Abstract
Background
Little is known about the effects of passive smoke exposures on the developing brain.
Objective
The purpose of the current study was to identify changes in gene expression in the murine hippocampus as a consequence of in utero exposure to sidestream cigarette smoke (an experimental equivalent of environmental tobacco smoke (ETS)) at exposure levels that do not result in fetal growth inhibition.
Methods
A whole body smoke inhalation exposure system was utilized to deliver ETS to pregnant C57BL/6J mice for six hours/day from gestational days 6–17 (gd 6–17) [for microarray] or gd 6–18.5 [for fetal phenotyping].
Results
There were no significant effects of ETS exposure on fetal phenotype. However, 61 “expressed” genes in the gd 18.5 fetal hippocampus were differentially regulated (up- or down-regulated by 1.5 fold or greater) by maternal exposure to ETS. Of these 61 genes, 25 genes were upregulated while 36 genes were downregulated. A systems biology approach, including computational methodologies, identified cellular response pathways, and biological themes, underlying altered fetal programming of the embryonic hippocampus by in utero cigarette smoke exposure.
Conclusions
Results from the present study suggest that even in the absence of effects on fetal growth, prenatal smoke exposure can alter gene expression during the “early” period of hippocampal growth and may result in abnormal hippocampal morphology, connectivity, and function.
Keywords: mouse, fetus, cigarette, sidestream smoke, hippocampus, microarray, environmental tobacco smoke, gene expression
1. INTRODUCTION
Despite worldwide attention highlighting the deleterious consequences of cigarette smoking on overall health and reproduction, over 20% of adults in the United States smoke [1]. It has been estimated that one third to one half of all pregnant women are exposed to cigarette smoke via passive or involuntary means in their homes, in public, or in the workplace [2–4]. Moreover, 43% of children in the United States −2 months to 11 years of age – live in homes with at least one smoker, rendering cigarette smoke exposure a significant environmental hazard for this cohort [5,6].
‘Passive’ smoking refers to exposure to environmental tobacco smoke (ETS), also known as secondhand smoke. ETS is comprised of the smoke released from the end of the smoldering cigarette and the portion of the mainstream smoke that is exhaled by the smoker. It is a complex mixture of over 4,000 compounds, many of which, such as nicotine, arsenic, and lead are possible human teratogens [7–9]. While numerous components of cigarette smoke are present in lower concentrations in sidestream smoke than in mainstream smoke, incomplete combustion of tobacco products from the smoldering end of the cigarette results in the release of higher concentrations of toxic constituents such as nitrosamines and formaldehyde [10–12].
Adverse pregnancy outcomes such as fetal growth restriction, increased rates of spontaneous abortion, premature placental abruption, perinatal lethality, congenital malformations, and cognitive impairments have been linked to maternal smoking during pregnancy [13–18]. While the deleterious consequences of maternal smoking on infant development have been well documented, less is known about the effects of ETS on developmental outcomes. Nevertheless, increased risk for respiratory illnesses, sudden infant death syndrome, middle ear disease, low birth weight and long-term cognitive and behavioral deficiencies have been linked to ETS exposure [19,20].
An extensive body of literature supports the notion that the brain, particularly regions associated with learning and memory, is a developmental target for the constituents of cigarette smoke. For example, prenatal nicotine exposure of rodents resulted in decreases in cell size, cell layer thickness, and cell density as well as alterations in dendritic morphology in the hippocampus [21,22]. In addition, long-term impairments in attention, learning and memory follow developmental exposures to nicotine [23–28]. Moreover, prenatal carbon monoxide exposure in rodents has also been shown to disrupt long-term potentiation and alter hippocampal-dependent behaviors [29,30].
Preliminary microarray analysis of whole brain tissue collected from murine fetuses exposed to sidestream smoke during gestational days 6–18.5 (gd 6–18.5), revealed significant alterations in gene expression profiles when compared to controls55. Included among the categories of genes whose expression was differentially altered by smoke exposure were those known to be involved in neurodevelopmental, neuro-behavioral, and cognitive processes. These data support the notion that subtle smoke-induced changes in the developing fetal brain can result in long-term behavioral and cognitive deficits. The purpose of the current study was to identify changes in gene expression in the fetal mouse hippocampus following in utero exposure to sidestream cigarette smoke, a model of ETS exposure.
2. MATERIALS AND METHODS
2.1 Experimental Animals
C57BL/6J mice (Jackson Laboratories; Bar Harbor, ME), were maintained in the American Association for Accreditation of Laboratory Animal Care (AAALAC) approved facility at the University of Louisville on a 12-hour light/dark cycle and ad libitum food and water. Timed pregnancies were obtained by overnight mating of a single mature male with two nulliparous females. The presence of a vaginal plug was considered to be evidence of mating and the time designated as gestational day 0 (gd 0). Pregnant mice were exposed to sidestream cigarette smoke (CSE) or ambient air (sham-exposed controls) for six hours per day on either gd 6–17 [for array] or gd 6–18.5 [fetal assessment]. Dams were weighed daily and monitored for overt signs of toxicity including weight loss, moribundity, mortality, ruffled fur, reluctance to ambulate, and chromodacryorrhea. Animals (dams and corresponding litters) were excluded from fetal outcome analysis if the dam was not pregnant (3 Sham and 1 CSE) or if the dam gave birth early (1 CSE).
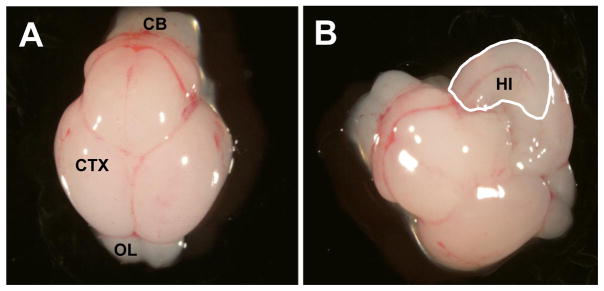
On gd 18.5, dams were anesthetized with Avertin (500 mg/kg, i.p.; Aldrich; (2,2,2 – tribromoethylalcohol; St. Louis, MO) [31] and then euthanized by carbon dioxide asphyxiation followed by cervical dislocation. Uteri were exteriorized and the number and location of implantations were recorded. Fetuses were then removed and the following outcomes were assessed: number of viable fetuses, fetal weight, number of abnormalities, and fetal crown-rump length. For microarray analysis, fetal brains were harvested, hippocampal tissue microdissected (Figure 1) pooled by litter, and processed as described below.
Figure 1.
Photographs of the dorsal view of the gestational day 17 murine brain. (A) The intact fetal brain on gestational day 17. (B) A unilateral view of the interior portion of the fetal cerebral cortex on gestational day 17. The cerebral cortex was opened along the midline and the hippocampal region was identified, removed and frozen on dry ice. The region demarcated by the white line represents the hippocampal tissue that was bilaterally excised from the cerebral cortex of the gestational day 17 murine fetus for RNA isolation. (CB) Cerebellum; (CTX) Cerebral cortex/Cerebrum; (OL) Olfactory lobes; (HI) Hippocampus.
2.2 Whole Animal Inhalation System
A Teague TE-10C whole body smoke inhalation exposure system (Teague Enterprises; Davis, CA) was used to generate and deliver sidestream cigarette smoke. The Teague, TE-10C [32] is a microprocessor controlled instrument that produces sidestream smoke or a combination of mainstream and sidestream smoke. Dams were exposed to sidestream cigarette smoke generated from Philip Morris Marlboro Red cigarettes™ (Philip Morris; Richmond, VA; 15 mg of tar/cigarette; 1.1 mg nicotine/cigarette; additives). Philip Morris Marlboro Red cigarettes™ were selected because they represent the most popular brand of cigarettes consumed among 18–25 year olds, the age group containing the majority of maternal smokers [33]. The cigarettes were stored at 4°C until 48 hours prior to use when they were brought to a relative humidity of 60%. Cigarettes were smoked using the standard Federal Trade Commission method: a two second, 35 cm3 puff, once a minute for a total of nine minutes [32]. For quality control purposes, paired exposure chambers (one receiving cigarette smoke and one receiving ambient air [sham]) were characterized twice during each daily exposure session for: total suspended particulates (TSP), temperature, carbon monoxide levels, and humidity. Tail blood was collected from each dam immediately following the six-hour exposure session on gestational day 6, 9, 12, and 15 for determination of plasma cotinine levels (described below).
2.3 Cotinine Assay
Cotinine, the principal metabolite of nicotine, is a well-documented marker of active tobacco smoking and passive/environmental tobacco smoke exposure [34]. Cotinine concentrations can be reliably measured in blood, urine, and hair [35–37]. At designated time points, tail blood was collected from each dam and plasma cotinine concentrations were determined utilizing a Cotinine One-Step ELISA Detection Kit (International Diagnostic Systems; St. Joseph, MI). In brief, 20 μl of plasma or cotinine standard was added to cotinine antibody-coated microtiter plates. Enzyme conjugate (100 μl) was added to each well and following a 30-minute incubation at room temperature the solution was removed and the plate washed. The substrate, tetramethylbenzidine, (150 μl) was added to each well and the plate further incubated at room temperature until the wells developed a medium blue color. The reaction was terminated by adding 150 μl of stop solution (3 N sulfuric acid) to each well and the absorbance was read at 450 nm. Cotinine concentrations were quantified using a standard curve (0–50 ng/ml) and the data reported as mean ng/ml cotinine ± standard error of the mean.
2.4 Isolation of RNA from Fetal Hippocampal Tissue
Total RNA was isolated from fetal hippocampal samples using the RNeasy Protect Mini Kit (Qiagen; Valencia, CA) following the manufacturer’s recommendations. The quality and quantity of the extracted total RNA were assessed by UV Absorbance (NanoDrop® ND1000 Spectrophotometer v3.1.2, NanoDrop Technologies, Wilmington, DE) and the RNA 6000 Nano Assay (Agilent Technologies, Santa Clara, CA). Hippocampal RNA was isolated from three sham-exposed and three sidestream cigarette smoke-exposed litters and each sample was applied to an Affymetrix high-density mouse genome 430 2.0 GeneChip® array.
2.5 cDNA Target Synthesis, Biotin-Labeling of cRNA, and GeneChip® Hybridization
Double stranded cDNA was prepared using the GeneChip® One cycle cDNA Synthesis kit (Affymetrix, Santa Clara, CA). In brief, 5 μg of total RNA was denatured and annealed to 2 μl (50 μM) T7- Oligo-dT primer for 10 minutes at 70°C. The reaction was cooled to 4 °C in a thermal cycler and the RNA reverse transcribed for 1 hour by adding 1 μl Superscript II at 42 °C in reaction buffer containing, 10 mM dithiothreitol, and 0.5 mM dNTP mix, in a total volume of 27 μl. Second-strand cDNA was synthesized by adding 4 μl DNA polymerase I, 1 μl E. coli DNA ligase, 1 μl RNase H, 30 μl 5X second-strand buffer, 3 μl 10 mM dNTP mix, and water to a total volume of 130 μl and incubating for 120 min at 16 °C. Subsequently, 2 μl of T4 DNA polymerase was added and the incubation continued at 16 °C for 5 additional minutes. Second-strand cDNA synthesis was stopped by the addition of 10 μl of 0.5 M EDTA. The resulting double stranded cDNA was purified using the cDNA clean-up module (Affymetrix).
Twelve microliters (12 μl) of double stranded cDNA was in vitro transcribed using a GeneChip® Expression 3′-Amplification Kit (Affymetrix) with biotinylated CTP and UTP according to the manufacturer’s instructions. Following a 16-hour incubation at 37°C, the resultant biotin-labeled cRNA was purified with the cRNA clean-up module (Affymetrix) and eluted in 21 μl of RNase-free water. The concentration of biotin-labeled cRNA was assessed by UV Absorbance (NanoDrop® ND1000 Spectrophotometer v3.1.2, NanoDrop Technologies, Wilmington, DE) and the RNA 6000 Nano Assay (Agilent Technologies, Santa Clara, CA). Twenty micrograms (20 μg) of labeled cRNA was then fragmented in 40 μl 1X fragmentation buffer (40 mM Tris-acetate pH 8.1, 100 mM potassium-acetate, 30 mM magnesium-acetate) for 35 min at 94°C and the effectiveness of fragmentation assessed by agarose gel electrophoresis. Fragmented cRNA was brought to a total volume of 300 μl with 1X hybridization buffer (100 mM MES, 1 M NaCl, 20 mM EDTA, 0.01% Tween 20), 100 μg/ml herring sperm DNA, 500 μg/ml acetylated BSA, 50 pM biotinylated control oligonucleotide B2 and 1X eukaryotic hybridization controls. cRNAs derived from fetal hippocampal tissue exposed to either sham or sidestream smoke conditions were hybridized for 16 hours to an individual GeneChip® from an identical lot of Affymetrix Mouse Genome 430 2.0 GeneChip® arrays. These arrays contain 45,101 genes and EST probes sets. GeneChip arrays were then washed and stained using antibody-mediated signal amplification and the Affymetrix Fluidics Station’s standard Eukaryotic GE Wash 5/ protocol.
2.6 Reduction, Statistical Analysis, and Biological Interpretation of Microarray Data
Individual GeneChip® arrays were scanned with the GeneChip® Scanner 3000 (Affymetrix) and images were processed using Affymetrix GCOS 1.2 software, according to Affymetrix protocols. The CEL files, containing individual raw chip data (probe intensities), were imported into GeneSpring GX 7.3.1 (Silicon Genetics, Inc.; Redwood City, CA) and pre-processed using Robust Multi-chip Average, with GC-content background correction (GC-RMA). The GC-RMA processed data were further normalized by the ‘per gene normalization’ step in which all the samples were normalized against the median of the control samples (i.e. the expression value for a gene across the different conditions is centered on 1. This is done by dividing the expression value of a given gene, by the median expression values for that gene across the conditions). This ensured that genes whose expression did not change across treatment conditions received a normalized expression value of 1, allowing for easy visual detection of differentially expressed genes. To define a set of statistically significant, differentially expressed genes, a one-way ANOVA (parametric test, assuming equal variances) was applied using the “Benjamini and Hochberg false discovery rate” as the multiple testing correction (p = 0.05). This restriction tested each of the 45,101 genes and ESTs on the microarray and generated a list of 3,109 genes with statistically significant expression values. A fold change “filter” (probes with fold differences ≥1.5 were considered significant) was then applied to the list of 3,109 genes. The gene/EST list resulting from this analysis includes those whose expression is either ≥1.5-fold up- (25 genes and ESTs) or down-regulated (36 genes and ESTs) in a statistically significant manner (p < 0.05) as a function of the treatment condition (sham versus sidestream smoke-exposed). Hierarchical clustering analysis was then performed using the GeneSpring 7.2 software (Silicon Genetics, Inc., Redwood City, CA) to generate a “condition tree” representing the functional category of genes based on their expression profile. A heat map (Figure 2) was generated by dividing each measurement by the 50th percentile of all the measurements in that sample. The average value of expression for each gene across the samples was then set to 1.0 and the normalized signal value for each sample was plotted (values less than 0.01 were set to 0.01). In order to establish an overview of different biological pathways impacted by in utero sidestream cigarette smoke, computational gene interaction predictions were made using Ingenuity Systems Pathway Analysis (Ingenuity Systems, Mountain View, CA; http://www.ingenuity.com). Several downregulated and upregulated genes from the study were used to construct gene association maps for predicting the effects of sidestream smoke on various cellular and molecular processes in the fetal mouse hippocampus.
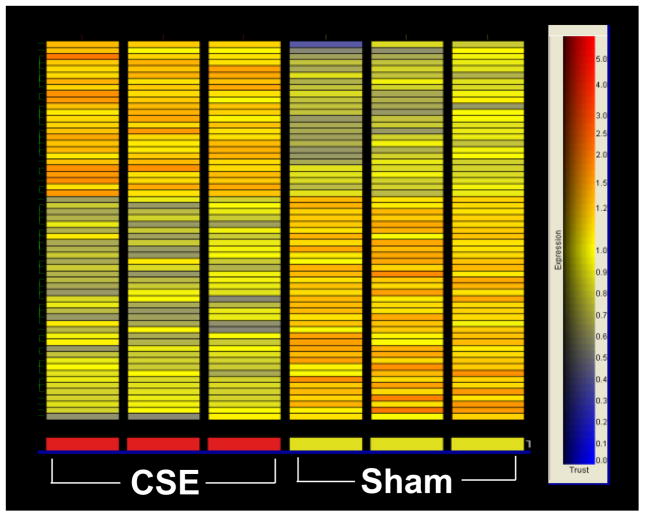
Figure 2.
Heat map illustrating the differentially regulated genes in the murine hippocampus following exposure to sham and sidestream smoke exposure. Each row of the heat map represents a gene while each column represents the experimental treatment (labeled at the bottom). The color saturation scale, shown to the right, represents the level of gene expression, with red indicating an increase in gene expression and blue indicating a decrease in gene expression. Only genes whose expression demonstrated a 1.5 fold or greater increase or decrease are depicted. The list of genes comprising the heat map is found in Table 3.
2.7 Quantitative Real-Time PCR
Quantitative Real-Time PCR analysis was performed using an ABI Prism 7000 Sequence Detector System (Applied Biosystems; Foster City, CA). Primers and their corresponding fluorescent probes for each of the genes that were examined were purchased from Applied Biosystems. In all cases, both forward and reverse primers were used at a concentration of 900 nM, while the concentration of the probe was 250 nM. For the PCR reaction, 1 pg of cRNA template was mixed with 0.2 mM each of dATP, dCTP, and dGTP, 0.4 mM dUTP, and 0.625 units of AmpliTaq Gold (Applied Biosystems) in a final volume of 25 μl. Cycling parameters were as follows: 50°C for 2 min for probe and primer activation, 95°C for 10 min of DNA strand denaturation, followed by 40 cycles of denaturation at 95°C for 15 s and primer extension at 60°C for 1 min. Raw data were acquired and processed with ABI Sequence Detector System software, v1.0 (Applied Biosystems; UK). Each cDNA sample was tested in triplicate and mean Ct values are reported. For each cDNA template reaction, a parallel reaction lacking template was performed as a negative control. Each determination of mRNA amount for the genes analyzed was normalized to GAPDH mRNA present in the sample by using TaqMan™ GAPDH PCR primers and probe.
2.8 Statistical Analysis
Outcome data were analyzed using a mixed-model ANOVA followed by t-tests with the Statistical Package for Social Sciences© version 14.0 (SPSS; Chicago, IL). Since the dam was the treated entity in the conducted studies, the litter rather than individual fetuses was regarded as the experimental unit [38]. Statistical significance was assigned a p < 0.05.
3. RESULTS
3.1 Exposure Conditions and Cotinine Levels
Chamber conditions (total suspended particulates, carbon monoxide levels, temperature, and humidity) were measured daily for quality control purposes and are reported in Table 1. Under the experimental paradigm utilized, the mean total suspended particulates (TSP) in the sidestream smoke chamber was 5.9 ± 0.2 mg/m3. Measured TSP resulted in exposure levels where dam plasma cotinine levels approximated those of pregnant women who are ‘passively’ exposed to cigarette smoke [34,39]. In order to monitor cigarette exposure levels, tail blood was collected for cotinine determination by ELISA following exposures on gd 6, 9, 12, and 15. Average maternal cotinine levels in dams exposed to sidestream smoke were 35.3 ± 3.5 ng/ml.
TABLE 1.
Chamber Conditions for Cigarette Smoke- and Sham-Exposed Dams1
| Sidestream Smoke Exposure | ||
|---|---|---|
| Condition | Sham Chamber2 | CSE3 Chamber2 |
| Carbon Monoxide (ppm) | ND4 | 32.9 ± 1.0 |
| Humidity (% RH) | 40.0 ± 1.3 | 46.9 ± 0.1 |
| Temperature (°C) | 22.5 ± 0.1 | 21.9± 0.1 |
| Total Suspended Particulates (mg/m3) | 0.2 ± 0.1 | 5.9 ± 0.2 |
Chamber measurements were monitored prior to daily animal exposures and then twice during the daily six-hour exposure period.
Data are reported as mean ± SEM.
CSE = Cigarette Smoke Exposure
ND = Not Detected
3.2 Fetal Outcomes
In an effort to avoid any confounding effects that exposure to cigarette smoke might have on murine embryo implantation, which is typically completed by gd 5 [40–42], dams were exposed beginning on gd 6 and continued throughout gestation. This exposure period represents post-implantation development and includes organogenesis, late term growth, and early hippocampal neurogenesis [43–45]. Notably, no dams died as a result of the exposure regimen, nor were there any observed signs of maternal toxicity.
Fetal outcomes are shown in Table 2. Exposure to sidestream smoke resulted in no observable effects on dam weight gain, fetal viability, or fetal morphometric measures. One fetus from a litter designated for array analysis, and exposed to sidestream cigarette smoke, exhibited micrognathia and cleft palate. This fetus was excluded from hippocampal tissue analysis.
TABLE 2.
Fetal Outcomes on Gestational Day 18.51
| Sidestream Smoke Exposure | ||
|---|---|---|
| Outcome | Sham2 | CSE2,3 |
| Number of Implantations | 8.2 ± 0.4 | 8.3 ± 0.5 |
| Number of Resorptions | 0.2 ± 0.1 | 0.5 ± 0.2 |
| Number of Viable Fetuses | 8.0 ± 0.4 | 7.7 ± 0.5 |
| Number of Abnormalities | 0.5 ± 0.3 | 0.6 ± 0.2 |
| Fetal Weight (g) | 1.15 ± 0.02 | 1.12 ± 0.03 |
| Crown-Rump Length (mm) | 27.5 ± 0.5 | 26.2 ± 0.4 |
Fetal outcomes were assessed on gestational day 18.5.
Data are reported as mean ± SEM for each measure (n = 10 – 11 litters).
CSE = Cigarette Smoke Exposure
There were no significant effects of gestational sidestream smoke exposure on any of the fetal outcomes measured.
3.3 “Transcriptional Profiling” of the Fetal Mouse Hippocampal tissue
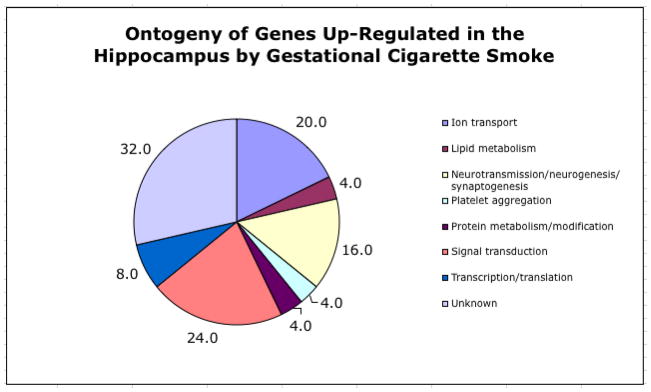
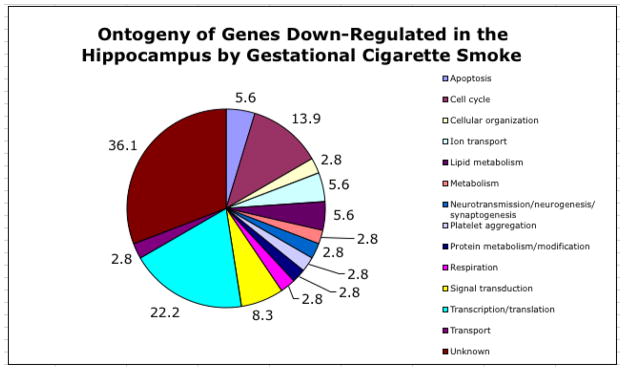
Of the 45,101 gene/EST probes on the Affymetrix high-density GeneChip® array, 3,109 demonstrated a detectable level of expression. Results indicated that 61 genes were differentially expressed in the hippocampus following in utero sidestream smoke exposure with 25 genes/ESTs being upregulated at least 1.5 fold and 36 genes/ESTs being down-regulated 1.5 fold or greater (Table 3). As shown in the heat map in Figure 2, expression of these genes was reproducible in the three biological samples taken from each treatment condition (smoke- and sham-exposed). Extensive literature mining of the differentially expressed genes identified numerous functional categories including cell cycling, neuronal transmission, neurogenesis, synaptogenesis, cellular organization, ion transport, lipid metabolism, protein metabolism/modulation, signal transduction, transcription/translation, and platelet aggregation. Figures 3 and 4 depict the functional categorization of genes differentially expressed within the hippocampus following sidestream smoke exposure during gestation.
TABLE 3.
Genes/ESTs that are Differentially Regulated in the Fetal Hippocampus Following Sidestream Cigarette Smoke Exposure on Gestational Days 1–17
| Gene Probe ID | Gene Descriptor | Fold Change1 |
|---|---|---|
| 1436544_at | ATPase, Class V, type 10D | 2.00 |
| 1438193_at | Neurexin III | 1.99 |
| 1424995_at | Riken cDNA 6230410P16 gene | 1.93 |
| 1417150_at | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 (Slc6a4; SERT) | 1.85 |
| 1441429_at | Insulin receptor substrate 4 | 1.85 |
| 1430379_at | RIKEN DNA 5830411K21 gene | 1.79 |
| 1450344_a_at | Prostaglandin E receptor 3 | 1.72 |
| 1444195_at | Required for meiotic nuclear division 5 homolog A (S. cerevisiae) | 1.68 |
| 1440358_at | Rho guanine nucleotide exchange factor (GEF) 15 | 1.62 |
| 1429808_at | Hypothetical protein LOC68625 | 1.61 |
| 1429180_at | Guanosine monophosphate reductase 2 | 1.61 |
| 1455373_at | Transcribed locus | 1.60 |
| 1455493_at | Synaptic nuclear envelope 1 | 1.60 |
| 1436593_at | RIKEN cDNA 1700016K19 gene = hypothetical protein LOC 74230 | 1.59 |
| 1459385_at | Calmodulin regulated spectrin-associated protein 1-like 1 | 1.56 |
| 1419501_at | Polymerase (DNA directed), Kappa | 1.56 |
| 1427558_s_at | Asparagine-linked glycosylation 12 homolog | 1.56 |
| 1445459_at | Somatostatin receptor 5 | 1.54 |
| 1450370_a_at | Kv channel interacting protein 4 | 1.54 |
| 1455223_at | Insulin-like growth factor 2 mRNA binding protein 1 | 1.53 |
| 1416275_at | Solute carrier family 26, member 6 | 1.53 |
| 1459143_at | Coiled-coil-helix-coiled-coil-helix domain containing 3 | 1.53 |
| 1449269_at | Coagulation factor V | 1.52 |
| 1424989_at | Transmembrane protein 142A/ORAI calcium release-activated calcium modulator | 1.51 |
| 1428948_at | RIKEN cDNA 5730414M22 gene | 1.51 |
| 1423084_at | UDP-Gal:betaGlcNAcbeta 1,3- galactosyltransferase, polypeptide 2 | −1.51 |
| 1450515_at | Potassium inwardly rectifying channel, subfamily J, member 11 | −1.51 |
| 1438268_at | Ring finger and CCCH-type zinc finger domains 2 | −1.51 |
| 1455586_at | Ring finger protein 168 | −1.51 |
| 1458465_at | PAN3polyA specific ribonuclease subunit homolog (yeast) | −1.52 |
| 1444917_at | H3098E04-3 NIA Mouse 15K cDNA clone set Mus Musculus DNA clone H3098E04 3′, mRNA sequence | −1.52 |
| 1437372_at | Cleavage and polyadenylation specific factor 6 | −1.52 |
| 1424971_at | Coiled-coil domain containing 99 | −1.52 |
| 1443229_at | ATPase family, AAA domain containing 2 | −1.53 |
| 1440241_at | Melastatin 1/transient receptor potential cation channel, subfamily M, member 1 | −1.53 |
| 1445484_at | Cytochrome c, somatic | −1.54 |
| 1429470_at | EST: similar to Speedy homolog A | −1.54 |
| 1456492_at | RIKEN cDNA 9130404D08 gene | −1.55 |
| 1450459_at | Signal peptide peptidase-like 2A/intramembrane protease3 | −1.56 |
| 1429010_at | U1 small nuclear ribonucleoprotien polypeptide A | −1.56 |
| 1450188_s_at | Lipase, endothelial | −1.56 |
| 1421512_at | Centrosomal protein 250 | −1.56 |
| 1429880_at | RIKEN cDNA 4921531C22 gene | −1.57 |
| 1439327_at | Collagen and calcium binding EGF domains 1 | −1.58 |
| 1448885_at | RAP2B, (RAS oncogene family member) | −1.58 |
| 1449311_at | Basic Leucine zipper transcription factor 1 | −1.59 |
| 1417642_at | Aldehyde dehydrogenase family 1, A3 | −1.59 |
| 1453573_at | Histone cluster 1, H3d | −1.59 |
| 1458050_at | Transcribed Sequences | −1.60 |
| 1415927_at | Actin, alpha, cardiac | −1.60 |
| 1436904_at | Thyroid hormone receptor associated protein 1/mediator complex subunit 3 | −1.61 |
| 14557787_at | Transcribed sequences | −1.61 |
| 1440282_at | Tubby-like protein 4 | −1.64 |
| 1422167_at | Sema domain (semaphorin 5A) | −1.64 |
| 1439664_at | WD repeat domain 68 | −1.69 |
| 1454354_at | RIKEN cDNA 8030476L19 gene | −1.70 |
| 1431278_s_at | Phospholipase A2, group VI | −1.73 |
| 1439704_at | Histone deacetylase 2 | −1.74 |
| 1421230_a_at | Musashi Homolog 2 (Drosophila) | −1.74 |
| 1444564_at | Apolipoprotein D | −1.83 |
| 1439040_at | Centromere protein E | −1.90 |
Only genes whose expression increased or decreased at least 1.5 fold (smoke exposed versus sham exposed) in the hippocampus are shown in this table. Positive numbers indicate an increase in expression, whereas negative numbers indicate a decrease in expression.
Figure 3.
Ontogeny of up-regulated genes in the hippocampus following exposure to ETS. The pie chart depicts the predicted function of genes that demonstrated an increase in expression in the hippocampus following ETS exposure. Numbers beside each pie segment indicate the percentage of the total number of upregulated genes assigned to a given functional category. Functional gene category assignments were determined through an extensive literature search in PubMed. Note that individual genes may be present in more than one category.
Figure 4.
Ontogeny of down-regulated genes in the hippocampus following exposure to ETS. The pie chart depicts the predicted function of genes that demonstrated a decrease in expression in the hippocampus following ETS exposure. Numbers on each pie segment indicate the percentage of the total number of down-regulated genes assigned to a given functional category. Functional gene category assignments were determined through an extensive literature search in PubMed. Note that individual genes may be present in more than one category.
3.4 Identification of Relevant Biological Networks for Differentially Expressed Genes within the Hippocampus as a result of in utero ETS Exposure
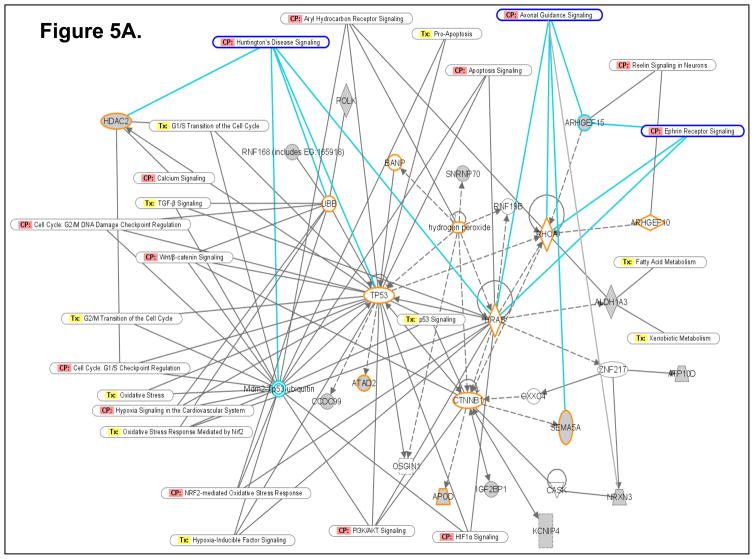
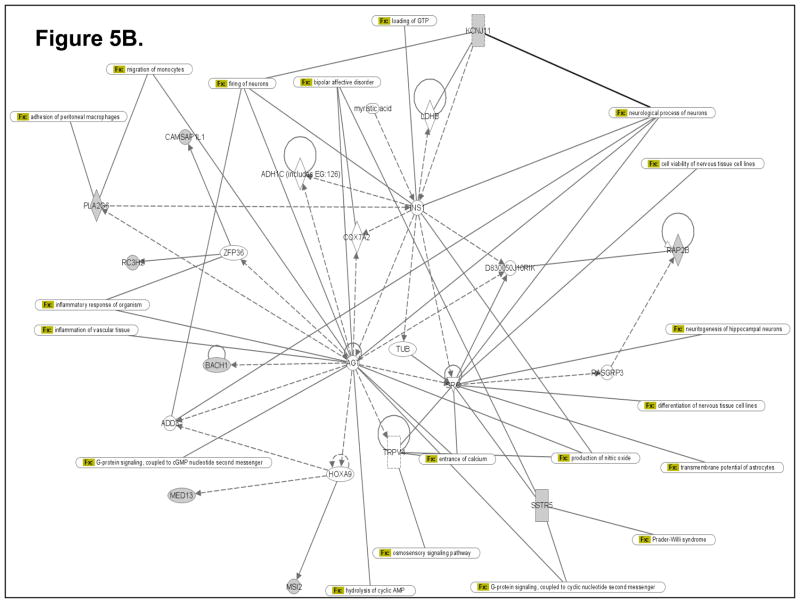
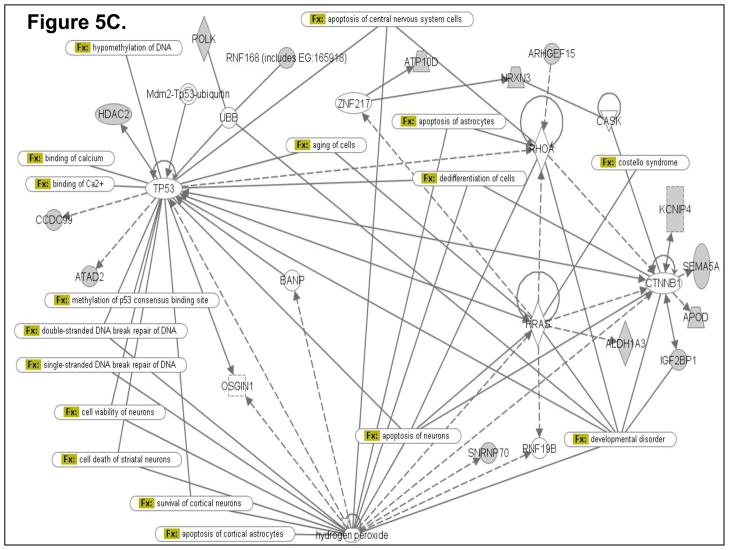
To determine how the differentially expressed genes might interact in the ETS-exposed hippocampus, an Ingenuity Pathway Analysis (IPA; Ingenuity Systems) was performed using the Ingenuity Systems program at www.ingenuity.com. IPA employs a knowledge base program to generate relevant biological networks. Four major networks (containing many genes already known to be expressed in the hippocampus) were discovered in the differentially-expressed gene list. All of these networks have significant p-values, indicating that it is highly implausible that these networks were detected by chance. Three of the four highest scoring networks (1st Network with p-value 1.0E-25; Fig. 5A; 2nd Network with p-value 1.0E-21; Fig. 5B; and 3rd Network with p-value 1.0E-20; Fig. 5C) are presented as examples in Figure 5. The pathway analysis data suggest that novel, differentially expressed genes (identified in the present gene expression profiling study) that cluster in these networks (containing several known hippocampus-expressed genes) may be essential for sustaining key hippocampal functions.
Figure 5.
Computational gene interaction predictions: selected gene networks (A, B, C) in the fetal hippocampus following ETS exposure. Gene networks were constructed with Ingenuity Systems Pathway Analysis (IPA) software. Several differentially regulated genes from the study were used to construct gene association maps for predicting effects of prenatal exposure to ETS on various cellular and molecular events in the developing mouse hippocampus. The first statistically significant network (Fig. 5A) that was generated, includes p53 tumor suppressor (TP53), Hras oncogene, β-catenin (CTNNB1) and Ubiquitin-B (UBB), and also consists of several genes such as those encoding HDAC2, Semaphorin-5A, Aldehyde dehydrogenase 1A3 (ALDH1A3), Rho guanine nucleotide exchange factor 15 (ARHGEF15), Apolipoprotein D, Polymerase kappa, and Neurexin-3 (NRXN3) among others, which demonstrated significant differential expression within the hippocampus as a consequence of prenatal ETS exposure (Table 3). The second statistically significant network (Fig. 5B) consists of – Rous sarcoma oncogene (SRC), Angiotensinogen (AGT), Zinc finger protein 36 (ZFP36) and Hoxa9, in addition to a number of genes encoding proteins such as Somatostatin receptor 5 (SSTR5), Potassium inwardly rectifying channel, subfamily J, member 11 (KCNJ11), Musashi-2 (MSI2), and Phospholipase A2, group VI (PLA2G6) among others, which are either up- or down-regulated in the developing hippocampus exposed to in utero sidestream tobacco smoke (Table 3). The third statistically significant network (Fig. 5C) is composed of – p53, RhoA, β-catenin, Hras, as well as several differentially expressed genes – detected in the present study – such as those encoding – Insulin-like growth factor 2 mRNA binding protein 1 (Igf2bp1), HDAC2, Apolipoprotein D (APOD), ATPase Class V, type 10D (ATP10D), Ring finger protein (RNF168), ATPase family, AAA domain containing 2 (ATAD2), etc. (Table 3). Solid lines specify direct relationships whereas dotted lines indicate indirect interactions.
3.5 TaqMan ™ Quantitative Real Time PCR Verification of Microarray Results
Twelve genes were randomly selected from the list of upregulated and downregulated genes (four upregulated and eight downregulated) and their expression patterns determined by quantitative real-time PCR using diluted cRNA as template. The overall expression profiles of the twelve mRNAs tested were found to be in agreement with the GeneChip microarray results (Table 4).
TABLE 4.
GeneChip® Microarray Verification by TaqMan™ Quantitative Real-Time PCR1
| Gene2 | Entrez Gene | Affymetrix Probe ID | Concordance3 |
|---|---|---|---|
| Downregulated Genes | |||
| Apolipoprotein D | 11815 | 1444564_at | +/+ |
| HDAC-2 | 15182 | 1439704_at | +/+ |
| Sema domain 5A (Semaphorin 5A) | 20356 | 1422167_at | +/+ |
| Musashi homolog 2 | 76626 | 1421230_a_at | +/+ |
| Centromere protein E | 229841 | 1439040_at | +/+ |
| UDP-Gal:betaGlcNAcbeta 1,3- galactosyltransferase, polypeptide 2 | 26878 | 1423084_at | +/+ |
| Aldehyde dehydrogenase family 1, A3 | 56847 | 1417642_at | +/+ |
| Phospholipase A2, group VI | 53357 | 1431278_s_at | +/+ |
| Upregulated Genes | |||
| Insulin Receptor Substrate 4 (IRS-4) | 16370 | 1441429_at | +/+ |
| Synaptic nuclear envelope 1 | 64009 | 1455493_at | +/+ |
| Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 15567 | 1417150_at | +/+ |
| Polymerase (DNA directed) Kappa | 27015 | 1419501_at | +/+ |
The differential expression of twelve genes in the murine hippocampus following exposure to sidestream cigarette smoke on gestational days 1–17, was compared using Affymetrix GeneChip® arrays and TaqMan™ quantitative real-time PCR.
Target genes were selected randomly.
Full concordance in the pattern or level of gene expression obtained using the Affymetrix GeneChip® arrays and the TaqMan™ quantitative real-time PCR is represented as ‘+/+’.
4. DISCUSSION
The purpose of the current study was to identify changes in gene expression in the murine hippocampus as a consequence of in utero exposure to sidestream cigarette smoke (an experimental equivalent of environmental tobacco smoke (ETS)). Exposures to ETS, also called secondhand smoke, are estimated to affect 60% of non-smokers [46]. These involuntary exposures pose a significant threat to the health of children and the developing fetus [47–50]. Cigarette smoke exposure has been shown to adversely affect implantation rates, [40,51,52] and exposure to low levels of cigarette smoke, typically through passive or involuntary means, has been linked to a number of adverse developmental outcomes [13–16,18,48–50]. Deleterious effects associated with exposure to ETS during development include impaired cognitive development and reduced vocabulary abilities in children [18,53,54]. Constituents of smoke exposure have been shown to target the developing brain [21,22,28,30]. Recent work within our laboratory has demonstrated significant changes in gene expression in the murine fetal brain following exposure to sidestream cigarette smoke during prenatal development [55]. In an attempt to better understand how the exposure to ETS during development modifies gene expression within the hippocampus, a brain region known to play a key role in learning and memory, microarray analysis was employed on hippocampal tissue isolated from fetuses exposed to ETS in utero.
As a consequence of exposure to ETS, expression of a number of genes regulating key processes within the hippocampus such as synaptic function, axon guidance, neurogenesis, and cell survival were significantly altered (Table 3 & Fig 5). These findings support the notion that prenatal exposure to sidestream smoke may negatively influence postnatal learning and memory tasks.
Serotonin and serotonergic innervations play a key role in central nervous system (CNS) development. Regulation of serotonin levels is, driven, in part, by serotonin uptake transporters, known as SERTs [56]. Serotonin has also been found in non-neuronal cells including platelets and in the placenta [57,58]. Altered platelet serotonin uptake and metabolism have been associated with Down Syndrome (DS), and SERT expression has been shown to be significantly increased in samples taken from the cortex of DS patients [59]. Increased SERT expression would result in altered serotonin signaling in the cortex of DS patients leading to abnormal neurogenesis (particularly within the cortex) which might be one of the responsible factors for learning disabilities seen in these patients [60]. Our results revealed a significant upregulation of the gene (slc6a4) encoding SERT, suggesting alteration of the serotonergic system in the hippocampus of mouse embryos following ETS exposure (Table 3). Since serotonin has been reported to reduce apoptosis during neurogenesis [61], ETS exposure may result in SERT-induced hippocampal apoptosis. Thus, altered serotonergic signaling may be hypothesized as contributing to ETS-associated impaired cognitive development.
The Klarsicht/ANC-1/Syne homologue (KASH)-domain-containing proteins execute key functions in nuclear positioning during various cellular and developmental processes [62]. Syne-1, Syne-2 and Nesprin-3 belong to the Nesprin family of proteins which mediate nuclear membrane localization, and binding to actin [63]. Syne-1 is expressed in multiple tissues, including the CNS [64]. Mutations in SYNE1 gene have been associated with autosomal recessive cerebellar ataxia type 1 [65]. Interestingly, nicotine has been reported to induce ataxia [66]. In the current study, significant upregulation of the Syne1 gene was observed in the hippocampus of mouse embryos following ETS exposure (Table 3). It can thus be postulated that in utero ETS exposure may render the embryos susceptible to ataxia and related neuropathies through enhanced hippocampal expression of Syne1 in conjunction with the ataxia-inducing effect of nicotine.
Several low fidelity mammalian polymerases, including Pol z, Pol h, Pol i, Pol k, and Rev1, function to evade unrepaired DNA lesions that otherwise inhibit replication by normal polymerases [67]. Pol-k is one such polymerase that is highly inaccurate when replicating undamaged DNA [68]. Indeed, transient expression of PolK in cultured mouse cells significantly increased the incidence of specific mutations [69]. Because of the inaccuracy of PolK in replicating undamaged DNA, overexpression may contribute to defective cell cycle control leading to anomalous embryogenesis [70]. Markedly enhanced expression of Polk within the hippocampus of in utero ETS exposed embryos (Table 3) may therefore lead to abnormal hippocampal neurogenesis via aberrant cell cycle control or anomalies in genome stability pathways, eventually affecting long-term behavioral and cognitive function.
It is becoming increasingly apparent that insulin and insulin-like growth factor-I (IGF-I) play previously unrecognized crucial roles in the developing and adult brain where they promote myelinization of neuronal axons, contribute to the formation, maintenance and repair of synaptic networks, and are involved in learning and memory, cell survival, neurogenesis and longevity [71,72]. Moreover, stimulation of neuronal nicotinic acetylcholine receptors (nAChRs) through exposure to nicotine resulted in time- and dose-dependent upregulation of insulin receptor substrate (IRS)-1 and IRS-2 mRNAs and proteins and enhanced insulin-induced activation of PI3K and ERK pathways [71]. This highlights the importance of the IRS family of proteins in regulating synaptic plasticity, learning, memory, and cell survival. Thus, significantly enhanced expression of the genes encoding Irs-4 and Insulin-like growth factor 2 mRNA binding protein 1 (Igf2bp1) in the hippocampus of mouse embryos following gestational exposure to ETS (Table 3), may result in functional compromise of hippocampal neuronal nicotinic acetylcholine receptors resulting in long-term cognitive impairments.
Apolipoprotein D (apoD), a member of the lipocalin family, is generally found in plasma associated with plasma high density lipoproteins (HDL) [73,74]. While tissue distribution of apoD mRNA is species-specific, all species investigated exhibit strong expression in the central and peripheral nervous systems [75]. It has been observed that apoD levels were significantly increased in the hippocampus and cerebrospinal fluid (CSF) of patients with Alzheimer’s disease (AD) [76]. Alterations in apoD expression within the brain have also been reported in a number of neurodegenerative diseases as well as patients with schizophrenic and bipolar disorders [77]. Exposure to cigarette smoke has also been reported to affect apoD expression [78]. Collectively, these findings suggest that altered apoD levels in neural tissues, including the hippocampus, could be considered as a marker of a range of neuropathologies associated with prenatal tobacco smoke exposure. Thus, significantly downregulated expression of ApoD in the embryonic hippocampal tissue observed in the present gene expression profiling study (Table 3), could be a contributing factor to generation of neuropathologies associated with in utero ETS exposure.
Epigenetic modifications of DNA or associated proteins are essential in controlling key processes such as cellular differentiation, development and behavior – including learning and memory [79]. Dysregulation of the epigenome has been widely accepted as a contributing factor towards the emergence of a range of neurodevelopmental disorders [80]. Inhibitors of class 1 HDACs (HDACs 1, 2, 3 and 8) and class 2 HDACs (HDACs 4, 5, 6, 7, 9 and 10) have been considered as targets for therapeutic intervention of neurodegenerative diseases and cognitive deficits associated with scores of neurodevelopmental disorders [80–82]. HDAC2 is highly expressed in the hippocampus and is also, a developmental target for the constituents of cigarette smoke [21,22]. We demonstrate, in the present study, that Hdac2 expression is significantly diminished in the hippocampus of murine embryos exposed in utero to ETS (Table 3). This observation is supported by the study of Yang et al. [83] which demonstrated reduced activity and levels of HDAC1, HDAC2, and HDAC3 in a human macrophage-like cell line exposed to cigarette smoke extract. These data implicate various histone deacetylases, including HDAC2, in the pathogenesis of a range of neurodegenerative and neurodevelopmental disorders, and emphasize that constituents present in cigarette smoke can modulate the expression and activity of HDAC2. Further, they highlight a possible linkage between prenatal exposure to cigarette smoke and postnatal development of certain neurodegenerative diseases with associated problems of depression, anxiety and cognitive deficits.
Semaphorins represent a large family of 20 different, secreted, GPI-linked, or transmembrane proteins characterized by the presence of a conserved 500-amino-acid- “Sema” domain at their amino-terminus. Semaphorins bind to two major families of receptors, neuropilins and plexins, to act as chemorepellents or inhibitors of growth cones [84,85] during regeneration of injured nerve fibers [86] and axon guidance [87]. These proteins are also thought to play a role in cell migration, morphogenesis and angiogenesis, during development [88], as well as contributing to distorted structural plasticity in neurodegenerative disorders such as Alzheimer’s disease [89]. Exposure to psychomotor-stimulant drugs induces changes in the expression of axon guidance molecules such as Sema5A [90] and this may contribute to cognitive deficits associated with drug abuse. Thus, the ETS-induced inhibition of Sema5A expression in the hippocampus (Table 3) could be a factor contributing to aberrant wiring of neural networks and axon guidance within the developing hippocampus leading to long-term postnatal deficiency in learning, memory and cognition.
Several neural-specific RNA-binding proteins (RBPs), such as Musashi1 (Msi1) and Musashi2 (Msi2), affect splicing, transport, translation and stability of target mRNAs [91]. Expression of Msi1 and Msi2 is developmentally regulated and coexpression of Msi1 and Msi2 has been detected in proliferating embryonic pluripotent neural precursors and in precursor cell populations of postnatal and adult CNS stem cells [92–94]. Indeed, it has been suggested that these two proteins contribute to the development and maintenance of CNS stem cells [95]. Inhibition of expression of msi1 and msi2 inhibited the ability of CNS stem cells to proliferate and form neurospheres [96]. Generation of specific neural populations takes place primarily in the subventricular zone (SVZ) of the lateral ventricles, and the subgranular zone (SGZ) of the dentate gyrus (DG) of the hippocampus [97]. Based on enhanced expression of Musashi-1 and -2 in proliferating neural progenitor cells in the brain of patients with subarachnoid hemorrhage, Sgubin et al., [98] concluded that Msi-2 may function as an activator of neural progenitor cell proliferation. In the present study, we demonstrate that the gene encoding Msi-2 was significantly downregulated in the embryonic murine hippocampus subsequent to ETS exposure (Table 3). It can thus be hypothesized that diminished expression of the Msi2 gene may result in altered hippocampal neurogenesis in fetuses exposed (in utero) to sidestream smoke, rendering them susceptible towards various neurological disorders associated with prenatal ETS exposure.
The Ingenuity Pathway Analysis (IPA; Ingenuity Systems) program allows generation of gene networks based on the biological associations of specific genes and various functions controlled by those genes. Based on the differentially expressed genes in the fetal hippocampus subsequent to maternal ETS exposure, pathway analysis revealed three statistically significant networks. One network (Fig. 5A) is noteworthy for including signaling pathways such as axon guidance, ephrin receptor signaling and calcium signaling, all crucial for hippocampal function. Other pathways include those regulating cell cycle and DNA damage, apoptosis, TGFβ-, Wnt/β-catenin-, PI3K/AKT-, Aryl hydrocarbon receptor- and p53- signaling, fatty acid and xenobiotic metabolism, NRF2-mediated oxidative stress and hypoxia-inducible factor signaling (Fig. 5A). Intriguingly, two neurological disease process interactomes, Huntington’s disease and Reelin signaling, are also present within this network (Fig. 5A). A second network (Fig. 5B) includes pathways crucial for neural cell formation, differentiation, viability, and transmembrane potential, as well as cell adhesion, cell migration, inflammatory response, nitric oxide production, calcium entry, G protein and osmosensory signaling. Similar to network 1, network 2 included the interactomes of two neurological disease process, bipolar affective disorder and Prader-Willi syndrome (Fig. 5B). A third network (Fig. 5C) includes pathways instrumental in regulating apoptosis and/or survival of various types of neurons and astrocytes, methylation of DNA, DNA repair and cellular aging. This network also links to a neurodevelopmental disease e.g. Costello Syndrome.
Pathway analysis thus reveals that genes differentially expressed within the hippocampus following gestational ETS exposure, may be functionally associated with several neurodegenerative diseases. Notably, a common theme underlying the pathologies of each of the aforementioned disorders is that the affected individuals have cognitive impairments and hippocampal dysfunction. Types of cognitive deficits common to all these disorders are attention deficits, and loss of short-term memory and sequential information processing. Furthermore, the neuroanatomical abnormalities seen in all such disorders localize to the hippocampus and cerebellum [99]. Collectively, our data strongly support the notion that prenatal ETS-exposure can trigger detrimental changes in normal hippocampal function leading to postnatal behavioral anomalies and cognitive deficits.
In the current study, pathway analysis enabled identification of a range of biological processes that, while indispensable for normal hippocampal function, could result in neuropathies when aberrantly regulated. For example, we showed ETS-induced downregulation in the fetal hippocampus of a number of genes that play key roles in cell survival and/or apoptosis (Tulp4, Pla2g6) [100,101]. Moreover, several p53-, Hras-, β-catenin-, RhoA- and Hydrogen peroxide-mediated neuronal apoptosis and/or cell survival pathways were detected within the networks generated with genes differentially-expressed within the prenatal ETS-exposed hippocampus (Fig 5A, 5B, 5C). Accordingly, these findings point to potential activation of various apoptosis and/or cell survival pathways within the hippocampus as a result of in utero sidestream cigarette smoke-exposure.
Several of the genes whose expression was altered in the fetal hippocampus by exposure to cigarette smoke are known to influence proliferation and growth. For example, genes encoding Centrosomal protein 250 (Cep 250) and Centromere-associated protein E (Cenp-E) are known regulators of cellular proliferation, and are significantly downregulated in the prenatal ETS-exposed hippocampus (Table 3). Decreased hippocampal cell proliferation has been linked to neurological diseases such as Huntington’s disease [102]. Thus, ETS-induced diminished expression of genes encoding the aforementioned proliferation markers could be associated with abnormal neurogenesis within the developing hippocampus.
The connection between cigarette smoke exposure and DNA damage has been established by a number of studies [103,104]. Our pathway analysis, wherein various p53- and hydrogen peroxide-mediated DNA repair pathways were detected (Fig 5A, 5B, 5C), provides further support for this connection.
Growth inhibition is the single most important factor affecting infant mortality in humans [105]. Maternal smoking decreases infant birth weight by ~150–200 grams and accounts for over 20% of the incidence of low birth weight [50,106,107]. Moreover, exposure to ETS is also associated with reduced birth weight in humans, albeit (30–100 g) less than that observed with ‘active’ maternal smoking [49,108]. We thus anticipated that our experimental paradigm of exposure to low levels of cigarette smoke during gestation would result in fetal growth restriction. However, in utero exposure to sidestream cigarette smoke during gd 6–18.5 had no significant effect on fetal weight or crown-rump length. Several studies are in agreement with our data, demonstrating that exposure to ETS during gestation failed to result in decreases in fetal weight [21,109–111]. In contrast, other studies demonstrate a link between exposure to sidestream smoke during gestation and reduced fetal weight [112–114]. These conflicting data may be in part due to differences in exposure parameters, including the period of gestational exposure. In the current study, exposures were begun on gestational day 6, and continued throughout gestation. Although this developmental period encompasses the period of organogenesis [115], it begins after implantation which is completed on gd 5 [40–42]. Recent studies in our laboratory have shown that exposure of murine dams to mainstream/sidestream tobacco smoke during the first five days of gestation (pre/peri-implantation period gd 1–5) resulted in decreases in fetal weight and crown-rump length, suggesting a temporal window of vulnerability for cigarette smoke-induced fetal growth restriction [112]. As women are typically exposed to environmental tobacco smoke both during and prior to pregnancy, the experimental paradigm used in this study may not adequately reveal true exposure risks.
However, it is important to note that even in the absence of observable effects on such phenotypic measures as term fetal-weight, -length and overall morphology, significant changes in the expression of genes associated with the development and function of the hippocampus were observed. These results are consistent with previous reports demonstrating alterations in brain development following prenatal exposure to nicotine, even in the absence of a low birth weight phenotype [22,116]. Collectively, data from the present study raise the possibility that subtle smoke-induced changes in the expression of genes within the developing hippocampus could affect long-term behavioral and cognitive function. Currently, studies are underway in our laboratory to determine whether exposure to low levels of cigarette smoke during gestation, in our animal model of in utero ETS exposure, induces permanent gene expression changes in the postnatal animal, and if such changes are accompanied by altered hippocampal-dependent behaviors.
Acknowledgments
These studies were supported, in part, by Philip Morris USA Inc. and Philip Morris International (KHH/MMP), the Centers for Disease Control and Prevention, CCU420170 (RMG/MMP), NIH grants HD053509 (RMG/MMP), DA027466 (MMP) and P20RR017702 from the COBRE Program of the National Center for Research Resources (RMG/MMP).
Footnotes
Conflict of Interest Statement:
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Centers for Disease Control and Prevention. Cigarette smoking among adults--United States, 2004. MMWR Morb Mortal Wkly Rep. 2005 Nov 11;54(44):1121–24. [PubMed] [Google Scholar]
- 2.Chen LH, Petitti DB. Case-control study of passive smoking and the risk of small-for-gestational-age at term. Am J Epidemiol. 1995 Jul 15;142(2):158–65. doi: 10.1093/oxfordjournals.aje.a117614. [DOI] [PubMed] [Google Scholar]
- 3.Jordan TR, Price JH, Dake JA, Shah S. Adolescent exposure to and perceptions of environmental tobacco smoke. J Sch Health. 2005 May;75(5):178–186. [PubMed] [Google Scholar]
- 4.Rudatsikira EM, Knutsen SF, Job JS, et al. Exposure to environmental tobacco smoke in the nonsmoking population of Cambodia. Am J Prev Med. 2008 Jan;34(1):69–73. doi: 10.1016/j.amepre.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Pirkle JL, Bernert JT, Caudill SP, Sosnoff CS, Pechacek TF. Trends in the exposure of nonsmokers in the U.S. population to secondhand smoke: 1988–2002. Environ Health Perspect. 2006 Jun;114(6):853–58. doi: 10.1289/ehp.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirkle JL, Flegal KM, Bernert JT, Brody DJ, Etzel RA, Maurer KR. Exposure of the US population to environmental tobacco smoke: the Third National Health and Nutrition Examination Survey, 1988 to 1991. Jama. 1996 Apr 24;275(16):1233–40. [PubMed] [Google Scholar]
- 7.Chang MJ, Walker K, McDaniel RL, Connell CT. Impaction collection and slurry sampling for the determination of arsenic, cadmium, and lead in sidestream cigarette smoke by inductively coupled plasma-mass spectrometry. J Environ Monit. 2005 Dec;7(12):1349–54. doi: 10.1039/b509048b. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, Zhao M, Kong H, et al. Characterization of cigarette smoke condensates by comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry (GC × GC/TOFMS). Part 2: basic fraction. J Sep Sci. 2004 Jan;27(1–2):101–9. doi: 10.1002/jssc.200301659. [DOI] [PubMed] [Google Scholar]
- 9.Stabbert R, Voncken P, Rustemeier K, et al. Toxicological evaluation of an electrically heated cigarette. Part 2: Chemical composition of mainstream smoke. J Appl Toxicol. 2003 Sep-Oct;23(5):329–39. doi: 10.1002/jat.924. [DOI] [PubMed] [Google Scholar]
- 10.Hammond SK, Sorensen G, Youngstrom R, Ockene JK. Occupational exposure to environmental tobacco smoke. Jama. 1995 Sep 27;274(12):956–60. [PubMed] [Google Scholar]
- 11.Schick S, Glantz S. Philip Morris toxicological experiments with fresh sidestream smoke: more toxic than mainstream smoke. Tob Control. 2005 Dec;14(6):396–404. doi: 10.1136/tc.2005.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schick S, Glantz SA. Sidestream cigarette smoke toxicity increases with aging and exposure duration. Tob Control. 2006 Dec;15(6):424–29. doi: 10.1136/tc.2006.016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein IM, Plociennik K, Stahle S, Badger GJ, Secker-Walker R. Impact of maternal cigarette smoking on fetal growth and body composition. Am J Obstet Gynecol. 2000 Oct;183(4):883–86. doi: 10.1067/mob.2000.109103. [DOI] [PubMed] [Google Scholar]
- 14.Cnattingius S. The epidemiology of smoking during pregnancy: smoking prevalence, maternal characteristics, and pregnancy outcomes. Nicotine Tob Res. 2004 Apr;6( Suppl 2):S125–40. doi: 10.1080/14622200410001669187. [DOI] [PubMed] [Google Scholar]
- 15.Cornelius MD, Taylor PM, Geva D, Day NL. Prenatal tobacco and marijuana use among adolescents: effects on offspring gestational age, growth, and morphology. Pediatrics. 1995 May;95(5):738–43. [PubMed] [Google Scholar]
- 16.Kharrazi M, DeLorenze GN, Kaufman FL, et al. Environmental tobacco smoke and pregnancy outcome. Epidemiology. 2004 Nov;15(6):660–70. doi: 10.1097/01.ede.0000142137.39619.60. [DOI] [PubMed] [Google Scholar]
- 17.Windham GC, Von Behren J, Waller K, Fenster L. Exposure to environmental and mainstream tobacco smoke and risk of spontaneous abortion. Am J Epidemiol. 1999 Feb 1;149(3):243–47. doi: 10.1093/oxfordjournals.aje.a009798. [DOI] [PubMed] [Google Scholar]
- 18.Yolton K, Dietrich K, Auinger P, Lanphear BP, Hornung R. Exposure to environmental tobacco smoke and cognitive abilities among U.S. children and adolescents. Environ Health Perspect. 2005 Jan;113(1):98–103. doi: 10.1289/ehp.7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goel P, Radotra A, Singh I, Aggarwal A, Dua D. Effects of passive smoking on outcome in pregnancy. J Postgrad Med. 2004 Jan-Mar;50(1):12–6. [PubMed] [Google Scholar]
- 20.Ward C, Lewis S, Coleman T. Prevalence of maternal smoking and environmental tobacco smoke exposure during pregnancy and impact on birth weight: retrospective study using Millennium Cohort. BMC Public Health. 2007;7:81. doi: 10.1186/1471-2458-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji CM, Royce FH, Truong U, Plopper CG, Singh G, Pinkerton KE. Maternal exposure to environmental tobacco smoke alters Clara cell secretory protein expression in fetal rat lung. Am J Physiol. 1998 Nov;275(5 Pt 1):L870–76. doi: 10.1152/ajplung.1998.275.5.L870. [DOI] [PubMed] [Google Scholar]
- 22.Roy TS, Seidler FJ, Slotkin TA. Prenatal nicotine exposure evokes alterations of cell structure in hippocampus and somatosensory cortex. J Pharmacol Exp Ther. 2002 Jan;300(1):124–33. doi: 10.1124/jpet.300.1.124. [DOI] [PubMed] [Google Scholar]
- 23.Genedani S, Bernardi M, Bertolini A. Sex-linked differences in avoidance learning in the offspring of rats treated with nicotine during pregnancy. Psychopharmacology (Berl) 1983;80(1):93–5. doi: 10.1007/BF00427504. [DOI] [PubMed] [Google Scholar]
- 24.Levin ED, Briggs SJ, Christopher NC, Rose JE. Prenatal nicotine exposure and cognitive performance in rats. Neurotoxicol Teratol. 1993 Jul–Aug;15(4):251–60. doi: 10.1016/0892-0362(93)90006-a. [DOI] [PubMed] [Google Scholar]
- 25.Martin JC, Becker RF. The effects of maternal nicotine absorption or hypoxic episodes upon appetitive behavior of rat offspring. Dev Psychobiol. 1971;4(2):133–47. doi: 10.1002/dev.420040205. [DOI] [PubMed] [Google Scholar]
- 26.Peters MA, Ngan LL. The effects of totigestational exposure to nicotine on pre- and postnatal development in the rat. Arch Int Pharmacodyn Ther. 1982 May;257(1):155–67. [PubMed] [Google Scholar]
- 27.Sorenson CA, Raskin LA, Suh Y. The effects of prenatal nicotine on radial-arm maze performance in rats. Pharmacol Biochem Behav. 1991 Dec;40(4):991–93. doi: 10.1016/0091-3057(91)90117-k. [DOI] [PubMed] [Google Scholar]
- 28.Vaglenova J, Birru S, Pandiella NM, Breese CR. An assessment of the long-term developmental and behavioral teratogenicity of prenatal nicotine exposure. Behav Brain Res. 2004 Apr 2;150(1–2):159–70. doi: 10.1016/j.bbr.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Giustino A, Cagiano R, Carratu MR, Cassano T, Tattoli M, Cuomo V. Prenatal exposure to low concentrations of carbon monoxide alters habituation and non-spatial working memory in rat offspring. Brain Res. 1999 Oct 9;844(1–2):201–5. doi: 10.1016/s0006-8993(99)01832-6. [DOI] [PubMed] [Google Scholar]
- 30.Mereu G, Cammalleri M, Fa M, et al. Prenatal exposure to a low concentration of carbon monoxide disrupts hippocampal long-term potentiation in rat offspring. J Pharmacol Exp Ther. 2000 Aug;294(2):728–34. [PubMed] [Google Scholar]
- 31.Nagy AGM, Vintersten K, Behringer R, editors. Manipulating the mouse Embryo: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Press; 2003. [Google Scholar]
- 32.Teague SVPK, Goldsmith M, Gebremichael A, Chang S. Inhalation Toxicology. Vol. 6. Taylor & Francis; 1994. Sidestream cigarette smoke generation and exposure system for environmental tobacco smoke studies; pp. 79–93. [Google Scholar]
- 33.Substance Abuse and Mental Health Services Administration. Office of Applied Studies, National Household Survey on Drug Abuse. 1999. [Google Scholar]
- 34.Society for Research on Nicotine and Tobacco Subcommittee on Biochemical Verification, Biochemical verification of tobacco use and cessation. Nicotine Tob Res. 2002 May;4(2):149–59. doi: 10.1080/14622200210123581. [DOI] [PubMed] [Google Scholar]
- 35.Koren G. Measurement of drugs in neonatal hair; a window to fetal exposure. Forensic Sci Int. 1995 Jan 5;70(1–3):77–82. doi: 10.1016/0379-0738(94)01623-d. [DOI] [PubMed] [Google Scholar]
- 36.Koren G, Klein J, Forman R, Graham K, Phan MK. Biological markers of intrauterine exposure to cocaine and cigarette smoking. Dev Pharmacol Ther. 1992;18(3–4):228–36. [PubMed] [Google Scholar]
- 37.Lackmann GM, Salzberger U, Tollner U, Chen M, Carmella SG, Hecht SS. Metabolites of a tobacco-specific carcinogen in urine from newborns. J Natl Cancer Inst. 1999 Mar 3;91(5):459–65. doi: 10.1093/jnci/91.5.459. [DOI] [PubMed] [Google Scholar]
- 38.Haseman JK, Hogan MD. Selection of the experimental unit in teratology studies. Teratology. 1975 Oct;12(2):165–171. doi: 10.1002/tera.1420120209. [DOI] [PubMed] [Google Scholar]
- 39.Jarvis MJ, Feyerabend C, Bryant A, Hedges B, Primatesta P. Passive smoking in the home: plasma cotinine concentrations in non-smokers with smoking partners. Tob Control. 2001 Dec;10(4):368–74. doi: 10.1136/tc.10.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Card JP, Mitchell JA. The effects of nicotine on implantation in the rat. Biol Reprod. 1979 Apr;20(3):532–39. doi: 10.1095/biolreprod20.3.532. [DOI] [PubMed] [Google Scholar]
- 41.Kaufman M. The Atlas of Mouse Development. San Diego, CA: Academic Press; 1992. [Google Scholar]
- 42.Surani MA. Zona pellucida denudation, blastocyst proliferation and attachment in the rat. J Embryol Exp Morphol. 1975 Apr;33(2):343–53. [PubMed] [Google Scholar]
- 43.Angevine JB., Jr Time of neuron origin in the hippocampal region. An autoradiographic study in the mouse. Exp Neurol Suppl. 1965 Oct;(Suppl 2):1–70. [PubMed] [Google Scholar]
- 44.Bayer SA. Development of the hippocampal region in the rat. I. Neurogenesis examined with 3H-thymidine autoradiography. J Comp Neurol. 1980 Mar 1;190(1):87–114. doi: 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- 45.Bayer SA. Development of the hippocampal region in the rat. II. Morphogenesis during embryonic and early postnatal life. J Comp Neurol. 1980 Mar 1;190(1):115–34. doi: 10.1002/cne.901900108. [DOI] [PubMed] [Google Scholar]
- 46.U.S. Department of Health and Human Services. The health consequences of involuntary exposure to tobacco smoke: A report of the Surgeon General. CDC, U.S. Department of Health and Human Services, Coordinating Center for Health Promotion, National Center for Chronic Disease Prevention and Health Promotion, Office of Smoking and Health; 2006. [Google Scholar]
- 47.Cook DG, Strachan DP. Health effects of passive smoking-10: Summary of effects of parental smoking on the respiratory health of children and implications for research. Thorax. 1999 Apr;54(4):357–66. doi: 10.1136/thx.54.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haddow JE, Knight GJ, Palomaki GE, Haddow PK. Estimating fetal morbidity and mortality resulting from cigarette smoke exposure by measuring cotinine levels in maternal serum. Prog Clin Biol Res. 1988;281:289–300. [PubMed] [Google Scholar]
- 49.Windham GC, Eaton A, Hopkins B. Evidence for an association between environmental tobacco smoke exposure and birthweight: a meta-analysis and new data. Paediatr Perinat Epidemiol. 1999 Jan;13(1):35–57. doi: 10.1046/j.1365-3016.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 50.Windham GC, Hopkins B, Fenster L, Swan SH. Prenatal active or passive tobacco smoke exposure and the risk of preterm delivery or low birth weight. Epidemiology. 2000 Jul;11(4):427–33. doi: 10.1097/00001648-200007000-00011. [DOI] [PubMed] [Google Scholar]
- 51.Neal MS, Hughes EG, Holloway AC, Foster WG. Sidestream smoking is equally as damaging as mainstream smoking on IVF outcomes. Hum Reprod. 2005 Sep;20(9):2531–35. doi: 10.1093/humrep/dei080. [DOI] [PubMed] [Google Scholar]
- 52.Yoshinaga K, Rice C, Krenn J, Pilot RL. Effects of nicotine on early pregnancy in the rat. Biol Reprod. 1979 Mar;20(2):294–303. doi: 10.1095/biolreprod20.2.294. [DOI] [PubMed] [Google Scholar]
- 53.Bauman KE, Flewelling RL, LaPrelle J. Parental cigarette smoking and cognitive performance of children. Health Psychol. 1991;10(4):282–8. doi: 10.1037//0278-6133.10.4.282. [DOI] [PubMed] [Google Scholar]
- 54.Eskenazi B, Trupin LS. Passive and active maternal smoking during pregnancy, as measured by serum cotinine, and postnatal smoke exposure. II. Effects on neurodevelopment at age 5 years. Am J Epidemiol. 1995 Nov 1;142(9 Suppl):S19–29. doi: 10.1093/aje/142.supplement_9.s19. [DOI] [PubMed] [Google Scholar]
- 55.Mukhopadhyay P, Horn KH, Esposito ER, Greene RM, Pisano MM. Maternal exposure to environmental tobacco smoke alters the gene expression profile of the developing murine brain. Molecular Brain. 2009 (manuscript in preparation) [Google Scholar]
- 56.Amara SG, Kuhar MJ. Neurotransmitter transporters: recent progress. Annu Rev Neurosci. 1993;16:73–93. doi: 10.1146/annurev.ne.16.030193.000445. [DOI] [PubMed] [Google Scholar]
- 57.Balkovetz DF, Tiruppathi C, Leibach FH, Mahesh VB, Ganapathy V. Evidence for an imipramine-sensitive serotonin transporter in human placental brush-border membranes. J Biol Chem. 1989 Feb 5;264(4):2195–98. [PubMed] [Google Scholar]
- 58.Amisten S, Braun OO, Bengtsson A, Erlinge D. Gene expression profiling for the identification of G-protein coupled receptors in human platelets. Thromb Res. 2008;122(1):47–57. doi: 10.1016/j.thromres.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 59.Gulesserian T, Engidawork E, Cairns N, Lubec G. Increased protein levels of serotonin transporter in frontal cortex of patients with Down syndrome. Neurosci Lett. 2000 Dec 15;296(1):53–7. doi: 10.1016/s0304-3940(00)01624-4. [DOI] [PubMed] [Google Scholar]
- 60.Smith L, von Tetzchner S. Communicative, sensorimotor, and language skills of young children with Down syndrome. Am J Ment Defic. 1986 Jul;91(1):57–66. [PubMed] [Google Scholar]
- 61.Persico AM, Baldi A, Dell’Acqua ML, et al. Reduced programmed cell death in brains of serotonin transporter knockout mice. Neuroreport. 2003 Mar 3;14(3):341–4. doi: 10.1097/00001756-200303030-00009. [DOI] [PubMed] [Google Scholar]
- 62.Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009 May;66(9):1518–33. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q, Skepper JN, Yang F, et al. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001 Dec;114(Pt 24):4485–98. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 64.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000 Oct 13;275(41):31986–95. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 65.Gros-Louis F, Dupre N, Dion P, et al. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet. 2007 Jan;39(1):80–5. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- 66.Kita T, Nakashima T, Shirase M, Asahina M, Kurogochi Y. Effects of nicotine on ambulatory activity in mice. Jpn J Pharmacol. 1988 Feb;46(2):141–6. doi: 10.1254/jjp.46.141. [DOI] [PubMed] [Google Scholar]
- 67.Lehmann AR. Replication of UV-damaged DNA: new insights into links between DNA polymerases, mutagenesis and human disease. Gene. 2000 Jul 25;253(1):1–12. doi: 10.1016/s0378-1119(00)00250-x. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Yuan F, Xin H, et al. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000 Nov 1;28(21):4147–56. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogi T, Kato T, Jr, Kato T, Ohmori H. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells. 1999 Nov;4(11):607–18. doi: 10.1046/j.1365-2443.1999.00289.x. [DOI] [PubMed] [Google Scholar]
- 70.Bavoux C, Hoffmann JS, Cazaux C. Adaptation to DNA damage and stimulation of genetic instability: the double-edged sword mammalian DNA polymerase kappa. Biochimie. 2005 Jul;87(7):637–46. doi: 10.1016/j.biochi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 71.Sugano T, Yanagita T, Yokoo H, Satoh S, Kobayashi H, Wada A. Enhancement of insulin-induced PI3K/Akt/GSK-3beta and ERK signaling by neuronal nicotinic receptor/PKC-alpha/ERK pathway: up-regulation of IRS-1/-2 mRNA and protein in adrenal chromaffin cells. J Neurochem. 2006 Jul;98(1):20–33. doi: 10.1111/j.1471-4159.2006.03846.x. [DOI] [PubMed] [Google Scholar]
- 72.Wada A, Yokoo H, Yanagita T, Kobayashi H. New twist on neuronal insulin receptor signaling in health, disease, and therapeutics. J Pharmacol Sci. 2005 Oct;99(2):128–43. doi: 10.1254/jphs.crj05006x. [DOI] [PubMed] [Google Scholar]
- 73.Bojanovski D, Alaupovic P, McConathy WJ, Kelly JL. Isolation and partial characterization of apolipoprotein D and lipoprotein D from baboon plasma. FEBS Lett. 1980 Apr 7;112(2):251–4. doi: 10.1016/0014-5793(80)80191-8. [DOI] [PubMed] [Google Scholar]
- 74.Weech PK, Camato R, Milne RW, Marcel YL. Apolipoprotein D and cross-reacting human plasma apolipoproteins identified using monoclonal antibodies. J Biol Chem. 1986 Jun 15;261(17):7941–51. [PubMed] [Google Scholar]
- 75.Rassart E, Bedirian A, Do Carmo S, et al. Apolipoprotein D. Biochim Biophys Acta. 2000 Oct 18;1482(1–2):185–98. doi: 10.1016/s0167-4838(00)00162-x. [DOI] [PubMed] [Google Scholar]
- 76.Terrisse L, Seguin D, Bertrand P, Poirier J, Milne R, Rassart E. Modulation of apolipoprotein D and apolipoprotein E expression in rat hippocampus after entorhinal cortex lesion. Brain Res Mol Brain Res. 1999 Jun 18;70(1):26–35. doi: 10.1016/s0169-328x(99)00123-0. [DOI] [PubMed] [Google Scholar]
- 77.Thomas EA, Dean B, Pavey G, Sutcliffe JG. Increased CNS levels of apolipoprotein D in schizophrenic and bipolar subjects: implications for the pathophysiology of psychiatric disorders. Proc Natl Acad Sci U S A. 2001 Mar 27;98(7):4066–71. doi: 10.1073/pnas.071056198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flatscher-Bader T, Wilce PA. Chronic smoking and alcoholism change expression of selective genes in the human prefrontal cortex. Alcohol Clin Exp Res. 2006 May;30(5):908–15. doi: 10.1111/j.1530-0277.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- 79.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005 Feb;6(2):108–18. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 80.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008 Feb;8(1):57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hahnen E, Hauke J, Trankle C, Eyupoglu IY, Wirth B, Blumcke I. Histone deacetylase inhibitors: possible implications for neurodegenerative disorders. Expert Opin Investig Drugs. 2008 Feb;17(2):169–84. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- 82.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008 Oct;7(10):854–68. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 83.Yang SR, Chida AS, Bauter MR, et al. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006 Jul;291(1):L46–57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 84.Skaliora I, Singer W, Betz H, Puschel AW. Differential patterns of semaphorin expression in the developing rat brain. Eur J Neurosci. 1998 Apr;10(4):1215–29. doi: 10.1046/j.1460-9568.1998.00128.x. [DOI] [PubMed] [Google Scholar]
- 85.Yu TW, Bargmann CI. Dynamic regulation of axon guidance. Nat Neurosci. 2001 Nov;4( Suppl):1169–76. doi: 10.1038/nn748. [DOI] [PubMed] [Google Scholar]
- 86.de Wit J, Verhaagen J. Role of semaphorins in the adult nervous system. Prog Neurobiol. 2003 Oct;71(2–3):249–67. doi: 10.1016/j.pneurobio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 87.He Z, Wang KC, Koprivica V, Ming G, Song HJ. Knowing how to navigate: mechanisms of semaphorin signaling in the nervous system. Sci STKE. 2002 Feb 12;119:RE1. doi: 10.1126/stke.2002.119.re1. [DOI] [PubMed] [Google Scholar]
- 88.Tamagnone L, Comoglio PM. Signalling by semaphorin receptors: cell guidance and beyond. Trends Cell Biol. 2000 Sep;10(9):377–83. doi: 10.1016/s0962-8924(00)01816-x. [DOI] [PubMed] [Google Scholar]
- 89.Good PF, Alapat D, Hsu A, et al. A role for semaphorin 3A signaling in the degeneration of hippocampal neurons during Alzheimer’s disease. J Neurochem. 2004 Nov;91(3):716–36. doi: 10.1111/j.1471-4159.2004.02766.x. [DOI] [PubMed] [Google Scholar]
- 90.Bahi A, Dreyer JL. Cocaine-induced expression changes of axon guidance molecules in the adult rat brain. Mol Cell Neurosci. 2005 Feb;28(2):275–91. doi: 10.1016/j.mcn.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 91.Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res. 2002 Apr 15;68(2):121–6. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- 92.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002 Apr 1;115(Pt 7):1355–59. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 93.Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001 Oct 15;21(20):8091–107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997 Nov 1;17(21):8300–12. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaneko Y, Sakakibara S, Imai T, et al. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22(1–2):139–53. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 96.Sakakibara S, Nakamura Y, Yoshida T, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):15194–99. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gage FH. Mammalian neural stem cells. Science. 2000 Feb 25;287(5457):1433–38. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 98.Sgubin D, Aztiria E, Perin A, Longatti P, Leanza G. Activation of endogenous neural stem cells in the adult human brain following subarachnoid hemorrhage. J Neurosci Res. 2007 Jun;85(8):1647–55. doi: 10.1002/jnr.21303. [DOI] [PubMed] [Google Scholar]
- 99.Pulsifer MB. The neuropsychology of mental retardation. J Int Neuropsychol Soc. 1996 Mar;2(2):159–76. doi: 10.1017/s1355617700001016. [DOI] [PubMed] [Google Scholar]
- 100.Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. J Lab Clin Med. 2005 Jan;145(1):47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 101.Shinzawa K, Sumi H, Ikawa M, et al. Neuroaxonal dystrophy caused by group VIA phospholipase A2 deficiency in mice: a model of human neurodegenerative disease. J Neurosci. 2008 Feb 27;28(9):2212–20. doi: 10.1523/JNEUROSCI.4354-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lazic SE, Grote H, Armstrong RJ, et al. Decreased hippocampal cell proliferation in R6/1 Huntington’s mice. Neuroreport. 2004 Apr 9;15(5):811–3. doi: 10.1097/00001756-200404090-00014. [DOI] [PubMed] [Google Scholar]
- 103.Howard DJ, Briggs LA, Pritsos CA. Oxidative DNA damage in mouse heart, liver, and lung tissue due to acute side-stream tobacco smoke exposure. Arch Biochem Biophys. 1998 Apr 15;352(2):293–7. doi: 10.1006/abbi.1998.0605. [DOI] [PubMed] [Google Scholar]
- 104.Wolz L, Krause G, Scherer G, Aufderheide M, Mohr U. In vitro genotoxicity assay of sidestream smoke using a human bronchial epithelial cell line. Food Chem Toxicol. 2002 Jun;40(6):845–50. doi: 10.1016/s0278-6915(02)00034-0. [DOI] [PubMed] [Google Scholar]
- 105.Das UG, Sysyn GD. Abnormal fetal growth: intrauterine growth retardation, small for gestational age, large for gestational age. Pediatr Clin North Am. 2004 Jun;51(3):639–54. doi: 10.1016/j.pcl.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 106.Meyer MB, Comstock GW. Maternal cigarette smoking and perinatal mortality. Am J Epidemiol. 1972 Jul;96(1):1–10. doi: 10.1093/oxfordjournals.aje.a121427. [DOI] [PubMed] [Google Scholar]
- 107.Ventura SJ, Hamilton BE, Mathews TJ, Chandra A. Trends and variations in smoking during pregnancy and low birth weight: evidence from the birth certificate, 1990–2000. Pediatrics. 2003 May;111(5 Part 2):1176–80. [PubMed] [Google Scholar]
- 108.Martin TR, Bracken MB. Association of low birth weight with passive smoke exposure in pregnancy. Am J Epidemiol. 1986 Oct;124(4):633–42. doi: 10.1093/oxfordjournals.aje.a114436. [DOI] [PubMed] [Google Scholar]
- 109.Gospe SM, Jr, Zhou SS, Pinkerton KE. Effects of environmental tobacco smoke exposure in utero and/or postnatally on brain development. Pediatr Res. 1996 Mar;39(3):494–8. doi: 10.1203/00006450-199603000-00018. [DOI] [PubMed] [Google Scholar]
- 110.Subramaniam S, Srinivasan S, Bummer PM, Gairola CG. Perinatal sidestream cigarette smoke exposure and the developing pulmonary surfactant system in rats. Hum Exp Toxicol. 1999 Apr;18(4):206–11. doi: 10.1191/096032799678839923. [DOI] [PubMed] [Google Scholar]
- 111.Witschi H, Lundgaard SM, Rajini P, Hendrickx AG, Last JA. Effects of exposure to nicotine and to sidestream smoke on pregnancy outcome in rats. Toxicol Lett. 1994 May;71(3):279–86. doi: 10.1016/0378-4274(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 112.Esposito ER, Horn KH, Greene RM, Pisano MM. An animal model of cigarette smoke-induced in utero growth retardation. Toxicology. 2008 Apr 18;246(2–3):193–202. doi: 10.1016/j.tox.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leichter J. Growth of fetuses of rats exposed to ethanol and cigarette smoke during gestation. Growth Dev Aging Autumn. 1989;53(3):129–34. [PubMed] [Google Scholar]
- 114.Rajini P, Last JA, Pinkerton KE, Hendrickx AG, Witschi H. Decreased fetal weights in rats exposed to sidestream cigarette smoke. Fundam Appl Toxicol. 1994 Apr;22(3):400–4. doi: 10.1006/faat.1994.1045. [DOI] [PubMed] [Google Scholar]
- 115.Lambers DS, Clark KE. The maternal and fetal physiologic effects of nicotine. Semin Perinatol. 1996 Apr;20(2):115–26. doi: 10.1016/s0146-0005(96)80079-6. [DOI] [PubMed] [Google Scholar]
- 116.Navarro HA, Seidler FJ, Schwartz RD, Baker FE, Dobbins SS, Slotkin TA. Prenatal exposure to nicotine impairs nervous system development at a dose which does not affect viability or growth. Brain Res Bull. 1989 Sep;23(3):187–92. doi: 10.1016/0361-9230(89)90146-9. [DOI] [PubMed] [Google Scholar]