Abstract
Developmentally regulated translation plays a key role in controlling gene expression during oogenesis. In particular, numerous mRNA species are translationally repressed in growing oocytes and become translationally activated during meiotic maturation. While many studies have focused on a U-rich sequence, termed the cytoplasmic polyadenylation element (CPE), located in the 3′-untranslated region (UTR) and the CPE-binding protein (CPEB) 1, multiple mechanisms likely contribute to translational control in oocytes. The stem-loop-binding protein (SLBP) is expressed in growing oocytes, where it is required for the accumulation of nonpolyadenylated histone mRNAs, and then accumulates substantially during meiotic maturation. We report that, in immature oocytes, Slbp mRNA carries a short poly(A) tail, and is weakly translated, and that a CPE-like sequence in the 3′-UTR is required to maintain this low activity. During maturation, Slbp mRNA becomes polyadenylated and translationally activated. Unexpectedly, proteasomal activity is required both to initiate and to sustain translational activation. This proteasomal activity is not required for the polyadenylation of Slbp mRNA during early maturation; however, it is required for a subsequent deadenylation of the mRNA that occurs during late maturation. Moreover, although CPEB1 is degraded during maturation, inhibiting its degradation by blocking mitogen-activated protein kinase 1/3 activity does not prevent the accumulation of SLBP, indicating that CPEB1 is not the protein whose degradation is required for translational activation of Slbp mRNA. These results identify a new role for proteasomal activity in initiating and sustaining translational activation during meiotic maturation.
Keywords: 3′-untranslated region, gamete biology, gametogenesis, gene regulation, meiotic maturation, oocyte development, oogenesis, ovum, proteasome, protein degradation, stem-loop-binding protein, translational control
The mRNA encoding the stem-loop-binding protein becomes polyadenylated and translationally activated during meiotic maturation, while blocking proteasomal activity at any time during maturation permits polyadenylation but inhibits translation.
INTRODUCTION
Growing oocytes synthesize and store mRNA that will become translationally activated at later stages of oogenesis or during early embryogenesis [1–4]. This reflects the fact that transcription declines substantially near the end of growth, and remains very low or undetectable throughout meiotic maturation and until the early cleavage stages of embryogenesis [5, 6]. The mRNAs encoding proteins required for development during the period of transcriptional quiescence must therefore be synthesized during oocyte growth, and regulatory mechanisms must ensure that these mRNAs are translated at the appropriate stage of oocyte or embryo development.
Considerable work has focused on mRNAs that become translationally activated during maturation [7–11]. In immature oocytes, these transcripts carry a short poly(A) tail of ∼25 nt and are translationally repressed; during maturation, the poly(A) tail lengthens to >100 nt and translation is activated. This translational control is mediated by specific sequences in the 3′-untranslated region (UTR) [2, 12–15]. Among the several regulatory sequences so far identified and characterized, the cytoplasmic polyadenylation elements (CPE) have been the most widely studied. CPE are U-rich sequences (URSs; consensus: UUUUUA[A]U) that typically are located <120 nt 5′ of the polyadenylation signal [16]. CPE-binding protein (CPEB) 1 binds to the CPE and nucleates the assembly of a complex of associated proteins [2, 17]. When the CPE sequences are deleted from the 3′-UTR, or competitor fragments containing the CPE are injected, mRNAs that normally are repressed in immature oocytes become translationally active [9, 18–20]. Conversely, when maturation is triggered, mRNAs lacking the CPE do not become polyadenylated nor translationally activated [7–9, 15]. Thus, the CPE plays a key role in regulating translational activity in immature and maturing oocytes.
Most studies have examined mRNAs encoding MOS, cyclin B1, and tissue plasminogen activator (tPA) [7–11]. These proteins are synthesized at very low or undetectable levels in immature oocytes, and play no known role in oogenesis prior to meiotic maturation. During maturation, translational activation leads to a significant increase in their quantity [21–23]. Synthesis of cyclin B1 is required to enter metaphase II following the first meiotic division [9, 24], and MOS is required to maintain meiotic arrest at metaphase II [25–27]. Thus, translational repression in immature oocytes and activation during maturation is consistent with the biological function of the proteins.
The stem-loop-binding protein (SLBP) is an RNA-binding protein that associates with a highly conserved stem-loop structure in the 3′-UTR of mRNAs encoding most histones, and is required for posttranscriptional processing and translation of the mRNAs [28]. SLBP is expressed in growing oocytes [29–32], where, in the mouse, it is required for the accumulation of mRNAs encoding histones H3 and H4 [33]. SLBP accumulates substantially during oocyte maturation, and regulates histone mRNA translation at this stage [30, 34]. Thus, like MOS and cyclin B1, SLBP accumulates during meiotic maturation, but, unlike these two proteins, it is also produced and functions in meiotically immature oocytes. Therefore, we sought to characterize the mechanisms that regulate its synthesis during these two stages of oogenesis. Our results indicate that, in immature oocytes, Slbp mRNA is translationally dampened through a CPE-dependent mechanism, and that, during maturation, proteasomal activity is required both to initiate and to sustain its translational activation.
MATERIALS AND METHODS
Oocyte Collection and Culture
All experiments were performed using CD1 mice (Charles River Canada, St.-Constant, QC, Canada) in compliance with the regulations and policies of the Canadian Council on Animal Care, and were approved by the Animal Care Committee of the Royal Victoria Hospital (protocol 1352). Fully grown oocytes were collected from 21-day-old female mice by puncture of the ovarian antral follicles, as previously described [35]. The oocytes were cultured in bicarbonate-buffered minimal essential medium (MEM) supplemented with sodium pyruvate, antibiotics, 3 mg/ml BSA and 0.1 mg/ml dibutyryl cyclic AMP (dbcAMP; this and the drugs listed below were from Sigma Chemicals, Windsor, ON) at 37°C, in 5% CO2 in air. Resumption of meiosis was initiated by transferring the oocytes into medium without dbcAMP. Dibutyryl cyclic AMP and puromycin were dissolved in water at 10 mg/ml, and used at 100 μg/ml and 50 μg/ml, respectively. MG132, N-acetyl-Leu-Leu-methioninal (LLM), and U0126 were dissolved in dimethyl sulfoxide (DMSO) at 50 mM and used at 25 μM. Epoxomycin was dissolved in DMSO at 5 mM and used at 10 μM.
Immunoblotting
Oocytes were collected and lysed in 10 μl of 2× Laemmli buffer. After denaturation, proteins were separated in a 10% polyacrylamide gel and transferred onto a polyvinylidene fluoride membrane (Amersham, Oakville, ON, Canada) under constant voltage. The membrane was subsequently blocked in 5% nonfat milk, in 0.1% Tween-PBS (PBST). The membrane was washed three times in PBST and incubated overnight at 4°C with primary antibody. After washing, the membrane was incubated in secondary antibody conjugated to horseradish peroxidase (Promega) at a dilution of 1:5000 for 1 h at room temperature. After the final washes, bound antibody was revealed using ECL+ (Amersham). For quantification, blots were scanned using a Storm phosphorimager (Amersham). Primary antibodies and dilutions were: anti-SLBP (1:5000 [34, 36]), anti-mitogen-activated protein kinase (MAPK) 1/3 (1:2000; Santa Cruz sc-94), anti-CDC2A (1:3000; Upstate Biotechnology 06–966), anti-CPEB (1:4000; Affinity Bioreagents PA1–1100).
RNA Ligation-Mediated Polyadenylation Test
Total RNA was extracted from 30 oocytes using a Picopure RNA isolation kit (Arcturus), including a DNase treatment, eluted in 10 μl of buffer, and stored at −80°C until use. Total RNA (10 μl), 4 μl of 20 μM RNA linker (5′-phosphate; 3′-ddC residue to prevent ligation to the 3′ end; Dharmacon, Lafayette, CO), 2 μl of 10× T4 RNA ligation buffer and 2 μl of T4 RNA ligase (5 U/μl; Ambion), and 2 ul of diethyl pyrocarbonate (DEPC)-treated water were combined in a final volume of 20 μl, and the solution was incubated for 2 h at room temperature. The next day, 20 μl of the ligation product was combined with 115 μl of DEPC-treated water, 15 μl of a solution of 5 M ammonium acetate and 100 mM EDTA, and 150 μl of phenol/chloroform (pH 8.0). This mixture was vortexed and centrifuged for 5 min at 13 000 rpm at room temperature, and the top layer was transferred to a new tube. Chloroform (150 μl) was added, the mixture was vortexed and centrifuged as above, and the top layer was transferred to a new tube. To this was added 2 μl of 20 mg/ml glycogen and 150 μl isopropanol, and the mixture was vortexed and then chilled at −20°C for 30 min. Centrifugation at 13 000 rpm for 20 min at 4°C produced a visible pellet at the bottom of tube. The pellet was washed using 0.5 ml of cold 70% ethanol and centrifuged for 5 min at 13 000 rpm, the ethanol was removed, and the pellet was allowed to dry at 37°C on a dry bath, and then resuspended in 10 μl of DEPC-treated water.
For the cDNA synthesis reaction, the resuspended ligation product was combined with 1 μl of 10 μM dNTPs and 1 μl of 20 μM of the RT primer complementary to the RNA linker. The mixture was heated to 65°C for 5 min, then chilled on ice for 5 min. An aliquot of 4 μl of 5× RT buffer, 2 μl of DTT (0.1 M), and 1 μl of RNaseOUT (Invitrogen) was added, and the mixture incubated at 42°C for 2 min. Superscript II (1 μl; Invitrogen) was added, and the mixture was incubated at 42°C for 50 min, then heated to 70°C for 15 min and stored at −20°C.
For the first PCR reaction, 1 μl of cDNA was amplified using High Fidelity Platinum Taq (Invitrogen) following the manufacturer's directions, using the RT primer and the appropriate gene-specific outer primer, for 20 cycles; 1 μl of the product was used for the nested PCR using the same conditions, except that the gene-specific inner primer was used, for 35 cycles. PCR products were separated on a 1.7% gel. The band corresponding to the expected size was cut out and purified by QIAquick Gel Extraction Kit (Qiagen). Purified products were cloned into pDrive vector with QIAGEN PCR Cloning Kit (Qiagen) and sequenced.
Primers were as follows: RNA linker, 5′-CGUAGGCUCAGCUCGGAAUC; linker RT primer, 5′-GGATTCCGAGCTGAGCCTACG; Slbp outer primer, 5′-GCCTTCAGTTGCCACTTTTC; Slbp inner primer, 5′-GCTCTGGACAAAGGATGCTAA; Ccnb1 outer primer, 5′-AAAAGAATTTGCCCCCAAGT; Ccnb1 inner PCR primer, 5′-CAGCATTCCTTTCAATGCCT.
Generation and In Vitro Transcription of Constructs
The Slbp mRNA 3′-UTR was cloned downstream of the luciferase coding sequence inserted into the Cs-2+ expression vector (gift of Dr. M. Featherstone). Deletion of different segments of SLBP 3′-UTR was performed by reverse PCR. The primers used for reverse PCR were designed such that the forward primer anneals downstream of the sequence to be deleted, while the reverse primer anneals upstream. Therefore, all vector and insert sequences will be amplified except the fragment to be deleted. Following PCR, the amplification product was circularized by ligation and transformed into bacteria. The clones obtained were confirmed by DNA sequencing. Following generation of the deletion constructs, an SP6 promoter was added at the 5′ end, and a sequence of 30 adenines at the 3′ end, of the reporter mRNA. In vitro transcription was performed using the mMessage mMachine kit (Ambion/Applied Biosystems, Austin, TX). The mRNA was purified by lithium chloride precipitation, followed by three washes using 70% ethanol. The pellet was resuspended in RNase-free water and stored at −80°C. About 10 pl of mRNA at 0.5 μg/μl in RNase-free water was microinjected, as previously described [34], into meiotically immature oocytes. After 1 h of recovery, half of the cells were transferred to medium without dbcAMP to allow resumption of meiosis, while the other half was kept in medium with dbcAMP.
Primer sequences were as follows (forward primer given first, followed by reverse primer, nucleotide positions in NM_009193 indicated): to generate the template for in vitro transcription, CCCAAGCTTGATTTAGGTGAC, (T)30CAGTTAAAGGGTCTTTAT-1613; to delete the terminal 54 nt, CCCAAGCTTGATTTAGGTGAC, (T)30CAGTTAAAGGGTCTTTATTTTTACTGGATACATGAACAG-1538; to delete URS-1, 1383-GGAACTTGAAATAGCATTAG, 1373-CTATATAACTTACATACAGCCC; to delete URS-2, 1484-GATCAGTTATTTACCATG, 1469-GTTCTTTCTAAGTCCCTCTC.
Oocyte Microinjection
Oocytes were microinjected in Hepes-buffered (pH 7.2) MEM supplemented with antibiotics, 3 mg/ml BSA, 0.1 mg/ml dbcAMP, and 3 mg/ml of polyvinyl pyrrolidone using a Leica inverted microscope equipped with Leica micromanipulators (Leica Canada, Montreal, QC, Canada), as previously described [34].
Luciferase Assay
Oocytes were lysed on ice in 45 μl of 1× lysis buffer (0.01% Triton X-100, 15 mM magnesium acetate, 4 mM EGTA, and 1 mM DTT) for 5 min. Immediately prior to the luciferase assay, 5 μl of assay buffer (1 mM ATP, 15 mM KH2PO4, 15 mM magnesium acetate) was added to each sample. Then 100 μl of luciferin solution (0.1 M KH2PO4, 15 mM magnesium acetate, 0.22 mM luciferin, 0.2 mg/ml coenzyme A) was added to each sample and activity was recorded using a EG&G Berthold LB96 luminometer.
RESULTS
Slbp mRNA Becomes Translationally Activated During Meiotic Maturation
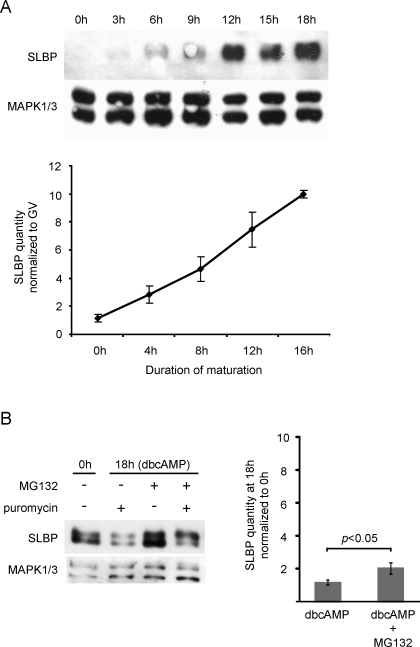
SLBP is present at a relatively low level in meiotically immature oocytes, and increases substantially during meiotic maturation [30]. To study the mechanism that controls this increase, we first determined the timing and extent of SLBP accumulation. We collected fully grown immature oocytes, allowed them to undergo maturation in vitro, and, at regular intervals, collected aliquots and subjected them to immunoblotting (Fig. 1A). SLBP had begun to accumulate by 3–4 h of incubation, shortly after germinal vesicle breakdown (GVBD), and continued to accumulate at an apparently steady rate throughout subsequent maturation. Note that samples were collected at slightly different time points for Figure 1, A and B. During a 16- to 18-h-period of maturation, the quantity of SLBP increased about 10-fold.
FIG. 1.
Accumulation of SLBP in maturing oocytes. A) Immature oocytes were collected immediately after isolation from the follicle (0 h) or were allowed to mature for the indicated period of time. They were immunoblotted using antibodies recognizing SLBP or MAPK1/3 (loading control). Only oocytes that had undergone GVBD were collected at the 3 h/4 h and later time points, and only those containing a polar body at 12 h and later time points. Fifty oocytes per lane. Upper panel shows a representative blot; lower panel shows cumulative results of a different series (note different time points) of six replicates. B) Immature oocytes were incubated for 18 h in the presence of dbcAMP to prevent maturation, and in the presence or absence of an inhibitor of protein synthesis, puromycin, or proteasomal activity, MG132, or both; 80 oocytes per lane. Left panel shows a representative blot; right panel shows cumulative results of 3 replicates using MG132. Means were compared using the t-test. Error bars indicate the standard error of the mean.
This accumulation could be due either to increased synthesis or to reduced degradation of SLBP during maturation. We collected immature oocytes and incubated them for 18 h in the presence of dbcAMP to maintain them in the immature state, together with either puromycin, which blocks protein synthesis, or MG132, which blocks proteasomal activity. SLBP diminished modestly in the presence of puromycin, and accumulated modestly in the presence of MG132 (Fig. 1B). In the presence of both drugs, the slower-migrating band, which likely represents a phosphorylated form targeted for degradation by the proteasome [37, 38], became relatively more intense. These results indicate that SLBP is synthesized by fully grown immature oocytes. However, the fact that it accumulated only very slightly when proteasomal activity was suppressed for 18 h in immature oocytes implies that the 10-fold accumulation that occurs in oocytes that are allowed to mature for the same period of time could not be due to reduced SLBP degradation during maturation. These results indicated that Slbp mRNA is translationally dampened in immature oocytes, and becomes translationally activated during maturation.
Translational Activation Is Accompanied by an Increase in the Length of the Poly(A) Tail of Slbp mRNA
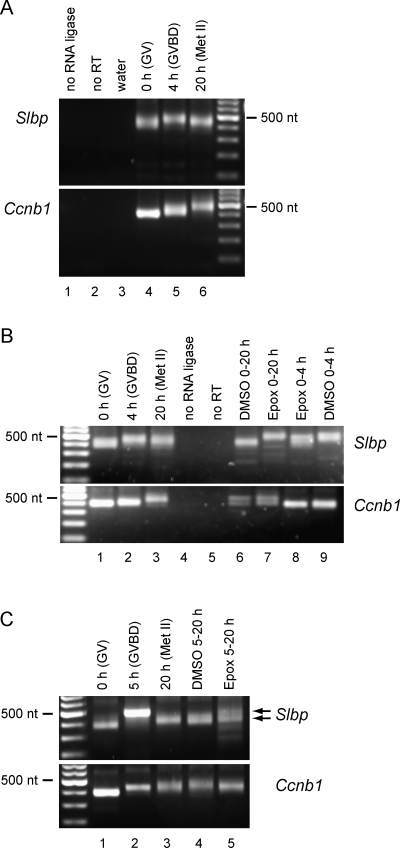
Translational activation of mRNAs during maturation is usually accompanied by a lengthening of the poly(A) tail [7–10, 39, 40]. We used an RNA ligation-based strategy, similar to that described by others [12, 41, 42], to determine the length of the Slbp mRNA poly(A) tail. Oocytes were collected before maturation (immature oocytes) and after 4 or 20 h of maturation, and the poly(A) tails of Slbp and Ccnb1 mRNAs were analyzed.
In immature oocytes, Slbp mRNA carried a relatively short poly(A) tail (Fig. 2A); sequencing of cloned PCR products indicated the range to be from 25 to 50 nt. By 4 h of maturation, the poly(A) tail had elongated to 100–125 nt. By 20 h of maturation, however, the poly(A) tail had shortened, indicating that it had become (partially) deadenylated. This deadenylation was not observed in the case of Ccnb1 mRNA, whose poly(A) tail continued to elongate throughout maturation. These results confirm that Slbp mRNA carried a short poly(A) tail in immature oocytes that became elongated during maturation.
FIG. 2.
Polyadenylation of Slbp mRNA during maturation. A) Oocytes were collected at the indicated stages of maturation, and the length of the poly(A) tails of Slbp and Ccnb1 (cyclin B1) were analyzed using a PCR-based assay. The PCR product comprises the portion of the mRNA that lies 3′ of the gene-specific primer, the poly(A) tail, and the ligated short RNA. The length of the poly(A) tail was determined by sequencing cloned products. B and C) Oocytes were treated with DMSO or epoxomycin for the indicated period of times. Arrows in C indicate the two products detected for Slbp.
Translational Dampening of Slbp mRNA in Immature Oocytes Requires a CPE-Like Sequence in the 3′-UTR
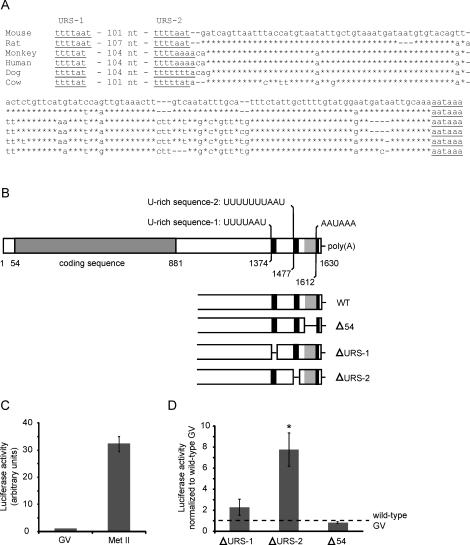
The distal region of the 3′-UTR of Slbp mRNA is highly conserved among mammals (Fig. 3A). Notably, two URSs that resemble CPE are present in all cases, are equally spaced from each other, and are at the same distance from the polyadenylation signal. We designated the proximal of these as URS-1 and the distal as URS-2, and tested whether either is required to dampen translation of Slbp mRNA in immature oocytes.
FIG. 3.
Identification and analysis of putative CPE sequences in the 3′-UTR of Slbp mRNA. A) The 3′-UTR of Slbp mRNA in different mammals. Sequences are from Mus musculus (Genbank NM_009193), Rattus norvegicus (XM_001060131), Pan troglodytes (NM_001144566), Homo sapiens (NM_006527), Canis lupus familiaris (XM_545928), and Bos taurus (NM_001045880). The position of first nucleotide and the sequence of URS-1 are shown, followed by the number of nucleotides that separate it from URS-2. The sequence between URS-2 and the polyadenylation signal is shown. Gaps have been introduced to maximize alignment. Nucleotides identical to the mouse are indicated by asterisks. B) The sequence of Slbp mRNA is shown together with the sequence and location of the two URSs identified as putative CPEs and the polyadenylation signal. The lower part of the panel shows the regions that were deleted to generate each mutant 3′-UTR. C) mRNA encoding luciferase linked to the wild-type 3′-UTR of SLBP RNA was synthesized and provided with a short poly(A) tail in vitro and then injected into immature oocytes. Injected oocytes were maintained at the immature (germinal vesicle [GV]) stage, or allowed to mature to metaphase II (Met II). Following a 16-h incubation, luciferase activity was measured; each group contained 15–25 oocytes. The experiment was performed three times. D) As in C, except that URS-1, URS-2, or the 54-nt sequence were deleted from the 3′-UTR. Injected oocytes were maintained at the GV stage, and activity was expressed relative to that in control oocytes injected with the wild-type construct and maintained at the GV stage in the same experiment. Asterisk indicates that the mean differed significantly from the control (t-test, P < 0.05). Error bars indicate the standard error of the mean.
First, to confirm that the Slbp mRNA 3′-UTR regulates its translation in oocytes, we linked the coding sequence of firefly luciferase to the Slbp mRNA 3′-UTR, followed by a short poly(A) sequence (Fig. 3B). A similar strategy was previously used to study the regulation of tPA mRNA translation in oocytes [20]. Messenger RNA was synthesized in vitro and injected into fully grown immature oocytes, which were either kept in the immature state or allowed to mature. After a 16-h incubation, the luciferase activity of the two groups was measured. Luciferase activity was ∼30-fold higher in mature oocytes than in immature oocytes (Fig. 3C). Thus, the 3′-UTR of Slbp mRNA was sufficient to confer the pattern of translation observed for the full-length mRNA; namely, a relatively low activity in immature oocytes and a high activity in maturing oocytes.
We then generated constructs lacking URS-1 or URS-2 or a 54-nt fragment that is located immediately 5′ to the polyadenylation signal, and possesses no CPE-like sequences (Fig. 3B). Messenger RNA was synthesized and injected into fully grown immature oocytes. For each experiment, control oocytes were injected with the construct containing the wild-type 3′-UTR. Injected oocytes were kept in the immature state for a 16-h incubation period, after which luciferase activity was measured. Neither deletion of the 54-nt fragment nor of URS-1 significantly altered luciferase activity as compared to the wild-type construct (Fig. 3D). In contrast, deletion of URS-2 produced a significant increase in luciferase activity, although the magnitude of the increase remained below that observed during maturation. These results indicate that URS-2 contributes to the translational dampening of Slbp mRNA in immature oocytes.
Proteasomal Activity Is Required Both to Initiate and to Sustain Translational Activation of Slbp mRNA During Maturation
We then turned to the mechanism that activates Slbp mRNA translation during maturation. In Xenopus, translational activation of Ccnb1 mRNA during oocyte maturation requires CPEB1 degradation [43]. This raised the possibility that translational activation of Slbp mRNA might require degradation of a protein that was bound to URS-2 and dampened translation in immature oocytes. If so, then blocking proteasomal activity at the beginning of maturation might prevent destruction of the putative inhibitor, and thus prevent the accumulation of SLBP. In contrast, blocking proteasomal activity at a later stage of maturation—following destruction of the putative inhibitor—would not prevent SLBP accumulation. To test this hypothesis, we transferred maturing oocytes to medium containing MG132 at different stages of maturation, and examined the effect on SLBP accumulation.
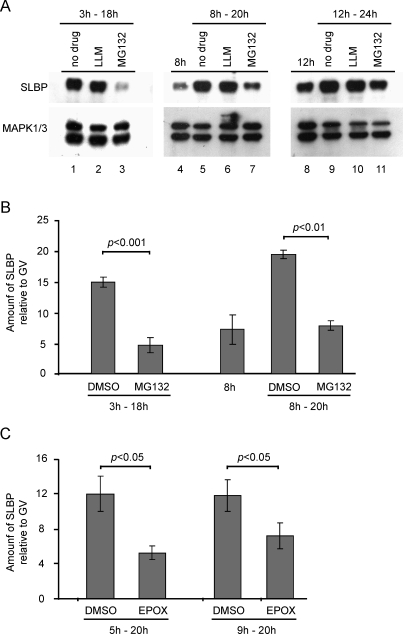
First, we added the MG132 or an inactive analogue (LLM) after 3 h of maturation, which is shortly after GVBD, and incubated the oocytes in its presence for an additional 15 h (i.e., until 18 h after the start of maturation). MG132 effectively inhibited the accumulation of SLBP (Fig. 4, A and B). This result indicates that proteasomal activity during early maturation was required for translational activation of Slbp mRNA.
FIG. 4.
Effect of proteasomal inhibition on the accumulation of SLBP during maturation. A) Immature oocytes were allowed to begin maturation, then, after 3 h (left), 8 h (middle), or 12 h (right), were transferred to medium containing no drug, LLM, or MG132, and incubated for an additional 12–15 h. Samples were also taken before the drug treatment for experiments shown in the middle and right panels. Samples were immunoblotted using antibodies recognizing SLBP and MAPK1/3 (loading control); 50 oocytes per lane. B) Results of a separate series of three experiments that were analyzed quantitatively. Oocytes were transferred to MG132 or DMSO after 3 or 8 h of maturation, as indicated. Mean values were compared using the t-test. C) Maturing oocytes were transferred to medium containing DMSO or epoxomicin at the indicated times and collected at 20 h. Results of four experiments are shown; 30 oocytes per group. Error bars indicate the standard error of the mean.
We then tested the effect of blocking proteasomal activity at later stages of maturation, after Slbp mRNA had become translationally activated. Immature oocytes were allowed to begin maturation. After 8 or 12 h, one sample was collected to measure the amount of SLBP present at that time point. The remaining oocytes were transferred to medium containing MG132 or left under control conditions, incubated for an additional 12 h, and then collected for immunoblotting. Oocytes incubated in the presence of DMSO or LLM contained substantially more SLBP than was present at the time of transfer (Fig. 4A, lanes 4 vs. 5/6 and 8 vs. 9/10; Fig. 4B, bars 3 vs. 4). This confirms that SLBP continued to be synthesized during the 12-h incubation following transfer. In contrast, oocytes transferred to MG132 contained little or no more SLBP than was present at the time of transfer (Fig. 4A, lanes 4 vs. 7 and 8 vs. 11; Fig. 4B, bars 3 vs. 5). Thus, when proteasomal activity was blocked midway through maturation, SLBP ceased to accumulate. To verify these results, we tested a different proteasomal inhibitor, epoxomycin. As observed using MG132, when epoxomycin was added shortly after GVBD (5 h) or at midmaturation (9 h), it inhibited the accumulation of SLBP (Fig. 4C). These results indicate that, even after Slbp mRNA had become translationally activated, proteasomal activity was required to sustain the state of translational activation.
Proteasomal Activity Is Not Required for Elongation of the Poly(A) Tail on Slbp mRNA
Because the length of a poly(A) tail is regulated by opposing activities that lengthen it, such as cytoplasmic poly(A) polymerase, or shorten it, such as poly(A)-specific ribonuclease [2, 17], we considered whether proteasomal activity might be required to degrade a factor that maintained a short poly(A) tail on Slbp mRNA. Immature oocytes were allowed to mature in the presence or absence of epoxomycin and, as described above, samples were collected before maturation (immature oocytes), after 4 h, and after ∼20 h.
In the presence of epoxomycin, the poly(A) tail of Slbp mRNA became elongated to the same extent as controls (Fig. 2B, lanes 7 vs. 2/9), although elongation may have occurred slightly more slowly (lanes 8 vs. 9). However, the poly(A) tail then failed to undergo the subsequent shortening observed under control conditions, but instead remained elongated in the presence of the drug (Fig. 2B, lanes 6 vs. 7). When oocytes were incubated in the presence of epoxomycin from 5 to 20 h of maturation, a doublet was observed, suggesting the presence of two populations of Slbp mRNAs, containing elongated and shortened poly(A) tails, respectively (Fig. 2C). In contrast to its effect on Slbp mRNA, epoxomycin did not detectably affect the length of the poly(A) tail of Ccnb1 mRNA. Thus, blocking proteasomal activity did not prevent the lengthening of the Slbp mRNA poly(A) tail at the beginning of maturation, but did prevent its subsequent shortening. These results indicate that the translational inhibition of Slbp mRNA observed when proteasomal activity was blocked was not due to shortening of its poly(A) tail.
Translational Activation of Slbp mRNA During Maturation Is Independent of CPEB1 Degradation
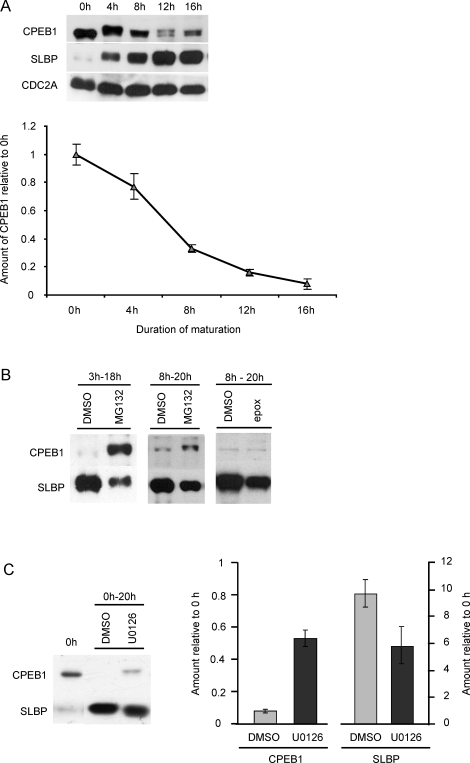
The link between CPEB1 degradation and translational activation of Ccnb1 mRNA in Xenopus [43], together with evidence that CPEB1 is degraded during oocyte maturation in the cow [9], suggest that CPEB1 degradation might be required for translational activation of Slbp mRNA. We tested this in two ways. First, we determined the timing of CPEB1 degradation during maturation. By 4 h, which is shortly after GVBD, CPEB1 had been converted to a slow-migrating form that could be resolved as a doublet in polyacrylamide gels (Fig. 5A). This electrophoretic mobility shift is likely due to phosphorylation [43, 44]. The amount of CPEB1 then decreased steadily during maturation. Importantly, after 12 h of maturation, when SLBP accumulation remained dependent on proteasomal activity (Fig. 4A), ∼80% of the CPEB1 present in immature oocytes had been degraded.
FIG. 5.
Degradation of CPEB1 during meiotic maturation. A) Immature oocytes were allowed to mature for the indicated periods of time and then immunoblotted using antibodies recognizing CPEB1, SLBP, and CDC2A (loading control). A representative immunoblot is shown together with the cumulative results of six experiments; 50 oocytes per lane. B) Immature oocytes were incubated in the presence of DMSO, MG132, or epoxomycin during the indicated periods of time, as described for Figure 3, and then immunoblotted using antibodies recognizing CPEB1 and SLBP; 50 oocytes per lane for MG132 groups, 30 for epoxomycin. The experiments were performed three times, and a representative immunoblot is shown. C) Immature oocytes were allowed to mature in the presence of DMSO or U0126, then immunoblotted as above. Representative blot and cumulative results of five experiments are shown. Error bars indicate the standard error of the mean.
It was possible that blocking proteasomal activity led to a reaccumulation of CPEB1, which, in turn, inhibited Slbp mRNA translation. We therefore examined the quantity of CPEB1 and SLBP in the same samples following exposure to MG132 or epoxomycin. Oocytes exposed to MG132 or epoxomycin beginning at 8 h of maturation contained only slightly more CPEB1 than control oocytes exposed to DMSO (Fig. 5B). Moreover, they contained much less CPEB1 than oocytes exposed to MG132 beginning at 3 h of maturation. These results indicate that CPEB1 did not accumulate when proteasomal activity was inhibited during maturation. Nonetheless, the inhibitor-treated oocytes contained less SLBP than controls. Therefore, the inhibition of Slbp mRNA translation that occurs when proteasomal activity is blocked in maturing oocytes is not due to an accumulation of CPEB1.
As a second approach, we inhibited MAPK1/3 activity during maturation, based on a recent report that this prevents the degradation of CPEB1 during bovine oocyte maturation [45]. Immature oocytes were allowed to mature for 18 h in the presence of the MAPK1/3 inhibitor, U0126. Although the quantity of CPEB1 diminished, it remained much more abundant than in control samples (Fig. 5C), indicating that blocking MAPK1/3 activity partially inhibited CPEB1 degradation. Despite the persistence of CPEB1, however, SLBP accumulated substantially (Fig. 5C). The slightly faster migration of the U0126-treated samples suggests that MAPK1/3 phosphorylates SLBP. Taken together, these results indicate that degradation of CPEB1 was not sufficient, and probably not required, for translational activation of Slbp mRNA during maturation.
DISCUSSION
We have studied the mechanism that regulates the translation of mRNA encoding SLBP in immature and maturing oocytes. We find that, in immature oocytes, the mRNA carries a short poly(A) tail, and is translationally dampened via a CPE-like sequence in the 3′-UTR. During maturation, it becomes polyadenylated and translationally activated. Unexpectedly, proteasomal activity is required both to initiate and to sustain translational activation. This proteasomal activity is not required for polyadenylation of the Slbp mRNA, and CPEB1 is probably not the protein whose destruction is required for translational activation. These results 1) indicate that CPE sequences, previously implicated in regulating the translation of mRNAs whose encoded proteins are required during maturation, also regulate expression of a protein required during growth, and 2) identify a new role for proteasomal activity in initiating and sustaining translational activation during meiotic maturation.
Studies of other mRNAs have highlighted the central role of the CPE in mediating translational repression in immature oocytes [2, 7–10, 40]. Three lines of evidence suggest that URS-2 acts as a CPE. First, it is present in the 3′-UTR and matches the CPE consensus. Second, it is located 128 nt from the polyadenylation signal, which is within the reported range of effective distance [16]. Third, deletion of URS-2 activates translation of a reporter mRNA in immature oocytes. These results suggest that URS-2 is a functional CPE that contributes to the translational dampening of Slbp mRNA in immature oocytes.
Although its mRNA is only weakly translationally active as compared with that in maturing oocytes, SLBP is present in immature oocytes [30, 33]. Moreover, the SLBP produced in growing immature oocytes is essential for the accumulation mRNAs encoding histones H3 and H4 that, in turn, are essential for embryonic development [33]. Thus, its mRNA is translationally active, albeit at a low level, in growing oocytes, despite the presence of a short poly(A) tail and the repressive activity of the URS-2. Although CPE sequences are generally linked to translational repression in immature oocytes [2, 7–10, 40], recent work has shown that CPEB1 is bound to some mRNA species that are translated in growing oocytes [46]. Thus, CPE sequences appear not to act exclusively to repress translation in immature oocytes, but instead, perhaps in concert with other sequences in the 3′-UTR [47], to precisely modulate translational activity at this stage.
Our observation, that Slbp mRNA becomes polyadenylated during maturation coincident with its translational activation, is consistent with previous reports in which other mRNAs were examined (see Introduction). However, the need for proteasomal activity has not previously been reported.
Proteasomal activity was not required in order to elongate the poly(A) tail of Slbp mRNA. Rather, in the presence of the proteasomal inhibitors, Slbp mRNA acquired a long poly(A) tail, yet remained translationally repressed. This rules out a model in which proteasomal activity during maturation is required to degrade a factor that maintains a short poly(A) tail on Slbp mRNA. Rather, the protein that must be degraded to activate translation of Slbp mRNA during maturation must be required for an event that follows polyadenylation.
The link between proteasomal activity and accumulation of SLBP during maturation recalls a previous report that translational activation of Ccnb1 mRNA during maturation in Xenopus requires degradation of CPEB1 [43]. Two lines of evidence suggest, however, that this is not the case for Slbp mRNA in mice. First, the dependence on proteasomal activity persisted even after most CPEB1 had been degraded. Second, when CPEB1 degradation was partially prevented by inhibiting MAPK1/3 activity during maturation, SLBP nonetheless accumulated. These results imply that CPEB1 is not the protein that must be degraded in order for Slbp mRNA to become translationally activated during maturation.
Unexpectedly, we found that, even after SLBP had begun to accumulate during maturation, proteasomal activity was required for its continued accumulation. This implies that mRNAs that had become translationally activated during maturation subsequently became, upon blocking of proteasomal activity, translationally inactive. Two models may be proposed. One possibility is that a translational inhibitor is continuously synthesized and degraded during maturation. When proteasomal activity is blocked, this protein accumulates and, perhaps by binding to the Slbp mRNA, prevents its translation. Alternatively, an inhibitor that is bound to Slbp mRNA in immature oocytes might be slowly degraded during maturation. Consequently, the inhibitor would be removed from individual mRNA molecules at different times during maturation. Once freed from the inhibitor, individual mRNAs would become translationally active. If each mRNA molecule was active for only a short period of time and then became translationally inactive, however, then continuous proteasomal activity would be required to replenish the pool of translationally active mRNA molecules.
In this context, it is intriguing that the poly(A) tail of Slbp mRNA, which becomes elongated during early maturation, subsequently shortens during late maturation. The same pattern has been reported for mRNA encoding PAPD4 (Gld-2) and EMI1 [12, 42], but does not occur in the case of Ccnb1 mRNA. Thus, Slbp mRNA is a member of a group of mRNA species whose poly(A) tail is initially elongated and subsequently shortened during maturation. The mechanism underlying this dynamic regulation of poly(A) tail length during maturation is unknown. There is evidence that mRNAs containing AU-rich elements (consensus: AUUUA) are deadenylated (and ultimately degraded) by a process linked to translation [48–50]. However, although the mRNAs encoding SLBP, PAPD4, and EMI1 each contain at least one AUUUA within the 3′-UTR, Ccnb1 mRNA contains the same element. Other sequences in the 3′-UTR may contribute to regulating the length of the poly(A) tail. Regardless of the mechanism of its shortening, the observation that the poly(A) tail of Slbp mRNA remained elongated when translation was repressed by blocking proteasomal activity raises the possibility that translation of Slbp mRNA during maturation leads to its deadenylation and translational inactivation. Such a process would be consistent with second model proposed above. In any case, studies can now be directed toward identifying the protein whose degradation enables translational activation of Slbp mRNA during maturation and understanding the mechanism and significance of its deadenylation during late maturation.
Acknowledgments
Heather Senn and Dana Murchison made important contributions to the development of the RNA ligation-mediated polyadenylation test assay. We thank W.F. Marzluff (University of North Carolina, Chapel Hill, NC) for purification of the anti-SLBP antibody and D. Arnold, D. Dufort, and M. Nagano for comments on the manuscript.
Footnotes
1Supported by the Canadian Institutes for Health Research through an operating grant to H.J.C. and the Program in Oocyte Health. P.A. was supported by the Fonds de Recherche en Santé du Québec. Infrastructural support for the Research Division of the Department of Obstetrics and Gynecology is generously provided by the Royal Victoria Hospital Foundation.
These authors contributed equally to this work.
REFERENCES
- Kashiwabara S-i, Nakanishi T, Kimura M, Baba T.Non-canonical poly(A) polymerase in mammalian gametogenesis. Biochim Biophys Acta 2008; 1779: 230–238. [DOI] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH.Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 2008; 1779: 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD.Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2001; 2: 521–529. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R.A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet 2002; 18: 150–157. [DOI] [PubMed] [Google Scholar]
- De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol 2006; 292: 1–12. [DOI] [PubMed] [Google Scholar]
- Latham KE.Mechanisms and control of embryonic genome activation in mammalian embryos. Int Rev Cytol 1999; 193: 71–124. [DOI] [PubMed] [Google Scholar]
- Dai Y, Newman B, Moor R.Translational regulation of MOS messenger RNA in pig oocytes. Biol Reprod 2005; 73: 997–1003. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Xu WH, Cooper GM, Richter JD.Translational control by cytoplasmic polyadenylation of c-Mos mRNA is necessary for oocyte maturation in the mouse. EMBO J 1994; 13: 5712–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J, Hodgman R, Richter JD.The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev Biol 2000; 221: 1–9. [DOI] [PubMed] [Google Scholar]
- Huarte J, Stutz A, Oconnell ML, Gubler P, Belin D, Darrow AL, Strickland S, Vassalli JD.Transient translational silencing by reversible mRNA deadenylation. Cell 1992; 69: 1021–1030. [DOI] [PubMed] [Google Scholar]
- Vassalli JD, Huarte J, Belin D, Gubler P, Vassalli A, O'Connell ML, Parton LA, Rickles RJ, Strickland S.Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev 1989; 3: 2163–2171. [DOI] [PubMed] [Google Scholar]
- Belloc E, Mendez R.A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature 2008; 452: 1017–1021. [DOI] [PubMed] [Google Scholar]
- Wang YY, Charlesworth A, Byrd SM, Gregerson R, MacNicol MC, MacNicol AM.A novel mRNA 3′ untranslated region translational control sequence regulates Xenopus Wee1 mRNA translation. Dev Biol 2008; 317: 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, Lissenden S.Mechanisms of translational control by the 3′ UTR in development and differentiation. Sem Cell Dev Biol 2005; 16: 49–58. [DOI] [PubMed] [Google Scholar]
- Richter JD.CPEB: a life in translation. Trends Biochem Sci 2007; 32: 279–285. [DOI] [PubMed] [Google Scholar]
- Richter JD.Breaking the code of polyadenylation-induced translation. Cell 2008; 132: 335–337. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD.Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 2006; 24: 173–183. [DOI] [PubMed] [Google Scholar]
- Barkoff AF, Dickson KS, Gray NK, Wickens M.Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev Biol 2000; 220: 97–109. [DOI] [PubMed] [Google Scholar]
- de Moor CH, Richter JD.Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J 1999; 18: 2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz A, Conne B, Huarte J, Gubler P, Volkel V, Flandin P, Vassalli J-D.Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev 1998; 12: 2535–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl A, Eppig JJ.Analysis of the mechanism(s) of metaphase I arrest in maturing mouse oocytes. Development 1995; 121: 925–933. [DOI] [PubMed] [Google Scholar]
- Huarte J, Belin D, Vassalli A, Strickland S, Vassalli JD.Meiotic maturation of mouse oocytes triggers the translation and polyadenylation of dormant tissue-type plasminogen activator mRNA. Genes Dev 1987; 1: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Paules RS, Buccione R, Moschel RC, Vande Woude GF, Eppig JJ.Mouse Mos protooncogene product is present and functions during oogenesis. Proc Natl Acad Sci U S A 1989; 86: 5395–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledan E, Polanski Z, Terret M-E, Maro B.Meiotic maturation of the mouse oocyte requires an equilibrium between cyclin B synthesis and degradation. Dev Biol 2001; 232: 400–413. [DOI] [PubMed] [Google Scholar]
- Colledge WH, Carlton MBL, Udy GB, Evans MJ.Disruption of c-mos causes parthenogenetic development of unfertilized mouse eggs. Nature 1994; 370: 65–68. [DOI] [PubMed] [Google Scholar]
- Hashimoto N, Watanabe N, Furuta Y, Tamemoto H, Sagata N, Yokoyama M, Okazaki K, Nagayoshi M, Takedat N, Ikawatll Y, Aizawai S.Parthenogenetic activation of oocytes in c-mos-deficient mice. Nature 1994; 370: 68–71. [DOI] [PubMed] [Google Scholar]
- Inoue D, Ohe M, Kanemori Y, Nobui T, Sagata N.A direct link of the Mos-MAPK pathway to Erp1/Emi2 in meiotic arrest of Xenopus laevis eggs. Nature 2007; 446: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Marzluff WF.Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol 2005; 17: 274–280. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Ingledue TC, Dominski Z, Sanchez R, Marzluff WF.Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol 1999; 19: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, Champigny MJ, Skoggard S, Erkmann JA, Whitfield ML, Marzluff WF, Clarke HJ.Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. J Cell Sci 2002; 115: 4577–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AJ, Howard JT, Dominski Z, Schnackenberg BJ, Sumerel JL, McCarthy JJ, Coffman JA, Marzluff WF.The sea urchin stem-loop-binding protein: a maternally expressed protein that probably functions in expression of multiple classes of histone mRNA. Nucleic Acids Res 2004; 32: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF.Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol Cell Biol 2002; 22: 2267–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold DR, Francon P, Zhang J, Martin K, Clarke HJ.Stem-loop binding protein expressed in growing oocytes is required for accumulation of mRNAs encoding histones H3 and H4 and for early embryonic development in the mouse. Dev Biol 2008; 313: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, Yang Q, Marzluff WF, Clarke HJ.The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Dev Biol 2005; 286: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke HJ, Oblin C, Bustin M.Developmental regulation of chromatin composition during mouse embryogenesis: somatic histone H1 is first detectable at the 4-cell stage. Development 1992; 115: 791–799. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF.Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol 2000; 20: 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu MM, Graves LM, Marzluff WF.Phosphorylation of threonine 61 by cyclin A/cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol Cell Biol 2008; 28: 4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LX, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF.Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol 2003; 23: 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B, Hwang SY, McLaughlin J, Solter D, Knowles BB.Timely translation during the mouse oocyte-to-embryo transition. Development 2000; 127: 3795–3803. [DOI] [PubMed] [Google Scholar]
- Stutz A, Conne B, Huarte J, Gubler P, Volkel V, Flandin P, Vassalli JD.Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev 1998; 12: 2535–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad CK, Mahadevan M, MacNicol MC, MacNicol AM.Mos 3prime UTR regulatory differences underlie species-specific temporal patterns of Mos mRNA cytoplasmic polyadenylation and translational recruitment during oocyte maturation. Mol Reprod Dev 2008; 75: 1258–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S-i, Miyado K, Baba T.Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev Biol 2006; 289: 115–126. [DOI] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD.Differential mNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 2002; 21: 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverte CG, Ahearn MD, Hake LE.CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev Biol 2001; 231: 447–458. [DOI] [PubMed] [Google Scholar]
- Uzbekova S, Arlot-Bonnemains Y, Dupont J, Dalbies-Tran R, Papillier P, Pennetier S, Thelie A, Perreau C, Mermillod P, Prigent C, Uzbekov R.Spatio-temporal expression patterns of aurora kinases A, B, and C and cytoplasmic polyadenylation-element-binding protein in bovine oocytes during meiotic maturation. Biol Reprod 2008; 78: 218–233. [DOI] [PubMed] [Google Scholar]
- Racki WJ, Richter JD.CPEB controls oocyte growth and follicle development in the mouse. Development 2006; 133: 4527–4537. [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R.A combinatorial code for CPE-mediated translational control. Cell 2008; 132: 434–448. [DOI] [PubMed] [Google Scholar]
- Lu J-Y, Bergman N, Sadri N, Schneider RJ.Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA 2006; 12: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen C-YA, Zhu W, He X, Shyu A-B.Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 2008; 182: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S-i.Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 2007; 21: 3135–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]