Abstract
Erg, a member of the ETS family of transcription factors, has been implicated by previous studies in endothelial and haematopoietic development. Deregulation of the human ERG locus is associated with acute myeloid leukaemia, prostate cancer and Ewing's sarcoma. To better understand the role of Erg during early development, we utilised the zebrafish as a model amenable to descriptive and functional studies in vivo. Zebrafish have a single erg gene that is expressed in mesoderm and its vascular derivatives during angioblast migration, vasculogenesis and early angiogenesis. Mutant and morphant expression analyses placed erg in a genetic pathway downstream of cloche, tal1/scl and etsrp during early angioblast migration. Furthermore, a combination of gain-of-function and loss-of-function studies suggested a redundant yet specific role for erg in both angioblast specification/proliferation and early angiogenesis, and a synergistic interaction with the critical ETS factor etsrp.
1. Introduction
During embryonic zebrafish development, vasculogenesis begins at the 6-somite stage with the specification of angioblasts from a putative haemangioblast subset in the lateral plate mesoderm (Liao et al., 1998). Between the 10- and 15-somite stages, angioblasts migrate to the midline of the embryo, where they coalesce to form the dorsal aorta and cardinal vein, the first functional embryonic blood vessels. At the 26-somite stage, angiogenesis begins, with secondary vessels sprouting from these primary vessels along the trunk of the embryo (reviewed in Childs et al., 2002).
Many of the transcriptional regulators directing zebrafish mesodermal specification, vasculogenesis and angiogenesis belong to the ETS family of transcription factors. This large family contains 26 members that share a common DNA-binding “ETS domain”, and is delineated into 9 subfamilies according to conservation of various other domains (for review see Oikawa and Yamada, 2003).
During mesodermal specification, the ETS factor spi1/pu.1 drives myeloid specification in the anterior mesoderm (Rhodes et al., 2005), while etsrp is required to drive haemangioblasts towards vascular fate (Sumanas and Lin, 2006). Expression of the closely-related ETS factors fli1, fli1b and ets1 overlaps that of etsrp during the specification and migration of angioblasts, and functional studies indicate redundant roles for these factors during vasculogenesis in zebrafish (Pham et al., 2007).
The human ets related gene (ERG) is an ETS family member 96% homologous to FLI1 (Maroulakou and Bowe, 2000) first identified in 1987 (Reddy et al., 1987). ERG is a transcription factor containing four functional domains, each responsible for sequence specific DNA-binding, transcriptional activation, and repression of transcriptional activation (Reddy and Rao, 1991; Siddique et al., 1993). Human ERG is also an important proto-oncogene with roles in a range of human malignancies, including Ewing's sarcoma, acute myeloid leukaemia, and more than half of all prostate cancers (Ichikawa et al., 1994; Sorensen et al., 1994; Tomlins et al., 2005). Malignancies are linked to chromosomal translocations fusing ERG to EWS, TLS/FUS or TMPRSS2, producing either transcriptionally volatile fusion proteins or ERG overexpression. High ERG expression levels are also an independent correlate of poor prognosis in acute myeloid leukaemia (Marcucci et al., 2007).
Deletion studies of ERG in vitro have identified two distinct domains that take part in formation of homodimers, heterodimers and ternary complexes, both with itself and other ETS proteins (Carrere et al., 1998). In vitro studies in mammalian cells have demonstrated ERG to be important in a number of developmental processes. These include early haematopoiesis; during which ERG is expressed highly in early myeloid cells (Murakami et al., 1993), and endothelial cell differentiation (McLaughlin et al., 2001). In the mouse, Erg is expressed in mesodermal tissues including precartilaginous, urogenital and endothelial cells, positioning it for functions in cell migration and differentiation, as well as for establishment of endothelial fate in mesenchymal cells (Vlaeminck-Guillem et al., 2000). Transient expression of Erg has also been described during murine T-cell lineage specification (Anderson et al., 1999). A missense mutation of a conserved amino acid in the DNA-binding ETS domain of ERG, resulting in a putative loss-of-function allele, has recently been shown to result in haematopoietic stem cell (HSC) deficiencies in mice (Loughran et al., 2008). Homozygosity of this mutation results in loss of definitive erythropoiesis and death by E13.5. Mice heterozygous for this mutation have lower numbers of lineage-negative Sca-1+c-kit+ (LSK) cells, which represent the long-term repopulating HSC compartment. Furthermore, remaining LSK cells appear functionally deficient when competitively transplanted with wild-type LSK cells into lethally irradiated mice. These studies suggest a role for ERG in either the specification or function of haematopoietic progenitor cells during development and/or steady-state production and survival of haematopoietic precursors during adult life.
Transient functional studies of ERG include knock-down of ERG in human umbilical vein endothelial cells (HUVECs) with GeneBloc antisense oligonucleotides/siRNA (McLaughlin et al., 2001; Huang et al., 2005; Birdsey et al., 2008) and overexpression studies in Xenopus (Baltzinger et al., 1999), both of which support a role for ERG in endothelial differentiation. (McLaughlin et al., 2001) described decreased expression of known regulators of angiogenesis, factors important in cellular remodelling and angiogenesis, von Willebrand factor (an adhesive glycoprotein for coagulation of platelets and a proposed factor involved in tumour angiogenesis) (Girma et al., 1987; Zanetta et al., 2000) and RhoA, a small GTPase involved in cellular adhesion pathways (Nobes et al., 1995). Furthermore, GenBloc treatment of HUVECs impaired the formation of tubular structures in vitro. ERG mediates angiogenesis in part by transcriptional regulation of VE-cadherin, overexpression of which rescues apoptosis in ERG-deficient HUVECs (Birdsey et al., 2008). In Xenopus, overexpression of XIerg by injection of mRNA led to ectopic endothelial cell accumulation and perturbations of the cardiovascular system (Baltzinger et al., 1999).
erg was first described in zebrafish as a vascularly-expressed gene by (Weber et al., 2005) based on differential expression in a morpholino microarray analysis, but this report did not present temporospatial expression patterns or any functional studies.
In this study, we detail the spatiotemporal expression pattern of erg during angioblast specification, vasculogenesis and early angiogenesis. From functional studies, we present data suggesting a redundant yet synergistic role for erg during blood vessel formation and its capacity to drive angioblast specification and/or proliferation in the lateral plate mesoderm during early somitogenesis.
2. Results
2.1. erg is highly conserved between vertebrate lineages
To identify the zebrafish erg locus, we utilised the BLAST tool at the Ensembl database to search the current zebrafish genome assembly (Zv7). The erg locus is located on chromosome 10 and encodes a predicted 2.8 kb mRNA translating into a 427 amino acid protein with two major functional domains: an N-terminal “pointed domain” implicated in protein-protein interactions; and an C-terminal ETS DNA-binding domain common to all ETS family members. Only one copy of the erg locus was identified in the zebrafish genome; unlike the fli1 locus (Zhu et al., 2005), the erg gene has not undergone duplication.
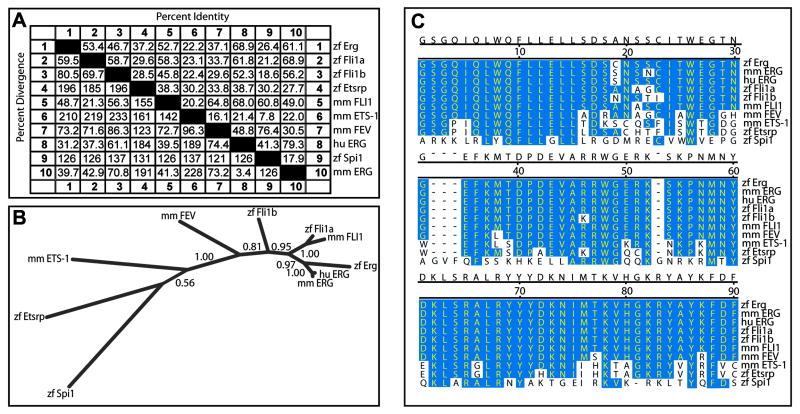
To investigate the degree of conservation between zebrafish Erg and various vertebrate homologues and paralogues, amino acid sequences were compared both over the full length of the protein and more specifically over the highly conserved ETS DNA-binding domain (Fig. 1). Zebrafish Erg is highly conserved between vertebrate lineages, sharing more than 60% identity with its murine and human homologues over the entire protein sequence, and more than 97% identity within the ETS domain.
Fig. 1. Sequence analysis of zebrafish erg.
(A) Percent identity and divergence of full-length protein sequences between various vertebrate ETS family members.
(B) Phylogenic tree describing evolutionary divergence of ETS family homologues and paralogues. Full-length protein sequences were aligned using MUSCLE, and phylogeny created using MrBayes. Branch labels are probabilities.
(C) Amino acid sequence alignment of highly conserved ETS DNA-binding domains from a number of vertebrate ETS family transcription factors. This important domain is almost completely conserved between zebrafish Erg and mammalian homologues, while significant identity is also observed with Fli1 paralogues.
Abbreviations: zf: zebrafish/Danio rerio; mm: mouse/Mus musculus and hu: human/Homo sapiens.
2.2. erg is expressed during vasculogenesis
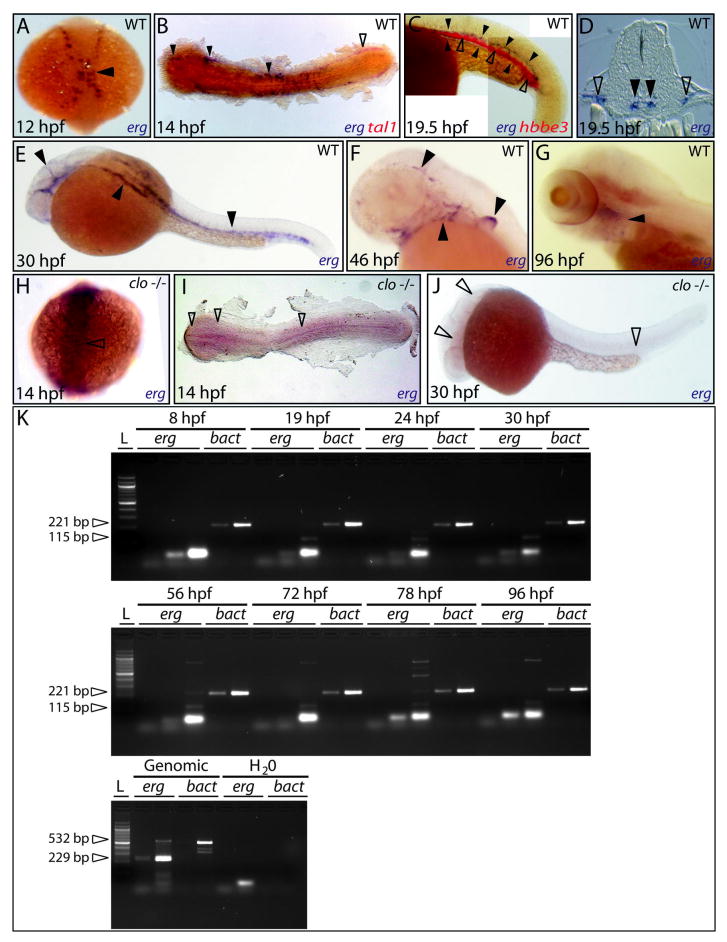
The basic spatiotemporal expression of zebrafish erg is included in an online database of high-throughput whole-mount in situ hybridisation (WISH) expression patterns and available through ZFIN (Thisse et al., 2004). Our studies expand on these observations with more detailed time-course and co-expression analyses by WISH and RT-PCR (Fig. 2, Supplementary Fig. 1). erg expression was first observed at the 6-somite stage of development (12 hpf) in anterior and posterior domains of the lateral plate mesoderm. A two-colour WISH of tal1 and erg at the 10-somite stage (14 hpf) confirmed that erg expression co-localised with the subset of tal1 labelled mesodermal haemangioblasts destined for vascular development, angioblasts (Fig. 2B). At the 15-somite stage (16.5 hpf), erg was expressed in cells positioned at the midline of the embryo, characteristic of precursors that form the primary trunk vessels between the 18- and 21-somite stages (18 - 19.5 hpf). Comparison of erg expression to that of the embryonic globin gene hbbe3 by two-colour WISH at 19.5 hpf (Fig. 2C) shows overlap of expression in the developing caudal vein but not the dorsal aorta, consistent with coalescence of the caudal vein around erythrocytes in the ICM prior to the onset of circulation. By the onset of circulation at around 26 hpf, erg was expressed throughout all developing primary vasculature (Supplementary Fig. 1H). Around 34 hpf, erg expression waned in most trunk vessels and became restricted to vascular endothelial cells in the head and developing aortic arches (Supplementary Fig. 1L). Interestingly, expression was also observed in a small subset of cells in the intermediate mesoderm region, reminiscent of those that express etsrp and have been suggested to mark precursors of pronephric and/or gut vessels (Sumanas and Lin, 2006). erg expression was still detectable by WISH and RT-PCR at 96 hpf, marking a subset of vasculature still developing in the aortic arches (Fig 2, Supplementary Fig. 1T).
Fig. 2. erg expression during early zebrafish development.
(A) Earliest expression of erg in lateral plate mesoderm at approximately 12 hpf (dorsal view, full arrowhead). (B) Flat-mount preparation shows overlap of early erg expression (blue) with tal1 expression (red) in vascular (filled arrowheads) but not haematopoietic progenitors (empty arrowhead). (C) Lateral view of tail displaying ICM expression of erg in developing vascular structures (blue, full arrowheads) partially overlapping that of the embryonic globin hbbe3 (red, empty arrowheads) as the developing caudal vein coalesces around erythroid cells prior to circulation. (D) Transverse section showing erg expression in bifurcated cental arterial (filled arrowheads) and lateral venous structures (empty arrowheads). (E) erg expression at 30 hpf in all developing vascular structures in the brain, trunk and tail of the embryos (full arrowheads). (F) Later erg expression restricted to vasculogenesis in the brain, aortic arches and developing fin bud (full arrowheads). (G) erg expression remains detectable in the aortic arches at 96 hpf (full arrowhead). (H-J) erg expression is absent in the cloche (clo) mutant during angioblast specification and during vasculogenesis (empty arrowheads indicate sites of loss of erg expression). (clo genotype recognised from the expected Mendelian ratio). All panels containing whole mount in situ hybridisation analyses; unless otherwise stated, anterior is to the left, and, except in flat-mounts, dorsal is to the top. (K) Semi-quantitative RT-PCR showing erg expression over the time period described (25, 30 and 35 cycles shown). β-actin (bact) expression was used as a positive control for each timepoint and to show linear amplification (20 and 25 cycles shown). Each primer pair was designed to span an intron, providing a genomic DNA-contamination internal negative control. For more details, see Methods.
2.3. erg expression is cloche-, tal1- and etsrp-dependent
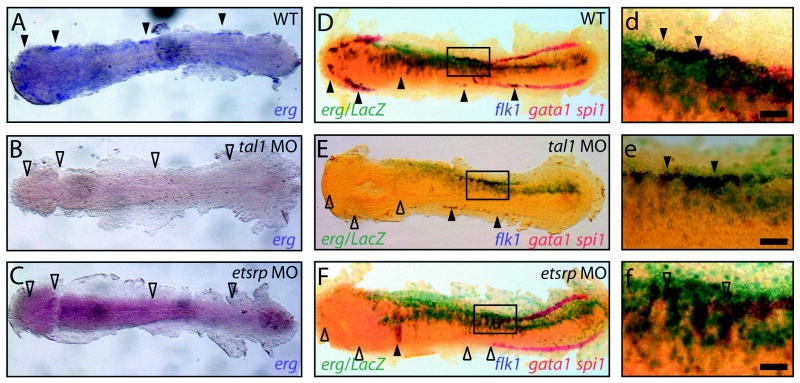
The earliest known factor controlling vascular development is thought to be the acyltransferase lycat, which is located within a deletion in an allele of cloche, a mutant lacking most haematopoietic and vascular mesodermal derivatives (Stainier et al., 1995; Xiong et al., 2008). Downstream of cloche, tal1 is also required for development of haematopoietic and myeloid lineages, as well as development and formation of the dorsal aorta (Stainier et al., 1995; Patterson et al., 2005). As described previously, the ETS factor etsrp has been shown to be a key regulator of vasculogenesis downstream of tal1 and cloche signalling (Sumanas and Lin, 2006). To dissect the position of erg in this functional hierarchy, we analysed the expression of erg at the 10-somite (14 hpf) stage in the lateral plate mesoderm in the absence of each of these factors by WISH. erg expression was absent in cloche (Fig. 2I), tal1-morphant (Fig. 3B) and etsrp-morphant embryos (Fig. 3C), suggesting that expression of erg at this time-point is dependant on the presence of these factors in the mesoderm. In contrast to tal1 morphants, absence of erg expression in cloche and etsrp morphant embryos may reflect the absence of flk1-expressing specified angioblasts in which erg is normally expressed, rather than indicate direct transcriptional regulation of the erg locus.
Fig. 3. erg overexpression drives mesodermal angioblast proliferation.
(A) Mesodermal erg expression defines angioblast populations at 14 hpf (full arrowheads) and is lost in etsrp (n = 20/20) and tal1 morphants (n = 20/20) (B and C respectively, empty arrowheads). (D-F/d-f) Triple in situ at 14 hpf shows spi1 and gata1 marking anterior myeloid and posterior erythroid progenitors respectively in red and flk1 marking angioblast populations in blue. Lateral erg overexpression is traced with LacZ mRNA in turquoise. (D/d, E/e) erg overexpression specifically expands existing flk1-expressing lateral angioblast populations present in WT (5/5) and tal1 morphant (n = 10/10) embryos (full arrowheads) but is unable to rescue absent flk1-expressing lateral angioblast populations in etsrp morphant (n = 5/5) embryos (F/f, empty arrowheads). Panels d,e,f are details of boxed areas in D,E,F respectively.
All panels are whole-mount in situ hybridisation analyses, flat-mounted, with anterior to the left, Scale bars: 50 μm.
2.4. erg overexpression drives mesodermal angioblast proliferation
Many ETS family members are expressed in the lateral plate mesoderm during somitogenesis in the zebrafish, where they have important functions during specification. To elucidate potential functions for erg in angioblast specification and proliferation, and to further dissect its position in transcriptional and biological pathways, Erg was overexpressed from injected mRNA on mutant and morphant backgrounds and the effect on various mesodermal progenitors at 14 hpf was assessed.
Widespread erg overexpression from 1-2 cell microinjections resulted in early, major developmental defects (e.g. 21/24 severely abnormal embryos at 24 hpf (12.5 ng/μl RNA); compared with 202/202 normal at 24 hpf (50 ng/μl) for a negative-control RNA encoding a carboxyl-truncated Erg lacking the DNA binding domain). However, erg overexpression consistently expanded the early expression domain of vascular markers flk1 (Supplementary Fig. 3) and fli1 (observed in Tg(fli1:EGFP) embryos, data not shown). This appeared to be at the expense of haematopoietic and myelopoietic progenitors, as these embryos displayed reduced extent of gata1 and spi1 expression respectively (Supplementary Fig. 3). To avoid confounding, potentially off-target effects of erg overexpression and to examine this effect more closely, erg mRNA delivery was compartmentalised by microinjection of 8-16 cell embryos and traced by co-injecting LacZ mRNA. Embryos with unilateral, mesodermally-restricted, LacZ-traced erg delivery were analysed by WISH, the normal contralateral side of such embryos serving as a stringent internal control.
Mesoderm-restricted overexpression of erg on a WT background resulted in specific expansion of flk1-expressing vascular progenitors in the lateral mesoderm (Fig. 3A,D). This outcome was also observed in the absence of tal1, in which flk1-expressing angioblast populations in the lateral mesoderm are still present, although populations marking the heart-field and anterior endothelial progenitors are lost (Dooley et al., 2005) (Fig. 3 B,E). In contrast, in an angioblast-deficient etsrp morphant background, in which anterior and lateral but not heart-field flk1-expressing populations are lost (Sumanas and Lin, 2006), erg overexpression was not sufficient to rescue this lateral population (Fig. 3 C,F). Collectively, these results suggest that during somitogenesis, erg may function to drive angioblast specification and/or proliferation via a tal1-independent, etsrp-dependent pathway.
2.5. erg acts synergistically with etsrp to regulate vascular development
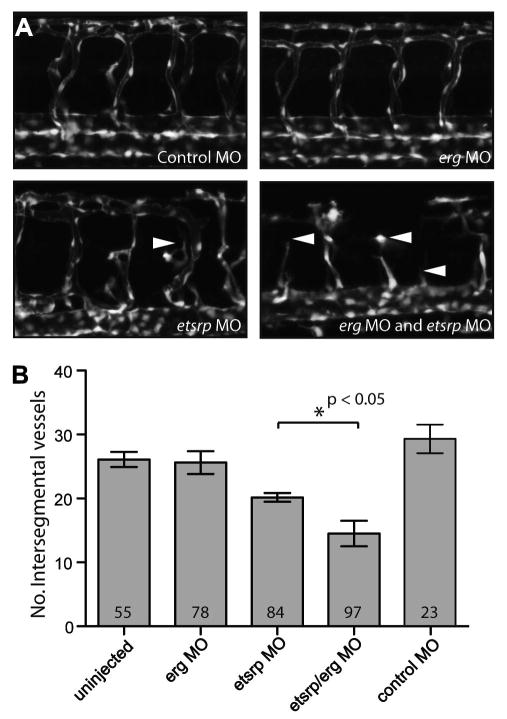
To knock-down erg function during early zebrafish development, we injected morpholino antisense oligonucleotides (MO) targeted to the erg translation-initiation site. To confirm that the ATG-targeting ergMO was biologically active, we employed an in vivo reporter assay. The MO target sequence was cloned upstream of EGFP and from this, mRNA was transcribed for injection. In the presence of ergMO, expression of EGFP from this construct was abolished completely, confirming that the ergMO strongly targets its complementary sequence to suppress translation (Supplementary Fig. 2). However, erg knockdown on the Tg(fli1:EGFP) background resulted in no discernable vascular phenotype (Fig. 4A).
Fig. 4. erg acts synergistically with etsrp to regulate vascular development.
(A) Panels show GFP-marked vasculature in wild-type and ergMO, etsrpMO and double morphants on the Tg(fli1:EGFP) line at 48 hpf. erg knock-down with an antisense morpholino oligonucleotide had no effect on vascular development unless introduced on a sensitised etsrp morphant background. Note the slight disruption of intersegmental vessel sprouting from the dorsal aorta to form the dorsal longitudinal anastomotic vessels in etsrpMO morphants, and the much more severe disruption in ergMO and etsrpMO double morphants (full arrowheads). (B) Vascular development was quantified by counting intersegmental vessels that fully connected the dorsal aorta to dorsal longitudinal anastomotic vessels in embryos at 48 hpf. Collated data show significant reduction of intersegmental sprouting in ergMO + etsrpMO versus ergMO morphants. Collectively the morphological and quantitative data indicate a synergistic but redundant role for erg in vascular development.
Error bars are standard errors (uninjected, morphant groups; n = 3 experiments) and standard deviation (control MO, n = 23 embryos from one experiment). Numbers within bars indicate collated number of embryos scored for each group.
To investigate a possible functional relationship between erg with etsrp, a sensitising dose of etsrp morpholino was administered at which a partial reduction of intersegmental vessel development was observed at 48 hpf (Fig. 4A). Scoring the number of intersegmental vessels fully joining the dorsal aorta to dorsal longitudinal anastomotic vessels provided a sensitive quantifiable readout for any further synergistic functional interaction between loss of erg and etsrp. Upon introduction of the ergMO into this system, a further reduction of intersegmental vessel development was observed, suggesting that erg may have a role, albeit redundant to etsrp, in zebrafish blood vessel development (Fig. 4).
3. Discussion
We present evidence suggesting a role for erg at multiple stages of vascular development in the zebrafish: (1) erg expression is spatiotemporally restricted to angioblasts and developing blood vessels during angioblast migration, vasculogenesis and early angiogenesis; (2) erg expression is lost in the absence of factors known to regulate angioblast specification, proliferation and vasculogenesis; (3) erg overexpression specifically drives specification/proliferation of angioblasts in the lateral plate mesoderm; (4) knock-down of erg in combination with etsrp significantly reduces formation of intersegmental vessels by angiogenesis.
Our data implicating erg in zebrafish vascular development are consistent with a recent independent study of erg biology based on expression data and a combinatorial ETS-family member loss-of-function analysis, but which did not include a gain-of-function analysis such as we also present (Liu and Patient, 2008). We observed a more subtle erg loss-of-function effect on intersegmental vessel morphology, in that we found the requirement for erg to discernable only in an etsrp-deficient context; this difference may be due to technical differences such as reagent dose and targeting efficiency, the baseline controls used, or the method of scoring. However, collectively these studies point to a role for erg in vascular development.
3.1. Redundant roles for ETS family members during vasculogenesis limit experimental approaches to functional characterisation
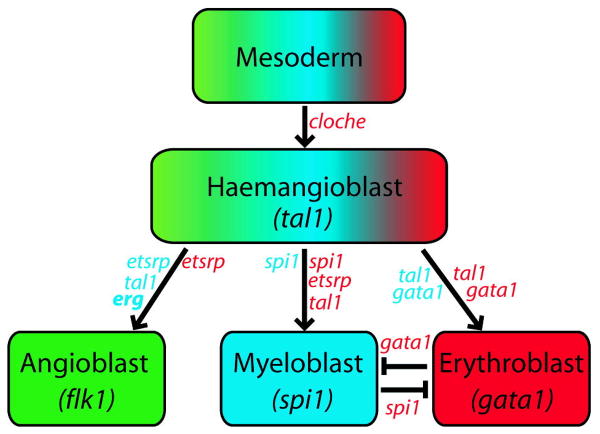
The cardiovascular system is the first fully functional system to develop during zebrafish embryogenesis and is integral for the correct development of many other organs and structures: it is therefore not surprising that such a crucial step in development is regulated by complex transcriptional networks with many integrated safeguards and redundancies. Past functional studies in zebrafish of blood-vessel development have strongly suggested combinatorial or redundant roles for members of the ETS family of transcription factors (Pham et al., 2007). Indeed, of the ETS factors investigated, etsrp alone is essential for the specification of angioblasts during early somitogenesis (Sumanas and Lin, 2006), where it has been implicated during specification of a common myeloid/vascular precursor (Sumanas et al., 2008). As described in knockdown studies of other vascular specific ETS factors fli1, fli1a and ets1 (Pham et al., 2007; Liu et al., 2008), we show that erg is also not essential for vasculogenesis. This systematic redundancy severely diminishes the utility of loss-of-function experiments for determining gene function. In this study, overexpression has proved a more useful tool, providing insight into the function of erg by focussing on genetic potential rather than developmental requirement, and has allowed positioning of erg in the genetic pathway regulating early vascular development (Fig. 5).
Fig. 5. A putative role for erg in the transcriptional regulation of vasculogenesis.
This highly simplified model represents published functional studies of transcriptional regulation of cellular populations in the zebrafish mesoderm. Compiled from (Stainier et al., 1995); (Liao et al., 1998; Galloway et al., 2005; Rhodes et al., 2005; Sumanas and Lin, 2006) (Lyons et al., 2002). Factors in red have been demonstrated as essential by mutant and knockdown experiments, while factors in blue have been implicated by overexpression studies. Factors within boxes are specifically expressed in that lineage. Sharp arrows represent positive regulation, flat arrows represent negative regulation. Ours studies suggest erg is positioned downstream of cloche, tal1 and etsrp, as a positive regulator of angioblast specification/proliferation.
3.2. Functional studies suggest combinatorial transcriptional regulation of flk1 by erg and etsrp
Unlike the murine system, in which the early expression of the tyrosine receptor kinase Flk1 drives Tal1 expression and haemangioblast specification, zebrafish flk1 (recently renamed kdr-like (Bussmann et al., 2008), but considered a functional homologue of murine Flk1) expression commences later, once haemangioblast specification has already occurred. Mutations in zebrafish flk1 result in normal specification of angioblasts and vasculogenesis, although later disruption of intersegmental angiogenesis is observed (Habeck et al., 2002). Furthermore, promoter deletion studies of murine Flk1 have identified regulatory binding sites for ETS factors able to be activated by co-expression of Ets1 and Ets2 (Kappel et al., 2000). The functional parallels between observations of zebrafish flk1 function and the phenotypic effects of erg knockdown and overexpression are striking. In this study, knockdown of erg alone did not affect early vasculogenesis, and its requirement in normal angiogenesis was demonstrable only in conjunction with knockdown of another ETS factor, etsrp. Furthermore, overexpression of erg only resulted in expansion of angioblasts where etsrp was present. Hence in zebrafish, redundant ETS factors such as erg may act in conjunction with etsrp to drive various stages of haemangioblast specification, vasculogenesis and angiogenesis, but the presence of etsrp is the limiting factor.
3.3 Practical applications of zebrafish erg biology
As the number of diseases associated with a various disruptions of the ERG locus increases, so does the need to develop an assay to identify whether these abnormalities are of functional significance. High functional conservation between vertebrates permits in vivo functional assays to be developed in zebrafish based on rescue of morphant or mutant zebrafish phenotypes by introduction of mRNA encoding human genes and their disease-related allelic variants, or even by injection of protein isolates (McWhorter et al., 2003; Winkler et al., 2005). Given the conservation between vertebrate Erg homologues, the overexpression bioassay we describe for zebrafish erg is potentially the basis for functional assessments of mutant human, or analogously-mutated zebrafish alleles. For example, overexpression of a truncated zebrafish erg lacking the ETS DNA-binding domain by mRNA injection had no effect on vasculogenesis even at high doses, indicating that this mutant Erg has been rendered functionally inactive.
Our studies suggest a synergistic yet redundant role for erg in zebrafish vasculogenesis, adding erg to the complex web of synergistically interacting ETS factors implicated in blood vessel development, and gained insight into the molecular regulation of the erg locus. It will be interesting to determine the degree to which the role of erg in vasculogenesis that we have discerned in zebrafish is conserved across species.
4. Methods
4.1. Zebrafish strains
All experiments were conducted on fish maintained at the Ludwig Institute of Cancer Research Aquarium Facility on protocols approved by the Walter & Eliza Hall Institute Animal Ethics Committee. All non-transgenic functional studies were conducted on AB wild-type or clom39 +/- mutant (Stainier et al., 1995) backgrounds, while the Tg(fli1:EGFP) strain (Lawson and Weinstein, 2002) was used to investigate synergistic interactions with etsrp.
4.2. Cloning of zebrafish erg
The erg 1.2 kb coding sequence was amplified from an adult kidney cDNA library using the Kozak optimised primers: (A) 5′-GCGGAATTCCATGACGGCGTCTGCAGCCGC-3′ and (B) 5′-GCGCCTCGAGGTTCTTCTAGTAGTATGAGC-3′. The 528 bp construct used for in situ hybridisation was amplified using (A) and (C) 5′-GCGCCTCGAGGTTGGGAAAGATGAAGTTGGC-3′. Constructs were cloned into the pCS2+ expression plasmid (Turner and Weintraub, 1994) using EcoRI and XhoI restriction sites and verified by sequencing.
4.3. Phylogenetic analysis
Full-length amino acid alignment was performed using MUSCLE, phylogeny created using MrBayes. Amino acid substitution prior: blosum62, equal rates among sites. All sequences were obtained from GenBank at NCBI (Accession Numbers: zf Erg: BC086811.1; mm Erg: BAC34461; hu ERG: BC040168; zf Fli1a: AAH66571, zf Fli1b: NP_001008780; mm Fli1: NP_032052; mm FEV: NP_694751; mm ETS-1: NP_035938; zf Etsrp: AAY89037 and zf Spi1: NP_932328). Full-length sequence divergence analysis and domain-restricted alignments were performed with DNAstar Lasergene MegAlign. ETS DNA-binding domain restrictions were defined by comparative alignment to the DNA-binding domain of zebrafish Erg: G(254)-F(339).
4.4. Whole-mount in situ hybridisation
Antisense and sense riboprobes for erg were generated from a 528 bp fragment targeting the 5′ end of erg mRNA, which lacks the highly conserved ETS domain. The template was linearised with EcoRI and StuI for transcription with T3 and SP6 polymerases respectively. A 2.6 kb antisense riboprobe for flk1 was included as a positive control for probe penetration for timecourse experiments, resulting in flk1 expression patterns as previously published (Bahary et al., 2007); Thisse et al., 2004; (Thompson et al., 1998) across the timecourse. In situ hybridisation experiments were performed as described previously (Lieschke et al., 2002); (Oates et al., 1999). Embryos presented in figures are representative of > 10 examples unless otherwise stated.
4.5. Semi-quantitative RT-PCR
cDNA for each timepoint was synthesised from mRNA collected from WT embryos using a SuperscriptIII RT-PCR kit (Invitrogen) according to manufacturer's instructions. erg primers (5′-GTGGGTTATGACGCTGTCAG-3′ and 5′-CTAACTGCGCTCTCTGCTC-3′) were designed to amplify from exon 1 to exon 2, spanning an intron, amplifying 115 bp from cDNA or 229 bp from genomic DNA. bact primers (5′-TGGCATCACACCTTCTAC-3′ and 5′-AGACCATCACCAGAGTCC-3′) were designed to amplify from exon 2 to exon 3, spanning an intron, amplifying 221 bp from cDNA or 532 bp from genomic DNA. PCR was performed using 1 μL of 1/10 dilution of stock cDNA, 500 nM of each primer and 0.2 μL of Phusion DNA polymerase (Finnzymes) in High Fidelity buffer. Cycle times and temperatures used: 95°C, 60′ (1 cycle), then 95°C, 15′; 70°C, 15′; 72°C, 12′ (20, 25, 30 or 35 cycles), then 16°C ∞. In Fig. 2K, erg is shown at 25, 30 and 35 cycles in test samples. bact positive control is shown at 20 and 25 cycles. Genomic and H2O controls are shown at 25 and 35 cycles.
4.6. erg overexpression constructs
Full length mRNA was generated from the 1.2 kb construct described above. The “ETS-less” control construct lacking the ETS DNA-binding domain was generated from the full-length clone by PCR exclusion using primers 5′-GGCTAGCATTGCGCAAGCCTGCAG-3′ and 5′-GGCTAGCACCAGAGCCTGGGTTGG-3′ that introduced an NheI site in place of the ETS domain. Capped, 3′ polyadenylated erg mRNA was generated from clones in PCS2+ following template linearisation with NotI using M Message M Machine Kit (Ambion) according to manufacturer's instructions.
4.7. Morpholino oligonucleotide design and validation
The erg morpholino antisense oligonucleotide ergMO (5′-CAGACGCCGTCATCTGCACGCTCAG -3′) was designed to target the ATG start-codon, all other morpholinos described are previously published: Control (Hogan et al., 2004; Rhodes et al., 2005; Sumanas and Lin, 2006), and tal1 (Dooley et al., 2005). The ergMO validation construct was generated by cloning oligonucleotides (5′-TTGCAGGATCCTGAGCGTGCAGATGACGGCGTCTGCAATCGATTCG -3′) and (5′-CGAATCGATTGCAGACGCCGTCATCTGCACGCTCAGGATCCTGCAA -3′) containing the morpholino target (underlined) in front of EGFP in pCS2+ using BamHI and ClaI restriction sites (bold).
4.8. Microinjection
etsrp morpholino was injected at 2 ng/μl in epistasis experiments and at 0.125 ng/μl as a sensitising dose in synergy experiments. The tal1 morpholino was injected at 4.19 ng/μl, erg morpholino was injected at 2.84 ng/μl and control morpholino at 2.085 ng/μl. Overexpression of erg traced by LacZ expression was performed using injectates containing 50 ng/μl of each mRNA. Average injection bolus for 1-2 cell stage injections was 1.8 nl, while 8-16 cell injections were 0.26 nl.
4.9. Statistical analyses
For data management and descriptive statistics, Microsoft Excel for Mac 2004 was used. Analytical statistics were derived using Prism 4 for Mac 2005: a one-tailed T-test was used expecting that knock-down of erg in conjunction with etsrp would result in a more severe phenotype, as observed for other Ets-proteins in previously published experiments (Pham et al., 2007).
4.10. Imaging and photography
Embryos were imaged using a Nikon SMZ 1500 dissecting microscope via a Nikon Coolsnap HQ camera, and a Nikon Optiphot-2 compound microscope via a Zeiss AxioCam Mrc 5 camera. Live confocal imaging was performed on a Bio-Rad inverted confocal microscope using a 40 × water-dipping lens. Confocal images were adjusted for subject movement where necessary using the Stackreg plugin in ImageJ. Photographs were manipulated (cropping and size only) using Adobe Suite CS2 programs Photoshop and Illustrator.
4.11. Nomenclature
Nomenclature for genes and proteins described throughout adhere to naming conventions as described at www.zfin.org/zf_info/nomen.html.
For example:
Species/gene/protein
Zebrafish/erg/Erg
Human/ERG/ERG
Mouse/Erg/ERG
Where referred to collectively, murine nomenclature is used.
Supplementary Material
Acknowledgments
We thank all members of the Lieschke laboratory for their support, particularly Sony Varma for technical assistance and Luke Pase for practical advice. We also thank Ben Hogan and Stephen Loughran for helpful discussions. We would like to thank Terry Speed and Toby Sargeant for help with bioinformatics and the construction of phylogenies, Stephen Cody and Adam Parslow for help with microscopy, Tania Tabone for cDNA preparation, Steven Mihajlovic for histological support and the reviewers for their constructive comments. Work in GL's laboratory is supported by the NHMRC (461222, 461208 and 516750) and NIH (HL079545). BK is supported by a QEII Fellowship from the Australian Research Council. FE was supported by a scholarship from the Dafydd Lewis Trust and an Australian Postgraduate Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- Bahary N, Goishi K, Stuckenholz C, Weber G, Leblanc J, Schafer CA, Berman SS, Klagsbrun M, Zon LI. Duplicate VegfA genes and orthologues of the KDR receptor tyrosine kinase family mediate vascular development in the zebrafish. Blood. 2007;110:3627–3636. doi: 10.1182/blood-2006-04-016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltzinger M, Mager-Heckel AM, Remy P. Xl erg: expression pattern and overexpression during development plead for a role in endothelial cell differentiation. Dev Dyn. 1999;216:420–433. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<420::AID-DVDY10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. The transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008 doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann J, Lawson N, Zon L, Schulte-Merker S. Zebrafish VEGF receptors: a guideline to nomenclature. PLoS Genet. 2008;4:e1000064. doi: 10.1371/journal.pgen.1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrere S, Verger A, Flourens A, Stehelin D, Duterque-Coquillaud M. Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene. 1998;16:3261–3268. doi: 10.1038/sj.onc.1201868. [DOI] [PubMed] [Google Scholar]
- Childs S, Chen JN, Garrity DM, Fishman MC. Patterning of angiogenesis in the zebrafish embryo. Development. 2002;129:973–982. doi: 10.1242/dev.129.4.973. [DOI] [PubMed] [Google Scholar]
- Dooley KA, Davidson AJ, Zon LI. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev Biol. 2005;277:522–536. doi: 10.1016/j.ydbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Girma JP, Meyer D, Verweij CL, Pannekoek H, Sixma JJ. Structure-function relationship of human von Willebrand factor. Blood. 1987;70:605–611. [PubMed] [Google Scholar]
- Habeck H, Odenthal J, Walderich B, Maischein H, Schulte-Merker S. Analysis of a zebrafish VEGF receptor mutant reveals specific disruption of angiogenesis. Curr Biol. 2002;12:1405–1412. doi: 10.1016/s0960-9822(02)01044-8. [DOI] [PubMed] [Google Scholar]
- Hogan BM, Hunter MP, Oates AC, Crowhurst MO, Hall NE, Heath JK, Prince VE, Lieschke GJ. Zebrafish gcm2 is required for gill filament budding from pharyngeal ectoderm. Dev Biol. 2004;276:508–522. doi: 10.1016/j.ydbio.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Huang MT, Mason JC, Birdsey GM, Amsellem V, Gerwin N, Haskard DO, Ridley AJ, Randi AM. Endothelial intercellular adhesion molecule (ICAM)-2 regulates angiogenesis. Blood. 2005;106:1636–1643. doi: 10.1182/blood-2004-12-4716. [DOI] [PubMed] [Google Scholar]
- Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- Kappel A, Schlaeger TM, Flamme I, Orkin SH, Risau W, Breier G. Role of SCL/Tal-1, GATA, and ets transcription factor binding sites for the regulation of flk-1 expression during murine vascular development. Blood. 2000;96:3078–3085. [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Paw BH, Thompson MA, Hall NE, Ward AC, Ho RK, Zon LI, Layton JE. Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274–295. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- Liu F, Patient R. Genome-Wide Analysis of the Zebrafish ETS Family Identifies Three Genes Required for Hemangioblast Differentiation or Angiogenesis. Circ Res. 2008 doi: 10.1161/CIRCRESAHA.108.179713. [DOI] [PubMed] [Google Scholar]
- Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, Hilton DJ, Alexander WS, Kile BT. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008 doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- Lyons SE, Lawson ND, Lei L, Bennett PE, Weinstein BM, Liu PP. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc Natl Acad Sci U S A. 2002;99:5454–5459. doi: 10.1073/pnas.082695299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Whitman SP, Vukosavljevic T, Paschka P, Langer C, Mrozek K, Baldus CD, Carroll AJ, Powell BL, Kolitz JE, Larson RA, Bloomfield CD. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. J Clin Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–42. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- McLaughlin F, Ludbrook VJ, Cox J, von Carlowitz I, Brown S, Randi AM. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. Blood. 2001;98:3332–3339. doi: 10.1182/blood.v98.12.3332. [DOI] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Mavrothalassitis G, Bhat NK, Fisher RJ, Papas TS. Human ERG-2 protein is a phosphorylated DNA-binding protein--a distinct member of the ets family. Oncogene. 1993;8:1559–1566. [PubMed] [Google Scholar]
- Nobes CD, Hawkins P, Stephens L, Hall A. Activation of the small GTP-binding proteins rho and rac by growth factor receptors. J Cell Sci. 1995;108(Pt 1):225–233. doi: 10.1242/jcs.108.1.225. [DOI] [PubMed] [Google Scholar]
- Oates AC, Brownlie A, Pratt SJ, Irvine DV, Liao EC, Paw BH, Dorian KJ, Johnson SL, Postlethwait JH, Zon LI, Wilks AF. Gene duplication of zebrafish JAK2 homologs is accompanied by divergent embryonic expression patterns: only jak2a is expressed during erythropoiesis. Blood. 1999;94:2622–2636. [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–3511. doi: 10.1182/blood-2004-09-3547. [DOI] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ES, Rao VN. erg, an ets-related gene, codes for sequence-specific transcriptional activators. Oncogene. 1991;6:2285–2289. [PubMed] [Google Scholar]
- Reddy ES, Rao VN, Papas TS. The erg gene: a human gene related to the ets oncogene. Proc Natl Acad Sci U S A. 1987;84:6131–6135. doi: 10.1073/pnas.84.17.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Siddique HR, Rao VN, Lee L, Reddy ES. Characterization of the DNA binding and transcriptional activation domains of the erg protein. Oncogene. 1993;8:1751–1755. [PubMed] [Google Scholar]
- Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Weinstein BM, Detrich HW, 3rd, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 http://zfin.org.
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Vlaeminck-Guillem V, Carrere S, Dewitte F, Stehelin D, Desbiens X, Duterque-Coquillaud M. The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech Dev. 2000;91:331–335. doi: 10.1016/s0925-4773(99)00272-5. [DOI] [PubMed] [Google Scholar]
- Weber GJ, Choe SE, Dooley KA, Paffett-Lugassy NN, Zhou Y, Zon LI. Mutant-specific gene programs in the zebrafish. Blood. 2005;106:521–530. doi: 10.1182/blood-2004-11-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JW, Yu Q, Zhang J, Mably JD. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ Res. 2008;102:1057–1064. doi: 10.1161/CIRCRESAHA.107.163907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetta L, Marcus SG, Vasile J, Dobryansky M, Cohen H, Eng K, Shamamian P, Mignatti P. Expression of Von Willebrand factor, an endothelial cell marker, is up-regulated by angiogenesis factors: a potential method for objective assessment of tumor angiogenesis. Int J Cancer. 2000;85:281–288. doi: 10.1002/(sici)1097-0215(20000115)85:2<281::aid-ijc21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Zhu H, Traver D, Davidson AJ, Dibiase A, Thisse C, Thisse B, Nimer S, Zon LI. Regulation of the lmo2 promoter during hematopoietic and vascular development in zebrafish. Dev Biol. 2005;281:256–269. doi: 10.1016/j.ydbio.2005.01.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.