Abstract
Purpose
In head and neck squamous cell carcinoma (HNSCC) cells, Rap1 shuttles between the nucleus and cytoplasm. Prior findings suggested that Rap1 may modulate the β-catenin-independent Wnt pathway in some settings, but the role of Rap1 in β-catenin-dependent Wnt signaling remains undefined.
Experimental Design and Results
We observed that β-catenin bound to active Rap1 in vitro and Rap1 activated β-catenin-TCF (T cell factor)-dependent transcription. Immunofluorescence studies showed that ectopic expression of Rap1 increased nuclear translocation of β-catenin. Overexpression of active Rap1 facilitated an increase in β-catenin-mediated transcription that was abrogated by dominant negative TCF4. Conversely, siRNA-mediated inhibition of endogenous Rap1 expression inhibited β-catenin/TCF-mediated transcription as well as invasion of HNSCC. Furthermore, inhibition of Rap1 expression downregulated the expresesion of MMP7, a transcriptional target of β-catenin/TCF. In HNSCC cells stably transfected with β-catenin or treated with lithium chloride or Wnt3A to stabilize endogenous β-catenin, inhibition of Rap1 expression led to decreases in the free pool of β-catenin. Immunohistochemical studies of tissue from HNSCC patients revealed that increased β-catenin intensity correlated with higher tumor stage. Furthermore, the prognostic effect of active Rap1 on tumor N-stage was found to depend on cytosolic β-catenin expression (p<0.013). When β-catenin is high, higher rap1GTP intensity is associated with more advanced N stage.
Conclusions
The findings suggest that Rap1 enhances β-catenin stability and nuclear localization. In addition to indicating that Rap1 has a significant role in regulating β-catenin and β-catenin-dependent progression to more advanced N-stage lesions, these data highlight Rap1 as a potential therapeutic target in HNSCC.
Keywords: nucleus, Wnt signaling, TCF transcription, small GTP-binding protein
Introduction
Rap1 is a Ras-like protein that has two isoforms, Rap1A and Rap1B, which are encoded by different genes on chromosomes 1 and 12, respectively. The ubiquitously expressed Rap1 protein is closely related to Ras proteins in the GTP-binding region, the effector domain, and the membrane attachment site. Rap1 switches from an inactive guanine diphosphate (GDP)-bound form to an active GTP-bound form. This switch is regulated by several guanine nucleotide exchange factors (GEFs), including C3G, Epac, CalDAG-GEF, smgGDS, PDZ, Dock-4, phospholipase Cε, and Mr-GEF (1, 2). Inactivation of Rap1A and Rap1B is regulated by GTPase-activating proteins (RapGAPs), which activate endogenous GTPase activity. Two Rap1GAP families have been identified: RapGAP (I and II) and SPA-1 (3–5). Rap1 is a key player in cell adhesion and migration. Active Rap1 modulates cellular functions by regulating the translocation of other proteins or by shuttling between intracellular compartments (6, 7). We previously demonstrated that Rap1 is linked to tumor growth and invasion in malignant oral keratinocytes (8, 9). In another study, we showed that active GTP-bound Rap1 is localized in the nucleus whereas inactive GDP-bound Rap1 was trapped in the cytoplasm (10).
Aberrant activation of the Wnt/β-catenin signaling cascade has been linked to tumor development and invasion in various settings (11–13). β-catenin, a central molecule in the Wnt pathway, translocates from a free cytosolic form to the nucleus where, as a co-factor with T-cell factor/lymphoid enhancer factor (TCF/LEF), it triggers gene transcription (14–16). The cytosolic pool of β-catenin is tightly regulated by interaction with a protein complex of adenomatous polyposis coli (APC), axin, and glycogen synthase kinase 3β (GSK3β) that facilitates phosphorylation and proteasomal degradation of β-catenin (17, 18). In contrast, activated Wnt signaling, which is initiated by specific ligands binding to their cognate frizzled receptors, inhibits phosphorylation and stabilizes cytosolic β-catenin (19). Wnt-induced signaling inhibits phosphorylation of β-catenin by inhibiting GSK3β (16).
In normal keratinocytes, E-cadherin binds β-catenin during the formation of adherens junctions, thereby sequestering a major fraction of the pool of β-catenin protein and ensuring low cytosolic concentrations (16, 20). Mutations in β-catenin’s N-terminal regulatory domain or in key components of the APC/Axin/GSK3β destruction complex lead to accumulation of free β-catenin in the cytosol (16, 20). In various types of cancer, mutations in key phosphorylation sites in the N-terminal region of β-catenin confer resistance to ubiquitination and subsequent degradation, thereby increasing the amount of free/cytosolic β-catenin available for nuclear translocation (20). Previous studies have investigated changes in the adherens junctions relative to Rap1 activation (21–24). However, the role of Rap1 in β-catenin-mediated transcription and invasion has not been investigated. In the current study we investigated the role of Rap1 in stabilization of β-catenin, induction of its transcriptional targets and in β-catenin-mediated invasion of HNSCC cells.
Materials and methods
Cell culture
UM-SCC (1, 5, 14A, 17B, 22B, 74A, and 81B), OSCC3, DLD-1 colon cancer cells, and human embryonic kidney (HEK) 293 cells were grown to 60–80% confluence in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, Inc., Grand Island, NY) containing 10% fetal bovine serum, penicillin 100 μg/ml, streptomycin 100μg/ml, and 50 μg/ml L-glutamine. UM-SCC-(1, 5, 14A, 17B, 22B, 74A, and 81B) were developed by and obtained from Dr. Thomas Carey, University of Michigan and were validated by genotyping in his laboratory during use for these studies. UM-SCC-1/Neo, UM-SCC-1/β-catenin and HEK293 cells were obtained from Dr. Cun-Yu Wang. UM-SCC-1/Neo and UM-SCC-1/β-catenin were transfected by Dr. CY Wang (13) and genotyped in Dr. Carey’s laboratory during this study. OSCC3 and DLD-1 colon cancer cells were obtained from Drs. Peter Polverini and Eric Fearon, University of Michigan and have not been genotyped during use for this study.
Vector constructs
The wild-type (Wt) FLAG-tagged β-catenin construct has been previously described (25). Hemagglutinin (HA)-tagged Rap1A G12V and Rac1 G12V were purchased from the University of Missouri-Rolla cDNA Resource Center (Rolla, MO). Dominant negative TCF4 was a generous gift from Dr. Benjamin Alman (Hospital for Sick Children, Toronto, Ontario, Canada). pTOPflash and pFOPflash were purchased from Upstate Biotechnology (Lake Placid, NY). TOPflash is a reporter construct that contains three repeats of the wildtype TCF4 binding site upstream of a thymidine kinase minimal promoter and the luciferase reporter gene. FOPflash, the negative control plasmid, has a mutated TCF4 binding site.
Immunoblot analysis
Whole-cell lysates were prepared as described previously (10). Proteins were quantified using the Bio-Rad protein assay (Bio-Rad, Richmond, CA) and electrophoresed and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH). Primary antibodies and concentrations were; mouse anti-β-catenin monoclonal antibody (1:1,000; BD Transduction Laboratories, Lexington, KY), mouse anti-Rap1 monoclonal antibody (1:1,000; BD Transduction Laboratories), anti-HA monoclonal antibody (1:1,000; Convance Research Products Inc, Berkeley, CA), anti-FLAG M2 monoclonal antibody (1: 5000; Sigma, St. Louis, MO), and mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (1:10,000; Chemicon Inc., Temecula, CA). Membranes were washed in TBS containing 0.1% Tween-20 (TBS-T). Horseradish peroxidase-conjugated, affinity-purified goat anti-mouse IgG (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA) was used for immunodetection. Immunoreactive proteins were visualized by SuperSignal West Pico chemiluminescence system (Pierce, Rockford, IL) and exposed to X-ray film.
Immunoprecipitation with HA was performed using ProFound HA Tag IP/Co-IP Assay system (Pierce) following the manufacturer’s instructions.
Precipitation of free β-catenin
The free β-catenin pool in cell lysates was quantified as described previously (26). Briefly, cell lysates were precleared with unbound glutathione-Sepharose 4B beads (Amersham Biosciences, Piscataway, NJ) followed by incubation with GST-E-cadherin fusion protein, which contains the C-terminal region of E-cadherin, bound to glutathionebeads. Free β-catenin was identified by Western blotanalysis.
Rap1 activation assay
Active Rap1 was retrieved by the Rap1-binding domain of ral-GDS, (gift from Dr. Johannes L. Bos, University Medical Center Utrecht, The Netherlands), as described previously (8, 10, 27). Equivalency of Rap1 protein expression was verified by immunoblot analysis of whole cell lysates.
RNAi-mediated knockdown of Rap1A
Rap1A was down-regulated by small interfering RNA (siRNA). ON-TARGETplus SMARTpool was used for endogenous Rap1A knockdown and ON-TARGETplus siCONTROL Non-targeting POOL (Dharmacon, Lafayette, CO) was used as a negative control. HNSCC cells were nucleofected with siRNAs using the Nucleofector Device (Amaxa Inc.) and Cell Line Nucleofector Kit V (Amaxa) according to the manufacturer’s protocol.
Luciferase-based transcription assay
HEK293 cells were seeded at a density of 5×105 cells/well (6-well plates) or 1×105 cells/well (24-well plates) in triplicate, and cultured in DMEM supplemented with 10% FBS. After 24h, the cells were transfected using FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. Cells were transfected with test DNA (total 2.0μg/well for 6-well plates and 0.5μg/well for 24-well plates), together with an internal control plasmid pRL-CMV (15ng/well for 6-well plates and 3ng/well for 24-well plates) and either pTOPflash or pFOPflash (0.5μg/well for 6-well plates and 0.1μg/well for 24-well plates).
For knockdown studies, HNSCC cells cultured in 100 mm dishes were nucleofected using the Nucleofector Device (Amaxa Inc., Gaithersburg, MD) and Cell Line Nucleofector Kit V (Amaxa Inc.) according to the manufacturer’s protocol. Briefly, cells were nucleofected with 2.0μg siRNAs, together with 15ng pRL-CMV and either 1.0μg pTOPflash or pFOPflash in 3×106 cells.
Twenty-four hours later, transcription was assayed according to the manufacturer’s protocol using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) with a luminometer, LMax II384 (Molecular Devices, Sunnyvale, CA). Results were expressed as an average of the firefly luciferase activity normalized to the renilla luciferase activity and presented as relative luciferase units. The error bars represent the standard deviation of triplicate samples. The data are representative of three independent experiments.
Immunostaining
UM-SCC cells cultured on Lab-tek slides were fixed in a mixture of methanol and acetone, and air dried. Slides were incubated with mouse anti-β-catenin monoclonal antibody (1:3,000, BD Transduction Laboratories). Primary antibody binding was detected using biotinylated goat anti-mouse IgG (Biocare Medical, Walnut Creek, CA), followed by incubation with biotin-streptavidin conjugated to peroxidase and the chromogen DAB500 (Biocare). Immunodetection was followed by hematoxylin counterstaining.
Formalin-fixed, paraffin-embedded human HNSCC tissue microarray sections (5 μm) were deparaffinized and rehydrated. Antigen retrieval was performed with 10mM sodium citrate buffer, pH 6.0, at 95°C for 15 minutes. Duplicate sections were stained with mouse anti-β-catenin and mouse anti-Rap1 monoclonal antibodies (BD Transduction Laboratories). Immunodetection was performed as described (10). Detection of active Rap1 was performed as described (9). Institutional Review Board approval was obtained prior to use of human tissue.
For immunofluorescence studies, HEK293 cells cultured on Lab-tek slides were transfected with FLAG-tagged β-catenin and/or HA-tagged Rap1A G12V or empty vector. After 24h, cells were fixed in ice cold methanol followed by blocking with 1% BSA. Immunodetection was performed with mouse anti-FLAG M2 monoclonal antibody (Sigma) followed by FITC (fluorescein isothiocyanate)-conjugated goat anti-mouse IgG (Jackson Immunoresearch, West Grove, PA). Nuclei were identified with 4′,6-diamidino-2-pheryl-indole, dihydrochloride (DAPI) (Invitrogen, Carlsbad, CA) and the cells were visualized on a laser scanning confocal microscope (Olympus FV-500 Confocal, Tokyo, Japan). Nuclear and cytoplasmic expression of exogenous (FLAG tagged) β-catenin was quantified in cells in twenty fields.
Cell invasion assay
Cell invasion was evaluated with the BD BioCoat Invasion Assay system (BD Biosciences, San Jose, CA) as described (9). Briefly, HNSCC cells stably transfected with HA-tagged β-catenin or control vector (NeoV) (13) were transfected with HA-tagged Rap1A G12V or control vector using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, the cells suspended in DMEM were seeded on Matrigel and Control inserts (5×104 cells/well) and incubated in DMEM/5% FBS as a chemoattractant in the lower chamber. After 24h, HNSCC cells that had migrated to the lower surface of the membrane were fixed in methanol and stained with hematoxylin. To verify protein expression in transfected cells, whole cell lysates were generated from duplicate plates.
University of Michigan Oral Cavity/Oropharyngeal Cancer OrganPreservation Trial
As described previously (9), after appropriate Institutional Review Board approval, a tissue microarray (TMA) was constructed from pretreatment tissue specimens of a randomized clinical trial (28). This was a clinical trial of stage III/IV squamous cell carcinoma of the oral cavity and oropharynx that compared concurrent chemotherapy/radiation in tumors that shrank >50% in response to induction chemotherapy with surgery/radiation in those whose tumors had <50% response to induction chemotherapy (28). Included in this group are squamous cell carcinomas with T-stage of 1 and 2 with nodal involvement (N1, N2, N3), which are considered clinical stage III or IV.
Data analysis
Statistical analysis of in vitro assays was performed using a Student’s t-test. For analysis of TMA data, interpretation and scoring were performed by a board certified pathologist, as described previously (9). The covariates of interest were T stage and N stage, which were analyzed as ordinal data. The outcomes of interest were overall survival, disease-specific survival, time to indication of surgery at primary site, and time to recurrence or second primary. For time to indication of surgery at primary site, the events were stable disease or disease progression after induction chemotherapy that required surgery at the primary site, local recurrence after chemoradiation therapy that required surgery at the primary site, and never disease free. For the outcome of time to recurrence or second primary, the events included recurrences, second primaries, and never disease free. The Spearman correlation coefficient was used to evaluate univariate associations between markers and ordinal variables of interest.
The Cox proportional hazards model was used to relate time-to-event outcomes to marker levels and other covariates. Proportional odds model was used to assess the association between N stage and the interaction of Rap1 and β-catenin.
Statistical analyses of TMA data were done using Statistical Analysis System version 9.0 (SAS). A two-tailed p value of 0.05 was considered to be statistically significant.
Results
Free β-catenin is present in human HNSCC
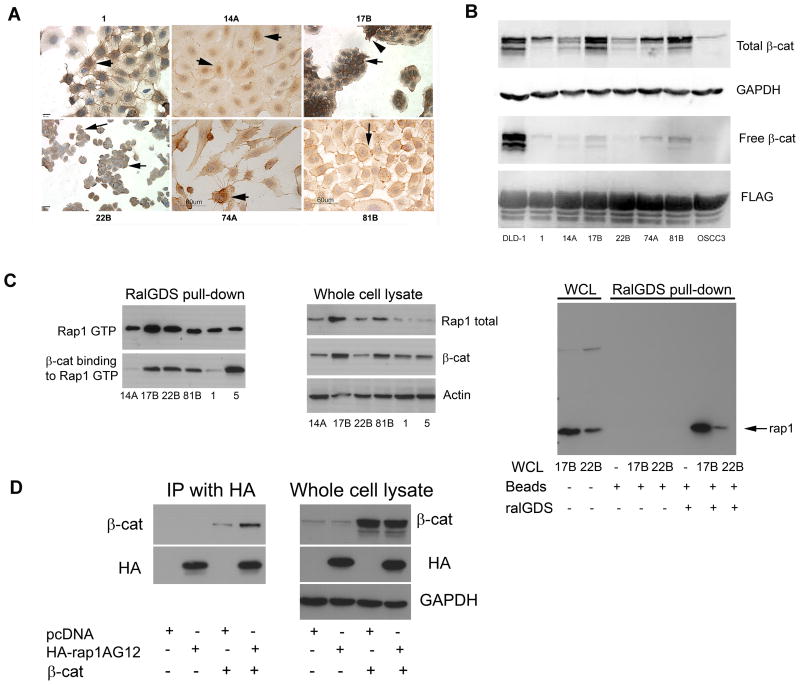
Due to conflicting reports regarding the levels and subcellular localization of β-catenin in HNSCC (29, 30), we investigated whether increased cytosolic and nuclear β-catenin are found in HNSCC. Immunohistochemical studies on HNSCC cell lines revealed β-catenin staining in the cytosol and nucleus of UM-SCC-1, UM-SCC-17B, UM-SCC-74A and UM-SCC-81B lines (Fig. 1A, arrows), whereas in UM-SCC-22B only membrane-bound β-catenin was observed (Fig. 1A, arrows). UM-SCC-14A exhibited primarily nuclear staining (arrows).
Figure 1.
Cytoplasmic and nuclear pools of free β-catenin are detectable in human SCC. A. UM-SCC-(1, 14A, 17B, 22B, 74A, and 81B) cells grown on Lab-tek slides were incubated with β-catenin monoclonal antibody (1:3000) followed by secondary antibody, DAB detection, and counterstaining with hematoxylin. IgG control was negative (not shown). Cytosolic and nuclear staining (1, 74A; arrows), cytosolic, membrane and nuclear (17B, 81B; arrows) or membrane staining (22B; arrows) were observed. (14A, 74A and 81B, bar = 60 μm; 17B and 22B, bar = 40 μm; 1, bar = 20 μm). B. Whole-cell lysates prepared from DLD-1, UM-SCC-(1, 14A, 17B, 22B, 74A, 81B) and OSCC3 were immunoblotted with anti-β-catenin (upper panel) and GAPDH (upper middle panel), as a loading control. Free β-catenin was retrieved with the FLAG-tagged cytoplasmic tail of E-cadherin and immunoblotted with anti-β-catenin (lower middle panel) and anti-Flag (lower panel), as a loading control. C.β-catenin binds to Rap1GTP. Cell lysates of human SCC cell lines UM-SCC-(14A, 17B, 22B, 81B, 1, 5) were evaluated for active Rap1 by the ralGDS pull down assay (upper left panel). The same filter was blotted with anti-β-catenin (lower left panel). Whole cell lysates were blotted to show total Rap1, β-catenin, and actin (middle panel). Control for ralGDS pull-down assay (right panel). For the ralGDS pull-down assay, beads were incubated alone or with whole cell lysates of 17B and 22B in the presence or absence of ralGDS as indicated (right panel, lanes 3–8). Total Rap1 in whole cell lysates from 17B and 22B was also immunoblotted (right panel, lanes 1 and 2). D. HEK293 cells were co-transfected with HA-tagged Rap1AG12 or pcDNA control vector and β-catenin as indicated. HA-Rap1G12 was immunoprecipitated (IP) and the immunoprecipitates were blotted with antiβ-catenin (upper left panel) and anti-HA (lower left panel) antibodies. Whole cell lysates were immunoblotted to show total β-catenin (upper right panel), HA (middle right panel), and GAPDH (lower right panel).
The presence of free β-catenin in HNSCC cells was verified with a pull-down assay using a recombinant protein containing glutathione S-transferase (GST) sequences fused upstream of the FLAG-tagged cytoplasmic tail of E-cadherin (25, 26). The samples were analyzed by immunoblotting for β-catenin, and a FLAG antibody against the epitope present in the GST-E-cadherin fusion protein was used as a control for loading. Among the HNSCC cell lines, the strongest signal for total β-catenin was observed in UM-SCC-17B followed by UM-SCC-81B, UM-SCC-74A and UM-SCC-1 (Fig. 1B, top panel). Free β-catenin was observed in all HNSCC cell lines except UM-SCC-22B and OSCC3 (Fig. 1B, lower middle panel). DLD-1, a colon cancer cell line carrying defects in the APC tumor suppressor gene, was used as a positive control. Relative to total β-catenin, the free β-catenin represented only a small fraction of the protein in whole cell lysates.
β-catenin binds to Rap1 in HNSCC cells
Since a previous study suggested that Rap1A may bind proteins with armadillo repeats (31, 32), we investigated whether Rap1 interacts with β-catenin. Proteins recovered from HNSCC cell lysates by binding to ralGDS, were electrophoresed and blotted first with Rap1 (Fig. 1C, upper left panel) and then with β-catenin antibodies (Fig. 1C, lower left panel). As shown in Fig. 1C (left panel), β-catenin bound to active, GTP-bound Rap1 was recovered with the RalGDS pull-down assay. A strong β-catenin signal was detected in UM-SCC-(17B, 22B, 81B and 5) (Fig. 1C, left panel). Weaker signals were detected in UM-(SCC-14A and 1). Total β-catenin and total Rap1 were variably expressed in these cell lines (Fig. 1C, middle panels). No cross-reactive signals are observed with either GST-beads alone or GST-beads coupled to ralGDS or GST-beads with whole cell lysates in the absence of ralGDS (Fig. 1C, right panel). Unlike immunoprecipitation assays, the pull-down assay uses a “bait” protein rather than IgG to specifically retrieve active Rap1 and its binding partners.
The protein interaction between β-catenin and Rap1 was also investigated by immunoprecipitation (IP) of Rap1. HEK293 cells were transfected with hemaglutinin (HA) epitope-tagged Rap1A and β-catenin. Extracts were immunoprecipitated with anti-HA agarose, and the immunoprecipitates were immunoblotted with anti-β-catenin and anti-HA antibodies. As shown in Fig. 1D, immunoprecipitated HA-tagged Rap1A interacted with β-catenin (left panel, lane 4). A faint cross-reactive signal is observed in cells transfected with β-catenin alone (left panel, lane 3). As shown in the immunoblot of the corresponding whole cell lysates, β-catenin was equivalently expressed in both samples (Fig 1D, right panel, lanes 3 and 4).
Rap1 enhances β-catenin mediated transcription
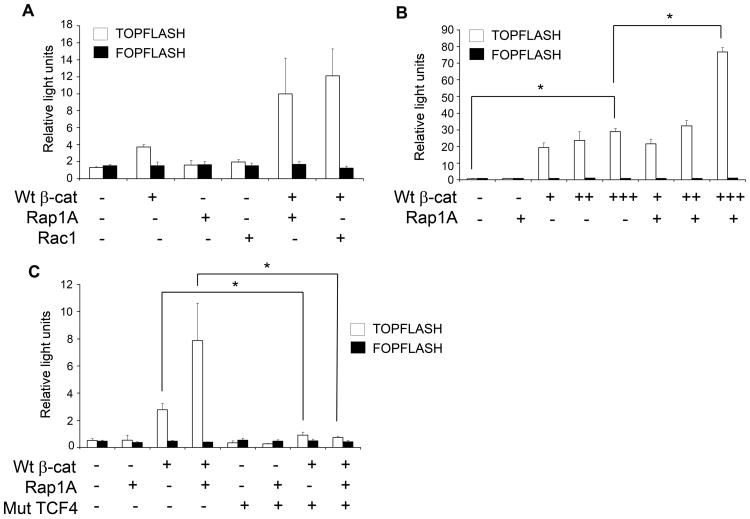
To determine if Rap1 induces nuclear translocation of β-catenin, 293 cells were co-transfected with a constitutively active form of Rap1A (Rap1A G12V) and β-catenin (Fig. 2A). Since these cells have a functionally intact but unstimulated β-catenin pathway, aberrant expression of exogenous β-catenin induces target gene transcription (33). The luciferase-based TOPflash/FOPflash assay system was used to quantify TCF4/LEF-dependent transcriptional activity. Rap1AG12V induced a two-fold increase in β-catenin-mediated transcription (Fig. 2A). Rac1, another small GTP-binding protein that promotes nuclear transport of β-catenin (33), was used as a positive control (Fig. 2A). The effect of Rap1A on β-catenin-mediated transcription was dependent on the concentration of β-catenin (Fig. 2B).
Figure 2.
Rap1 enhances β-catenin/TCF mediated transcription. A. HEK293 cells were co-transfected with pTOPflash or pFOPflash β-catenin mediated reporter genes, Renilla luciferase, wild type β-catenin (Wt β-cat; +: 0.1 μg), the active form of Rap1A (Rap1AG12V, +: 0.1 μg), and active form of Rac1 (Rac1G12V, +: 0.1μg) in a 24-well plate. β-catenin mediated transcription was assayed 24 h later. B. HEK293 cells were co-transfected with pTOPflash or pFOPflash reporter genes, Renilla luciferase, wild-type β-catenin (Wt β-cat, +: 0.2μg, ++: 0.4μg, +++: 0.6μg), and Rap1A G12V (+: 0.9μg), as indicated, in a 6-well plate. Luciferase was assayed 24 h later (*, p<0.05). C, HEK 293 cells were co-transfected with a dominant negative mutant form of TCF4 (Mut TCF4, +: 0.1μg) and pTOPflash, pFOPflash, Renilla luciferase, wild type β-catenin (Wt β-cat, +: 0.1 μg), and Rap1A G12V (+: 0.1 μg), as indicated. The data are the average of at least three independent experiments (*, p<0.05).
To determine if Rap1A induces β-catenin-dependent transcription via TCF4/LEF transcription factor, transcription was evaluated in the presence or absence of a dominant negative mutant form of TCF4 (DNTCF) (34). Rap1A-induced, β-catenin-dependent transcription was completely blocked by a mutant TCF4 protein lacking its β-catenin-binding domain (Fig. 2C). As expected, the transcriptional activity of ectopically expressed β-catenin was also completely blocked by the mutant TCF4 protein. Thus, Rap1A-induced, β-catenin-dependent transcription occurs via a TCF-dependent pathway.
Rap1 enhances nuclear translocation of β-catenin and upregulates β-catenin mediated invasion of HNSCC
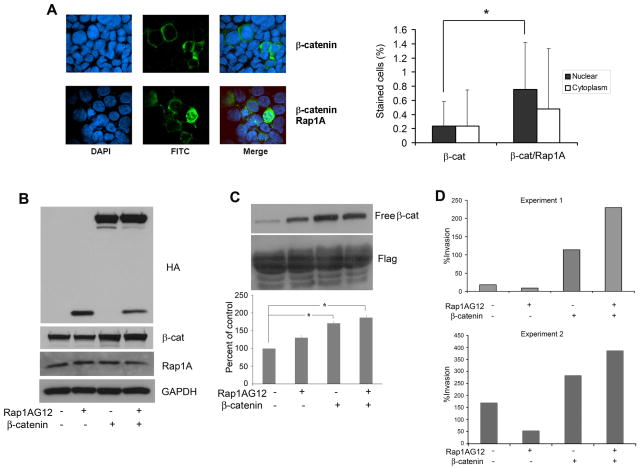
To verify whether Rap1 promotes nuclear translocation of β-catenin, immunofluorescence studies were performed (Fig. 3A). 293 cells were transfected with FLAG-tagged β-catenin alone or with Rap1A G12V and β-catenin. As shown in Fig. 3A (right panel), Rap1 increased the nuclear localization of β-catenin by more than 35% compared to cells transfected with β-catenin and vector control.
Figure 3.
Effects of Rap1 on β-catenin and cell invasion in vitro. A. Rap1 enhances nuclear translocation of β-catenin. HEK 293 cells were transfected with FLAG-tagged wild-type β-catenin (left upper three panels) or co-transfected with wild type β-catenin and Rap1AG12V (left lower three panels). Cells were fixed with methanol and incubated with anti-FLAG antibody (FITC) to detect FLAG-tagged β-catenin (green fluorescence). The slides were counterstained with DAPI nuclear stain (x40 magnification). Quantification of FITC-labeled β-catenin in the nucleus or cytoplasm of 293 cells co-transfected with β-catenin and pcDNA or β-catenin and Rap1A G12 (*, p<0.05) (right panel). Data are representative of two independent experiments. B. SCC cells stably transfected wth HA-tagged β-catenin or empty vector were transfected with HA-tagged Rap1A G12V or pcDNA. Expression of Rap1A G12V and β-catenin were verified by immunoblot analysis with anti-HA antibody as well as β-catenin and Rap1 antibodies. GAPDH was used as a loading control. C. Free β-catenin was retrieved with the cytoplasmic domain of E-cadherin and evaluated by immunoblot analysis (upper panel). Free β-catenin, quantified by densitometry, was normalized to FLAG and expressed as percent of control (lower panel). D. Rap1 promotes β-catenin-mediated invasion in SCC cells. UM-SCC-1 cells stably transfected with HA-tagged β-catenin or empty vector were transfected with HA-tagged Rap1A G12V or pcDNA. Transfected cells suspended in DMEM were added to the upper chamber of a matrigel insert and DMEM/FBS was introduced in the lower chamber. After overnight incubation, the invasive cells were stained and quantified. Data from two independent experiments are shown.
In order to investigate whether Rap1 promotes the functional effects of β-catenin in HNSCC, we determined whether Rap1 enhanced β-catenin-induced invasion. HNSCC cells stably expressing HA-tagged β-catenin (UM-SCC-1/β-cat) or empty vector (UM-SCC-1/NeoV) were transfected with HA tagged Rap1AG12V or pcDNA vector control (Fig. 3B). Overexpression of exogenously expressed β-catenin and Rap1 were confirmed by blotting with HA antibody (Fig. 3B, top panel). Total β-catenin and Rap1 expression were confirmed by blotting with the respective antibodies (Fig. 3B, middle panels). GAPDH confirmed equivalency of loading.
To determine whether Rap1’s effects may be attributable to an increase in free β-catenin, the latter was evaluated by binding to the C-terminal domain of E-cadherin. As expected, cells transfected with β-catenin showed significantly more free-β-catenin than control cells in the presence or absence of Rap1 (Fig. 3C, top and lower panels, p<0.05). However, there was no difference in free β-catenin in HNSCC cells stably expressing HA-tagged β-catenin (Fig. 3C, top and lower panels) in the presence or absence of Rap1AG12V. Rap1AG12 alone increased free β-catenin in HNSCC cells. FLAG was used as a loading control for the pull down protein (C-terminal domain of E-cadherin).
Transfected cells were subsequently seeded on Matrigel and invasion was assayed. Overexpression of Rap1A G12V upregulated invasion in UM-SCC-1/β-catenin by roughly 50% compared to UM-SCC-1/β-catenin cells (Fig. 3D, Experiments 1 and 2). As reported previously (13), UM-SCC-1/β-catenin cells were more invasive than UM-SCC-1/NeoV cells (Fig. 3D). Of note, the effect of Rap1A G12V on β-catenin-mediated invasion was dependent on a threshold level of free β-catenin since they were not observed in control cells transfected with Rap1AG12 alone, even though there was an increase in free β-catenin in these cells (Fig. 3C, lanes 2 versus 4). A similar threshold effect was observed in Fig. 2B which showed that the effect of Rap1A on β-catenin-mediated transcription was dependent on the concentration of β-catenin.
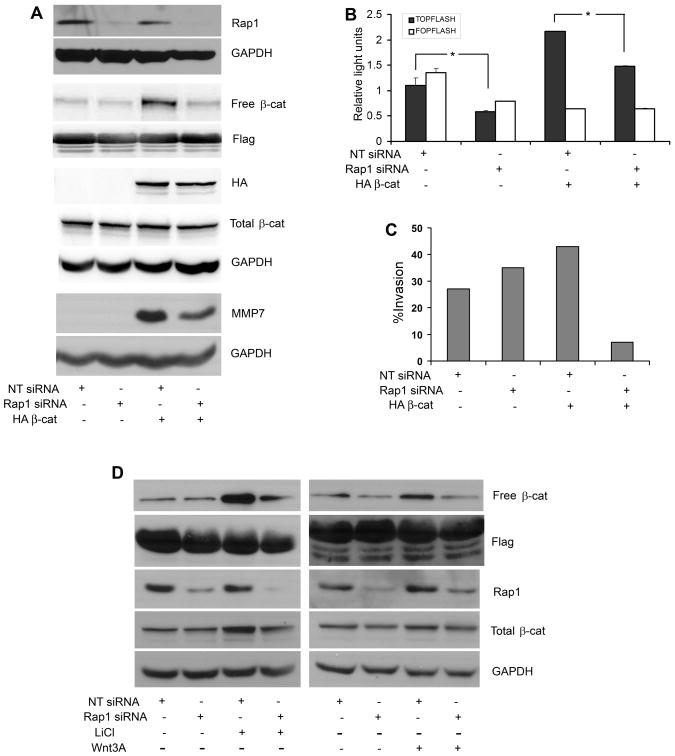
siRNA knockdown of Rap1A inhibits β-catenin-mediated invasion and transcription
To determine whether inhibition of endogenous Rap1 downregulates β-catenin-mediated invasion, UM-SCC-1/Neo and UM-SCC-1/β-catenin cells were nucleofected with siRNAs. As verified in whole cell lysates, Rap1 expression was reduced in UM-SCC-1/Neo and UM-SCC-1/β-catenin cells nucleofected with siRNA Rap1A compared to the corresponding cells nucleofected with non targeting (control) siRNA (Fig. 4A). Moreover, knockdown of Rap1A led to markedly reduced levels of free β-catenin in UM-SCC-1/β-catenin cells (Fig. 4A). FLAG was used as a loading control for the pull down protein (C-terminal domain of E-cadherin), used to retrieve free β-catenin. Immunoblots with HA antibody confirmed exogenously expressed β-catenin in stably transfected cells (Fig. 4A). Total β-catenin remained unchanged. GAPDH was used as a loading control. There was increased expression of MMP7, a transcriptional target of β-catenin/TCF (35), in cells transfected with β-catenin, and levels of MMP-7 expression were reduced in the presence of siRNA Rap1 (Fig. 4A). MMP7 expression was not detected in the absence of HA-β-catenin.
Figure 4.
Rap1A-β-catenin mediated transcription and invasion of SCC. A. UM-SCC-1 cells stably transfected with HA-tagged β-catenin or empty vector were nucleofected with siRNA Rap1A or Non-target siRNA. Cell lysates were immunoblotted with Rap1, HA, FLAG, GAPDH, β-catenin and MMP7 antibodies. Free β-catenin was retrieved with the cytoplasmic domain of E-cadherin. B. UM-SCC-1 cells stably transfected with HA-tagged β-catenin or empty vector were nucleofected with siRNA Rap1A or Non-target siRNA concurrently with pTOPflash, pFOPflash, and Renilla luciferase. Luciferase activity was assayed 24h later. Data are representative of three independent experiments, each in triplicate. C. UM-SCC-1 cells stably transfected with HA-tagged β-catenin or empty vector were nucleofected with siRNA Rap1A or Non-target siRNA. Transfected cells suspended in DMEM were added to the upper chamber of a matrigel insert and DMEM/FBS was introduced in the lower chamber. After overnight incubation, the invasive cells were stained and quantified. Data are representative of three independent experiments. D. UM-SCC-1 cells nucleofected with siRNA Rap1A or non target siRNA were treated with lithium chloride (40 mM, 6h; two independent experiments, each in duplicate) or Wnt3A (50 ng/ml, 6h; one experiment in duplicate) as indicated. Free β-catenin and whole cell lysates were immunoblotted with Flag, Rap1, β-catenin and GAPDH antibodies.
In corresponding luciferase assays, siRNA Rap1A inhibited TCF-dependent luciferase activity in the presence or absence of overexpressed β-catenin (Fig. 4B, p<0.03). As predicted, and as shown in Fig. 4C, invasion was upregulated in UM-SCC-1/β-catenin cells compared to corresponding UM-SCC-1/Neo control cells. Furthermore, siRNA Rap1A inhibited invasion in UM-SCC-1/β-catenin cells compared to UM-SCC-1/β-catenin cells nucleofected with non targeting siRNA (Fig. 4C). To confirm whether downregulation of Rap1 reduces free β-catenin, HNSCC cells nucleofected with siRNA Rap1A were treated with lithium chloride. HNSCC cells treated with 40 mM lithium chloride (Alexis Biochemicals) for 6h showed an increase in free β-catenin (Fig. 4D, left panel). This increase was abrogated in the presence of siRNA Rap1A. Similar results were observed in HNSCC cells treated with Wnt3A (50 ng/ml, R&D Systems) for 6h (Fig. 4D, right panel). The Wnt3A induced increase in free β-catenin was reduced in cells transduced with siRNA Rap1A (Fig. 4D, right panel). As determined by densitometry of lithium chloride treated sample, the total β-catenin normalized to GAPDH, is decreased by ~21%, in the presence of siRNA Rap1A. In contrast, the free β-catenin is decreased ~54%, in the presence of siRNA Rap1A. In the presence of Wnt3A, the total β-catenin normalized to GAPDH, shows a slight increase of ~7%. In contrast, the free β-catenin is decreased ~42%, in the presence of siRNA Rap1A. Together these studies support a decrease of at least 30% in free β-catenin in cells transduced with siRNA Rap1A.
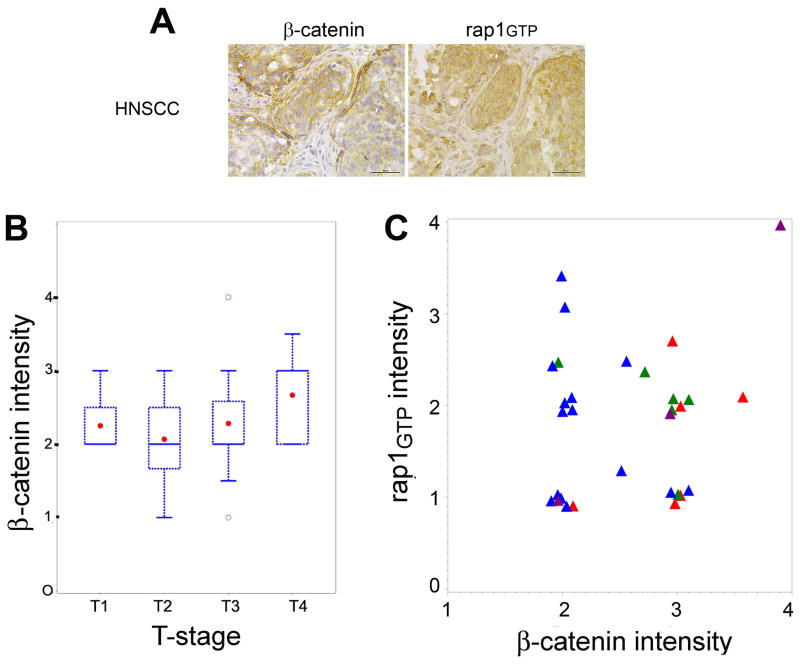
Immunohistochemical studies using a tissue microarray of pre-treatment tissue specimens from primary oropharyngeal SCCs showed that increased β-catenin intensity was correlated with higher tumor stage, as assessed by the Spearman correlation coefficient (Fig. 5B, p<0.05). Membrane and cytoplasmic staining for β-catenin were observed in tumor cells (Fig. 5A, left panel). Multivariate analysis (proportional odds model) showed that the association of active Rap1 on tumor N-stage depended on the expression of cytosolic β-catenin. When β-catenin expression was high, higher active Rap1 intensity (Fig. 5A,) was associated with more advanced N-stage; when β-catenin expression is low, lower active Rap1 intensity is associated with more advanced N-stage (Fig. 5C, p<0.013). These finding are consistent with our in vitro data showing that HNSCC cells expressing high β-catenin and high active Rap1 were more invasive (Fig. 3D; Rap1AG12 +, β-catenin + ) than cells expressing high β-catenin and low Rap1 (Fig. 3D; Rap1AG12 -, β-catenin + ). Also consistent were the in vitro findings showing that HNSCC cells expressing low β-catenin and low active Rap1 (Fig. 3D; Rap1AG12 -, β-catenin - ) were more invasive than cells expressing low β-catenin and high active Rap1 (Fig. 3D; Rap1AG12 +,β-catenin - ). There were no significant immunohistochemical correlations between β-catenin intensity and other survival outcomes such as disease-specific survival, and time to recurrence or second primary tumor, as assessed by a univariate Cox proportional hazard model..
Figure 5.
The prognostic effect of active Rap1 on tumor N-stage depends on the expression of cytosolic β-catenin. A. A 5μm tissue section of a human HNSCC tissue microarray was incubated with β-catenin antibody (left panel) or IgG control (not shown) or Rap1 antibody (right panel) as described in the Methods section. After DAB reaction, the slides were counterstained with hematoxylin (bars = 50 μm). High cytosolic β-catenin and rap1GTP staining are shown for the same tumor specimen. B. High β-catenin intensity is associated with more advanced T-stage as assessed by the Spearman Correlation Coefficient and represented in box and whisker plots. Patient groups: T1, tumor size <2 cm (n = 4); T2, tumor size 2–4 cm (n = 10); T3, tumor size >4cm (n = 16), T4, tumor invades through cortical bone, inferior alveolar nerve, floor of mouth or skin of face (n = 13). C. The proportional odds model was used to assess the association between N stage and the interaction of Rap1 and β-catenin. When cytosolic β-catenin was high, higher active Rap1 intensity was prognostic of more advanced N-stage; when β-catenin intensity is low, lower active Rap1 is associated with more advanced N-stage. Early N-stage is N0 (red triangles, n=6) and N1 (green triangles, n=6); advanced N-stage is N2 (blue triangles, n=15) and N3 (purple triangles, n=3).
Discussion
In HNSCC, invasion of tumor cells leads to extension into the underlying tissues and metastasis to regional lymph nodes and distant sites, which impact adversely on patient survival (36). Various Wnt ligands, increase the levels of the free pool of β-catenin (16). β-catenin, a central molecule in the Wnt pathway, translocates to the nucleus where it triggers the transcription of genes that facilitate invasion and migration (20). Many β-catenin-regulated target genes have been identified and include matrix metalloproteinases (MMP) (35, 37–41), cyclin D1 and c-myc (42, 43). We have previously shown that downregulation of active Rap1 by rap1GAP, a regulatory protein, promoted invasion of HNSCC cells via effects on secretion of MMP9 and MMP2 (9). Furthermore, Rap1 translocates between the nucleus and cytoplasm in HNSCC cells. In the present study, we show that upregulation of active, GTP-bound Rap1 promotes invasion, perhaps via β-catenin-mediated effects on MMP7 secretion.
Using the pull-down assay to retrieve active, GTP-bound Rap1, we observed that β-catenin was also retrieved. We also demonstrated by immunoprecipitation of exogenously expressed, HA-tagged Rap1, that β-catenin binds to Rap1. To investigate whether this association might affect β-catenin function in the nucleus, we performed transcriptional assays with TOPflash and FOPflash luciferase reporter genes. The TOPflash reporter construct contains three repeats of the wildtype TCF4 binding site upstream of a thymidine kinase minimal promoter and the luciferase gene. Together with β-catenin, TCF4 induces transcription of specific genes. Consistent with a possible role in nuclear translocation of β-catenin, we observed that Rap1 significantly enhanced β-catenin mediated transcription. The findings were supported by immunofluorescence studies showing nuclear translocation of β-catenin. Furthermore, we showed that Rap1’s effects on β-catenin/TCF transcription were inhibited by a dominant-negative mutant of TCF4.
Matrix metalloproteinase 7 (MMP7, matrilysin), which has a TCF binding site in its promoter, is a transcriptional target of β-catenin (35, 44). Consistent with this role, MMP7 was upregulated in HNSCC cells stably overexpressing β-catenin and was downregulated in the presence of siRNA Rap1. MMPs are zinc-dependent proteolytic enzymes that remodel the extracellular matrix. MMPs facilitate invasion and tumor progression by destroying the basement membrane (45). Using tissue microarrays, MMP7 has been previously shown to be upregulated in HNSCC (45).
Rap1 was originally identified as a tumor suppressor gene in ras-transformed NIH 3T3 fibroblasts (46). Rap1A was later shown to facilitate cell proliferation by inducing DNA synthesis (47). In Drosophila and in osteosarcoma cell lines, Rap1 facilitates cell adhesion, thereby inhibiting cell migration (21–23). In the case of work in Drosophila, adherens junctions were uniformly distributed around the circumference of cells with wildtype Rap1, but were clustered on one side of cells with inactivating Rap1 mutations (48). The mutant cell clones randomly invaded surrounding tissues, suggesting a role for Rap1 in appropriate maintenance of cell-cell contacts and inhibition of invasion. In a recent study, we showed that overexpression of Rap1GAP, which inactivates Rap1, promotes invasion in HNSCC cells (9). In the present study, HNSCC cells with siRNA-mediated downregulation of Rap1A were more invasive than control cells. Consistent with these findings, osteosarcoma cells with inactive DOCK4 and reduced levels of active Rap1 do not form adherens junctions and are invasive (49). Similarly, overexpression of Rap1 in MDCK epithelial cells facilitates cell-cell junction formation (24).
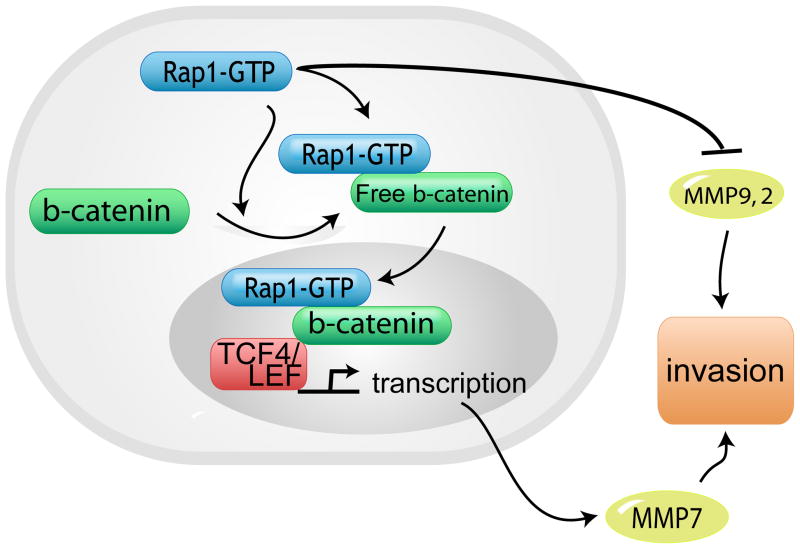
In contrast, in human breast cancer and mouse thyroid cells, disruption of cell contacts with endocytosis of E-cadherin initiates Rap1 activation (22, 23, 50) via rap1GEFs (GTP-GDP exchange factors) (50). These apparently contradictory effects in Rap1 function may be related to concurrent expression of other significant regulatory proteins. Our findings support this notion. Previously (9) and in the current study, downregulation of Rap1 promotes invasion of HNSCC cells. However, in the presence of free β-catenin, Rap1 promotes invasion in HNSCC cells and β-catenin-mediated transcription in 293 cells. Furthermore, Rap1 stabilizes free β-catenin that is increased by lithium chloride or Wnt3A, a secreted factor. Taken together, these findings are consistent with a dual role for Rap1 in invasion (Fig. 6).
Figure 6.
Proposed Model for interaction between Rap1, β-catenin and MMP7 in SCC progression. Rap1 promotes invasion via β-catenin mediated MMP7 secretion and inhibits invasion via inhibition of MMP9 and MMP2 secretion. Tumors with high β-catenin expression are associated with more advanced tumor stage. Rap1 promotes invasion via β-catenin mediated effects and inhibits invasion via inhibition of MMP9 and MMP2.
There are conflicting reports regarding nuclear β-catenin in HNSCC (29, 30). In one study with formalin-fixed HNSCC tissue (29), β-catenin was localized in the nucleus and cytoplasm of cells at the invasive front. In another study on frozen tissue with an antibody from a different source, nuclear β-catenin staining was not observed (30). In the current study we demonstrated cytosolic and nuclear localization of β-catenin in HNSCC cell lines and human tissue specimens. We show that high β-catenin intensity is prognostic of late tumor stage. Additionally, the association between active Rap1 and tumor progression was dependent on the level of cytosolic β-catenin. Together, high β-catenin and high active Rap1 are associated with a more advanced N-stage. Taken together, these results from a small group of patients suggest that pre-treatment screening for active Rap1 and free β-catenin in early stage lesions may identify those patients who are more likely to show progression to more advanced N-stage lesions.
The exact mechanism of Rap1-mediated effects on β-catenin is unknown but may in part be due to an increase in free-β-catenin to a threshold level. In the luciferase assay, when overexpression of β-catenin is constant (0.6 ug of cDNA), an increase in Rap1 facilitated a significant increase in luciferase activity (Fig. 2B, lanes 5 and 8). However, overexpression of Rap1 did not enhance β-catenin mediated transcription in cells transfected with 0.2 or 0.4 ug of β-catenin cDNA. In β-catenin overexpressing cells, transfection with Rap1 did not further increase free β-catenin (Fig. 3C), but it did enhance the invasion (Fig. 3D). Together the results presented in Figs. 3C, 3D and 2B, suggest a threshold ratio for the interaction between Rap1 and β-catenin.
Since free β-catenin translocates to the nucleus to induce target genes that regulate invasion, proliferation and metastasis (16, 20), elucidation of the mechanism of nuclear transport would identify an important therapeutic target. Rac1, another Ras-like protein, binds armadillo proteins, such as smgGDS, in the cytosol transporting them to the nucleus (32). Rac1 has a C-terminal polybasic region, a series of adjacent lysines and arginines that act as a canonical nuclear localization signal (Lys-Arg/Lys-x-Arg/Lys where x is Pro, Lys, Arg, Val, Ala) (32, 51). Our previous studies showing localization of active Rap1GTP primarily in the nucleus and inactive Rap1GDP primarily in the cytoplasm of HNSCC suggest that Rap1 shuttles between these two compartments (10). Given Rap1’s prominent nuclear localization in HNSCC, and its putative nuclear localization sequence (Lys-Lys-Pro-Lys), it is reasonable to speculate that Rap1 regulates nuclear transport of β-catenin in addition to upregulating free β-catenin. Consistent with this hypothesis, a recent study has shown that Rac1 regulates the nuclear translocation of β-catenin and upregulates β-catenin mediated transcription via the polybasic region (33). Future studies will explore whether Rap1 mediates nuclear transport of β-catenin.
Acknowledgments
The authors wish to thank Wei Ao for excellent technical assistance.
Financial support: This work was supported by NIDCR DE019513-01, DE018512-01, and University of Michigan Head and Neck Specialized Program of Research Excellence project and developmental project grants (NJD).
Footnotes
Translational Relevance
In the current study we investigated the role of Rap1 in stabilization of β-catenin, induction of its transcriptional targets and in β-catenin-mediated invasion of head and neck squamous cell carcinoma. We show that increased β-catenin intensity correlated with higher tumor stage. Furthermore, the association between active Rap1 and tumor N-stage was dependent on the level of cytosolic β-catenin. High β-catenin and high active Rap1 are associated with a more advanced N-stage. Taken together, these results from a small group of patients suggest that pre-treatment screening for active Rap1 and free β-catenin in early stage lesions may identify those patients who are more likely to show progression to more advanced N-stage lesions.
References
- 1.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001;2:369–77. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 2.Stork PJ, Dillon TJ. Multiple roles of Rap1 in hematopoietic cells: complementary versus antagonistic functions. Blood. 2005;106:2952–61. doi: 10.1182/blood-2005-03-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mochizuki N, Ohba Y, Kiyokawa E, et al. Activation of the ERK/MAPK pathway by an isoform of rap1GAP associated with G alpha(i) Nature. 1999;400:891–4. doi: 10.1038/23738. [DOI] [PubMed] [Google Scholar]
- 4.Rubinfeld B, Munemitsu S, Clark R, et al. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–42. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- 5.Su L, Hattori M, Moriyama M, et al. AF-6 controls integrin-mediated cell adhesion by regulating Rap1 activation through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem. 2003;278:15232–8. doi: 10.1074/jbc.M211888200. [DOI] [PubMed] [Google Scholar]
- 6.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 7.Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Mitra RS, Henson BS, et al. Rap1GAP inhibits tumor growth in oropharyngeal squamous cell carcinoma. Am J Pathol. 2006;168:585–96. doi: 10.2353/ajpath.2006.050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitra RS, Goto M, Lee JS, et al. Rap1GAP promotes invasion via induction of matrix metalloproteinase 9 secretion, which is associated with poor survival in low N-stage squamous cell carcinoma. Cancer Res. 2008;68:3959–69. doi: 10.1158/0008-5472.CAN-07-2755. [DOI] [PubMed] [Google Scholar]
- 10.Mitra RS, Zhang Z, Henson BS, Kurnit DM, Carey TE, D’Silva NJ. Rap1A and rap1B ras-family proteins are prominently expressed in the nucleus of squamous carcinomas: nuclear translocation of GTP-bound active form. Oncogene. 2003;22:6243–56. doi: 10.1038/sj.onc.1206534. [DOI] [PubMed] [Google Scholar]
- 11.Leethanakul C, Patel V, Gillespie J, et al. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19:3220–4. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- 12.Rhee CS, Sen M, Lu D, et al. Wnt and frizzled receptors as potential targets for immunotherapy in head and neck squamous cell carcinomas. Oncogene. 2002;21:6598–605. doi: 10.1038/sj.onc.1205920. [DOI] [PubMed] [Google Scholar]
- 13.Yang F, Zeng Q, Yu G, Li S, Wang CY. Wnt/beta-catenin signaling inhibits death receptor-mediated apoptosis and promotes invasive growth of HNSCC. Cell Signal. 2006;18:679–87. doi: 10.1016/j.cellsig.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Behrens J, von Kries JP, Kuhl M, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 15.Molenaar M, van de Wetering M, Oosterwegel M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–9. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 16.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–81. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 18.Sakanaka C, Weiss JB, Williams LT. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc Natl Acad Sci U S A. 1998;95:3020–3. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–90. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 20.Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer. 2002;34:255–68. doi: 10.1002/gcc.10083. [DOI] [PubMed] [Google Scholar]
- 21.Fujita Y, Hogan C, Braga VM. Regulation of cell-cell adhesion by Rap1. Methods Enzymol. 2006;407:359–72. doi: 10.1016/S0076-6879(05)07030-8. [DOI] [PubMed] [Google Scholar]
- 22.Hogan C, Serpente N, Cogram P, et al. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price LS, Hajdo-Milasinovic A, Zhao J, Zwartkruis FJ, Collard JG, Bos JL. Rap1 regulates E-cadherin-mediated cell-cell adhesion. J Biol Chem. 2004;279:35127–32. doi: 10.1074/jbc.M404917200. [DOI] [PubMed] [Google Scholar]
- 24.Asuri S, Yan J, Paranavitana NC, Quilliam LA. E-cadherin dis-engagement activates the Rap1 GTPase. J Cell Biochem. 2008;105:1027–37. doi: 10.1002/jcb.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winer IS, Bommer GT, Gonik N, Fearon ER. Lysine residues Lys-19 and Lys-49 of beta-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J Biol Chem. 2006;281:26181–7. doi: 10.1074/jbc.M604217200. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, Bommer GT, Zhai Y, et al. Drosophila split ends homologue SHARP functions as a positive regulator of Wnt/beta-catenin/T-cell factor signaling in neoplastic transformation. Cancer Res. 2007;67:482–91. doi: 10.1158/0008-5472.CAN-06-2314. [DOI] [PubMed] [Google Scholar]
- 27.Franke B, Akkerman JW, Bos JL. Rapid Ca2+-mediated activation of Rap1 in human platelets. Embo J. 1997;16:252–9. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf GT, Urba S, Hazuka M. Induction chemotherapy for organ preservation in advanced squamous cell carcinoma of the oral cavity and oropharynx. Recent Results Cancer Res. 1994;134:133–43. doi: 10.1007/978-3-642-84971-8_15. [DOI] [PubMed] [Google Scholar]
- 29.Uraguchi M, Morikawa M, Shirakawa M, Sanada K, Imai K. Activation of WNT family expression and signaling in squamous cell carcinomas of the oral cavity. J Dent Res. 2004;83:327–32. doi: 10.1177/154405910408300411. [DOI] [PubMed] [Google Scholar]
- 30.Tsuchiya R, Yamamoto G, Nagoshi Y, Aida T, Irie T, Tachikawa T. Expression of adenomatous polyposis coli (APC) in tumorigenesis of human oral squamous cell carcinoma. Oral Oncol. 2004;40:932–40. doi: 10.1016/j.oraloncology.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL. The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J Biol Chem. 2004;279:44197–210. doi: 10.1074/jbc.M404977200. [DOI] [PubMed] [Google Scholar]
- 32.Lanning CC, Ruiz-Velasco R, Williams CL. Novel mechanism of the co-regulation of nuclear transport of SmgGDS and Rac1. J Biol Chem. 2003;278:12495–506. doi: 10.1074/jbc.M211286200. [DOI] [PubMed] [Google Scholar]
- 33.Esufali S, Bapat B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of beta-catenin and TCF/LEF-mediated transcriptional activation. Oncogene. 2004;23:8260–71. doi: 10.1038/sj.onc.1208007. [DOI] [PubMed] [Google Scholar]
- 34.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 35.Crawford HC, Fingleton BM, Rudolph-Owen LA, et al. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–91. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 36.Werner JA, Dunne AA, Ramaswamy A, et al. Sentinel node detection in N0 cancer of the pharynx and larynx. Br J Cancer. 2002;87:711–5. doi: 10.1038/sj.bjc.6600445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Angelis T, Noe A, Chatterjee M, Mulholland J. Stromelysin-1 activation correlates with invasiveness in squamous cell carcinoma. J Invest Dermatol. 2002;118:759–66. doi: 10.1046/j.1523-1747.2002.01755.x. [DOI] [PubMed] [Google Scholar]
- 38.Impola U, Jeskanen L, Ravanti L, et al. Expression of matrix metalloproteinase (MMP)-7 and MMP-13 and loss of MMP-19 and p16 are associated with malignant progression in chronic wounds. Br J Dermatol. 2005;152:720–6. doi: 10.1111/j.1365-2133.2005.06447.x. [DOI] [PubMed] [Google Scholar]
- 39.Impola U, Uitto VJ, Hietanen J, et al. Differential expression of matrilysin-1 (MMP-7), 92 kD gelatinase (MMP-9), and metalloelastase (MMP-12) in oral verrucous and squamous cell cancer. J Pathol. 2004;202:14–22. doi: 10.1002/path.1479. [DOI] [PubMed] [Google Scholar]
- 40.Margulis A, Zhang W, Alt-Holland A, Crawford HC, Fusenig NE, Garlick JA. E-cadherin suppression accelerates squamous cell carcinoma progression in three-dimensional, human tissue constructs. Cancer Res. 2005;65:1783–91. doi: 10.1158/0008-5472.CAN-04-3399. [DOI] [PubMed] [Google Scholar]
- 41.Pelosi G, Scarpa A, Veronesi G, et al. A subset of high-grade pulmonary neuroendocrine carcinomas shows up-regulation of matrix metalloproteinase-7 associated with nuclear beta-catenin immunoreactivity, independent of EGFR and HER-2 gene amplification or expression. Virchows Arch. 2005;447:969–77. doi: 10.1007/s00428-005-0044-x. [DOI] [PubMed] [Google Scholar]
- 42.Chen YJ, Lin SC, Kao T, et al. Genome-wide profiling of oral squamous cell carcinoma. J Pathol. 2004;204:326–32. doi: 10.1002/path.1640. [DOI] [PubMed] [Google Scholar]
- 43.Ginos MA, Page GP, Michalowicz BS, et al. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 44.Takeda K, Kinoshita I, Shimizu Y, et al. Clinicopathological significance of expression of p-c-Jun, TCF4 and beta-Catenin in colorectal tumors. BMC Cancer. 2008;8:328. doi: 10.1186/1471-2407-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber A, Hengge UR, Stricker I, et al. Protein microarrays for the detection of biomarkers in head and neck squamous cell carcinomas. Hum Pathol. 2007;38:228–38. doi: 10.1016/j.humpath.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida Y, Kawata M, Miura Y, et al. Microinjection of smg/rap1/Krev-1 p21 into Swiss 3T3 cells induces DNA synthesis and morphological changes. Mol Cell Biol. 1992;12:3407–14. doi: 10.1128/mcb.12.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science. 2002;295:1285–8. doi: 10.1126/science.1067549. [DOI] [PubMed] [Google Scholar]
- 49.Yajnik V, Paulding C, Sordella R, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–84. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 50.Balzac F, Avolio M, Degani S, et al. E-cadherin endocytosis regulates the activity of Rap1: a traffic light GTPase at the crossroads between cadherin and integrin function. J Cell Sci. 2005;118:4765–83. doi: 10.1242/jcs.02584. [DOI] [PubMed] [Google Scholar]
- 51.Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol Cell Biol. 1989;9:2487–92. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]