinase-A regulates kinetochore/chromatin associated microtubule assembly in human cells. Cell Cycle. 2008;7:2691–704. doi: 10.4161/cc.7.17.6460. [DOI] [PubMed] [Google Scholar]
- 75.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–10. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 76.Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–7. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeLuca M, Brunetto L, Asteriti IA, Giubettini M, Lavia P, Guarguaglini G. Aurora-A and ch-TOG act in a common pathway in control of spindle pole integrity. Oncogene. 2008;27:6539–49. doi: 10.1038/onc.2008.252. [DOI] [PubMed] [Google Scholar]
- 78.Jang CY, Wong J, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J Cell Biol. 2008;181:255–67. doi: 10.1083/jcb.200711032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banks JD, Heald R. Adenomatous polyposis coli associates with the microtubule-destabilizing protein XMCAK. Curr Biol. 2004;14:2033–8. doi: 10.1016/j.cub.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 80.Barth AI, Caro-Gonzalez HY, Nelson WJ. Role of adenomatous polyposis coli (APC) and microtubules in directional cell migration and neuronal polarization. Semin Cell Dev Biol. 2008;19:245–51. doi: 10.1016/j.semcdb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rusan NM, Peifer M. Original CIN: Reviewing roles for APC in chromosome instability. J Cell Biol. 2008;181:719–26. doi: 10.1083/jcb.200802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dikovskaya D, Newton IP, Nathke IS. The adenomatous polyposis coli protein is required for the formation of robust spindles formed in CSF Xenopus extracts. Mol Biol Cell. 2004;15:2978–91. doi: 10.1091/mbc.E03-08-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee T, Langford KJ, Askham JM, Bruning-Richardson A, Morrison EE. MCAK associates with EB1. Oncogene. 2008;27:2494–500. doi: 10.1038/sj.onc.1210867. [DOI] [PubMed] [Google Scholar]
- 84.Su LK, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, et al. APC binds to the novel protein EB1. Cancer Res. 1995;55:2972–7. [PubMed] [Google Scholar]
- 85.Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–8. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 86.Niethammer P, Kronja I, Kandels-Lewis S, Rybina S, Bastiaens P, Karsenti E. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 2007;5:e29. doi: 10.1371/journal.pbio.0050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct Kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–45. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- 88.Honnappa S, Gouveia S, Weisbrich A, Damberger F, Bhavesh N, Jawhari H, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–76. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 89.Jiang K, Wang J, Liu J, Ward T, Wordeman L, Davidson A, et al. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 2009 doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]