Abstract
The hnRNP C heterotetramer [(C13)C2] binds RNA polymerase II transcripts in the nucleus, along with other proteins of the core hnRNP complex, and plays an important role in mRNA biogenesis and transport. Infection of HeLa cells with poliovirus causes hnRNP C to re-localize from the nucleus, where it is normally retained during interphase, to the cytoplasm. We have proposed that in the cytoplasm, the protein isoforms of hnRNP C participate in the recognition of viral specific RNAs by the poliovirus replication proteins and/or in the assembly of membrane-bound RNA replication complexes. In SK-OV-3 cells, which express reduced levels of hnRNP C compared to HeLa cells or 293 cells, the kinetics of poliovirus replication are delayed. hnRNP C is also re-localized from the nucleus to the cytoplasm in SK-OV-3 cells infected with poliovirus. Increased expression of hnRNP C in SK-OV-3 cells by transient transfection increases the rate of virus production and overall yield over that seen in mock-transfected cells. We propose that hnRNP C interacts with poliovirus RNA and replication proteins to increase the efficiency of viral genomic RNA synthesis.
Keywords: Poliovirus, Picornavirus, hnRNP C proteins, RNA replication, SK-OV-3 cells, Virus–cell interactions
Introduction
Identifying host factors that function in the replication of positive-strand RNA viruses is a major goal in molecular virology. Members of Picornaviridae, such as the well-studied poliovirus, with their relatively limited coding capacity must rely on available host factors to participate in the macromolecular interactions required for viral replicative processes. Cellular proteins functionally linked to viral RNA replication have been initially defined by their interaction with viral RNAs or replication polypeptides, or both. Compared to the study of host proteins involved with the viral proteins and RNA determinants required for picornavirus negative-strand RNA synthesis, much less work has been aimed at determining the factors necessary for positive-strand RNA synthesis. Not surprisingly, detailed studies of positive-strand RNA synthesis could only be undertaken once the process of negative-strand RNA synthesis had been somewhat unraveled. Further, the role of nuclear RNA-binding proteins in picornavirus replication has not been comprehensively investigated due, in part, to the original report by Pollack and Goldman (Pollack and Goldman, 1973) demonstrating that BSC-1 cells enucleated by treatment with cytochalasin B retained the ability to support the production of infectious poliovirus. However, it is possible that the treatment utilized in these early studies released nuclear proteins into the cytoplasm of the treated cells due to disruption of the nuclear envelope. In addition, it is noteworthy that following two rounds of cytochalasin B treatment and centrifugation, yields of poliovirus were reduced between five- and ten-fold compared to infection of untreated cells (Pollack and Goldman, 1973).
In the nucleus of the uninfected cell, primary transcripts of RNA polymerase II associate with a large number of nuclear RNA-binding proteins to form hnRNP complexes. These nuclear proteins assist with mRNA biogenesis and transport of nuclear pre-mRNA. Purified hnRNP complexes contain over 20 proteins, named according to size, from A1 to U. The most abundant protein in the complex, hnRNP C1, is expressed at approximately 100 million copies per cell, levels comparable to those for core histones (Dreyfuss et al., 1993). hnRNP C2, a splicing variant with an extra 13 amino acids, is expressed at approximately one-third the level of hnRNP C1. These two isoforms form stable heterotetramers [(C1)3C2] that bind cooperatively to RNA. Each isoform has an RNA-recognition motif (RRM), a basic leucine zipper-like motif (bZLM), a nuclear localization signal, a nuclear retention signal, and an acidic auxiliary domain thought to be involved in protein–protein interactions that serve to increase the specificity of RNA binding (Gorlach et al., 1992, McAfee et al., 1996, Lai, 1998). The hnRNP C proteins are highly conserved among vertebrates, both immunologically and in their amino acid sequence, which suggests essential cellular functions (Dreyfuss et al., 1993). The major constituents of the core hnRNP complex, A1, A2, B1, B2, C1, and C2, are assembled into hetero-tetrameric complexes of A1 and B2, B1 and A2, and C1 and C2 that together occupy approximately 700 nucleotides of RNA (Conway et al., 1988). The aggregation and disaggregation of the core hnRNP complex have been shown to be associated with RNA trafficking and nuclear export. hnRNP C is the only member of the core complex that does not appear to shuttle between the nucleus and cytoplasm during interphase, due to the presence of a nuclear retention signal (Pinol-Roma and Dreyfuss, 1992). This suggests that removal of hnRNP C may be required for RNA to be exported. Because hnRNP C is the most tightly bound component of the core ribonucleoprotein complex, remodeling may be a regulated, energy-dependent process (Beyer et al., 1977). Moreover, hnRNP C is preferentially phosphorylated during mitosis, suggesting that its functions are regulated during the cell cycle (Pinol-Roma and Dreyfuss, 1993, Schepens et al., 2007).
Investigators have reported that several nuclear RNA-binding proteins re-localize to the cytoplasm as a result of poliovirus infection. In addition to hnRNP A1, C, and K (Gustin and Sarnow, 2001), these include nucleolin, a primarily nuclear RNA-binding protein thought to be involved in rRNA metabolism, DNA replication, and apoptosis (Waggoner and Sarnow, 1998), Sam 68, an SH domain-binding protein (McBride et al., 1996), and the La autoantigen, a protein that binds to the poliovirus IRES (Meerovitch et al., 1993). We have previously described the interaction of hnRNP C with the 3′ end of poliovirus negative-strand RNA and with poliovirus RNA replication polypeptides (Roehl and Semler, 1995, Brunner et al., 2005). A mutated form of hnRNP C1, with a truncation of the protein–protein interaction auxiliary domain, impaired RNA synthesis in an in vitro replication assay. We demonstrated an interaction between hnRNP C and poliovirus RNA of both polarities recovered from extracts prepared from poliovirus-infected HeLa cells (Brunner et al., 2005). To further understand the involvement of hnRNP C in poliovirus RNA synthesis in infected cells, we utilized a human cell line (SK-OV-3) that expresses decreased levels of hnRNP C compared to other established human cell lines (e.g., 293 cells) (Holcik et al., 2003). SK-OV-3 cells, derived from an ovarian adenocarcinoma, are variably hypo-diploid (42 to 45 chromosome number), which could explain their modified expression of hnRNP C. Here we report that the concentration of hnRNP C proteins is substantially lower in SK-OV-3 cells compared to HeLa or 293 cells, in agreement with published findings (Holcik et al., 2003). Following infection of SK-OV-3 cells with poliovirus, we discovered that the kinetics of viral replication in these cells are significantly slower than the kinetics of replication in HeLa cells, especially during the first 8 h of infection. We provide evidence that this replication defect is due, in part, to reduced levels of positive-strand RNA produced during infection of SK-OV-3 cells. In addition, immunofluorescence studies, undertaken to examine hnRNP C distribution in SK-OV-3 cells during poliovirus infection, demonstrate that hnRNP C re-localizes to the cytoplasm, indicating an alteration in protein trafficking similar to that seen in poliovirus-infected HeLa cells (Gustin and Sarnow, 2001). Expression of hnRNP C1, hnRNP C2, or both simultaneously by transient transfection of recombinant expression vectors in SK-OV-3 cells increased the kinetics of poliovirus replication compared to vector alone. These studies provide new evidence for a functional role of hnRNP C in poliovirus replication and further indicate that the protein may be involved in increasing the efficiency of genomic RNA synthesis.
Results
hnRNP C is less abundant in SK-OV-3 cells than in HeLa cells
Holcik et al. reported that SK-OV-3 cells express decreased levels of hnRNP C compared to H661, H520, and 293 cell lines (Holcik et al., 2003). We evaluated the levels of endogenous hnRNP C in HeLa, SK-OV-3, and 293 cell lines by Western blot analysis (Fig. 1 ). In accordance with earlier studies, we observed that hnRNP C expression in SK-OV-3 cells was decreased approximately 3- to 4-fold compared to 293 cells. HeLa cells express higher levels of hnRNP C protein than SK-OV-3 cells (by ∼ 1.5- to 2-fold), although expression is still lower in HeLa cells than in 293 cells. However, poliovirus growth kinetics in infected 293 cells are comparable to those in HeLa cells (Campbell et al., 2005). Thus, the levels of hnRNP C expression in HeLa cells must be sufficient for poliovirus RNA synthesis and overall replication functions.
Fig. 1.
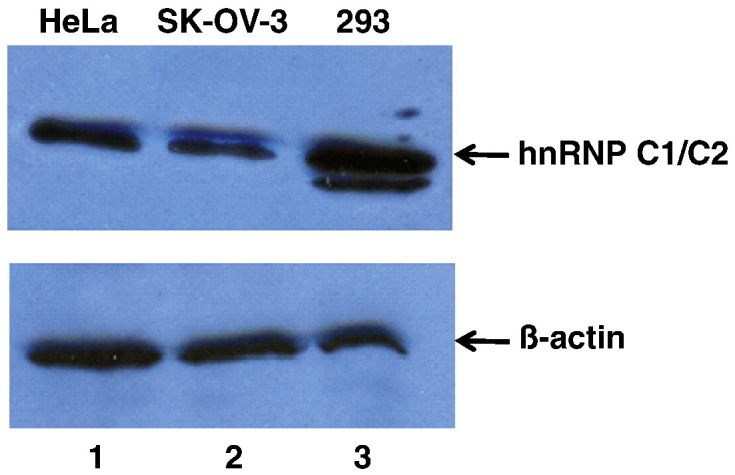
Western blot analysis of hnRNP C protein levels in three different cell lines. Protein samples from whole cell extracts generated from HeLa cells (lane 1), SK-OV-3 cells (lane 2), or 293 cells (lane 3) were subjected to SDS-polyacrylamide gel electrophoresis. Proteins were then transferred to a PVDF membrane by electro-blotting and probed with a monoclonal antibody to hnRNP C1/C2 (top panel) or β-actin (bottom panel) and then a secondary anti-mouse antibody conjugated to horseradish peroxidase (HRP). Due to the abundance of hnRNP C1/C2 in cells and differential antibody sensitivities, less total protein was loaded (10 µg) for hnRNP C1/C2 detection than for β-actin. To detect β-actin as the loading/transfer control, 50 µg of total protein from whole cell extracts was loaded on adjacent lanes of the gel prior to electrophoresis. All samples were loaded and subjected to electrophoresis on the same gel. After electro-blotting, the PVDF membrane was cut into two sections for separate protein detection with the two different monoclonal antibodies. A chemiluminescence substrate (Pierce) was utilized to develop the protein bands detected by antibodies. Band intensities were quantitated using Quantity One software (Bio-Rad).
Kinetics of poliovirus replication are decreased in SK-OV-3 cells compared to HeLa cells
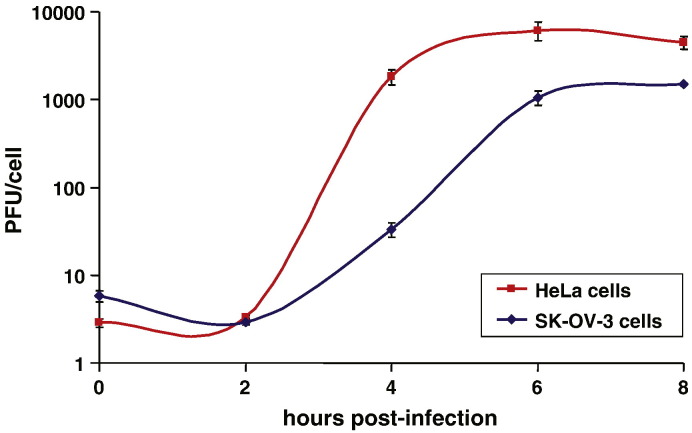
Having confirmed that SK-OV-3 cells express reduced levels of hnRNP C compared to HeLa or 293 cells, we wanted to determine if such a reduction had an effect on poliovirus replication. We expected that the kinetics of replication might be delayed in these cells if they were capable of serving as a permissive host for the virus. Monolayers of SK-OV-3 cells or HeLa cells were infected with poliovirus at a multiplicity of infection (MOI) of 25 to carry out a single-cycle growth analysis. The one-step growth curves generated from the data are shown in Fig. 2 . During the first 8 h after infection by wild type poliovirus, the kinetics of replication of poliovirus in SK-OV-3 cells are significantly delayed when compared to replication in HeLa cells. Maximum yields of virus were seen at 6 to 8 h post-infection in HeLa cells (refer to Fig. 2) compared to about 24 h post-infection in SK-OV-3 cells (data not shown).
Fig. 2.
Single-cycle growth analysis of poliovirus infection of HeLa or SK-OV-3 cells. Monolayers of either HeLa cells (■) or SK-OV-3 (♦) cells were infected with poliovirus at an MOI of 25, and then the total PFU/cell was determined at different times post-infection.
Decreased strand-specific RNA replication in poliovirus-infected SK-OV-3 cells
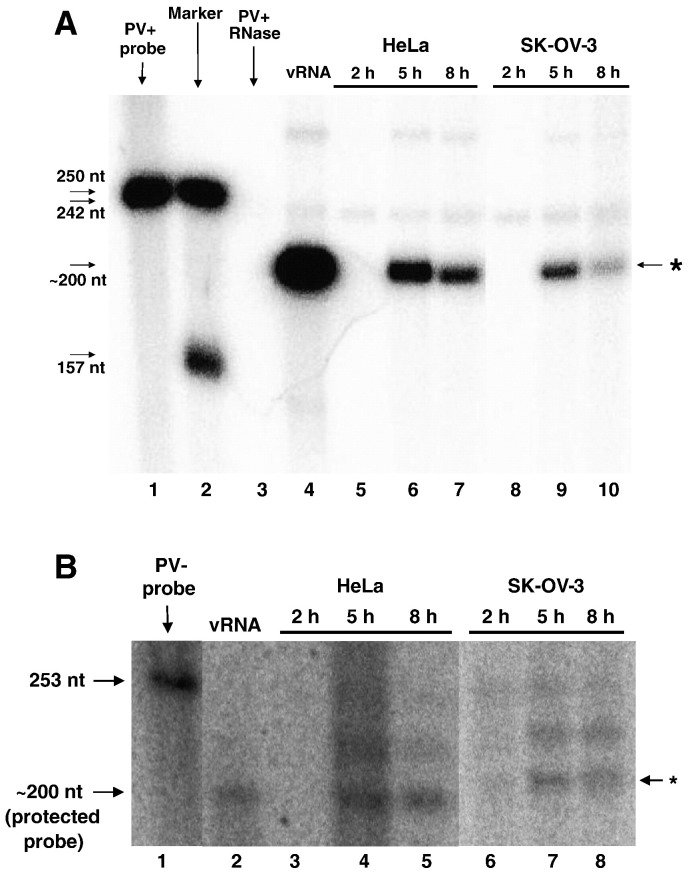
To determine the origin of the virus growth defect in SK-OV-3 cells that was observed in the single-cycle growth assay, HeLa or SK-OV-3 cell monolayers were infected with poliovirus and total cellular RNA was harvested at specific times post-infection (Fig. 3 ). We employed an RNase protection assay to detect virus positive-strand RNA in both cell lines during the course of a poliovirus infection. Using phosphorimager quantitation, the maximum level of positive-strand RNA synthesis in HeLa cells (Fig. 3A, lane 6) compared to SK-OV-3 cells (Fig. 3A, lane 9) was analyzed. Positive-strand viral RNA accumulation in poliovirus-infected SK-OV-3 cells at 5 h post-infection was approximately 45% of the level observed in HeLa cells, suggesting a positive-strand RNA synthesis defect during poliovirus infection of the SK-OV-3 cell line. To further investigate the difference in RNA synthesis for poliovirus negative-strand RNAs (using the total cellular RNA prepared in the same manner described above), we carried out additional RNase protection assays for poliovirus negative-strand RNA using the two-cycle method that has been described previously (Novak and Kirkegaard, 1991). Interestingly, the maximum level of negative-strand RNA accumulation in poliovirus-infected SK-OV-3 cells was approximately 85% of the amount observed for infected HeLa cells (Fig. 3B), suggesting that poliovirus negative-strand RNA synthesis in infected SK-OV-3 cells remains largely intact. Therefore, these data indicate that the low virus yield observed in SK-OV-3 cells infected with poliovirus stems, in part, from an RNA replication defect primarily at the level of positive-strand RNA synthesis.
Fig. 3.
Viral RNA accumulation during poliovirus infection of HeLa or SK-OV-3 cell lines. A. Positive-strand viral RNA accumulation was determined using an RNase protection assay as described in Materials and methods. Untreated radiolabeled probe is shown in lane 1. Marker RNAs (with sizes indicated in nucleotides on the left side of the panel) are shown in lane 2. To ensure the RNase used in the assay was active, probe was digested with RNase in the absence of cellular RNA as a control in lane 3. As a positive control for protection of the target virus RNA, 100 ng of purified virion RNA was used for lane 4. Equal amounts (100 ng) of total cellular RNA were protected by 25 fmol of probe complementary to positive-sense poliovirus RNA sequences 5601 to 5809 (lanes 5 to 10). HeLa cells (lane 5 to lane 7) or SK-OV-3 cells (lane 8 to lane 10) were infected with wild type poliovirus. The RNA harvest times (hours post-infection) are indicated at the top of each lane. The protected probe is indicated by the arrow and asterisk on the right side of the panel. These data are representative of three independent experiments. Gel lanes not relevant to this experiment were cropped from this figure, resulting in a spliced image (between lanes 7 and 8). B. Negative-strand viral RNA accumulation was determined using an RNase protection assay as described in Materials and methods. Untreated radiolabeled probe is shown in lane 1. As a positive control for protection of the target virus RNA, 100 ng of purified pT7NPV1 negative-strand RNA was used for lane 2. Equal amounts (1.3 µg) of total cellular RNA were protected by 75 fmol of probe corresponding to positive-sense poliovirus RNA sequences 5601 to 5809 (lanes 3 to 8). HeLa cells (lane 3 to lane 5) or SK-OV-3 cells (lane 6 to lane 8) were infected with wild type poliovirus. The RNA harvest times (hours post-infection) are indicated at the top of each lane. The protected probe is indicated by the arrow and asterisk on the right side of the panel. These data are representative of three independent experiments. Gel lanes not relevant to this experiment were cropped from this figure, resulting in a spliced image (between lanes 1 and 2, and lanes 5 and 6).
hnRNP C is re-localized from the nucleus to the cytoplasm in SK-OV-3 cells infected with poliovirus
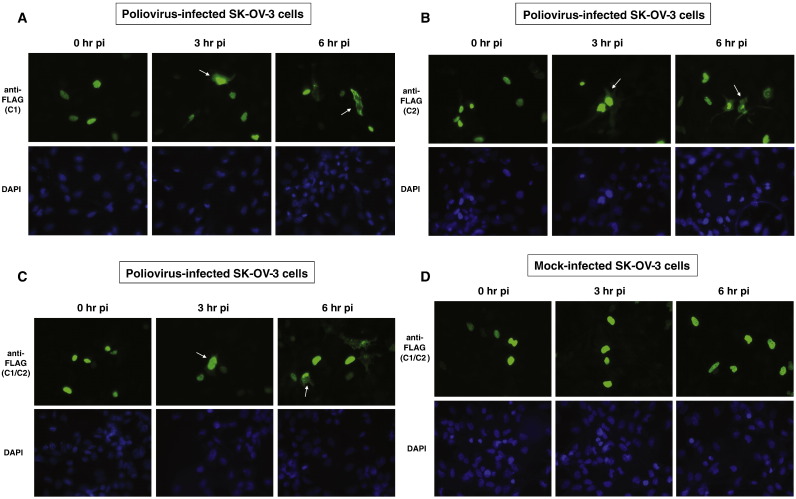
To determine if hnRNP C re-localizes from the nucleus to the cytoplasm during poliovirus infection of SK-OV-3 cells, we used immunofluorescence assays to examine the sub-cellular distribution of FLAG-tagged versions of hnRNP C in transiently transfected/poliovirus-infected SK-OV-3 cells. Expression vectors FLAG-hnRNP C1, FLAG-hnRNP C2, FLAG-hnRNP C1 and C2, or pcDNA 3.1 (vector only) were transfected into SK-OV-3 cells growing on cover slips prior to infection with poliovirus at an MOI of 25. In uninfected cells, endogenous hnRNP C is the only component of the core hnRNP complex that does not shuttle between the nucleus and cytoplasm during interphase, due to the presence of a nuclear retention signal. To calculate transfection efficiency, we determined the number of cells expressing FLAG-tagged protein divided by the total number of cells in several representative fields. Cells were counterstained with DAPI to visualize nuclei and determine total cells per field. Transfection efficiency was determined to be approximately 25%.
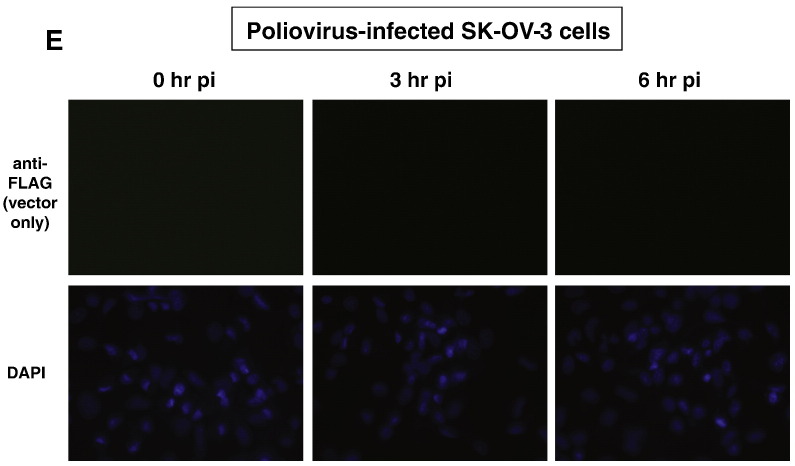
In Fig. 4 , panels A and B, the distributions of FLAG-tagged hnRNP C1 and C2, respectively, were shown to be altered in SK-OV-3 cells as the result of poliovirus infection. At 0 h post-infection, the localization of FLAG-tagged hnRNP C1 and C2 was strictly nuclear by immunofluorescence detection (Fig. 4, panels A, B, and C). This indicates that the hnRNP C proteins expressed in the cytoplasm from the transfected expression vectors were imported into the nucleus and were retained there due to the presence of intact nuclear localization and retention signals. However, at 3 h post-infection, FLAG-tagged hnRNP C1 and C2 were present in the cytoplasm of the poliovirus-infected SK-OV-3 cells, albeit at much lower levels than in the nucleus (Fig. 4, panels A, B, and C; white arrows). By 6 h post-infection, these proteins were almost completely re-localized to the cytoplasm in some cells (Fig. 4, panels A, B, and C; white arrows). In mock-infected SK-OV-3 cells, no change in sub-cellular distribution of hnRNP C was seen at either 3 h or 6 h post-infection (Fig. 4, panel D). This suggests that some disruption of nucleo-cytoplasmic trafficking of proteins occurred in the transiently transfected SK-OV-3 cells as a result of poliovirus infection similar to what has been demonstrated in HeLa cells (Gustin and Sarnow, 2001). The results of our experiments indicated that by 3 h post-infection, transiently-expressed hnRNP C proteins were available in the cytoplasm of approximately 25% of the cells for utilization in poliovirus RNA replication complexes.
Fig. 4.
Sub-cellular localization of FLAG-tagged hnRNP C proteins transiently expressed in SK-OV-3 cells infected with poliovirus or mock-infected. Cells were processed and examined by fluorescence microscopy using a FITC filter at 0, 3, and 6 h post-infection. Green fluorescence indicates localization of FLAG-tagged hnRNP C1 in poliovirus-infected cells (A), FLAG-tagged hnRNP C2 in poliovirus-infected cells (B), FLAG-tagged hnRNP C1 and C2 in poliovirus-infected cells (C), and FLAG-tagged hnRNP C1 and C2 in mock-infected cells (D). Background fluorescence in cells transfected with empty vector pcDNA 3.1 is seen in (E). All cells were counterstained with DAPI to visualize nuclei.
Expression of hnRNP C in SK-OV-3 cells increases the kinetics of poliovirus replication
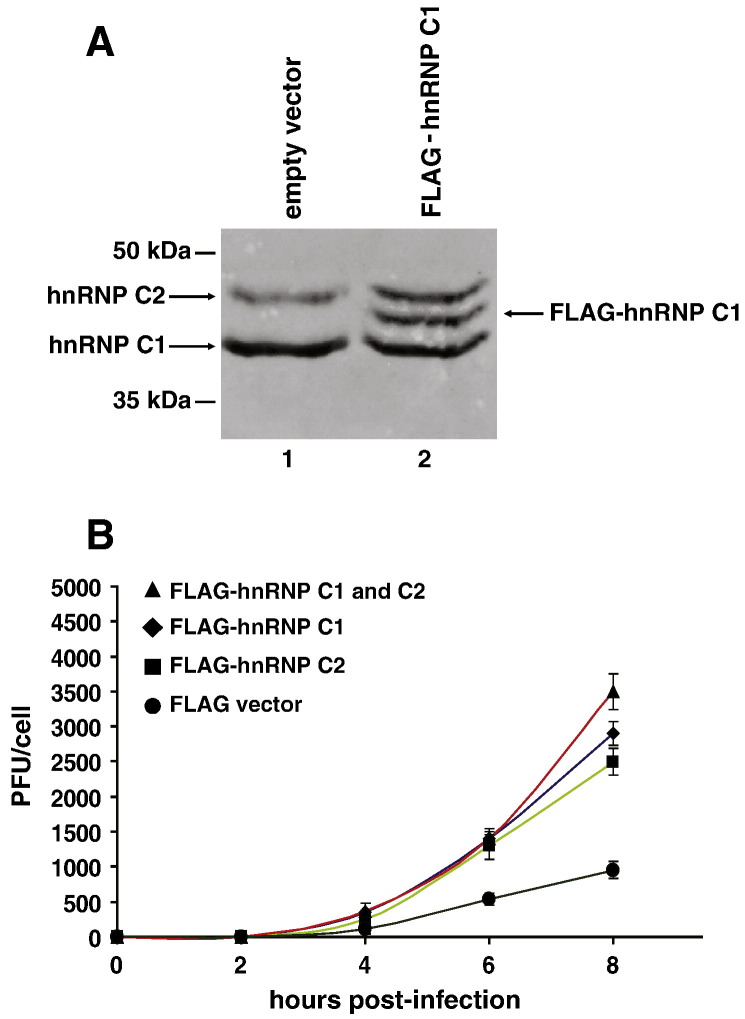
Because we determined that the expression of hnRNP C is decreased in SK-OV-3 cells and then showed that the kinetics of poliovirus replication are delayed in these cells, we hypothesized that increasing the expression of hnRNP C in the SK-OV-3 cells would increase the rate of poliovirus production. We transfected monolayers of SK-OV-3 cells at 50 to 60% confluency with the expression vectors FLAG-hnRNP C1, FLAG-hnRNP C2, FLAG-hnRNP C1 and C2, or pcDNA 3.1 (empty vector). An example of the increased levels of hnRNP C expression following transient transfection of SK-OV-3 cells with a FLAG-hnRNP C1 expression vector is shown in Fig. 5A. Thirty-six hours after transfection, we infected these same cells with poliovirus at an MOI of 25 and then harvested cells and media to determine total virus yield per cell at specific time points post-infection. From this single-cycle growth analysis of poliovirus in SK-OV-3 cells, we generated one-step growth curves (Fig. 5B).
Fig. 5.
Single-cycle growth analysis of poliovirus in transiently transfected SK-OV-3 cells. Monolayers of SK-OV-3 cells were transfected using Fugene (Roche) transfection reagent and plasmid DNAs corresponding to either FLAG-hnRNP C1, FLAG-hnRNP C2, FLAG-hnRNP C1 and C2, or pcDNA 3.1 (empty vector). A. To demonstrate the increased levels of hnRNP C proteins following transient transfection, monolayers of SK-OV-3 cells were transfected with plasmid DNA corresponding to pcDNA3.1 (empty vector), lane 1, or FLAG-hnRNP C1 expression vector, lane 2. At 36 h post-transfection, cells were harvested. Equal amounts of protein (5 µg) taken from whole cell lysates were subjected to SDS-polyacrylamide gel electrophoresis. Proteins were then electro-blotted to a PVDF membrane which was subsequently probed with a monoclonal antibody to hnRNP C (Abcam) and a monoclonal antibody recognizing the FLAG epitope (Sigma-Aldrich). Secondary antibody was goat anti-mouse IgG-alkaline phosphatase diluted 1:5000 in blotting buffer. The blot was developed at room temperature with 1-STEP NBT/BCIP (Pierce). Arrows indicate hnRNP C1 and C2 and the FLAG-tagged hnRNP C1 protein. B. Thirty-six hours after transfection, cells were infected with poliovirus (MOI of 25) and the total PFU/cell was subsequently determined at different times post-infection.
We compared the virus yield in cells transfected with empty vector pcDNA 3.1 to cells transfected with vectors expressing hnRNP C1, C2, or C1 and C2. As shown in Fig. 5B, transient expression of the hnRNP C proteins increases the yield of poliovirus at 6 h and 8 h after infection by approximately two- and four-fold, respectively. This represents an average increase in plaque-forming units per cell (PFU/cell), taking into account that the transfection efficiency was approximately 25%. A ten-fold increase in PFU/cell in one quarter of the cells would equate to an approximate overall average of three-fold increase in PFU/cell.
Discussion
The RNA-dependent RNA polymerase of the positive-strand RNA bacteriophage Q beta consists of four subunits, three of which are host-derived proteins [ribosomal protein S1, protein elongation factor EF-Tu, and protein elongation factor EF-Ts (Blumenthal and Carmichael, 1979, Miranda et al., 1997)]. In a prokaryotic cell, a virus such as Q beta has access to nucleic acid-binding proteins in the absence of a nuclear envelope. Gene-poor cytoplasmic positive-strand RNA viruses (like poliovirus) that infect eukaryotic cells may have evolved successfully due, in part, to an ability to utilize the abundant cellular RNA-binding proteins that are primarily localized in the nucleus. There is accumulating evidence demonstrating that viruses not only use components of nuclear import–export machinery for shuttling of viral macromolecules, but that viral infections disrupt normal host cell nucleo-cytoplasmic trafficking pathways (Schneider and Shenk, 1987, von Kobbe et al., 2000, Petersen et al., 2000, Belov et al., 2000, Gustin and Sarnow, 2001, Gustin and Sarnow, 2002). A block in nuclear import combined with perturbed nuclear export is predicted to result in cytoplasmic accumulation of cellular proteins that could be recruited for viral processes, including amplification of the viral genome. Infection with picornaviruses has been shown to cause this type of trafficking disruption and re-localize nuclear proteins to the cytoplasm that have been implicated in functional interactions with poliovirus RNA (Belov et al., 2000, Gustin and Sarnow, 2001, Meerovitch et al., 1993, McBride et al., 1996, Waggoner and Sarnow, 1998). Particularly relevant to the results of the present study is the finding that hnRNP C is re-localized to the cytoplasm during infection of HeLa cells by poliovirus (Gustin and Sarnow, 2001).
In this manuscript, we have shown that the kinetics of replication of poliovirus are delayed in SK-OV-3 cells, which express decreased levels of hnRNP C compared to HeLa cells. The delay primarily involves a defect in positive-strand RNA synthesis, although a slight defect in negative-strand synthesis was also detected. We have also demonstrated cytoplasmic re-localization of hnRNP C during poliovirus infection in SK-OV-3 cells that we propose results from a disruption of nucleo-cytoplasmic trafficking similar to that shown in poliovirus-infected HeLa cells (Gustin and Sarnow, 2001). The re-localization appears to occur to a lesser extent or perhaps at a slower rate in SK-OV-3 cells than in HeLa cells. Increased expression of hnRNP C, by transient transfection, in SK-OV-3 cells increases the rate of poliovirus production compared to the rate seen in vector-only transfected cells. Unless there are other as yet undetected genetic alterations in SK-OV-3 cells, we would expect that cells stably transfected with these expression vectors would produce yields of poliovirus approaching those seen in HeLa cells for the same times after infection. We plan to develop stable SK-OV-3 cell lines with increased expression levels for hnRNP C and carry out such experiments.
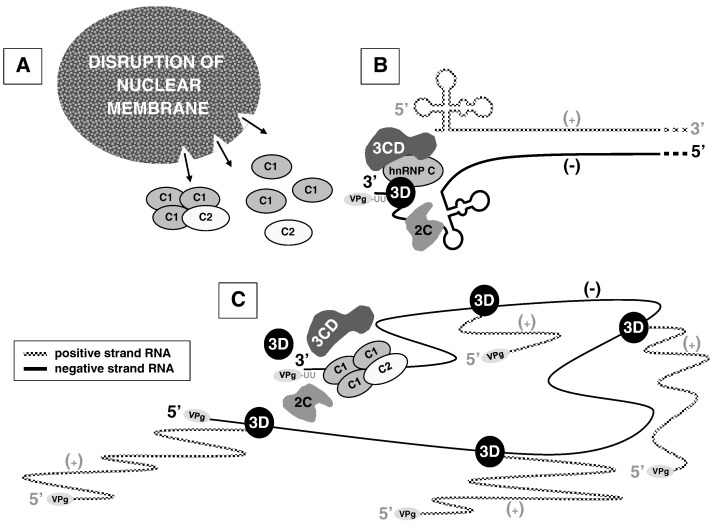
Previously, we demonstrated a functional interaction of hnRNP C with components of poliovirus RNA synthesis initiation complexes (Brunner et al., 2005). Based on the binding of hnRNP C to the 3′ ends of poliovirus negative-strand RNAs and the genetic/functional data presented in the current study, we have proposed a model in which hnRNP C functions as part of a multi-component RNP complex or as an RNA chaperone that facilitates multiple initiations on the template for synthesis of poliovirus positive-strand RNA, as shown in Fig. 6 . For simplicity, we have depicted the template for positive-strand RNA synthesis as single-stranded, although it may be a partially or completely double-stranded template RNA. Indeed, one possible role for hnRNP C is to facilitate or maintain strand separation of the partially double-stranded template as depicted in Fig. 6B. Once a relatively small number of poliovirus negative-strand RNA templates have been synthesized in the cytoplasm of the infected cell, the concentration of hnRNP C1 or C2 (or both) in the cytoplasm may be high enough to allow its binding to the high-affinity site, identified by SELEX (Soltaninassab et al., 1998; Brunner et al., 2005), at the 3′ end. The viral RNA replication machinery and host cell functions could then be recruited to initiate positive-strand RNA synthesis. We propose that hnRNP C proteins contribute to the assembly, remodeling, and re-assembly of the complex that allows for initiation and rapid re-initiations of positive-strand RNA synthesis, especially at mid-to-late times after infection when they are significantly re-localized to the cytoplasm (Fig. 6C).
Fig. 6.
Model for efficient poliovirus positive-strand RNA synthesis. A. hnRNP C proteins are re-localized to the cytoplasm due to disruption of nucleo-cytoplasmic trafficking as the result of poliovirus infection. B. Initiation of positive-strand RNA synthesis occurs on the replicative form involving hnRNP C bound to the 3′ end of negative-strand template RNA, possibly in complexes with the viral 3D polymerase and other viral replication proteins (e.g., 3CD and/or 2C). VPg-UU depicts the genome-linked protein covalently linked to two uridine residues, which serves as a primer for RNA synthesis. Note that in this part of the model, we have shown hnRNP C without designating the specific involvement of hnRNP C1 or C2 (or both). C. As RNA replication proceeds, the efficiency of multiple re-initiations of poliovirus positive-strand RNA synthesis may be facilitated by the binding of hnRNP C proteins to negative-strand template RNA and poliovirus replication proteins (e.g., 3D polymerase, 3CD, and/or 2C). Here we have depicted hnRNP C as a (C1)3C2 heterotetramer, although there are currently no data regarding the multimeric state of hnRNP C bound to poliovirus RNAs.
For picornaviruses and other positive-strand RNA viruses, RNA synthesis is asymmetric. Although we suspect that the double-stranded replicative form (RF) may be the primary template for assembly of the complex that initiates poliovirus positive-strand RNA synthesis, multiple initiations on the subsequently freed single-stranded negative-strand RNA are the means to efficient positive-strand (genomic) RNA synthesis. This leads to the accumulation of large numbers of positive-strand RNA molecules compared to negative-strand RNAs. Such template selection may require the functions of host cell proteins, given that many viral RNA-dependent RNA polymerases (RdRps) require cellular proteins for full replication activity (Lai, 1998, Ahlquist et al., 2003). Since viral RdRps must replicate only viral RNA, but not any of the numerous cellular RNAs present in the infected cell, selection of the appropriate template for RNA-dependent RNA synthesis is critically important for successful viral replication. Cellular proteins may serve as part of the RdRp holoenzyme. Alternatively, host cell factors may bind directly to the viral RNA template, thus directing the RdRp to the template. Our data suggest that this latter possibility is the mechanism of action for hnRNP C in stimulating positive-strand RNA synthesis by the poliovirus RNA replication complex. Presumably, synthesis of RNA strands of each polarity requires unique cellular factors. Interestingly, the RdRp holoenzyme of Q beta can synthesize positive-strand RNA from the negative-strand RNA template, but requires an additional cellular factor, HF1, to direct the synthesis of negative-strand RNA from the phage genome (Barrera et al., 1993).
Brown et al. proposed that an intact 3′ NCR of poliovirus genomic RNA contributes to the efficiency of positive-strand synthesis through sequence and/or structural elements conserved at the 5′ end of the poliovirus negative-strand RNA (Brown et al., 2004). It is hypothesized that RNA viruses may assemble RNA transcription or replication complexes which bring non-contiguous regions of viral RNA together, similar to the interaction between enhancer and promoter elements in DNA-dependent RNA synthesis. One end of the viral RNA could function as an enhancer while the other end could serve as a promoter (Lai, 1998). Long-range interactions bridging the 5′ and 3′ termini of the viral genome via protein–protein contacts have been studied in the context of initiation of negative-strand and positive-strand RNA synthesis. For mouse hepatitis virus, a coronavirus, published data suggest that there is protein-mediated 5′ to 3′ cross talk between the termini of the RNA of both polarities which may be important for RNA transcription and replication (Huang and Lai, 2001). Protein-mediated circularization of the poliovirus positive-strand RNA template has been proposed to occur, thereby increasing the efficiency of negative-strand RNA synthesis (Barton et al., 2001, Herold and Andino, 2001). It is possible that once sufficient concentrations of both hnRNP C1 and C2 proteins are present in the cytoplasm, stable tetramers could form and link the 3′ and 5′ ends of the negative-strand RNA to make re-initiations and positive-strand RNA synthesis more efficient (Fig. 6C).
Intuitively, a lytic cytoplasmic RNA virus with a small genome, such as poliovirus, must rely on cellular factors to increase the efficiency of replication. Cellular factors localized to the cytoplasm may potentiate RNA replication so that viral egress from the infected cell can occur prior to an anti-viral cellular response induced by the transient presence of double-stranded RNA in the cytoplasm. This concept is further supported for poliovirus by our finding that the kinetics of viral replication are delayed in a cell line that expresses decreased levels of hnRNP C.
Materials and methods
Cell culture and cell extracts
HeLa cells were grown in monolayers in DME with 8% newborn calf serum. SK-OV-3 cells (obtained from American Type Culture Collection) were grown in monolayers in RPMI with 12% fetal calf serum. 293 cells were grown in monolayers in DME with 10% fetal calf serum. To prepare whole cell extracts, equal numbers of cells (1 × 107) scraped from monolayers were washed three times in PBS and suspended in 100 μl of protein extraction buffer [50 mM Tris–HCl (pH 8), 5 mM EDTA, 150 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride]. The extract protein concentration was determined by Bradford assay (Bradford, 1976).
Western blot analysis
Total protein samples from whole cell extracts in protein extraction buffer [50 mM Tris–HCl (pH 8), 5 mM EDTA, 150 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride] were boiled in an equal volume of two times Laemmli sample buffer and resolved by SDS-polyacrylamide gel electrophoresis. The resolved proteins were electro-blotted in transfer buffer [25 mM Tris (pH 8.0), 10 mM glycine] onto PVDF membranes. The membranes were blocked by incubating in blotting buffer (2.5% nonfat dry milk, 0.5% Tween 20 in PBS). The blots were then probed with 4F4 monoclonal antibody to hnRNP C (Choi and Dreyfuss, 1984; Abcam), β-actin (Abcam), or the FLAG epitope (Sigma-Aldrich) diluted in blotting buffer.
Transient transfection/infection
SK-OV-3 cells were seeded onto 12 mm glass cover slips in six-well culture plates or grown in monolayers in 60 mm tissue culture dishes. At 50 to 60% confluency, the cells were transfected with FLAG-hnRNP C1, FLAG-hnRNP C2, FLAG-hnRNP C1 and C2, or pcDNA 3.1 DNA (Invitrogen) in Fugene-OptiMEM (6:1 Fugene μl:DNA μg). The expression plasmids coding for the FLAG-tagged hnRNP C proteins were obtained from Dr. Martin Holcik (Holcik et al., 2003). Thirty-six hours post-transfection, cells on cover slips were washed with PBS and infected with a passage three (P3) stock of poliovirus type 1 at an MOI of 25. Following a 30 min adsorption period at room temperature, cells on cover slips were overlaid with four ml of medium (RPMI with 12% fetal calf serum) and then treated as described below.
Immunofluorescence assays
At specific time points post-infection, the transfected/infected cells on cover slips were washed with PBS and fixed/permeabilized with ice-cold methanol. The cells were washed with PBS and incubated with anti-FLAG monoclonal antibody diluted 1:1000 in PBS-BSA (1× PBS with 10 mg/ml BSA), washed three times with PBS and incubated with goat anti-mouse antibody bound to Alexa 488 (Molecular Probes) diluted 1:5000 in PBS-BSA. The cells were counterstained with DAPI (1:1000) in PBS-BSA. The cover slips were inverted into permanent mounting fluid on slides and examined with a Zeiss AxioVert 200 M inverted fluorescence microscope. Images were processed in AxioVision, release 4.2.
Single-cycle growth analysis
Monolayers of SK-OV-3 cells (transfected or mock-transfected) in 60 mm culture dishes at 80 to 90% confluency in RPMI with 12% fetal calf serum were washed with PBS and infected with a P3 stock of poliovirus type 1 at an MOI of 25. Following a 30 min adsorption period at room temperature, unadsorbed virus was removed. The cells were washed three times with PBS and then overlaid with 4 ml of medium (RPMI with 12% fetal calf serum). At specific times after infection, the cell monolayers were scraped off the plates and frozen together with the medium at − 70 °C. Following five freeze–thaws of the samples to release intracellular virus particles, total plaque-forming units were determined by plaque assay on HeLa cell monolayers.
Total cellular RNA extraction
Total cellular RNA was harvested using TriReagent (Molecular Research Center, Inc.). Briefly, HeLa or SK-OV-3 monolayers were infected with poliovirus at an MOI of 25 at 37 °C. At the indicated times post-infection, 1 ml of TriReagent was added to the cell monolayers and the cell lysates were harvested. Total cellular RNA was extracted from the lysates by chloroform extraction of the aqueous phase. The RNA was precipitated with ethanol, pelleted, dried, and resuspended in diethylpyrocarbonate-treated H2O.
RNase protection assays
RNase protection assays to determine positive-strand and negative-strand RNA accumulation were performed as previously described (Novak and Kirkegaard, 1991, Brunner et al., 2005). A 100 ng aliquot of total RNA from infected cells was hybridized with 25 fmol of [32P]UTP-labeled RNA probe, designated the (+) probe, containing nucleotides complementary to positive-sense sequences 5601 to 5809 of poliovirus viral RNA. In addition to virus-specific sequences, the probe contained 49 nucleotides of vector sequence. For negative-strand RNA accumulation analysis, 1.3 µg of total RNA from infected cells was hybridized with 75 fmol of [32P]UTP-labeled RNA probe, designated the (−) probe, containing approximately 50 nucleotides of vector sequence in addition to virus-specific sequence complementary to negative-sense sequences 5601 to 5809 of poliovirus viral RNA. Hybridization reactions were denatured for 10 min at 85 °C in hybridization buffer (1 mM EDTA, 0.4 M NaCl, 80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) [PIPES]; pH 6.4) and then incubated overnight at 60 °C. The mixture was then cooled to room temperature, 300 µl of RNase digestion cocktail (0.5 M NaCl, 5 mM EDTA, 4.5 µg of RNase A/ml, 350 U of T1/ml, 10 mM Tris; pH 7.5) was added, and the reaction was incubated for 1 h at 7 °C. 20 µl of 10% sodium dodecyl sulfate (SDS) and 100 µg of proteinase K were then added, and samples were incubated for 30 min at 37 °C. Protected RNAs were then phenol–chloroform extracted, ethanol precipitated in the presence of 25 µg of carrier tRNA, dried using a lyophilizer, resuspended in 25 µl of formamide loading buffer, and resolved on an 8% polyacrylamide gel containing 7 M urea. For negative-strand RNase protection assays after samples were precipitated in ethanol and dried, samples were resuspended in 50 µl hybridization buffer and the hybridization protocol was repeated. As a positive control for negative-strand RNA accumulation, transcript RNA of the poliovirus negative-strand cDNA (pT7NPV1) was used (Ertel et al., in press). Gels were subjected to phosphorimager analysis on a Personal Molecular Imager FX (Bio-Rad).
Acknowledgments
We thank Martin Holcik for the gift of hnRNP C1/C2 expression plasmids and Gideon Dreyfuss for an initial gift of hnRNP C monoclonal antibodies. We are grateful to Annie Lee and Alan Goldin for help with immunofluorescence microscopy and to Sarah Daijogo and Kerry Fitzgerald for critical comments on the manuscript.
This study was supported by Public Health Service grant AI 22693. J.E.B. was a pre-doctoral trainee supported by Public Health Service training grant GM 07311. J.M.R. was supported, in part, by a grant from the American Asthma Foundation Research Program.
References
- Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera I., Schuppli D., Sogo J.M., Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J. Mol. Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- Barton D.J., O'Donnell B.J., Flanegan J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Evstafieva A.G., Rubtsov Y.P., Mikitas O.V., Vartapetian A.B., Agol V.I. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology. 2000;275:244–248. doi: 10.1006/viro.2000.0427. [DOI] [PubMed] [Google Scholar]
- Beyer A.L., Christensen M.E., Walker B.W., LeStourgeon W.M. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Carmichael G.G. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown D.M., Kauder S.E., Cornell C.T., Jang G.M., Racaniello V.R., Semler B.L. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 2004;78:1344–1351. doi: 10.1128/JVI.78.3.1344-1351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner J.E., Nguyen J.H.C., Roehl H.H., Ho T.V., Swiderek K.M., Semler B.L. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 2005;79:3254–3266. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S.A., Lin J., Dobrikova E.Y., Gromeier M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 2005;79:6281–6290. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.D., Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 1984;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway G., Wooley J., Bibring T., LeStourgeon W.M. Ribonucleoproteins package 700 nucleotides of pre-mRNA into a repeating array of regular particles. Mol. Cell. Biol. 1988;8:2884–2895. doi: 10.1128/mcb.8.7.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Ertel, K.J., Brunner, J.E., Semler, B.L., in press. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J. Virol. [DOI] [PMC free article] [PubMed]
- Gorlach M., Wittekind M., Beckman R.A., Mueller L., Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.E., Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.E., Sarnow P. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 2002;76:8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Gordon B.W., Korneluk R.G. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Lai M.M. Heterogeneous nuclear ribonucleoprotein a1 binds to the 3′-untranslated region and mediates potential 5′–3′-end cross talks. J. Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- McAfee J.G., Shahied-Milam L., Soltaninassab S.R., LeStourgeon W.M. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA. 1996;2:1139–1152. [PMC free article] [PubMed] [Google Scholar]
- McBride A.E., Schlegel A., Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Svitkin Y.V., Lee H.S., Lejbkowicz F., Kenan D.J., Chan E.K., Agol V.I., Keene J.D., Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda G., Schuppli D., Barrera I., Hausherr C., Sogo J.M., Weber H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- Novak J.E., Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J.M., Her L.S., Varvel V., Lund E., Dahlberg J.E. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol. 2000;20:8590–8601. doi: 10.1128/mcb.20.22.8590-8601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S., Dreyfuss G. Cell cycle-regulated phosphorylation of the pre-mRNA-binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol. Cell. Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack R., Goldman R. Synthesis of infective poliovirus in BSC-1 monkey cells enucleated with cytochalasin B. Science. 1973;179:915–916. doi: 10.1126/science.179.4076.915. [DOI] [PubMed] [Google Scholar]
- Roehl H.H., Semler B.L. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative- strand RNA. J. Virol. 1995;69:2954–2961. doi: 10.1128/jvi.69.5.2954-2961.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens B., Tinton S.A., Bruynooghe Y., Parthoens E., Haegman M., Beyaert R., Cornelis S. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.J., Shenk T. Impact of virus infection on host cell protein synthesis. Annu. Rev. Biochem. 1987;56:317–332. doi: 10.1146/annurev.bi.56.070187.001533. [DOI] [PubMed] [Google Scholar]
- Soltaninassab S.R., McAfee J.G., Shahied-Milam L., LeStourgeon W.M. Oligonucleotide binding specificities of the hnRNP C protein tetramer. Nucleic Acids Res. 1998;26:3410–3417. doi: 10.1093/nar/26.14.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C., van Deursen J.M., Rodrigues J.P., Sitterlin D., Bachi A., Wu X., Wilm M., Carmo-Fonseca M., Izaurralde E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Waggoner S., Sarnow P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 1998;72:6699–6709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]