Abstract
Glucocorticoids (GCs) are used in combination therapy for treating acute lymphoblastic leukemia (ALL). In T-cell (CEM-C7) and pre-B-cell (697) ALL cell lines, dexamethasone (Dex) treatment causes an auto-upregulation of glucocorticoid receptor (GR) mRNA transcripts and protein. We hypothesized that there is a threshold level of GR transcripts/protein needed for cells to respond to the apoptosis-inducing effects of hormone. GR knock down using a doxycycline-controllable shRNAmir indicated that the apoptotic response changes from sensitive to resistant with changing GR levels. Titration of the 697 cell GR to equal that of the CEM-C7 T-cell ALL line caused a shift in sensitivity to that seen in CEM-C7 cells. While the same level of GR is required to trigger apoptosis in both T-cell and pre-B-cell ALL lineages, similarities and differences were observed for the regulation of target genes in these lineages. These preliminary gene regulation patterns may lead to the development of a molecular signature for GC-sensitive and GC-resistant leukemia cells.
Keywords: glucocorticoid receptor, threshold, acute lymphoblastic leukemia, apoptosis, auto-regulation
1. Introduction
Glucocorticoids (GCs) are steroid hormones that affect virtually all cell types, and their effects are mediated by the ubiquitously expressed glucocorticoid receptor (GR). The binding of steroid ligand activates the GR, which then functions as a transcription factor to either activate or repress transcription of GC responsive genes (Webster and Cidlowski, 1999; Wright et al., 1993).
Although GC actions are widespread, they are tissue-specific and may elicit opposite responses in different cell types (Gross and Cidlowski, 2008; Keith, 2008). The pro-differentiation and apoptotic effects of glucocorticoids are particularly important in the treatment of acute lymphoblastic leukemia (ALL). Indeed, GCs cause rapid reduction in ALL tumor burden without causing myelosuppression (Langebrake et al., 2002), and numerous studies have shown that a patient’s initial response to GC-therapy is a strong prognostic indicator (Dordelmann et al., 1999; Kato et al., 1993; Pui and Costlow, 1986).
The precise mechanism and molecular trigger for GC-mediated apoptosis are unclear. Elucidating why some ALL patients respond to GC-therapy and others do not remains the focus of many studies, details of which have previously been reviewed (Kofler et al., 2003; Schaaf and Cidlowski, 2002; Sionov et al., 2008; Tissing et al., 2003). One facet of this mechanism in ALL cells is whether GC-mediated GR auto-upregulation is required and important for GC-mediated apoptosis. Many studies show that auto-upregulation of the GR is required for GC sensitivity (Ashraf et al., 1991; Miller et al., 2007; Pedersen and Vedeckis, 2003; Ramdas et al., 1999; Riml et al., 2004) while others suggest that it is not (Bachmann et al., 2007; Tissing et al., 2006). Another point of controversy is whether the absolute GR transcript and/or protein level is important for triggering GC-mediated apoptosis. At least two published studies (Gruber et al., 2009; Tissing et al., 2005) suggest that the absolute level of GR may be the trigger that induces GC-mediated apoptosis, whereas other studies suggest that in some cell types the absolute level of GR does not affect the cell’s sensitivity to hormone (Gomi et al., 1990; Kfir et al., 2007; Wiegers et al., 2001).
In the present study, we aimed to determine whether the GC-mediated auto-upregulation of GR, the absolute expression level of GR, or a combination of both act as the trigger for GC-mediated apoptosis. We used controllable, lentiviral-based shRNA specific for GR knock down as a tool to manipulate GR levels in both T-cell and pre-B-cell ALL model systems to test the hypothesis that there is a certain threshold level of GR that must be reached in order to trigger GC-mediated apoptosis. Further, we used this system to determine if the level of GR needed to trigger apoptosis in T-cell and pre-B-cell lines was the same, and to identify genes that are coordinately regulated and dependent upon GR levels. These studies characterize the threshold level of GR needed for steroid mediated leukemic cell apoptosis. Preliminary studies using patient samples suggest a mechanism that could resolve the question regarding the need for auto-upregulation of GR levels for hormone-mediated apoptosis, and they reveal genes that may be candidates for inclusion in a molecular signature for T-cell ALL hormone-sensitivity.
2. Materials and methods
2.1 Cell lines and cell culture
The human T-ALL cell line, CEM-C7, was a gift from Dr. E. Brad Thompson, University of Texas Medical Branch, Galveston, TX. The human pre-B-ALL cell line, 697, was obtained from Dr. Noreen M. Robertson, Drexel University School of Medicine, Philadelphia, PA. Both of these cell lines and their transduced derivatives (see below) were grown in RPMI 1640 culture medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 2 mM L-glutamine, and they were incubated at 37 °C, at 5% CO2. Cells were treated with dexamethasone (Dex) for 18 hours at a final concentration of 1 μM or an equivalent volume of the ethanol (EtOH) vehicle (control). Transduced controllable cells were pre-treated with varying concentrations (0.5 ng/ml to 1000 ng/ml) of doxycycline (Doxy) before the addition of Dex. HEK-293T cells (ATCC, CRL-11268, Manassas, VA) used for lentiviral production were grown in DMEM culture medium supplemented with 10% fetal bovine serum and 4 mM L-glutamine, and they were incubated at 37 °C, at 5% CO2.
2.2 Lentivirus production, titering, and transduction of target cell lines
The Expression Arrest™ TRIPZ lentiviral shRNAmir system (Open Biosystems, Huntsville, AL) was used to produce lentiviral particles containing a Doxy-controllable GR-specific shRNA construct, according to the manufacturer’s protocol, via HEK-293T transfection. HEK-293T cells were plated in a 100 mm culture dish at a density of 5.5 × 106 cells/plate. Twenty-four hours following initial plating, serum-containing media was aspirated and replaced with serum-free media containing the transfection mixture. The transfection mixture is composed of the transfer plasmid that contains the specific shRNAmir sequence, a mixture of packaging plasmids (supplied by manufacturer), and the Arrest-In™ transfection reagent in a 1:5 DNA:Arrest-In™ ratio. The transfection duration in serum-free media was 6 hours. Following transfection, the serum-free media with the transfection reagents was aspirated and replaced with media containing serum. Lentiviral particles were collected at 2 time intervals (48 and 72 hours) following the addition of fresh serum-containing media. Media containing the lentiviral particles was combined, centrifuged, filtered, and concentrated via ultracentrifugation (70,000 × g for 1.5 hours at 4 °C). The parental HEK-293T cells were used to calculate a functional titer of the viral stocks using the red fluorescent protein (RFP) within the transfer construct as a marker for microscopic visualization. After serially diluting an aliquot of concentrated virus, HEK-293T cells were transduced with each dilution of virus. Forty-eight hours following transduction the number of RFP-positive colonies per well were counted via fluorescent microscopy. Transducing units per milliliter (TU/ml) were calculated by the following equation: (# RFP+ colonies) × (dilution factor) × (40); multiplying by a factor of 40 converts microliters of virus used in the transduction to milliliters for TU/ml. CEM-C7 and 697 parental cell lines were transduced with a multiplicity of infection (MOI) between 0.25 and 0.15, and they were subsequently selected for via puromycin treatment (shRNA-containing provirus contains a puromycin resistance gene) to produce 2 new cell lines, CEM-C7-shGR1-F11 and 697-shGR2-F5. Puromycin killing curves were completed on both parental cell lines to determine the correct puromycin selection dosage: CEM-C7 = 0.35 μg/ml and 697 = 1.0 μg/ml. Clonal populations of each new cell line were obtained by either limited dilution cloning or flow cytometric cell sorting based on RFP positivity using the BD FACSAria instrument (BD Biosciences, San Jose, CA).
2.3 Western blotting
Cells were collected, washed with PBS, and lysed with 1x Laemmli sample buffer containing 1/100 volume of protease inhibitor cocktail (Sigma, St. Louis, MO) and 1 mM PMSF. Proteins were resolved by SDS-PAGE using 10% separating and 4% stacking gels and transferred to 0.45 μm nitrocellulose membranes. The membranes were blocked with 5% non-fat milk and blotted with protein specific antibodies. The antibodies, anti-GAPDH (sc-25778) and anti-hGR (sc-8992) were from Santa Cruz Biotechnologies, Santa Cruz, CA. The anti-TurboRFP (AB321) antibody was from Evrogen, Moscow, Russia. Quantification of western blots was performed with band intensity data obtained from the resident software on the BioRad (Herculaes, CA) Versa Doc instrument, insuring that the pixels were below saturation levels.
2.4 Real time quantitative reverse transcription polymerase chain reaction (QRT-PCR)
The PCR DNA primer and Taqman® probe sequences specific for GR transcripts containing the exon 5/6 junction were previously reported (Pedersen and Vedeckis, 2003). QRT-PCR reactions were normalized to the concentration of their internal 18S rRNA concentration. Real time analysis was completed with the iScript™ One-Step RT-PCR Kit for Probes on the Bio-Rad (Hercules, CA) CFX96 system following the manufacturer’s instructions. Total RNA used in QRT-PCR reactions was isolated from cell samples using the Qiagen RNeasy Mini RNA isolation kit. All RNA samples were treated with DNase I according to the manufacturer’s (Qiagen, Valencia, CA) on-column digestion instructions.
2.5 Cell viability assays
Cell viability was measured via either Trypan Blue exclusion and hemacytometry or by flow cytometry (FACSCalibur, BD Biosciences, Franklin Lakes, NJ) with the Vybrant® Apoptosis Assay Kit #4 (Invitrogen) or by analyzing the sub-G1 DNA content (LSRII, BD Biosciences, Franklin Lakes, NJ) in individual cells. A sub-G1 DNA content is observed in late apoptosis. The Leukemic Cell Survival (LCS) was calculated by the following equation: LCS = (% ViableDex/% ViableEtOH) × 100.
2.6 GeXP multiplex analysis
Multiplex analysis was completed using a custom multiplex containing primers designed using the GenomeLab™ eXpress Profiler software (Beckman-Coulter, Fullerton, CA). Custom multiplex optimization (e.g., primer validation and attenuation) was completed according to the manufacturer’s instructions. The reverse transcription reactions, PCR reactions, and GeXP capillary electrophoresis were completed according to the manufacturer’s protocol using the GeXP Start Kit reagents. Relative RNA expression levels are calculated relative to a pooled RNA standard using the GeXP Quant Tool software and are normalized to 18S rRNA, α-tubulin, β-actin, and HPRT expression levels.
2.7 Patient samples
All patient bone marrow samples were obtained at initial diagnosis by the Hematology/Oncology Department at Children’s Hospital of New Orleans (CHNO) with IRB-approved informed consent. Lymphoblasts were isolated via centrifugation over Ficoll-Paque™ (GE Healthcare, Piscataway, NJ). Isolated lymphoblasts were cultured in RPMI 1640 media supplemented with 10% heat-inactivated FBS and 2 mM L-glutamine. Samples were treated with the same dosages of Dex and EtOH as the cell lines above and incubated at 37 °C, at 5% CO2 for 18 hours before isolating total RNA using Trizol® Reagent (Invitrogen).
3. Results
3.1 GR knock down is controllable via doxycycline treatment in pre-B-cell and T-cell ALL cell lines
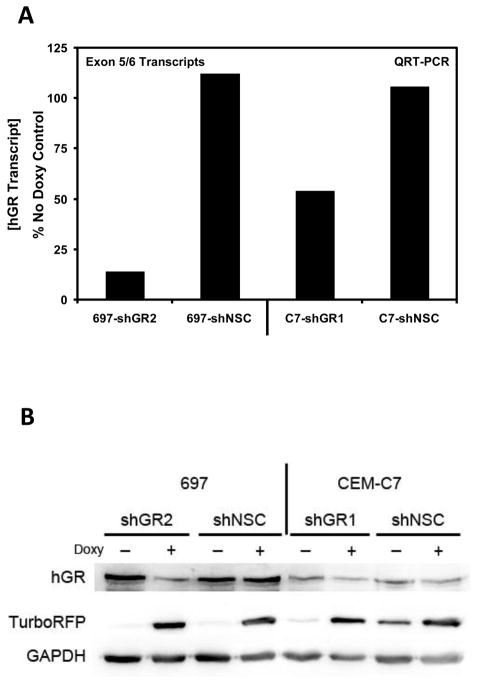
A total of 5 GR-specific shRNAmir transfer plasmids were obtained from the manufacturer (Open Biosystems). Two sequences were chosen based upon their binding locations on the GR transcript. These sequences (Cat. No. V2THS_82725/RHS4696-99634103 and V2THS_239399/RHS4696-99361104) bind to locations near others known to successfully knock down GR (J.R. Schwartz and W.V. Vedeckis, unpublished); they were renamed hGR1 and hGR2, respectively. Transduction of parental 697 and CEM-C7 cells with lentiviral particles containing GR-specific Doxy-controllable shRNAmir sequences resulted in 2 new, stable cell lines. The cell lines, 697-shGR2 and C7-shGR1, exhibited controllable reduction in the levels of both GR mRNA transcripts and protein. In addition to the cell lines with controllable GR knockdown, 2 other cell lines were produced that contained an integrated provirus with a controllable shRNAmir sequence that was not specific for any human gene, the non-silencing control (NSC) cell lines: 697-shNSC and C7-shNSC. When 697-shGR2 and C7-shGR1 were pre-treated for 48 hours with Doxy the GR mRNA transcript level (Fig. 1A) and protein level (Fig. 1B) were knocked down, whereas in the NSC cell lines, Doxy did not change the GR expression levels.
Figure 1.
Controllable GR knock down in ALL cell lines. A) GR mRNA transcript measurements were performed via QRT-PCR with a probe and primer set specific for all GR RNA transcripts (containing the exon 5/6 junction); the resulting measurement is representative of the total GR transcript level. The data are normalized to endogenous 18S rRNA levels in each respective sample and expressed as the percent of the total GR measured in the matched non-Doxy treated sample. Total RNA was isolated following 48 hours of Doxy treatment. The concentration of Doxy used for treating 697-shGR2 and C7-shGR1 cells was 250 ng/ml and 500 ng/ml, respectively; at these concentrations RFP expression was saturated. B) Western blot analysis showing the expression levels of GR, TurboRFP, and GAPDH in 697-shGR2 and C7-shGR1 with and without Doxy treatment (concentrations the same as in A).
Biological analysis of 697 and CEM-C7 cells transduced with either hGR1 or hGR2 showed that, for reasons that are not understood, hGR2 worked better in 697 cells and hGR1 worked better in CEM-C7 cells (data not shown). Furthermore, as indicated in Figure 1, hGR2 in 697 cells reduced GR levels more efficiently than hGR1 did in CEM-C7 cells. It is our experience that the integrated provirus behaves more predictably and as expected in 697 cells than in CEM-C7 cells. One indication for this is seen in Figure 1B in the immunoblot for TurboRFP, a marker for Doxy induction. In 697 cells there is a very pronounced on/off relationship between Doxy treatment and RFP expression in both the experimental and control cell lines. The same relationship is not present in the CEM-C7 cells, as some leakiness is evident. Based upon biological response assays of a number of CEM-C7 clones transduced with a number of different shRNAmir sequences, we feel that there is a higher propensity for recombination, resulting in a disordered provirus, in CEM-C7 cells (data not shown).
3.2 Controllable GR knock down results in controllable GC-resistance in sensitive ALL cell lines
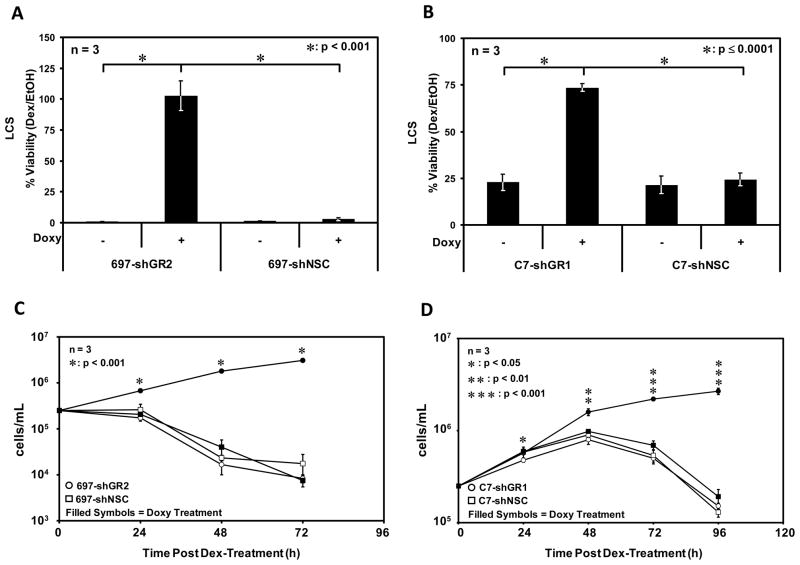
The controllable knock down of GR has important implications related to the biological response of these ALL cell lines to GCs. The fact that corticosteroids kill these cell lines via apoptosis has been validated previously using caspase 3 activation, a DNA fragmentation assay, and annexin V staining in 697 cells (Yamada et al., 2003) and caspase 3 activation, a TUNEL assay, and the accumulation of cells containing a sub-G1 DNA content in CEM-C7 cells (Medh et. al. 2001). We have further confirmed a substantial increase in cells containing a sub-G1 DNA content in these cell lines (see below). In 697 (Fig. 2A & C) and CEM-C7 (Fig. 2B & D) cells treated with 1 μM Dex in the absence of Doxy, an cell death response that replicates that of parental cells is observed, whereas in cell lines transduced with GR specific shRNA and treated with Dex in the presence of Doxy the cell death response is completely abolished. In control cell lines transduced with the NSC shRNA, Doxy treatment did not result in GC-resistance, as the level of GR was not decreased in these cells. The LCS values for C7-hGR1 cells not treated with Doxy and the control cell lines (+/− Doxy) are elevated compared to those in 697 cells. This is not surprising as in the non-transduced/parental state 697 cells apoptose at a faster rate than CEM-C7 cells (see below).
Figure 2.
GR knock down results in resistance to GC-mediated apoptosis. A & C) 697-shGR2 and 697-shNSC cells were pre-treated or not with Doxy and subsequently treated with 1 μM Dex or an equal volume of EtOH. Apoptosis assays were quantitated via Trypan Blue exclusion and hemacytometry 48 hours after treatment. B & D) Apoptosis assays were performed as in A & C with C7-shGR1 and C7-shNSC cells except the data were obtained 72 hours after hormone or vehicle treatment. Error bars represent the standard error of the mean of 3 separate experiments. A 2-sample t-test was used to test for statistically significant differences between GR knock down cell lines +/− Doxy.
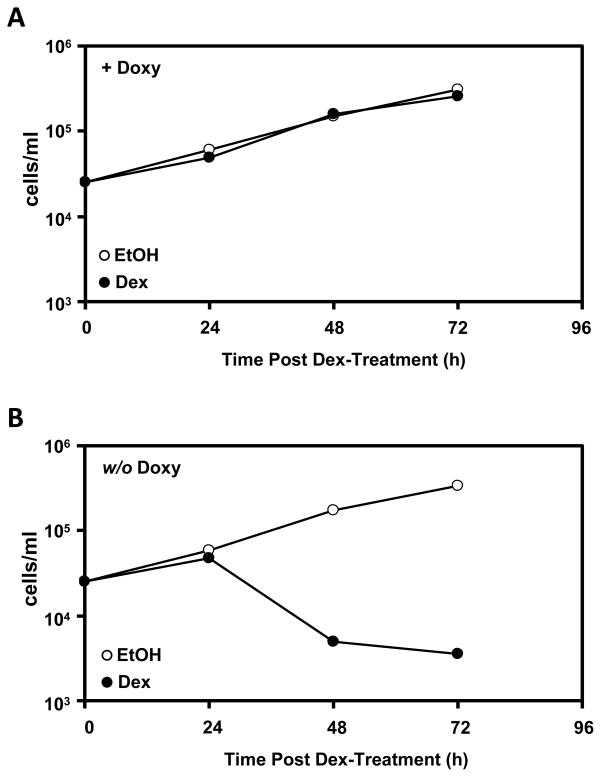
The pTRIPZ lentiviral system for GR knock down is not only controllable, but also reversible (Figure 3). The transduced 697 cells that were pre-treated with Doxy are resistant to GC-mediated apoptosis. When the Doxy is washed out and fresh media not containing Doxy is used to culture this same resistant 697 clone, these cells regain their GC-sensitivity.
Figure 3.
GR resistance is reversible. A) The 697-shGR2-F5 clone was pre-treated with Doxy for 48 hours and subsequently treated with 1 μM Dex or an equal volume of EtOH. After 72 hours of treatment, the Dex-treated cells were washed with PBS (4–5 times over 72 hours) and cultured in fresh media without Doxy. B) After 72 hours RFP expression had nearly ceased and the washed and re-cultured cells were re-treated with either 1 μM Dex or an equal volume of EtOH. Assays were quantitated via Trypan Blue exclusion and hemacytometry.
3.3 Titration with doxycycline results in a titrated GR knock down and a corresponding shift in sensitivity to GC-mediated apoptosis
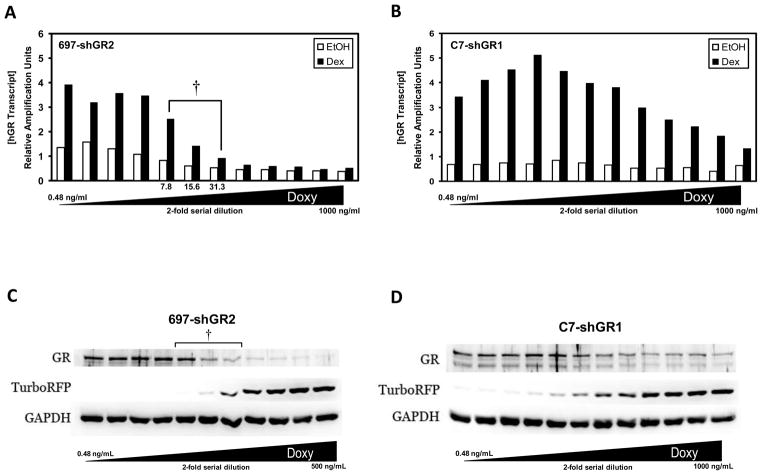
GR transcripts are knocked down in a titrated fashion in 697-shGR2 (Fig. 4A) and C7-shGR1 (Fig. 4B) cells after a 48 hour pre-treatment with a 2-fold serial dilution of Doxy and a subsequent 18 hour Dex treatment. The auto-upregulation of GR transcripts in the 697-shGR2 cell lines seems to be abolished when treated with Doxy concentrations ≥62.5 ng/mL while GR transcript up-regulation in the C7-shGR1 cell line is not lost even when treated with a Doxy concentration of 1000 ng/mL. We hypothesize that because the GR knock down in the CEM-C7 cells is less efficient than in 697 cells (Fig. 1) enough receptor always remains to bind Dex and drive GR auto-upregulation of its own gene transcription. Western blots using cellular lysates from 697-shGR2 (Fig. 4C) and C7-shGR1 (Fig. 4D) cells indicate that GR protein levels are also knocked down in a titrated fashion after pre-treatment with increasing Doxy concentrations and an 18 hour Dex-challenge.
Figure 4.
GR specific shRNAmir sequences reduce both mRNA and protein expression. 697 and CEM-C7 cells stably transduced with a GR-specific shRNAmir lentivirus were treated with 2-fold serial dilutions of Doxy, starting at a concentration of 1 μg/mL (rightmost sample). The total GR (containing the exon 5/6 junction) transcript concentration was measured with QRT-PCR in 697-shGR2 (A) and C7-shGR1 (B) cells after 48 hours of Doxy pre-treatment followed by 18 hours of Dex treatment. (†) notes, in A, the Dex-induced transcript level after pre-treatment with Doxy that corresponds to the same Doxy concentrations within which the biological response switches (see Fig. 5A ). C & D) Expression levels of GR, turboRFP, and GAPDH proteins were analyzed by western blotting following the same treatment regimen as in A and B. During these studies one 697-shGR2 sample was lost; therefore, in C, the highest Doxy concentration analyzed was 500 ng/mL. This does not affect the interpretation of the data.
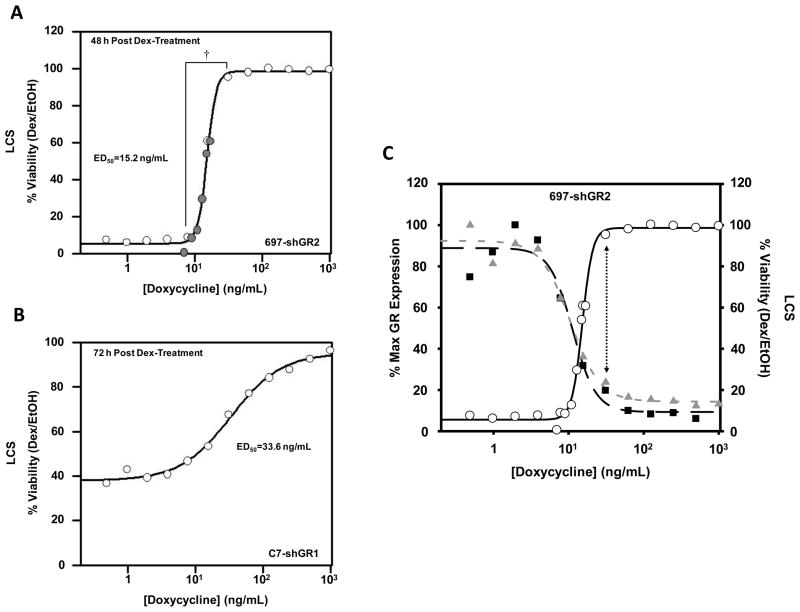
Cell viability was measured either 48 hours (697-shGR2) or 72 hours (C7-shGR1) following 1 μM Dex-treatment of cell lines pre-treated with various Doxy concentrations. The apoptotic response to GC-treatment in 697-shGR2 cells switches from GC-sensitivity to GC-resistance within a narrow range (~7.8–31.3 ng/mL) of Doxy concentration and, therefore, GR level (compare bracketed range in Figs. 4A, 4C, and 5A). In C7-shGR1 cells (Fig. 5B) this biological switch was not as abrupt. Furthermore, the calculated ED50 (Doxy concentration at which cell viability is 50%) was approximately 2-fold higher in the C7-hGR1 cells than in the 697-shGR2 cells. Two factors may explain the difference in apoptosis sensitivity between the 697-shGR2 and C7-shGR1 cells with the Doxy titration: 1) CEM-C7 cells contain one wild-type allele and one mutated GR allele that results in a non-functional protein unable to bind ligand (Ashraf and Thompson, 1993; Palmer et al., 1992; Powers et al., 1993); and 2) as shown in Figure 1, the shGR1 sequence knocks down GR less efficiently in CEM-C7 cells than the shGR2 sequence does in 697 cells, and the amount of Dex-induced up-regulation is not affected to the same degree in the middle range of the Doxy dilution in the C7-shGR1 cells (Fig. 4B). Therefore, in the C7-shGR1 cell line, the apoptotic response after pre-treatment with the full 2-fold Doxy dilution series looks similar to that in the 697-shGR2 cell line when pre-treated with a Doxy concentration range from 7.8 to 31.3 ng/mL, perhaps because the shRNAmir sequences are not specific for functional transcripts, and, consequently, knock down both functional and non-functional transcripts (and therefore protein). This, in addition to a less efficient shRNA, results in less overall functional GR knock down, therefore, probably requiring the wider range of Doxy concentration to achieve the same apoptotic response to Dex. A mathematical analysis (Fig. 5C) of the Dex-induced GR transcript and protein expression and cell viability in the 697-shGR2 cells suggests that a GR transcript and protein level above ~20–25% of maximum Dex-induced GR expression levels is the amount of receptor needed to trigger some degree of apoptosis; if GR expression falls below this level the 697-shGR2 cells are completely resistant to GC-mediated apoptosis.
Figure 5.
C) Compilation of the 697 cell data. Cell viability (open circles) and Dex-induced GR transcript (gray triangles) and protein expression (black squares) are plotted versus the pre-treatment Doxy concentration used in the experiment with 697-shGR2 cells. Expression levels of GR transcripts and protein shown are plotted as the calculated percentages of the maximum Dex-induced expression values from QRT-PCR (transcripts) and densitometery (protein) performed on the western blot shown in Figure 4. The vertical dotted arrow indicates that a GR expression level above ~20–25% is required for triggering some degree of GC-mediated apoptosis in these cells. The curve fits are theoretical calculations via the same equation used in Figure 5A. The calculated Doxy ED50 values for GR transcripts, GR protein, and cell viability are 10.5 ng/mL, 11.3 ng/mL, and 15.2 ng/mL, respectively.
3.4 GC-sensitivity differences in 697 pre-B-ALL cells and CEM-C7 T-ALL cells can be abrogated by equalizing functional GR levels
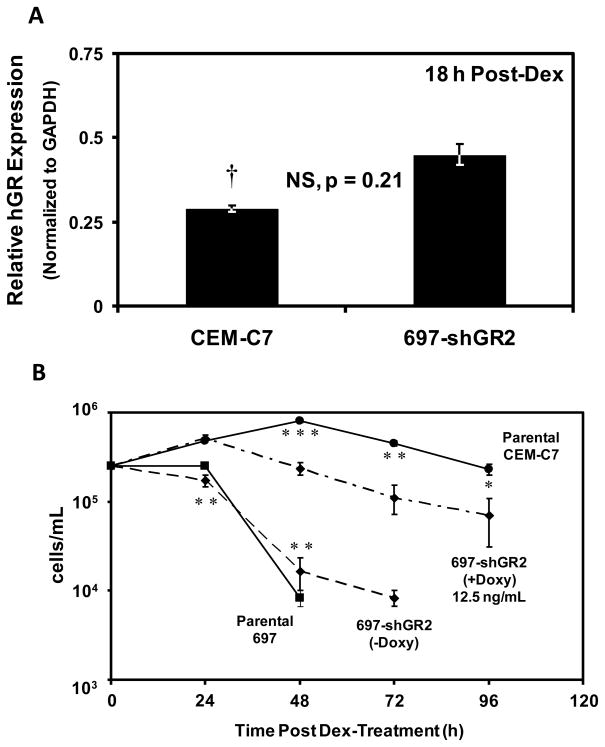
As discussed previously, 697 cells contain 2 functional GR alleles and, therefore, the full diploid complement of functional GR protein, while CEM-C7 cells contain only a haploid level of functional GR protein, as one allele is mutated. The delay in the appearance of apoptosis in CEM-C7 cells as compared to 697 cells may be attributed to the difference in functional GR levels and the requirement of a greater amount of auto-upregulation in CEM-C7 cells to reach the putative GR threshold level needed to trigger GC-mediated apoptosis. We hypothesized that if the levels of functional GR in 697 cells were manipulated such that they were equal to those in CEM-C7 cells, then the 2 cell lines would respond to Dex-treatment similarly. Figure 6A shows densitometric analyses of western blots comparing GR levels in 697-shGR2 cells pre-treated with 12.5 ng/ml Doxy followed by induction with 1 μM Dex to the GR levels in parental CEM-C7 cells following Dex induction. The densitometry values measured from the CEM-C7 cell samples were divided by 2 in order to account for their functional GR haploinsufficiency. There was not a statistically significant difference between these levels. The apoptosis analysis seen in Figure 6B shows that, in support of our hypothesis, the apoptotic response of 697-shGR2 cells treated with 12.5 ng/ml Doxy now more closely resembles that of parental CEM-C7 cells than 697-shGR2 cells not pre-treated with Doxy or parental 697 cells, particularly in the first 24–48 hours following Dex treatment. Thus, the GR threshold required to trigger apoptosis appears to be the same in T- and pre-B- ALL cell lines.
Figure 6.
Manipulation of functional GR level changes the apoptosis rate. A) Densitometry values of 3 biological replicates of Dex-treated 697-shGR2 cells pre-treated with 12.5 ng/mL Doxy and of Dex-treated parental CEM-C7 cells. Values are expressed as relative GR expression, normalized to GAPDH protein expression. The p value is that of a 2-tailed t-test assuming equal variances. (†) indicates that the CEM-C7 values were divided by 2 because CEM-C7 cells are functionally haploinsufficient for GR. B) Cell viability was measured by Trypan Blue exclusion and hemacytometry of cells treated as in A. Error bars represent the SEM; n=3 for all cell lines except 697-shGR2 (12.5 ng/mL Doxy) where n=4. The parental 697 cell line is shown to demonstrate that the cell death rate of the 697-shGR2 derivative cell line is phenotypically comparable to the parental cell line in the absence of Doxy (no GR knock down). Using the same statistical analysis as in A, 697-shGR2 (12.5 ng/mL Doxy) was compared to either parental CEM-C7 or 697-shGR2 (-Doxy). (*) p<0.05. (**) p<0.01. (***) p<0.001.
3.5 An intracellular GR threshold level must be reached in order for steroid-induced apoptosis to occur
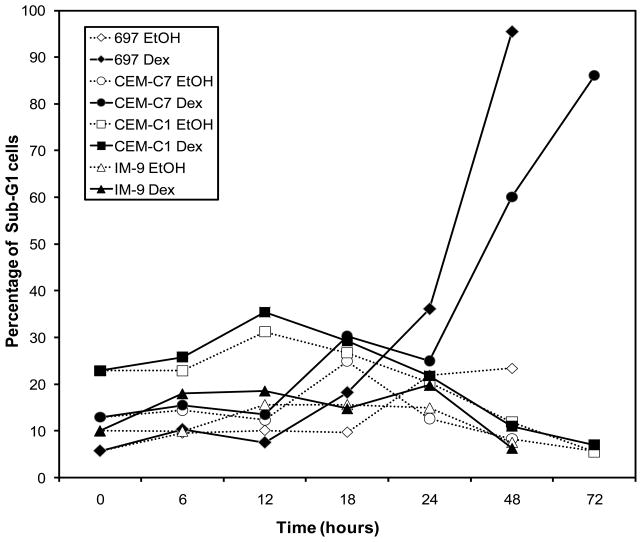
The above results agree with previous studies demonstrating that a certain level of GR is needed for a particular cell to respond (undergo apoptosis) to GC treatment (Gruber et al., 2009; Ramdas et al., 1999; Tissing et al., 2005). To explore this further, we used four established cell lines and four fresh T-ALL patient samples. Two cell lines, CEM-C7 and 697, underwent apoptosis as evidenced by an increase in cells containing a sub-G1 content of DNA after Dex treatment, while this was not seen in two cell lines that are resistant to GC-mediated cell death, the CEM-C1 TALL line (derived from the sensitive CEM-C7 line) and the IM-9 lymphoblastoid cell line (Fig. 7). The cell lines and the four T-ALL patient samples were also evaluated by examining the number of living cells after Dex treatment using the Vybrant #4 Apoptosis Assay kit (Table 1). The cell lines exhibited the same hormone sensitivity seen using the DNA content flow cytometry assay, and all four patient samples were also sensitive to steroid-mediated killing.
Figure 7.
Cell cycle analysis of cell lines. Cells were treated for the indicated times with either 1 μM Dex or an equal volume of the EtOH vehicle. After staining with propidium iodide, cell cycle analysis was performed to analyze the DNA content in individual cells. The percentage of sub-G1 cells as compared to the total number of cells analyzed for each sample is shown.
Table 1.
Dex-mediated Killing of Cell Lines And Patient Samples (LCS*)
| Cell Line or Patient Sample Time of Treatment (Hours) | 48 | 72 |
|---|---|---|
| 697 | 4.2% | 1.6% |
| CEM-C7 | 85.8% | 22.2% |
| CEM-C1 | 92.9% | 95.3% |
| IM-9 | 99.1% | 98.4% |
| **TALL1 | 22.1% | - |
| **TALL2 | - | 34.9% |
| **TALL3 | - | 10.2% |
| **TALL4 | - | 18.2% |
Leukemic Cell Survival was calculated as described in Materials and Methods
To insure a clear result, instead of analyzing the LCS after 48 hours of Dex-treatment of the patient samples (as was done initially), it was decided to use 72 hours of treatment to assay sensitivity to steroid-mediated cell death.
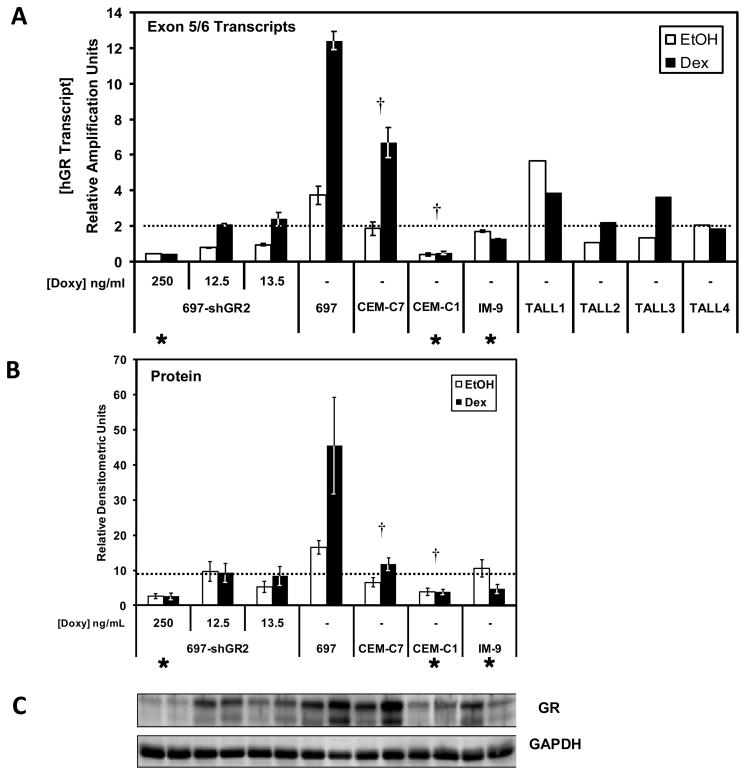
Results from the cell viability assays and QRT-PCR of the 4 fresh T-cell ALL patient samples suggest that not only is the basal level of GR important, but, depending on basal GR levels, autoregulation of the GR can be equally important for predicting the responsiveness of lymphoblasts to GC-therapy. Shown in Figure 8A are the basal and GC-induced levels of total (Exon 5/6-containing) GR transcript levels in various cell lines and in the 4 patient samples. In all cells that undergo cell death after GC treatment, the proposed GR mRNA threshold is surpassed. Conversely, the resistant cell lines analyzed (697-shGR2 cells in 250 ng/ml Doxy; CEM-C1; IM-9) did do not reach the GR transcript threshold basally or after Dex induction and were resistant. However, the 697-shGR2 cells in which the GR was titrated with Doxy (12.5 and 13.5 ng/ml) to resemble CEM-C7 cell hormone-sensitivity, did reach this threshold, but only after Dex-mediated auto-upregulation of the GR transcript levels. Obviously, the actual effector of steroid-mediated cell death is the GR protein, not the mRNA transcripts. This can be seen in the titration of GR transcript levels in the 697-shGR2 cells, where the intracellular GR protein levels closely follow the GR transcript levels (Fig. 5C). We have also assayed the GR protein levels in the same cell lines, and under the same conditions, as those used for the transcript measurements. While the correlation is not identical, the overall GR protein patterns (Fig. 8B) are remarkably similar to the transcript profile, and a threshold line that distinguishes sensitive from resistant samples can be drawn, which closely mimics that obtained when mRNA transcript levels (Fig. 8A) are used. Because of limited amounts of sample, not all of the patient samples could be analyzed at the protein level, and so these data could not be presented. Taken together, data obtained with parental cell lines (697; CEM-C7; CEM-C1; IM-9), derivative cell lines in which the GR level can be experimentally reduced (697-shGR2), and 4 T-ALL patient samples suggest that a minimal level of GR mRNA and protein, which we refer to as a threshold, must either be present basally or must be reached via GR auto-upregulation in order to trigger apoptosis, and this threshold appears to be similar in a variety of ALL blasts, including those of the pre-B-cell and T-cell lineages.
Figure 8.
Basal versus GC-induced GR transcript and protein levels in cell lines and patient samples. A) QRT-PCR analysis was performed using the probe specific for GR transcripts containing the exon 5/6 junction in various cell lines and 4 fresh, ALL patient samples. All cells were treated for 18 hours with either 1 μM Dex or an equal volume of the EtOH vehicle. Data are expressed as relative amplification units (RAU), and all samples were normalized to their respective internal 18S rRNA expression level. Symbols: (✱) denotes cells that are resistant to GC-mediated apoptosis; all other cells are sensitive. (†) indicates that both CEM-C7 and CEM-C1 RAU values were divided by 2 as both are functionally haploinsufficient for GR. The dotted line marks the apparent GR threshold that must be reached to trigger GC-mediate apoptosis. Error bars represent the SEM of 3 separate biological replicates where possible (i.e., there are no error bars for the patient samples as the RNA samples were from a single treatment experiment due to the minimal amount of patient material received). B) Quantitative western blotting of the GR proteins bands after SDS-PAGE. All cells were treated with 1 μM Dex or an equal volume of the EtOH vehicle for 18 hours prior to extraction. Three separate experiments were performed and the data are the means ± the SEM. C) Representative western blot of the cell line samples. For B) and C), there was not enough material from the patient samples to allow quantitative western blotting to be performed.
3.6 Multiplex analysis identifies several genes affected in a coordinate manner in response to titrated GR knock down in cell lines and patient sample analysis identifies candidate genes of a molecular signature for hormone-sensitivity
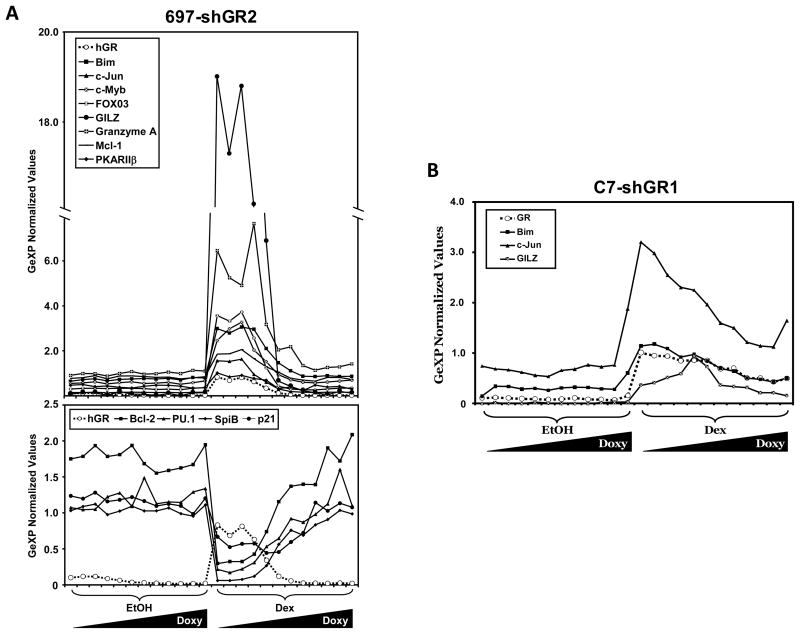
We have customized a 28 gene multiplex using the GenomeLab™ eXpress Profiler software. Twenty-four genes (Table 2) were chosen because of their known or potential importance in GC-mediated apoptosis. Also present in our custom multiplex are 4 typical normalization genes: hypoxanthine guanine phosphoribosyltransferase (HPRT), β-actin, 18S rRNA, and α-tubulin. GeXP gene expression data were normalized to all 4 reference genes simultaneously using the GeXP Quant Tool software. RNA was isolated from 697-shGR2 and C7-shGR1 cells following 48 hours of pre-treatment with the 2-fold Doxy dilution series and 18 hours after EtOH or Dex treatment. The normalized expression values for the genes for which the regulation was substantially altered by the titration of GR levels in the 697-shGR2 cell line are shown in Figure 9A. The genes that were up-regulated following Dex treatment were Bim, c-Jun, c-Myb, FOXO3a, GILZ, granzyme A, Mcl-1, and PKARIIβ. The genes that were down-regulated following Dex treatment were Bcl-2, PU.1, SpiB, and p21. Interestingly, these effects were seen in a titrated fashion which corresponded to the titrated knock down of GR. For 3 genes (Bim, c-Jun, and GILZ) the titrated up-regulation behavior was common between 697-shGR2 and C7-hGR1 cells (Fig. 9B).
Table 2.
Glucocorticoid-Mediated Changes in Gene Expression in T-Cell Acute Lymphoblastic Leukemia Patient Samples
| Gene | TALL1 | TALL2 | TALL3 | TALL4 |
|---|---|---|---|---|
| Bim | 19.87 | 1.44 | 9.08 | 3.66 |
| GILZ | 5.79 | 11.60 | 27.91 | 11.92 |
| c-Jun | 3.29 | 1.54 | 3.20 | 4.88 |
| PKARIIβ | 19.18 | 1.88 | 1.58 | 1.57 |
| Total GR | 1.19 | 2.26 | 3.78 | 1.58 |
| PU.1 | 1.55 | 0.88 | 1.44 | 2.74 |
| JunB | 1.03 | 1.02 | 2.49 | 2.71 |
| p21 | 0.76 | 0.46 | 1.84 | 1.97 |
| FOXO3a | 1.41 | 1.22 | 1.29 | 1.32 |
| Granzyme A | 1.33 | 0.64 | 1.28 | 2.42 |
| Bcl-2 | 1.36 | 0.97 | 2.51 | 0.83 |
| c-Myb | 1.66 | 0.74 | 1.07 | 0.90 |
| Mcl-1 | 1.02 | 0.95 | 1.25 | 1.54 |
| MKK3 | ✘ | 0.63 | 1.40 | 0.89 |
| Granzyme B | 1.59 | 0.46 | 0.94 | 0.62 |
| p16INK | ✘ | 0.30 | 1.40 | 0.38 |
| AIF1 | 0.79 | 0.65 | 0.95 | 0.75 |
| PUMA | 1.16 | 0.16 | 1.17 | 0.74 |
| p38 MAPK | 0.68 | 0.96 | 1.23 | 1.20 |
| c-Abl | 1.02 | 0.90 | 1.24 | 1.12 |
| Bcl-XL | 0.86 | 1.14 | 1.10 | 0.94 |
| ERK2 | 1.15 | 0.86 | 0.99 | 1.17 |
| Spi-B | 1.09 | 0.90 | 0.94 | 0.99 |
| PGC-1 | ✘ | ✘ | ✘ | ✘ |
The absence of an entry (✘) indicates that the expression of the transcript was not detectable. All values are normalized to a set of 4 normalization genes (see text), and the expression of the transcript in the presence of hormone is divided by its expression in cells treated with the ethanol vehicle alone (control). The numbers, thus, are fold changes in dexamethasone-treated samples compared to control values. The cut-off values are as follows: up-regulated transcripts (≥1.3-fold; red); no change in transcript levels (> 0.7-fold and <1.29-fold; yellow); down-regulated (<0.7-fold; green).
Figure 9.
Multiplex analysis of the gene expression in 697-shGR2 and C7-shGR1 cells titrated with a 2-fold serial dilution of Doxy, followed by EtOH or Dex. A) Top panel: genes that are up-regulated with GR following 1 μM Dex treatment in 697-shGR2 cells. Bottom panel: genes that are down-regulated oppositely to the upregulation of GR following 1 μM Dex treatment in 697-shGR2 cells. Irrespective of the direction, the regulation of the genes shown is in a titrated manner that corresponds to the titrated GR levels. B) Genes that are up-regulated with GR following 1 μM Dex treatment in C7-shGR1 cells. Data are expressed as GeXP normalized expression values; they are normalized to HPRT, β-actin, 18S rRNA, and α-tubulin simultaneously and relative to a total RNA standard curve.
Finally, we examined the 4 T-ALL patient samples in a preliminary attempt to identify genes that might be able to be used as a molecular signature to predict sensitivity to glucocorticoid-mediated apoptosis (Table 2). Four transcripts were up-regulated above the cutoff value of 1.3-fold in all 4 patient samples: Bim, GILZ, c-Jun, and PKARIIβ. Two other transcripts were up-regulated in three of the four patient samples: the GR itself and PU.1. There was no consistent pattern of up- or down-regulation for the remaining genes in the multiplex. Thus, there are 6 genes whose expression may be potentially helpful in determining if a T-ALL will be responsive to steroid-mediated apoptosis.
4. Discussion
In 2 acute lymphoblastic leukemia (ALL) model cell lines, 697 (pre-B-cell) and CEM-C7 (T-cell), stable integration of Doxy-controllable GR-specific shRNA sequences allows for controllable resistance to GC-mediated apoptosis. These results show that the absolute threshold level of GR is important for the biological response in at least these 2 cell lines, most dramatically seen in 697 cells where the switch from GC-sensitivity to GC-resistance occurs within a very narrow range of GR. This is the first time that such studies have been performed in a pre-B-ALL cell line system. These results are consistent with a previous study, where controllable overexpression of GR converted GC-resistance in a T-ALL, CEM-C7 derivative cell line to GC-sensitivity (Gruber et al., 2009). Thus, by complementary means (introduction of the GR into a resistant ALL cell and the knock down of the GR in a sensitive ALL cell line) a threshold for GR that is required for steroid-mediated apoptosis has been demonstrated. An important unanswered question was whether or not the threshold level of GR required for triggering apoptosis is the same in acute lymphoblastic leukemia cells of different lineages. Indeed, 697 pre-B-ALL cells have more rapid cell death kinetics than CEM-C7 T-ALL cells. By manipulating the GR level in 697 pre-B-cell ALL cells we have successfully changed the GC-responsive phenotype to more closely resemble that of CEM-C7 T-ALL cells, which contain only a haploid amount of functional GR (Ashraf and Thompson, 1993; Palmer et al., 1992; Powers et al., 1993). Thus, it appears that the same amount of GR is sufficient to trigger apoptosis in ALL cells of different lineages (T-cell, Pre-B-cell), at least for the 2 cell lines tested. Even though the panel of genes that are regulated in these 2 cell types is not identical (see below), the same amount of activated receptor seems to be sufficient in order to interact with the GR-binding sites in the hormone-regulated genes involved in hormone-mediated apoptosis in both cell types.
A major controversy still exists regarding whether or not the amount of GR present in ALL blasts is important for steroid sensitivity in ALL patients and if there is a necessity for steroid-mediated up regulation of GR gene expression (auto-upregulation). While parental 697 cells appear to have a basal level of GR above the required threshold and should not require GR upregulation (even though it occurs) to trigger apoptosis (Fig. 8), the basal GR in CEM-C7 cells appears to fall just below the threshold, thus requiring the observed auto-upregulation. This is in accord with previous findings (Ramdas et al., 1999) in which a CEM-C7 cell derivative cell line was resistant to GC-mediated apoptosis when the GR was held at the basal, uninduced level. Interestingly, auto-downregulation, or the absence of upregulation, may not affect a cell’s GC-sensitivity if the basal GR amount already exceeds the required threshold level (e.g., TALL1 and TALL4). These results seem to contradict those of Tissing et al. (2006), where neither the baseline level of the 5 different GR transcripts nor the up-regulation of GR transcripts by steroid seemed to be related to resistance. This apparent contradiction may be due to the fact that, in nearly all instances in their study, the percentages of upregulation of the GR transcripts (rather than the absolute level of GR) were compared between sensitive and resistant patient samples. An analysis only taking into consideration fold-change will miss cases similar to TALL1 and TALL4, where the basal level already exceeds the threshold. In 697 cells, this threshold appears to occur within a relatively narrow range of intracellular GR levels (Fig. 5A). A decrease in GR protein to about one-fourth to one-fifth of the basal level (see Figs. 4 and 5) is sufficient to cause a complete conversion of the cell from hormone-sensitivity to steroid-resistance with regard to apoptosis. These results, in both cell lines and fresh patient samples, strongly suggest that a threshold level must be reached to trigger GC-mediated apoptosis, and in some (but not all) cases, GC-mediated GR auto-upregulation may be the mechanism by which that level is achieved. Thus, we believe that there is no contradiction in the published results – the important criterion is that the GR must exceed the required threshold level to trigger apoptosis and this may result from the elevated basal expression of GR or from steroid-mediated auto-upregulation of sub threshold, basal GR levels to amounts exceeding the threshold level.
Using GeXP multiplex analysis of 697-hGR2 and C7-hGR1 cells, we have shown that the level of GR either directly or indirectly affects several genes (some known to be important in the apoptotic pathway), importantly, in a GR-titrated fashion. The upregulated genes that are common in the two cell lines, c-jun, Bim, and GILZ, have previously been implicated in steroid-mediated apoptosis. The coordinate upregulation of c-jun and the GR was demonstrated many years ago (Zhou and Thompson, 1996; Barrett et al., 1996), and the suppression of c-jun upregulation using an antisense c-jun construct blocked steroid-mediated apoptosis (Zhou and Thompson, 1996). Glucocorticoid induction of Bim (a BH3-only member of the bcl-2 family) expression has been implicated in GR-mediated apoptosis in both T-cell ALL (Wang et al., 2003; Zhang and Insel, 2004) and pre-B-cell ALL (Abrams, et al., 2004) tissue culture cell systems. GILZ, a gene that is often induced by GCs, especially in lymphoid cells, can also promote apoptosis, at least in certain cellular contexts (Delfino et al., 2004; Grugan et al., 2008). For the 697-shGR2 cell line, the involvement of other upregulated genes in apoptosis include: granzyme A in GC-treated 697 cells and pre-B-cell ALL patient samples (Yamada et al., 2003; Myomoto et al., 2007); FOXO3a in mouse splenocytes and a myeloma cell line (Ma et al., 2008); and, Protein Kinase A Regulatory Subunit IIβ (PKARIIβ) in GC-medicated apoptosis in CEM-C7 T-ALL cells (Ji et al. 2008). Upregulation of c-Myb could presumably contribute to the auto-upregulation of GR gene expression via the molecular switch mechanism that we have proposed (Geng and Vedeckis, 2005; Geng et al., 2008). Mcl-1 is actually anti-apoptotic, so its up-regulation is an enigma at the present time. The down-regulation of the well known anti-apoptotic protein, bcl-2, would also tend to promote steroid mediated cell death. The inhibition of the 2 Ets family members, PU.1 and SpiB, could also potentially contribute to increased auto-upregulation of GR gene expression, again, via the previously proposed molecular switch mechanism (Geng and Vedeckis, 2005; Geng et al., 2008). The down-regulation of the cell cycle inhibitor, p21, cannot be easily explained; however, hormonal effects on its expression are widely variable in the T-ALL patient samples, and the cell cycle stage in which the cells are may contribute to this heterogeneity.
Preliminary results indicate that some of these genes are regulated similarly in T-cell ALL patient samples (Table 2). It seems likely that hormone-sensitivity cannot be predicted solely upon GR levels, as some will be very close to the threshold level needed to trigger apoptosis (e.g., TALL4). However, by measuring the levels and regulation of transcripts for the target genes identified here (and perhaps additional ones) it might be possible to generate a “molecular signature” that can be used to stratify ALL patients into steroid-responsive and – resistant populations. At present, for T-ALL, it appears that Bim, GILZ, c-Jun, and PKARIIβ are potential candidate genes. TALL1 and TALL4 showed no, or marginal upregulation of the GR gene in this assay, emphasizing the fact that GR upregulation alone is not a good measure of hormone-sensitivity, because basal levels may be high, above the necessary threshold (see above). T-cell ALL is rare, and the number of fresh patient samples (which yield better results than cryopreserved samples; unpublished results) available to us is limited. Thus, the results here are still of limited scope. It is hoped that additional patient samples, and perhaps more candidate genes, can be analyzed by us and others in order to develop the necessary confidence in a molecular signature to predict hormone-responsiveness. The need for additional patient samples is emphasized by the fact the PKARIIβ was expressed at a low level in the established CEM-C7 cell line, which made it difficult to categorize this gene as an upregulated target in this cell line. It should be noted that any GR-regulated gene, even if it is not directly involved in apoptosis (although those are most desirable), could be used in a molecular signature for hormone-sensitivity in patient samples. Only 3 of 12 GC-responsive genes in 697, pre-B-cells were also responsive in CEM-C7, T-cells. This raises the possibility that the molecular signature may be different for these 2 lymphoid lineages and perhaps for other steroid responsive leukemias, such as multiple myeloma and non-Hodgkin’s lymphoma. Ultimately, this could lead to a more tailored targeted therapy for these patients. Another goal (J. R. Schwartz and W. V. Vedeckis, submitted for publication) is to develop a facile, ex vivo clinical assay that would predict GC-sensitivity. The strategy is that by monitoring the level and regulation of, perhaps, 4 to 6 transcripts (actual number to be determined) a predictive assay may be developed that would result in treatments that completely omit or simply increase the steroid dosage (Schwartz et al., 2001) depending on the degree of resistance that the ALL exhibits. This would spare patients who are totally resistant from the undesirable side effects of long-term, high-dose, corticosteroid treatment. Obviously, achieving this goal will require a much larger number of patient samples and the analysis of an expanded scope of regulated genes to identify a potentially therapeutically useful molecular signature for steroid-responsive acute lymphoblastic leukemia.
Acknowledgments
We thank Drs. Lily E. Leiva, Maria C. Velez, Tammuella C. Singleton, Renee V. Gardner, and Lolie C. Yu for their help with obtaining the patient samples and Dr. Beatriz E. Finkel-Jimenez for flow cytometry assistance. This work was supported by NCI grant CA116042 (to W.V.V.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams MT, Robertson NM, Yoon K, Wickstrom E. Inhibition of glucocorticoid-induced apoptosis by targeting the major splice variants of BIM mRNA with small interfering RNA and short hairpin RNA. J Biol Chem. 2004;279:55809–55817. doi: 10.1074/jbc.M411767200. [DOI] [PubMed] [Google Scholar]
- Ashraf J, Kunapuli S, Chilton D, Thompson EB. Cortivazol mediated induction of glucocorticoid receptor messenger ribonucleic acid in wild-type and dexamethasone-resistant human leukemic (CEM) cells. J Steroid Biochem Mol Biol. 1991;38:561–568. doi: 10.1016/0960-0760(91)90313-t. [DOI] [PubMed] [Google Scholar]
- Ashraf J, Thompson EB. Identification of the activation-labile gene: a single point mutation in the human glucocorticoid receptor presents as two distinct receptor phenotypes. Mol Endocrinol. 1993;7:631–642. doi: 10.1210/mend.7.5.8316249. [DOI] [PubMed] [Google Scholar]
- Bachmann PS, Gorman R, Papa RA, Bardell JE, Ford J, Kees UR, Marshall GM, Lock RB. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 2007;67:4482–4490. doi: 10.1158/0008-5472.CAN-06-4244. [DOI] [PubMed] [Google Scholar]
- Barrett TJ, Vig E, Vedeckis WV. Coordinate regulation of glucocorticoid receptor and c-jun gene expression is cell-type specific and exhibits differential hormonal sensitivity for down- and up-regulation. Biochemistry. 1996;35:9746–9753. doi: 10.1021/bi960058j. [DOI] [PubMed] [Google Scholar]
- Delfino DV, Agostini M, Spinicelli S, Vito P, Riccardi C. Decrease of Bcl-X and augmentation of thymocyte apoptosis in GILZ overexpressing transgenic mice. Blood. 2004;104:4134–4141. doi: 10.1182/blood-2004-03-0920. [DOI] [PubMed] [Google Scholar]
- Dordelmann M, Reiter A, Borkhardt A, Ludwig WD, Gotz N, Viehmann S, Gadner H, Riehm H, Schrappe M. Prednisone response is the strongest predictor of treatment outcome in infant acute lymphoblastic leukemia. Blood. 1999;94:1209–1217. [PubMed] [Google Scholar]
- Geng C-d, Vedeckis WV. c-Myb and members of the c-Ets family of transcription factors act as molecular switches to mediate opposite steroid regulation of the human glucocorticoid receptor 1A promoter. J Biol Chem. 2005;280:43264–43271. doi: 10.1074/jbc.M508245200. [DOI] [PubMed] [Google Scholar]
- Geng, Schwartz JR, Vedeckis WV. A conserved molecular mechanism is responsible for the auto-up-regulation of glucocorticoid receptor gene promoters. Mol Endocrinol. 2008;22:2624–2642. doi: 10.1210/me.2008-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi M, Moriwaki K, Katagiri S, Kurata Y, Thompson EB. Glucocorticoid effects on myeloma cells in culture: correlation of growth inhibition with induction of glucocorticoid receptor messenger RNA. Cancer Res. 1990;50:1873–1878. [PubMed] [Google Scholar]
- Gross KL, Cidlowski JA. Tissue-specific glucocorticoid action: a family affair. Trends Endocrinol Metab. 2008;19:331–339. doi: 10.1016/j.tem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber G, Carlet M, Turtscher E, Meister B, Irving JA, Ploner C, Kofler R. Levels of glucocorticoid receptor and its ligand determine sensitivity and kinetics of glucocorticoid-induced leukemia apoptosis. Leukemia. 2009 doi: 10.1038/leu.2008.360. [DOI] [PubMed] [Google Scholar]
- Grugan KD, Ma C, Singhal S, Krett NL, Rosen ST. Dual regulation of glucocorticoid-induced leucine zipper (GILZ) by the glucocorticoid receptor and the PI3-kinase/AKT pathways in multiple myeloma. J Steroid Biochem Mol Biol. 2008;110:244–254. doi: 10.1016/j.jsbmb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Mei FC, Miller AL, Thompson EB, Cheng X. Protein kinase A (PKA) isoform RIIβ mediates the synergistic killing effect of cAMP and glucocorticoid in acute lymphoblastic leukemia cells. J Biol Chem. 2008;283:21920–21925. doi: 10.1074/jbc.M803193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato GJ, Quddus FF, Shuster JJ, Boyett J, Pullen JD, Borowitz MJ, Whitehead VM, Crist WM, Leventhal BG. High glucocorticoid receptor content of leukemic blasts is a favorable prognostic factor in childhood acute lymphoblastic leukemia. Blood. 1993;82:2304–2309. [PubMed] [Google Scholar]
- Keith BD. Systematic review of the clinical effect of glucocorticoids on nonhematologic malignancy. BMC Cancer. 2008;8:84. doi: 10.1186/1471-2407-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfir S, Sionov RV, Zafrir E, Zilberman Y, Yefenof E. Staurosporine sensitizes T lymphoma cells to glucocorticoid-induced apoptosis: role of Nur77 and Bcl-2. Cell Cycle. 2007;6:3086–3096. doi: 10.4161/cc.6.24.5023. [DOI] [PubMed] [Google Scholar]
- Kofler R, Schmidt S, Kofler A, Ausserlechner MJ. Resistance to glucocorticoid-induced apoptosis in lymphoblastic leukemia. J Endocrinol. 2003;178:19–27. doi: 10.1677/joe.0.1780019. [DOI] [PubMed] [Google Scholar]
- Langebrake C, Reinhardt D, Ritter J. Minimising the long-term adverse effects of childhood leukaemia therapy. Drug Saf. 2002;25:1057–1077. doi: 10.2165/00002018-200225150-00002. [DOI] [PubMed] [Google Scholar]
- Ma J, Xie Y, Shi Y, Qin W, Zhao B, Jin Y. Glucocorticoid-induced apoptosis requires FOXO3A activity. Biochem Biophys Res Commun. 2008;377:894–898. doi: 10.1016/j.bbrc.2008.10.097. [DOI] [PubMed] [Google Scholar]
- Medh RD, Wang A, Zhou F, Thompson EB. Constitutive expression of ectopic c-Myc delays glucocorticoid-evoked apoptosis of human leukemic CEM-C7 cells. Oncogene. 2001;20:4629–4639. doi: 10.1038/sj.onc.1204680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Garza AS, Johnson BH, Thompson EB. Pathway interactions between MAPKs, mTOR, PKA, and the glucocorticoid receptor in lymphoid cells. Cancer Cell Int. 2007;7:3. doi: 10.1186/1475-2867-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myoumoto A, Nakatani K, Koshimizu T-a, Matsubara H, Adachi S, Tsujimoto G. Glucocorticoid-induced granzyme A expression can be used as a marker of glucocorticoid sensitivity for acute lymphoblastic leukemia therapy. J Hum Genet. 2007;52:328–333. doi: 10.1007/s10038-007-0119-4. [DOI] [PubMed] [Google Scholar]
- Palmer LA, Hukku B, Harmon JM. Human glucocorticoid receptor gene deletion following exposure to cancer chemotherapeutic drugs and chemical mutagens. Cancer Res. 1992;52:6612–6618. [PubMed] [Google Scholar]
- Pedersen KB, Vedeckis WV. Quantification and glucocorticoid regulation of glucocorticoid receptor transcripts in two human leukemic cell lines. Biochemistry. 2003;42:10978–10990. doi: 10.1021/bi034651u. [DOI] [PubMed] [Google Scholar]
- Powers JH, Hillmann AG, Tang DC, Harmon JM. Cloning and expression of mutant glucocorticoid receptors from glucocorticoid-sensitive and -resistant human leukemic cells. Cancer Res. 1993;53:4059–4065. [PubMed] [Google Scholar]
- Pui CH, Costlow ME. Sequential studies of lymphoblast glucocorticoid receptor levels at diagnosis and relapse in childhood leukemia: an update. Leuk Res. 1986;10:227–229. doi: 10.1016/0145-2126(86)90046-9. [DOI] [PubMed] [Google Scholar]
- Ramdas J, Liu W, Harmon JM. Glucocorticoid-induced cell death requires autoinduction of glucocorticoid receptor expression in human leukemic T cells. Cancer Res. 1999;59:1378–1385. [PubMed] [Google Scholar]
- Riml S, Schmidt S, Ausserlechner MJ, Geley S, Kofler R. Glucocorticoid receptor heterozygosity combined with lack of receptor auto-induction causes glucocorticoid resistance in Jurkat acute lymphoblastic leukemia cells. Cell Death Differ. 2004;11(Suppl 1):S65–72. doi: 10.1038/sj.cdd.4401413. [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Cidlowski JA. Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem Mol Biol. 2002;83:37–48. doi: 10.1016/s0960-0760(02)00263-7. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Thompson EB, Gelber RD, Young ML, Chilton D, Cohen HJ, Sallan SE. Improved response with higher corticosteroid dose in children with acute lymphoblastic leukemia. J Clin Oncol. 2001;19:1040–1046. doi: 10.1200/JCO.2001.19.4.1040. [DOI] [PubMed] [Google Scholar]
- Sionov RV, Spokoini R, Kfir-Erenfeld S, Cohen O, Yefenof E. Mechanisms regulating the susceptibility of hematopoietic malignancies to glucocorticoid-induced apoptosis. Adv Cancer Res. 2008;101:127–248. doi: 10.1016/S0065-230X(08)00406-5. [DOI] [PubMed] [Google Scholar]
- Tissing WJ, Meijerink JP, den Boer ML, Pieters R. Molecular determinants of glucocorticoid sensitivity and resistance in acute lymphoblastic leukemia. Leukemia. 2003;17:17–25. doi: 10.1038/sj.leu.2402733. [DOI] [PubMed] [Google Scholar]
- Tissing WJ, Lauten M, Meijerink JP, den Boer ML, Koper JW, Sonneveld P, Pieters R. Expression of the glucocorticoid receptor and its isoforms in relation to glucocorticoid resistance in childhood acute lymphocytic leukemia. Haematologica. 2005;90:1279–1281. [PubMed] [Google Scholar]
- Tissing WJ, Meijerink JP, Brinkhof B, Broekhuis MJ, Menezes RX, den Boer ML, Pieters R. Glucocorticoid-induced glucocorticoid-receptor expression and promoter usage is not linked to glucocorticoid resistance in childhood ALL. Blood. 2006;108:1045–1049. doi: 10.1182/blood-2006-01-0261. [DOI] [PubMed] [Google Scholar]
- Wang Z, Malone MH, He H, McColl KS, Distelhorst CW. Microarray analysis uncovers the induction of the proapoptotic BH3-only protein Bim in multiple model of glucocorticoid-induced apoptosis. J Biol Chem. 2003;278:23861–23867. doi: 10.1074/jbc.M301843200. [DOI] [PubMed] [Google Scholar]
- Webster JC, Cidlowski JA. Mechanisms of Glucocorticoid-receptor-mediated Repression of Gene Expression. Trends Endocrinol Metab. 1999;10:396–402. doi: 10.1016/s1043-2760(99)00186-1. [DOI] [PubMed] [Google Scholar]
- Wiegers GJ, Knoflach M, Bock G, Niederegger H, Dietrich H, Falus A, Boyd R, Wick G. CD4(+)CD8(+)TCR(low) thymocytes express low levels of glucocorticoid receptors while being sensitive to glucocorticoid-induced apoptosis. Eur J Immunol. 2001;31:2293–2301. doi: 10.1002/1521-4141(200108)31:8<2293::aid-immu2293>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Wright AP, Zilliacus J, McEwan IJ, Dahlman-Wright K, Almlof T, Carlstedt-Duke J, Gustafsson JA. Structure and function of the glucocorticoid receptor. J Steroid Biochem Mol Biol. 1993;47:11–19. doi: 10.1016/0960-0760(93)90052-x. [DOI] [PubMed] [Google Scholar]
- Yamada M, Hirasawa A, Shiojima S, Tsujimoto G. Granzyme a mediates glucocorticoid-induced apoptosis in leukemia cells. FASEB J. 2003;17:1712–1714. doi: 10.1096/fj.02-1116fje. [DOI] [PubMed] [Google Scholar]
- Zhang L, Insel PA. The pro-apoptotic protein Bim is a convergence point for cAMP/protein kinase A- and glucocorticoid-promoted apoptosis of lymphoid cells. J Biol Chem. 2004;279:20858–20865. doi: 10.1074/jbc.M310643200. [DOI] [PubMed] [Google Scholar]
- Zhou F, Thompson EB. Role of c-jun induction in the glucocorticoid-evoked apoptotic pathway in human leukemic lymphoblasts. Mol Endocrinol. 1996;10:306–316. doi: 10.1210/mend.10.3.8833659. [DOI] [PubMed] [Google Scholar]