Abstract
Mouse embryos display a strain-dependent propensity for blastomere cytofragmentation at the two-cell stage. The maternal pronucleus exerts a predominant, transcription-dependent effect on this phenotype, with lesser effects of the ooplasm and the paternal pronucleus. A parental origin effect has been observed as an inequality in the cytofragmentation rate of embryos produced through genetic crosses of reciprocal F1 hybrid females. To understand the basis for this, we conducted an extensive series of pronuclear transfer studies employing different combinations of inbred and F1 hybrid maternal and paternal genotypes. We find that the parental origin effect is the result of a transgenerational epigenetic modification, whereby the inherited maternal grandpaternal contribution interacts with the fertilizing paternal genome and the ooplasm. This result indicates that some epigenetic information related to grandparental origins of chromosomes (i.e., imprinting of chromosomes in the mother) is retained through oogenesis and transmitted to progeny, where it affects gene expression from the maternal pronucleus and subsequent embryo phenotype. These results reveal for the first time that mammalian embryonic development can be affected by the epigenotype of at least three individuals. Additionally, we observe a significant suppression of fragmentation by F1 hybrid ooplasm when it is separated from the F1 hybrid maternal pronucleus. This latter effect is a striking example of heterosis in the early mammalian embryo, and it provides a new opportunity for examining the molecular mechanisms of heterosis. These results are relevant to our understanding of the mechanisms of epigenetic effects on development and the possible fertility effects of genetic and epigenetic interactions in reproductive medicine.
Keywords: apoptosis, cytofragmentation, embryo, gene regulation, genomic imprinting, heterosis, mitochondria, nuclear transfer, oocyte development, parental origin effects, superovulation, transgenerational effect
Summary: Cytofragmentation in two-cell mouse embryos is controlled by strain-specific factors, epigenetic information from the maternal grandfather, and a striking hybrid vigor effect mediated by the ooplasm
INTRODUCTION
Life begins as a cooperative venture between two gametes, each fated separately to die. Once the gametes are united, the first requirement on the path to normal development is to suppress or override the molecular and cellular predisposition toward death. As a truly cooperative venture, one can hypothesize that both gametes may contribute information in the form of various macromolecules as well as developmental programming of the parental genomes, and that such information plays a vital role in diverting the zygote from a pathway leading to death to one leading to the creation of a new life.
Subsequent processes in the early embryo promote DNA replication, cell cycle progression, transcriptional activation, translation and degradation of maternal mRNAs, and the correct and timely activation of gene transcription. The interactions between maternal and paternal components of the zygote are, therefore, highly complex, affecting essentially every cellular process. The molecular basis of these interactions is poorly understood, and little is known about how some of these early interactions affect long-term developmental potential.
Defects in the suppression of cell death can lead to apoptosis in early embryos [1–5]. Apoptotic processes in early embryos include cytofragmentation and cytoplasmic blebbing, DNA fragmentation, changes in mitochondrial membrane polarity, cytochrome c release, annexin V exteriorization, and cellular degeneration [6–14]. Such processes are frequent among human embryos. More than 88% (203 of 229) of “spare” human embryos made by in vitro fertilization displayed fragmentation in one study, and half fail to develop to blastocyst stage [7]. Recent studies revealed that fragmentation reduced competence to form blastocysts in culture [15]. Fragmentation also occurs in other species, including cow [11, 12, 16] and mouse [2, 3, 5, 8], providing animal models for understanding the mechanisms that either promote or inhibit the process.
Apoptosis and cytofragmentation are most often observed around the time of transcriptional activation of the embryonic genome [2, 3]. It is believed that early gene transcription promotes the accumulation of antiapoptotic gene products that suppress apoptosis and cytofragmentation, and that failure to do so constitutes one mechanism of early embryo quality control [2, 3, 6]. Not all of these processes must necessarily accompany one another, however, and cytofragmentation has been observed in the absence of other hallmarks of apoptosis [17]. One study reported that chemical inhibition of caspase activity did not affect cytofragmentation [5], indicating that cytofragmentation may not be obligatorily linked to other apoptotic events. Moreover, the various components of molecular pathways mediating apoptosis may not be expressed equally between embryonic blastomeres and somatic cells. The early embryo may lack some components of the apoptotic pathways, or express a unique array or alternative forms of such proteins.
The embryo has three potential sources of molecules that could affect the incidence of apoptosis and cytofragmentation: maternal protein stores, maternal mRNAs that can be recruited for translation, and embryonic transcripts produced from either maternal or paternal chromosomes. As a result, the incidence of cytofragmentation may be influenced by components of the ooplasm or by products encoded by either maternal or paternal genomes. The information contributed by each of the two gametes must be encoded in the genome, and therefore may be subject to genetic variability. One would predict, therefore, that certain genetic combinations might favor early embryo survival, whereas other genetic combinations might predispose the embryo to early demise. Consequently, the cellular integrity and viability of the embryo during the immediate postfertilization period should be affected by the genotype of both parents (i.e., both gametes). Available data indicate that, indeed, both maternal and paternal genetic factors control the frequency at which embryos undergo apoptotic processes. Mathematical modeling reveals that, although cell loss in the human embryo does not begin until the eight-cell stage, the propensity to undergo cell loss and arrest is most likely established in the one-cell embryo [18]. Extrinsic factors, such as culture conditions, and maternal age may also affect the incidence of cell loss in a subpopulation of embryos [1, 3,19–21].
Genetic systems provide powerful tools for understanding early embryogenesis and the clinical aspects of infertility. In the mouse, apoptosis is most often observed at advanced cleavage stages and in blastocysts, where it is influenced strongly by growth factors [1, 19]. However, blastomere fragmentation also occurs at the one-cell and two-cell stages, and is subject to genetic effects [2, 3, 17, 22]. A significant effect of paternal genotype on fragmentation in mouse one-cell embryos was reported [2, 3]. More recently, we observed a significant effect of paternal genotype on fragmentation at the two-cell stage in crosses involving C57BL/6 and C3H/HeJ strains [17, 22]. The maternal genotype exerts an even stronger effect. Embryos from C3H/HeJ mothers display a higher incidence of cytofragmentation at the two-cell stage than embryos from C57BL/6 mothers, and maternal pronuclear (PN) transfer studies revealed that the maternal PN exerts the predominant effect on cytofragmentation within the context of inbred genetic backgrounds [22].
Interestingly, reciprocal F1 hybrid females made with these two strains yield embryos that display unequal rates of cytofragmentation [17]. Because the oocytes of the reciprocal F1 hybrid females transmit both maternal pronuclei and mitochondria of different genetic origins, this result signifies a parental origin effect related either to genomic imprinting or to mitochondrial genotype. Either explanation for the parental origin effect would provide crucial new insight into the biology of early embryos and the potential causes of cytofragmentation clinically.
Collectively, these results indicate that all components of the fertilized embryo, that is, maternal and paternal pronuclei, ooplasm, and mitochondria, may contribute to the control of blastomere cytofragmentation in the early embryo. The details of these interactions, however, have remained uncharacterized. Paternal PN transfers testing whether ooplasmic effects during PN formation contribute to later fragmentation rates have not been performed. Whether a mixture of ooplasm can affect fragmentation has not been tested. Most importantly, the basis for the parental origin effect has not been examined. To address these questions, we conducted an extensive series of PN transfer and ooplasm transfer studies using many different combinations of inbred strain and F1 hybrid genotypes in order to test systematically the contributions of each component (maternal PN, paternal PN, ooplasm) to embryo phenotype. We report here that early interactions between the ooplasm and paternal PN revealed by ooplasm transfer and paternal PN transfer can exert subtle effects on the rate of fragmentation. More importantly, we provide new results related to the parental origin effect. We find that the parental origin effect is not the result of mitochondrial origin, but rather is the result of epigenetic modifications that act transgenerationally. The maternal PN retains predominant control over cytofragmentation, but the grandpaternal origin of chromosomes transmitted via the maternal PN influences embryonic phenotype strongly. The maternal transmission of grandpaternal epigenetic information indicates that paternal epigenetic information is retained by the maternal genome through oogenesis. This information is able to affect gene expression from the maternal genome in the zygote. Additionally, we observe a striking suppression of cytofragmentation rate by F1 hybrid ooplasm when separated from the F1 hybrid maternal PN, indicating that the ooplasm is subject to heterosis. These results lead us to a new model of genetic and epigenetic control of cytofragmentation in the early embryo.
MATERIALS AND METHODS
Mice, Embryos, and Embryo Culture
Mice of the C57BL/6 strain (hereafter referred to as “B6”) were purchased from Harlan Sprague Dawley (Indianapolis, IN). Adult C3H/HeJ (hereafter referred to as “C3H”) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Unless otherwise indicated, females (8–15 wk of age) were superovulated by injection of 5 IU eCG (Calbiochem, San Diego, CA) followed 48 h later by 5 IU hCG (Sigma-Aldrich, St. Louis, MO), and then mated (males >10 wk of age). Some control studies employed either spontaneous ovulation or a dosage of 10 IU of each gonadotropin. All studies adhered to procedures consistent with the National Research Council Guide for the Care and Use of Laboratory Animals.
Embryos were isolated at the one-cell stage, approximately 19 h after hCG. Cumulus cells were removed by digestion with hyaluronidase (500 IU/mg, 120 IU/ml; ICN Pharmaceuticals, Costa Mesa, CA) in M2 medium at room temperature. Embryos were cultured in CZB medium as in the previous studies [17, 22]. Any unfertilized eggs or morphologically abnormal embryos were removed. Maternal and paternal PN transfers were performed as described [22]. Ooplasm transfer was performed using identical procedures, except that cytoplasts (10 μm diameter) were transferred from donor embryos to recipient embryos. Schematic diagrams of breeding crosses, pronuclear transfer, and ooplasm strategies (>7000 total PN and ooplasm transfers completed) are provided (Figs. 1–5). The polar bodies were removed during PN transfer. Electrofusion (900 V/cm, 10 μsec, one pulse) was performed in fusion medium (275 mM mannitol, 0.05 mM CaCl2, 0.10 mM MgSO4, 0.3% BSA). Only constructs that fused after a single pulse and yielded single-cell, intact embryos were retained for the analysis. After PN transfer, embryos were returned to CZB medium and cultured overnight as described [17, 22].
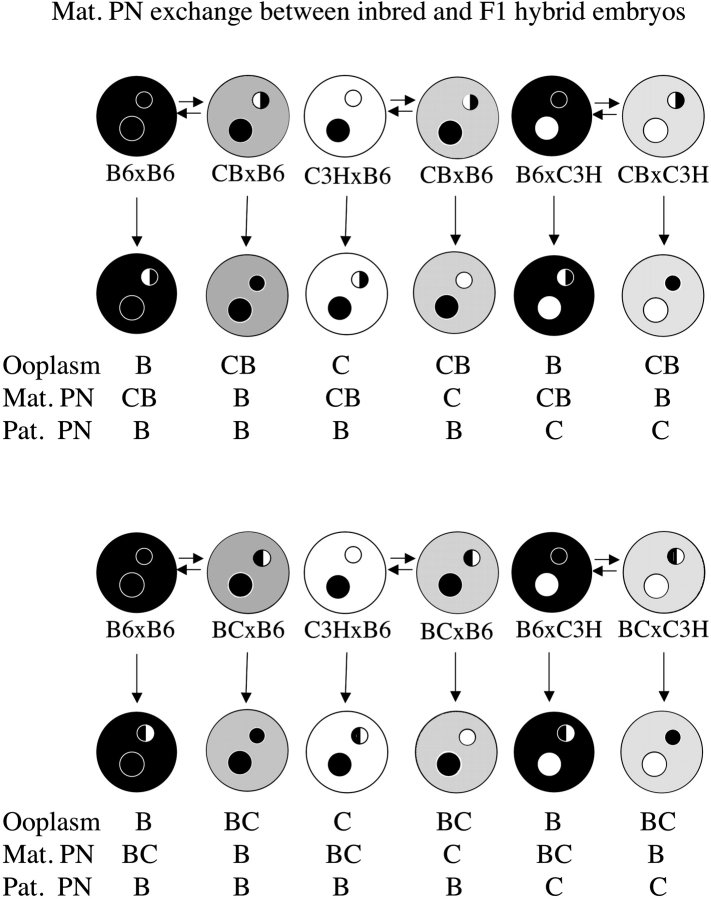
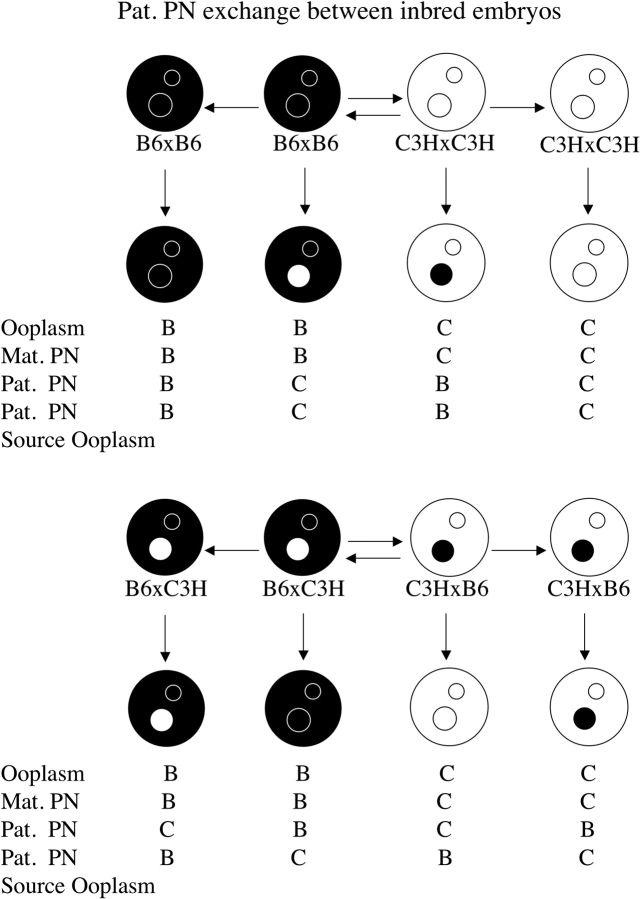
FIG. 1. Schematic illustration of maternal PN transfers performed using inbred strains of mice. Black ooplasm and black PN represent B6 (denoted B), and white ooplasm and white PN represent C3H (denoted C). The combinations of ooplasm, maternal PN (Mat. PN), and paternal PN (Pat. PN) are shown below the embryos, and are those used in Table 1.
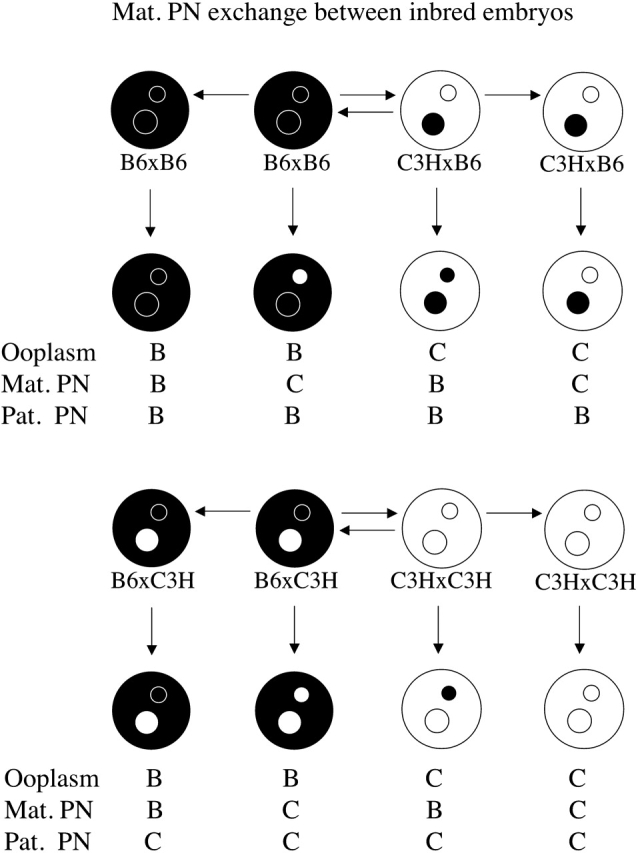
FIG. 5. Schematic illustration of maternal PN transfers performed using inbred and F1 hybrid strains of mice. Black ooplasm and black PN represent B6 (denoted B), and white ooplasm and white PN represent C3H (denoted C). Light stippled ooplasm are C3H×B6 (denoted CB), heavy stippled ooplasm are B6×C3H (denoted BC). Maternal PNs with left half white and right half black are from C3H×B6 (denoted CB) donors. Maternal PNs with left half black and right half white are from B6×C3H (denoted BC) donors. The combinations of ooplasm, maternal PN, and paternal PN produced are shown below the embryos, and are those used in Tables 6 and 7.
Analysis of Cytofragmentation
Embryos were examined using 320× magnification, phase-contrast microscopy, and were scored as either unfragmented or with fragmentation graded into three categories as described [22]. Briefly, grade 1 fragmentation referred to embryos with one or more small cytofragments smaller than the size of a polar body and often clustered at one or both poles or in the crevice between blastomeres; grade 2 fragmentation referred to embryos having many more fragments accounting for an equivalent of less than one half the volume of a blastomere; and grade 3 referred to embryos with a total volume of fragments being approximately one half of one blastomere or greater. The proportion of embryos falling into each category was recorded.
Statistical Analysis
Each experimental manipulation or breeding cross was conducted numerous times to produce a statistically large number of embryos of each class. The numbers of embryos thus analyzed provide sufficient statistical power for discerning small differences in overall fragmentation rates. The overall proportion of embryos of each fragmentation grade and the total proportion of fragmented embryos were calculated for each embryo class. The significance of differences in the incidence of fragmentation between classes of embryos was evaluated using a chi-square test of independence applied to the total number of embryos obtained for each class.
RESULTS
Role of Maternal Pronucleus and Epigenetic Mechanisms in Controlling Cytofragmentation
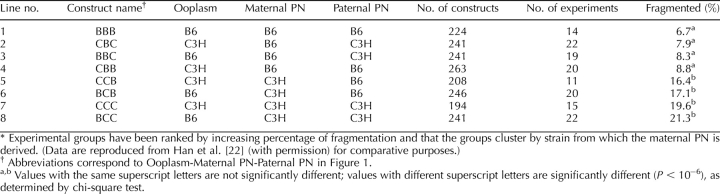
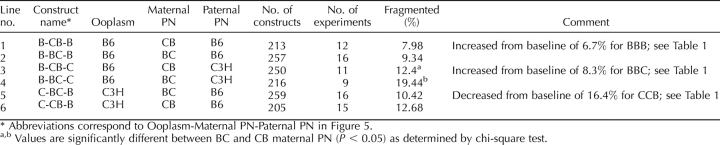
We reported that the incidence of cytofragmentation is predominantly controlled by the maternal pronucleus (PN; Han et al. [22], reproduced here in Fig. 1 and Table 1). The presence of a B6 maternal PN leads to cytofragmentation rates between 6.7% and 8.8%, whereas the presence of a C3H maternal PN leads to much higher cytofragmentation rates between 16.4% and 21.2% (P < 10−6). These studies with inbred mice demonstrated little or no effect of the ooplasm (note that the equivalent of complete ooplasm exchange has little effect on fragmentation rate—compare line 1 with line 4, and line 7 with line 8 in Table 1) or the paternal PN strain of origin. We also reported that this early effect of the maternal PN is sensitive to α-amanitin, and thus is transcription dependent [22]. Alpha-amanitin sensitivity indicates that the cytofragmentation phenotype is controlled by one or more products encoded by the maternal PN during the one-cell or early two-cell stage, making this one of the earliest documented effects of the embryonic genome on embryo phenotype.
TABLE 1.
Cytofragmentation of mouse two-cell stage embryos produced by maternal PN transfer using inbred strains of mice.*
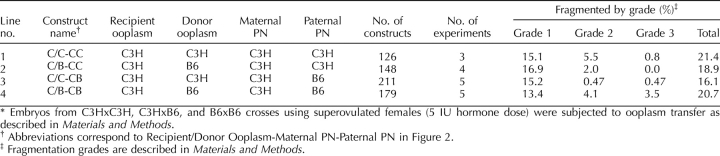
We conducted additional studies employing ooplasm transfer and paternal pronuclear transfer to test whether a mixture of different ooplasm affects outcome and whether ooplasmic effects on paternal PN could affect outcome (Tables 2 and 3). These studies confirmed the predominant role of the maternal PN. Although subtle effects of these manipulations on the severity of the cytofragmentation phenotype were seen in some instances, indicative of interactions between the ooplasm and the paternal pronucleus, the total rates of cytofragmentation were generally unaffected (Figs. 2 and 3; Tables 2 and 3). With paternal PN transfer, the total G2 + G3 cytofragmentation rates were affected significantly in both experiments (CCC/B vs. CCC/C and CCB/B vs. CCB/C) by whether the paternal PN formed within a B6 or C3H ooplasm within same embryo genotype (Table 3). No consistent effect was seen on total rate of fragmentation; CCB/B was greater than CCB/C; however, CCC/B was not greater than CCC/C. These results reveal a subtle effect of the ooplasm on paternal PN function, leading to a limited degree of paternal PN effect on cytofragmentation phenotype.
TABLE 2.
Effect of ooplasm transfer on cytofragmentation in two-cell stage mouse embryos.*
TABLE 3.
Effect of paternal pronuclear transfer on cytofragmentation in two-cell stage mouse embryos.
FIG. 2. Schematic illustration of ooplasm transfers performed using inbred strains of mice. Black ooplasm and black PN represent B6 (denoted B), and white ooplasm and white PN represent C3H (denoted C). Cytoplasts were approximately the size of a polar body. The combinations of recipient ooplasm, donor ooplasm, maternal PN, and paternal PN are shown below the embryos, and are those used in Table 2.
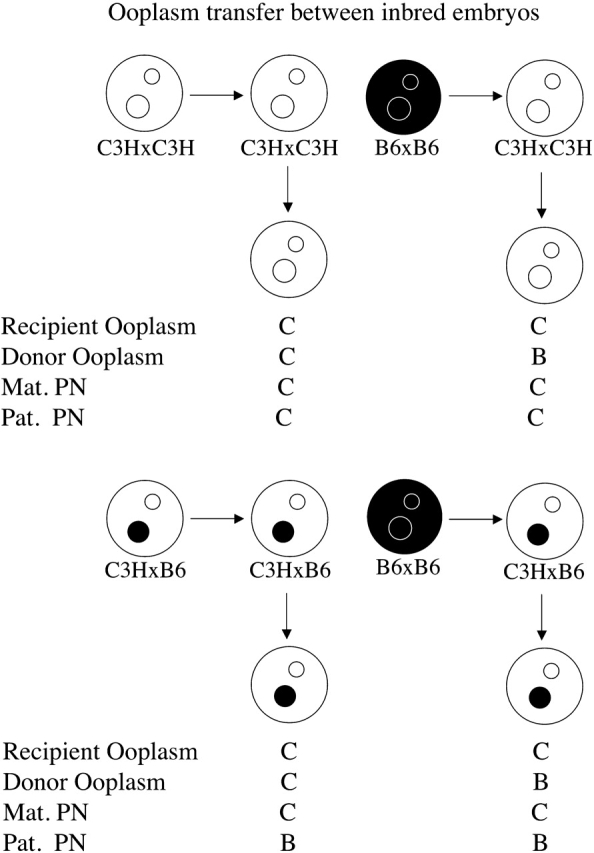
FIG. 3. Schematic illustration of paternal PN transfers performed using inbred strains of mice. Black ooplasm and black PN represent B6 (denoted B), and white ooplasm and white PN repersent C3H (denoted C). The combinations of ooplasm, maternal PN, paternal PN, and paternal PN source ooplasm are shown below the embryos, and are those used in Table 3.
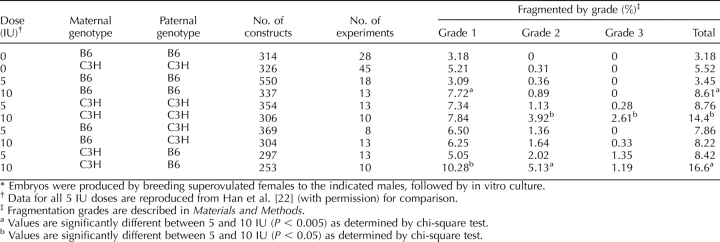
Another factor that could contribute to the effect of the maternal genotype is the response to the superovulation protocol. We evaluated this possibility by employing two different doses of hormones and then comparing rates of cytofragmentation among embryos produced with different combinations of maternal and paternal strains (Table 4). Neither B6 nor C3H females displayed any statistically significant difference in the total incidence of cytofragmentation following spontaneous ovulation compared with superovulation with 5 IU hormones (the dose employed for all previous studies). For both strains of female, the rate of cytofragmentation was elevated significantly to a similar degree (2.7-fold and 2.6-fold for B6 and C3H homozygous matings, respectively) using 10 IU hormones for superovulation compared with spontaneous ovulation. This difference between 5- and 10-IU doses was seen in all but one combination of maternal and paternal strains (B6×C3H). To avoid the effect of the higher hormone dose and to maintain consistency with previous studies, only the 5-IU hormone dosage was used for all other experiments employing superovulation.
TABLE 4.
Effects of parental genotype and gonadotropin dosage on cytofragmentation of two-cell stage mouse embryos.*
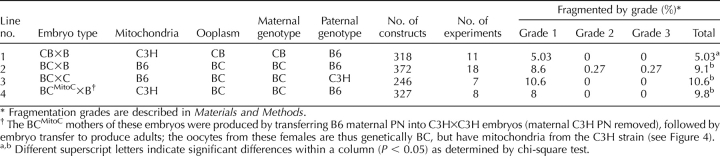
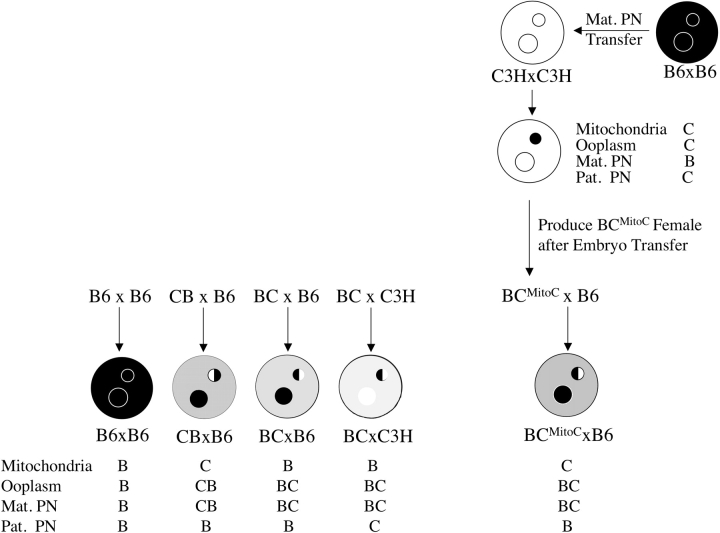
We also reported a significant parental origin effect on cytofragmentation, with reciprocal F1 hybrid maternal genotypes yielding significantly different rates [17]. This effect could be the result of either an epigenetic effect within the embryonic genome, or an effect of the mitochondria. To discriminate between these possibilities, we constructed by PN transfer females that were B6×C3H genetically but possessed C3H mitochondria, and we compared the rates of cytofragmentation among embryos from these females to normal F1 hybrid females (Table 5). We can exclude an effect of maternal mitochondrial origin, because nuclear transfer F1 hybrid females possessing the maternal genome of one strain but the mitochondria of the opposite strain yield embryos that fragment according to the maternal nuclear genotype (Fig. 4 and Table 5). Specifically, females that are genetically BC but possess C3H mitochondria yield embryos fragmenting at the same rate as normal BC females.
TABLE 5.
Effects of mitochondrial origin and maternal F1 hybrid genotype on cytofragmentation at the two-cell stage.
FIG. 4. Schematic illustration of the embryo types with different mitochondria origin and maternal F1 hybrid genotype. Black ooplasm and black PN represent C57BL/6 (B6, denoted B), and white ooplasm and white PN represent C3H (denoted C). Light stippled ooplasm is from C3H×B6 (denoted CB), heavy stippled ooplasm is from B6×C3H (denoted BC), and medium stippled ooplasm is from B6×C3H ooplasm with C3H mitochondria origin (denoted BCMitoC). Maternal PNs with left half white and right half black are from C3H×B6 (denoted CB) donors. Maternal PNs with left half black and right half white are from B6×C3H (denoted BC) donors. The combinations of mitochondria, ooplasm, maternal PN, and paternal PN are shown below the embryos, and are those used in Table 5.
Nuclear Versus Ooplasmic Basis for Parental Origin Effect
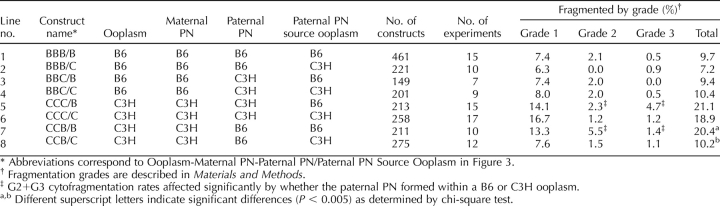
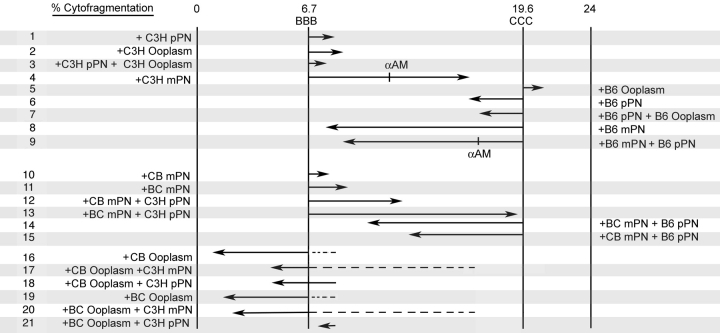
The parental origin effect, therefore, combined with the dominant effect of the maternal PN on cytofragmentation, indicated a novel and important role for an epigenetic mechanism, that is, genomic imprinting, in controlling the cytofragmentation rate. Our objective here was to define the specific interactions through which these epigenetic effects are mediated. We wanted to determine whether the parental origin effect is a property of the ooplasm or of the maternal PN; that is, whether the product of the imprinted gene was present in the ooplasm, expressed from the embryonic genome, or both. We produced a series of embryo constructs possessing different combinations of F1 ooplasm, F1 or inbred maternal PN, and inbred paternal PN, including combinations that separated the F1 ooplasm from the F1 maternal PN (>2800 maternal PN transfer constructs total, employing F1 hybrid oocytes). The effects of F1 maternal PN are presented in Table 6, and the effects of F1 ooplasm are presented in Table 7 (Fig. 5). In addition, a summary of the effects of each manipulation on the expected cytofragmentation rate is presented in Figure 6, where the origin and direction of each vector are the departures from the expected cytofragmentation rate, and the length of each vector is proportional to the strength of the effect of each manipulation. It should be noted that all of the data provided in Tables 1, 6, and 7 and in Figure 6 are from maternal PN transfer embryos, allowing these experiments to be compared directly to each other and eliminating the microsurgical procedure itself as an experimental variable.
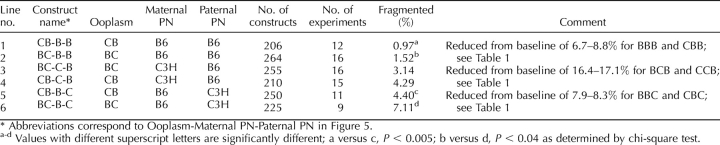
TABLE 6.
Effects of F1 maternal PN on cytofragmentation rates in mouse two-cell stage embryos produced by maternal PN transfer.
TABLE 7.
Effects of F1 hybrid ooplasm on cytofragmentation rates in mouse two-cell stage embryos produced by maternal PN transfer.
FIG. 6. Quantitative effects of pronucleus and ooplasm strain of origin on cytofragmentation in maternal pronucleus transfer studies. The parental rates of cytofragmentation for B6 and C3H strains are indicated (6.7% and 19.6%). The effect of each manipulation is represented by a vector, where the origin and direction of each vector is the observed departure from the cytofragmentation rate expected and the length of each vector is directly proportional to the strength of the effect of each manipulation. In general, addition of C3H alleles into the pronuclei enhances the rate of fragmentation above the B6 parental level, whereas addition of B6 alleles suppresses fragmentation below the C3H parental level. Note the synergistic effect of a BC maternal pronucleus and C3H paternal pronucleus (vector no. 13), which is greater than the sum of individual effects of BC maternal PN (vector no. 11) and C3H paternal PN (vector no. 1). This degree of synergy is not seen with a CB maternal pronucleus (vector no. 12), revealing the novel transgenerational effect of the maternal grandpaternal genome. Note also the hybrid vigor effect observed with F1 ooplasm when separated from F1 maternal PN (vector nos. 16–21), which suppresses fragmentation.
A significant parental origin effect is observed in constructs possessing reciprocal F1 hybrid maternal PN. This effect is most apparent when the paternal PN is of C3H origin and the ooplasm of B6 origin (lines 3 and 4, Table 6, and vectors 12 and 13, Fig. 6). In these constructs, the combination of a BC maternal PN and a C3H paternal PN yields a synergistic increase in the total rate of fragmentation, with a value greater than the sum of individual effects of a BC maternal PN and a C3H paternal PN (compare line 4, Table 6, with the sum of line 2, Table 6 and line 3, Table 1; or compare sum of vectors 1 and 11 with vector 13, Fig. 6). Overall, the effect of the combination is to raise the rate of cytofragmentation by nearly 3-fold over the rate seen with all B6 components (Table 1, line 1), and to produce an overall difference in fragmentation rate that is more than three times greater than the sum of differences seen with the C3H paternal PN and C3H ooplasm alone. If there were no grandparental origin effect (i.e., maternal and paternal alleles functioned identically in the oocyte), there would be no difference between the two reciprocal F1 hybrid maternal PN. Contrary to this prediction, we note that the synergy between the combination of B6 ooplasm, CB maternal PN, and C3H paternal PN is much reduced (compare line 3, Table 6, with the sum of line 1, Table 6 and line 3, Table 1; or compare the sum of vectors 1 and 10 with vector 12, Fig. 6) and that there is a significant difference between BC and CB maternal PN effects in combination with B6 ooplasm and C3H paternal PN, indicating that the synergistic increase in cytofragmentation specifically requires the transmission of C3H alleles from the maternal grandfather. We note that there is a trend for the reciprocal effect, wherein a CB maternal PN interacts more strongly with a B6 paternal PN within a C3H ooplasm (compare lines 5 and 6, Table 6); however, this does not reach statistical significance. We also note that F1 hybrid maternal PN can direct higher rates of fragmentation (lines 1 and 2, Table 6) than B6 maternal PN in BBB constructs (Table 1), and lower rates of fragmentation (lines 5 and 6, Table 6) than C3H maternal PN in CCB constructs (Table 1), once again attesting to the major role played by the maternal PN in controlling cytofragmentation. The combined interactions between ooplasm and paternal PN and between maternal grandpaternal alleles and the paternal PN, however, clearly modulate the otherwise dominant effect of the maternal PN.
The dependence of the grandparental origin effect on the ooplasm strain of origin indicates that the ooplasm influences the overall effect of a maternal F1 PN, and that one would predict that a C3H component in the ooplasm could cooperate with the maternal PN to increase cytofragmentation. An F1 hybrid ooplasm combined with a C3H maternal PN indeed yields a higher rate of fragmentation than a B6 maternal PN (compare lines 3 and 4 with lines 1 and 2, Table 7; compare vectors 17 and 18 with vectors 16 and 19, Fig. 6). The effect of F1 ooplasm, however, is tempered dramatically in three ways. First, F1 ooplasm has a strongly suppressive effect on cytofragmentation rate, regardless of inbred strain origin of maternal and paternal PN (compare Tables 1 and 7, and note the magnitude and direction of vectors 16–21, Fig. 6). For example, with B6 maternal and B6 paternal PN, F1 ooplasm reduces the rate of cytofragmentation to less than 2%, compared with 6.7% seen with B6 ooplasm (4.4-fold to 6.9-fold reduction). This indicates that there is a nonadditive effect of an F1 genotype on ooplasm properties indicative of heterosis; in other words, the F1 phenotype is not characteristic of either parental strain and is not intermediate between the two parental strains. Second, there is an apparent interaction between the F1 hybrid ooplasm and the paternal PN. Either CB or BC ooplasm, combined with a C3H paternal PN, yields a significantly greater (4.5-fold, and 4.7-fold, P < 0.005 and P < 0.04, respectively) rate of cytofragmentation than the same F1 ooplasm combined with a B6 paternal PN (compare vector 16 with 18 and vector 19 with 21, or lines 1 and 5, and lines 2 and 6, Table 7). The most unfavorable combinations tested, F1 hybrid ooplasm with a C3H paternal PN, in fact yield the greatest rates of fragmentation, even with a B6 maternal PN present (lines 5 and 6, Table 7; vectors 18 and 21, Fig. 6). Thus, the normally dominant effect of the maternal PN is overcome by the F1 ooplasm suppressive effect, so that the ooplasm-paternal PN interaction can be revealed (again, compare vector 16 with 18 and vector 19 with 21, Fig. 6). Moreover, a maternal C3H PN does not direct the typical high rate of fragmentation with F1 ooplasm as seen with inbred ooplasm, with F1 ooplasm suppressing fragmentation by as much as 5.4-fold (compare vectors 17 and 20 with vector 4, Fig. 6; see also lines 3–4, Table 7, and lines 5–8, Table 1). Third, the BC ooplasm displays 1.5-fold to 1.6-fold greater rates of cytofragmentation than the CB ooplasm in two of the three manipulations (compare vector 19 with 16, vector 21 with 18, and vector 20 with 17, Fig. 6), but this effect does not reach statistical significance in any of the three maternal-paternal genetic combinations. Thus, the parental origin effect observed in crosses with reciprocal F1 mothers is masked by the F1 ooplasm suppressive effect.
DISCUSSION
The studies presented here encompass an extensive series of pronuclear transfer experiments to test the specific effects of genetic origin and composition of maternal and paternal PN, mitochondria, and ooplasm on the blastomere integrity of mouse two-cell stage embryos. The data reveal a number of novel epigenetic and genetic effects on this early embryonic phenotype not observed previously, most notably synergistic interactions between both PN and the ooplasm, a parental origin effect dependent upon the genetic origin of the embryo's maternal grandfather, and a hybrid vigor effect of the ooplasm.
Predominant Effect of the Maternal PN
We demonstrated previously that the maternal PN exerts the greatest control over cytofragmentation rate, and that this effect is sensitive to α-amanitin, indicating that it depends on gene expression from the maternal genome at the late one-cell or early two-cell stage (Han et al. [22] and vectors 1–9, Fig. 6). This result indicates that one or more genes expressed exclusively or predominately from the maternal PN control cytofragmentation in the early embryo. These genes may either be imprinted or reside on the X chromosome. The strain difference that is observed between C3H and C57BL/6 maternal PN may thus reflect strain-specific or genetically polymorphic imprinting, as has been seen in other situations (Xu et al. [23]; review, Latham [24]), or a difference in allele activity or function between the two. Resolution of this point awaits the eventual identification of the affected gene(s) and characterization of their chromosomal locations and mechanisms of regulation.
Transgenerational Contribution to the Maternal PN Effect
The effect of the maternal PN is further dependent upon a transgenerational epigenetic effect, wherein genes derived from the maternal grandfather comprise critical determinants of cytofragmentation (vectors 10–15, Fig. 6). An F1 hybrid maternal PN shifts the phenotype toward values intermediate between the two parental strains. This effect is modest when the paternal PN is of B6 origin, but much more pronounced when the paternal PN is of C3H origin. The selective ability of the maternal grandpaternal allele to exert these effects implies transmission of epigenetic information (imprinting) through the oocyte to the embryo. Consequently, it is clear that the phenotype of early mouse embryos is determined by at least three distinct epigenomes: maternal, paternal, and maternal grandpaternal.
Transgenerational effects via epigenetic mechanisms affecting specific genes have been described elsewhere. For example, the DNA methylation state of a retrotransposon within the Axin1Fu allele can be transmitted to progeny via either oocyte or sperm, and this epigenetic state is sensitive to genetic strain [25]. Similarly, the DNA methylation state of the endogenous IAP retrotransposon at the mouse agouti viable yellow (Avy) locus, also sensitive to genetic strain, can be transmitted to progeny via the oocyte [26, 27]. Maternal dietary supplementation of methyl donors can affect the DNA methylation state of the Avy and Axin1Fu loci [26, 28]; however, this dietary effect is not cumulative across generations [29]. In humans, paternal cigarette smoking and diet exert sex-dependent transgenerational effects [30], and grandparental effects related to undernutrition are seen in the grandchildren of victims of the Dutch famine and in rat studies [31], which likely involve one or more epigenetic mechanisms. Other transgenerational epigenetic effects, likely heritable epimutations [32] or possible failures to reestablish the proper epigenetic state [33], have been postulated in some human disease phenotypes [34–36] and with the mouse metastable epialleles discussed above. The unexpected phenotypes observed in both the human disease examples and with metastable epialleles have been suggested to appear as a result of failure to reset genome imprints completely; that is, as a result of an abnormal process. It is also noteworthy that some epigenetic distinction is maintained between maternal and paternal alleles long after differential methylation differences have been eliminated during both normal spermatogenesis [37] and oogenesis [38], so that one parental allele consistently undergoes its final DNA methylation modification ahead of the other, again indicating that some epigenetic information endures throughout the entire process of imprint erasure and reestablishment, fertilization, and early development. The data presented here reveal for the first time that epigenetic information unrelated to maternal diet or other exogenous factors and transmitted from the maternal grandfather affects embryonic phenotype at the earliest stage when the embryonic genome becomes transcriptionally active.
Our data significantly extend observations of transgenerational effects. First, our results demonstrate a novel transgenerational effect of the maternal grandpaternal epigenome, yielding a quantitative trait affecting early embryo phenotype. We note that the maternal grandpaternal strain of origin can yield visible effects on the phenotype at a rate of up to 20% of embryos. We note, further, that reciprocal effects of the alternate grandpaternal alleles could also exist but may not be evident, potentially affecting an equivalent or greater fraction of additional embryos in each generation. This effect could be mediated by a single gene neighboring a retrotransposon, as seen with Axin1Fu and Avy, or could reflect direct epigenetic effects on one or more endogenous genes. Second, our results demonstrate that such transgenerational effects can be manifested from the very earliest time of embryonic gene expression, and do not require further epigenetic changes as development progresses. Third, our results demonstrate interactions between the maternal grandpaternal epigenome and the ooplasm and fertilizing sperm pronucleus. The dependence of transgenerational epigenetic effects on interactions between the two parental genomes and nuclear-cytoplasmic interactions is novel and adds a significant new aspect to understanding how transgenerational effects are mediated.
Ooplasm Hybrid Vigor Effect
We also observe a novel and striking example of hybrid vigor, with F1 ooplasm of either type (BC or CB) markedly suppressing the incidence of fragmentation when separated from the maternal F1 PN. This effect is highly significant, in that it eliminates the normally predominant effect of the maternal PN (note magnitude and direction of vectors 17 and 20, Fig. 6) and unmasks interactions between the ooplasm and the paternal PN (compare vector 16 with 18 and vector 19 with 21, Fig. 6). This result suggests that the normal function of the maternal PN is to suppress an inherent predisposition of the oocyte and zygote toward cytofragmentation via a transcription-dependent mechanism.
The F1 hybrid vigor effect observed with the ooplasm constitutes a striking example of heterosis, a process whereby genetic differences between parents can produce synergistic, favorable effects in offspring (hybrid vigor) or unfavorable (outbreeding depression) effects. Hybrid vigor is observed in both plants and animals, but it has not typically been described for the early stages of animal development. One study of interspecies breeding effects in embryos of bovine species failed to disclose a consistent effect; however, it appeared that interbreeding favored the in vitro development of purebred embryos (i.e., akin to outbreeding depression), an effect that may have resulted from differential effects of in vitro culture on the two species of embryos [39]. In mice, F1 hybrid embryos tolerate in vitro culture better than embryos from many inbred strains, and F1 hybrid ooplasm is superior to inbred strain ooplasm for cloning by nuclear transfer [40]. Additionally, F1 hybrid donor cell genomes more readily produce cloned embryos than outbred or inbred donor nucleus strains [40, 41]. Collectively, these observations indicate that hybrid vigor effects can significantly affect oocyte quality and early embryogenesis.
What mechanisms may underlie such effects of F1 genotypes? Overdominance, wherein different alleles interact more productively with one another than is seen in parental genotypes, is widely thought to contribute to heterosis [42]. In chickens, for example, multiple genes contribute to heterosis in fat content as a quantitative trait [43]. However, other mechanisms may apply. In maize, small-scale deletions or duplications that alter genic content for specific fragments, as well as variations in transposons and repetitive DNA around genes can play a role [44]. Such differences can lead to novel F1 hybrid gene expression patterns that are nonadditive and outside of the range displayed by the two parental strains. Allelic interactions are required to maintain a repressed state at the purple (pl) locus in maize, and some alleles lack the necessary sequences for this interaction [45]. In Arabidopsis, differential heterozygosity at specific loci is critical for heterosis [46]. The results presented here offer a discrete phenotype in a mammalian system that can be employed for identifying the genes that provide the basis of hybrid vigor, and subsequently identifying the molecular mechanism underlying the process. It is tempting to speculate that heterozygosity may lead to allelic interactions affecting the expression of genes that promote or enhance cytofragmentation and apoptotic processes in the oocyte.
Interactions with the Paternal PN
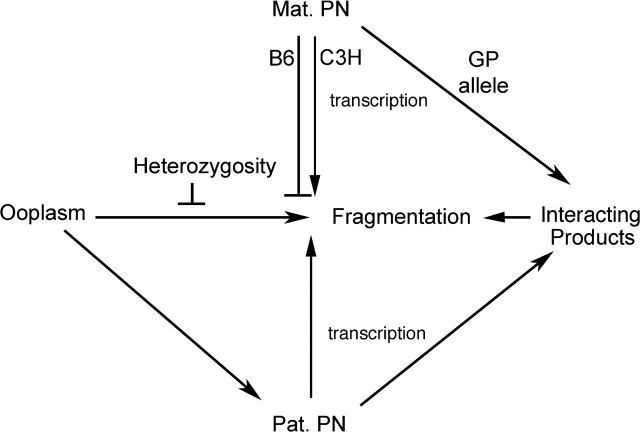
The transgenerational effect requires interactions between the maternal grandpaternal genetic contribution and the fertilizing paternal genome, with a synergistic interaction between C3H maternal grandpaternal and C3H paternal contributions (compare magnitude of difference between vectors 12 and 13 with magnitude of difference between vectors 14 and 15, Fig. 6). This synergy is affected by the ooplasm strain of origin, indicating that all three components of the embryo contribute to the overall rate of cytofragmentation. Additional effects of paternal genotype may exist to account for differences in outcomes of embryo crosses with inbred strains, specifically the increased cytofragmentation in crosses of B6 females with C3H males versus B6 males with no pronuclear transfer [17, 22]. Other results also reveal significant interactions between the ooplasm and paternal PN when this maternal PN effect is ablated. We note that the ooplasm transfer data and the paternal pronucleus transfer data also indicated the existence of ooplasm-paternal PN interactions that were largely masked by the maternal PN effect (Tables 2 and 3). The demonstration of ooplasm-paternal PN effects by multiple approaches indicates that these effects are real, and that they likely contribute to the paternal genotype effect seen with crosses employing the C3H and B6 inbred strains (i.e., enhanced fragmentation in embryos from B6 females mated to C3H males, compared with B6 males) [17, 22]. It should also be noted that, although the ooplasmic hybrid vigor effect only became apparent when the F1 ooplasm was separated from the F1 maternal PN, the F1 ooplasm must nevertheless suppress fragmentation, even in the absence of microsurgery, because we see that the synergistic profragmentation effect of a BC maternal PN and a C3H paternal PN is not realized in simple crosses that maintain the F1 maternal PN-ooplasm combination, and we see that the parental origin effect with a B6 paternal PN is masked when the ooplasm is from an inbred strain instead of from an F1 strain. A complex set of interactions thus regulates cytofragmentation in the early embryo. These interactions are summarized schematically in Figure 7.
FIG. 7. Schematic diagram summarizing interactions between maternal and paternal pronuclei and ooplasm affecting cytofragmentation in two-cell stage mouse embryos. The initial predisposition toward cytofragmentation is established in the ooplasm. This can occur either before or after fertilization, but is dependent upon the oocyte strain of origin. The predisposition toward fragmentation can be attenuated by heterozygosity during oogenesis, accounting for the observed hybrid vigor effect. This hybrid vigor effect, however, is seen only when the ooplasm is separated from its normal F1 maternal PN partner. This is most likely because C3H alleles inherited via the maternal PN promote fragmentation, or fail to suppress it fully, when combined with ooplasmic C3H products. The predominant effect of the maternal PN on cytofragmentation, and the absence of this effect when F1 ooplasm is separated from the F1 maternal PN, indicate that the primary function of the maternal PN may be to suppress the overall rate of cytofragmentation directed by the ooplasm. This effect is transcription dependent, and it may involve direct effects of maternal PN-encoded gene products on the pathway leading to cytofragmentation. A B6 maternal PN suppresses cytofragmentation much more than a C3H maternal PN, and the latter may actually promote cytofragmentation. The maternal PN effect, however, must also require interactions either between products encoded by the zygotic maternal and paternal PN, or between maternal PN-encoded products and the paternal PN, the overall effect of which is to promote cytofragmentation. This requirement arises from the finding that the parental origin effect seen with CB and BC maternal PN is sensitive to the paternal PN genotype and the ooplasm strain of origin. For example, we observe a strong synergistic interaction between a BC maternal PN and a C3H paternal PN, which clearly indicates interactions between products expressed from these two genomes in the early embryo. Additionally, the ooplasm itself appears to modify the paternal PN to promote cytofragmentation, a subtle effect that is seen in the severity of fragmentation following one combination of paternal PN transfer (Table 3, compare rows 7 and 8). Additional effects of ooplasm strain of origin are seen in that the ooplasm of one strain tends to increase cytofragmentation when combined with maternal plus paternal PN of the opposite strains (compare lines 1 and 4, and lines 7 and 8, Table 1), and also in the effect of transient exposure of the paternal PN to one kind of ooplasm followed by a need to interface with ooplasm of the opposite strain (compare grades 2 and 3 fragmentation, lines 5 and 6, and lines 7 and 8, Table 3). GP, grandpaternal.
Understanding Origins of Cytofragmentation
The results presented here also have significant implications for understanding the origin of cytofragmentation phenotypes in human patients undergoing assisted reproduction, where more than 80% of embryos are subject to cytofragmentation [7]. First, our results reveal that high doses of hormones for inducing ovulations can lead to increased rates of cytofragmentation (Table 4). This may reflect an increased recruitment of oocytes of lesser quality that are predisposed to fragment when the more extreme doses of hormone are employed. Additionally, our results illustrate the importance of maternal and paternal genotype on phenotype, as well as a novel epigenetic transgenerational effect. The latter finding further raises the possibility of environmental factors, such as maternal diet (i.e., availability of methyl group donors) on the cytofragmentation phenotype. Through detailed molecular and genetic studies it should be possible to identify the genes that control the rate of cytofragmentation, and hence develop novel diagnostic tools with which to identify patients that may be at risk for increased cytofragmentation, thus enabling this parameter to be addressed prospectively during assisted reproduction procedures.
Overall, these results demonstrate a central role for the maternal genotype in the control of early embryo cellular integrity. A severe deficiency in maintaining cellular integrity compromises embryo developmental potential. The interactions between maternal and paternal PN and the ooplasm shown here are likely to be highly relevant to other biological processes such as speciation and evolution. The synergistic interaction between a BC maternal PN and a C3H paternal PN, for example, indicates that certain combinations of alleles can lead to unfavorable consequences. This is reminiscent of the polar lethal Ovum mutant phenotype manifested in the DDK mouse strain, wherein DDK ova fertilized by an “alien” non-DDK strain of sperm display >95% lethality during cleavage [47]. This outcome has been accounted for, in part, by the alleles expressed in the ovum that interact negatively with alleles in the paternally inherited genome [48–52]. The hybrid vigor effect that we observe for F1 ooplasm appears to counteract such negative ooplasm-paternal PN interactions. In a population of randomly breeding individuals, the specific combinations of oocyte-expressed alleles and paternally inherited alleles could influence heritability of these alleles and thereby favor the reproductive success of particular maternal and paternal genetic combinations. The predominant effect of the maternal PN in controlling blastomere integrity and the increased fragmentation with the presence of C3H alleles would seem to favor the reproductive success of favorable hybrid allelic combinations, which would complement the hybrid vigor effect observed for the ooplasm.
Acknowledgments
We thank Sue Varmuza for helpful comments on the manuscript.
Footnotes
1Supported in part by grants from the National Institutes of Health, National Institute for Child Health and Human Development and National Center for Research Resources (HD41440 and RR15253 to K.E.L., and HD48730 and HD34508 to C.S.).
REFERENCES
- Brison D, Schultz RM. Apoptosis during mouse blastocyst formation: evidence for a role for survival factors including transforming growth factor alpha. Biol Reprod 1997; 56: 1088 1096 [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Latham KE, Casper RF, Varmuza SL. Expression and regulation of genes associated with cell death during murine preimplantation embryo development. Mol Reprod Dev 1998; 51: 243 253 [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Rogers I, Fasciani A, Casper RF, Varmuza S. Effect of maternal age and conditions of fertilization on programmed cell death during murine preimplantation embryo development. Mol Hum Reprod 1998; 4: 139 145 [DOI] [PubMed] [Google Scholar]
- Brewster JL, Martin SL, Toms J, Goss D, Wang K, Zachrone K, Davis A, Carlson G, Hood L, Coffin JD. Deletion of Dad1 in mice induces an apoptosis-associated embryonic death. Genesis 2000; 2: 271 278 [PubMed] [Google Scholar]
- Xu J, Cheung T, Chan ST, Ho P, Yeung WS. The incidence of cytoplasmic fragmentation in mouse embryos in vitro is not affected by inhibition of caspase activity. Fertil Steril 2001; 75: 986 991 [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Varmuza S, Casper RF. Involvement of programmed cell death in preimplantation embryo demise. Hum Reprod Update 1995; 1: 558 566 [DOI] [PubMed] [Google Scholar]
- Jurisicova A, Varmuza S, Casper RF. Programmed cell death and human embryo fragmentation. Mol Hum Reprod 1996; 2: 93 98 [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis PW. DNA strand break and phosphatidylserine redistribution in newly ovulated cultured mouse and human oocytes: occurrence and relationship to apoptosis. Hum Reprod 1998; 13: 1317 1324 [DOI] [PubMed] [Google Scholar]
- Antczak M, Van Blerkom J. Temporal and spatial aspects of fragmentation in early human embryos: possible effects on developmental competence and association with the differential elimination of regulatory proteins from polarized domains. Hum Reprod 1999; 14: 429 447 [DOI] [PubMed] [Google Scholar]
- Hardy K. Apoptosis in the human embryo. Rev Reprod 1999; 4: 125 134 [DOI] [PubMed] [Google Scholar]
- Byrne AT, Southgate J, Brison DR, Leese HJ. Analysis of apoptosis in the preimplantation bovine embryo using TUNEL. J Reprod Fertil 1999; 117: 97 105 [DOI] [PubMed] [Google Scholar]
- Otoi T, Yamamoto K, Horikita N, Tachikawa S, Suzuki T. Relationship between dead cells and DNA fragmentation in bovine embryos produced in vitro and stored at 4 degrees C. Mol Reprod Dev 1999; 54: 342 347 [DOI] [PubMed] [Google Scholar]
- Liu L, Trimarchi JR, Keefe DL. Involvement of mitochondria in oxidative stress-induced cell death in mouse zygotes. Biol Reprod 2000; 62: 1745 1753 [DOI] [PubMed] [Google Scholar]
- Matwee C, Betts DH, King WA. Apoptosis in the early bovine embryo. Zygote 2000; 8: 57 68 [DOI] [PubMed] [Google Scholar]
- Alikani M, Calderon G, Tomkin G, Garrisi J, Kokot M, Cohen J. Cleavage anomalies in early human embryo and survival after prolonged culture in vitro. Hum Reprod 2000; 15: 2634 2643 [DOI] [PubMed] [Google Scholar]
- Watson AJ, De Sousa P, Caveney A, Barcroft LC, Natale D, Urquhart J, Westhusin ME. Impact of bovine oocyte maturation media on oocyte transcript levels, blastocyst development, cell number, and apoptosis. Biol Reprod 2000; 62: 355 364 [DOI] [PubMed] [Google Scholar]
- Hawes SM, Chung YG, Latham KE. Genetic and epigenetic factors affecting blastomere fragmentation in preimplantation stage mouse embryos. Biol Reprod 2001; 65: 1050 1056 [DOI] [PubMed] [Google Scholar]
- Hardy K, Spanos S, Becker D, Iannelli P, Winston RM, Stark J. From cell death to embryo arrest: mathematical models of human preimplantation embryo development. Proc Nat Acad Sci U S A 2001; 98: 1655 1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brison DR, Schultz RM. Increased incidence of apoptosis in transforming growth factor alpha-deficient mouse blastocysts. Biol Reprod 1998; 59: 136 144 [DOI] [PubMed] [Google Scholar]
- Moley KH. Hyperglycemia and apoptosis: mechanisms for congenital malformations and pregnancy loss in diabetic women. Trends Endocrinol Metab 2001; 12: 78 82 [DOI] [PubMed] [Google Scholar]
- Chi MM, Hoehn A, Moley KH. Metabolic changes in the glucose-induced apoptotic blastocyst suggest alterations in mitochondrial physiology. Am J Physiol Endocrinol Metab 2002; 283: E226 E232 [DOI] [PubMed] [Google Scholar]
- Han Z, Chung YG, Gao S, Latham KE. Maternal factors controlling blastomere fragmentation in early mouse embryos. Biol Reprod 2005; 72: 612 618 [DOI] [PubMed] [Google Scholar]
- Xu Y, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun 1993; 197: 747 754 [DOI] [PubMed] [Google Scholar]
- Latham KE. Epigenetic modification and imprinting of the mammalian genome during development. Curr Top Dev Biol 1999; 43: 1 49 [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin (Fu) allele occurs after maternal and paternal transmission. Proc Nat Acad Sci U S A 2003; 100: 2538 2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 1998; 12: 949 957 [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 1999; 23: 314 318 [DOI] [PubMed] [Google Scholar]
- Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis 2006; 44: 401 406 [DOI] [PubMed] [Google Scholar]
- Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J 2007; 21: 3380 3385 [DOI] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, Golding J. ALSPAC Study Team. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 2006; 14: 159 166 [DOI] [PubMed] [Google Scholar]
- Barker DJ, Gluckman PD, Robinson JS. Conference report: fetal origins of adult disease—report of the First International Study Group, Sydney, 29–30 October 1994. Placenta 1995; 16: 317 320 [DOI] [PubMed] [Google Scholar]
- Chong S, Youngson NA, Whitelaw E. Heritable germline epimutation is not the same as transgenerational epigenetic inheritance. Nat Genet 2007; 39: 574 575 [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI. Reply to “Heritable germline epimutation is not the same as transgenerational epigenetic inheritance.” Nat Genet 2007; 39: 575 576 [DOI] [PubMed] [Google Scholar]
- Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, el-Maarri O, Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet 2003; 72: 571 577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 2003; 34: 157 165 [DOI] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 2006; 38: 1178 1183 [DOI] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 2000; 9: 2885 2894 [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 2004; 13: 839 849 [DOI] [PubMed] [Google Scholar]
- Fischer AE, Bernal DP, Gutierrez-Robayo C, Rutledge JJ. Estimates of heterosis for in vitro embryo production using reciprocal crosses in cattle. Theriogenology 2000; 54: 1433 1442 [DOI] [PubMed] [Google Scholar]
- Gao S, Czirr E, Chung YG, Han Z, Latham KE. Genetic variation in oocyte phenotype revealed through parthenogenesis and cloning: correlation with differences in pronuclear epigenetic modification. Biol Reprod 2004; 70: 1162 1170 [DOI] [PubMed] [Google Scholar]
- Kishigami S, Bui HT, Wakayama S, Tokunaga K, Van Thuan N, Hikichi T, Mizutani E, Ohta H, Suetsugu R, Sata T, Wakayama T. Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J Reprod Dev 2007; 53: 165 170 [DOI] [PubMed] [Google Scholar]
- Baack EJ, Rieseberg LH. A genomic view of introgression and hybrid speciation. Curr Opin Genet Dev 2007; 17: 513 518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abasht B, Lamont SJ. Genome-wide association analysis reveals cryptic alleles as an important factor in heterosis for fatness in chicken F2 population. Anim Genet 2007; 38: 491 498 [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM. Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res 2007; 17: 264 275 [DOI] [PubMed] [Google Scholar]
- Hollick JB, Chandler VL. Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 1998; 150: 891 897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NH, Chen ZJ. Molecular marker genotypes, heterozygosity and genetic interactions explain heterosis in Arabidopsis thaliana. Heredity 2005; 94: 295 304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi N, Tomita T, Kondo K. Differences of fertility in reciprocal crosses between inbred strains of mice. DDK, KK and NC. J Reprod Fertil 1967; 13: 41 50 [DOI] [PubMed] [Google Scholar]
- Renard JP, Baldacci P, Richoux-Duranthon V, Pournin S, Babinet C. A maternal factor affecting mouse blastocyst formation. Development 1994; 120: 797 802 [DOI] [PubMed] [Google Scholar]
- Baldacci PA, Richoux V, Renard JP, Guenet JL, Babinet C. The locus Om, responsible for the DDK syndrome, maps close to Sigje on mouse chromosome 11. Mamm Genome 1992; 2: 100 105 [DOI] [PubMed] [Google Scholar]
- Sapienza C, Paquette J, Pannunzio P, Albrechtson S, Morgan K. The polar-lethal Ovum mutant gene maps to the distal portion of mouse chromosome 11. Genetics 1992; 132: 241 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Naumova AK, Verner AE, Jin WH, Sapienza C. Confirmation of maternal transmission ratio distortion at Om and direct evidence that the maternal and paternal “DDK syndrome” genes are linked. Mamm Genome 1997; 8: 642 646 [DOI] [PubMed] [Google Scholar]
- Bell TA, de la Casa-Esperon E, Doherty HE, Ideraabdullah F, Kim K, Wang Y, Lange LA, Wilhemsen K, Lange EM, Sapienza C, de Villena FP. The paternal gene of the DDK syndrome maps to the Schlafen gene cluster on mouse chromosome 11. Genetics 2006; 172: 411 423 [DOI] [PMC free article] [PubMed] [Google Scholar]