Abstract
The lateral mobility of cell membranes plays an important role in cell signaling, governing the rate at which embedded proteins can interact with other biomolecules. The past two decades have seen a dramatic transformation in understanding of this environment, as the mechanisms and potential implications of nanoscale structure of these systems has become accessible to theoretical and experimental investigation. In particular, emerging micro- and nano-scale fabrication techniques have made possible the direct manipulation of model membranes at the scales relevant to these biological processes. This review focuses on recent advances in nanopatterning of supported lipid bilayers, capturing the impact of membrane nanostructure on molecular diffusion and providing a powerful platform for further investigation of the role of this spatial complexity on cell signaling.
1. Introduction
Lipid membranes are powerful organizational components of living cells, delineating and separating the various components of these systems. Somewhat paradoxically, lipid membranes are largely fluid, allowing motion of membrane biomolecules along the structure. This fluidity arises from the concept that membranes are held together by hydrophobic/hydrophilic interactions between lipid molecules, an idea that was the foundation for the Fluid Mosaic model formalized by Singer and Nicolson (Singer and Nicolson, 1972). Since that time, new experimental tools and conceptual frameworks have allowed exploration of the more subtle regions of this model, revealing short-range organization within membranes of living cells. What has emerged is a complex and not completely understood mix of interactions that give rise to a membrane nanostructure that modulates diffusion and mobility of membrane biomolecules. These effects are often invoked as modulators of cell signaling over a range of spatial scales. Development of sufficiently quantitative models of these effects would be greatly accelerated by the ability to capture the nanoscale complexity of the proposed membrane structure in controllable, reductionist systems. In this direction, substrate-supported lipid bilayers have provided elegant insights into the role of membranes as matrices for cell signaling by capturing this system in format that is compatible with contemporary, high-resolution microscopy (Chan et al., 1991; Dean et al., 2003; Grakoui et al., 1999; Groves and Dustin, 2003). Techniques for micropatterning these lipid systems hold much promise for capturing nanoscale membrane organization (Cremer et al., 1999; Groves and Boxer, 2002; Groves et al., 1997; Kam and Boxer, 2000; Kung et al., 2000a; Kung et al., 2000b; Ulman et al., 1997). However, the underlying mechanisms that give rise to membrane structure in cells are dynamic, fugitive, and very different than the manipulations that are commonly done to pattern supported lipid bilayers. We review here recent progress in the design of supported lipid bilayers systems that seek to capture this nanoscale complexity.
2. Nanoscale membrane structure
The recognition that cell membranes exhibit nanoscale structure has had a profound impact on the study of cell signaling and membrane physiology. Current research focuses on the formulation of general, guiding principles that govern membrane structure and identification of how these rules act in specific signaling proteins and pathways. Within this wide field, we focus here on capturing two general types of nanoscale organization in substrate-supported lipid bilayers.
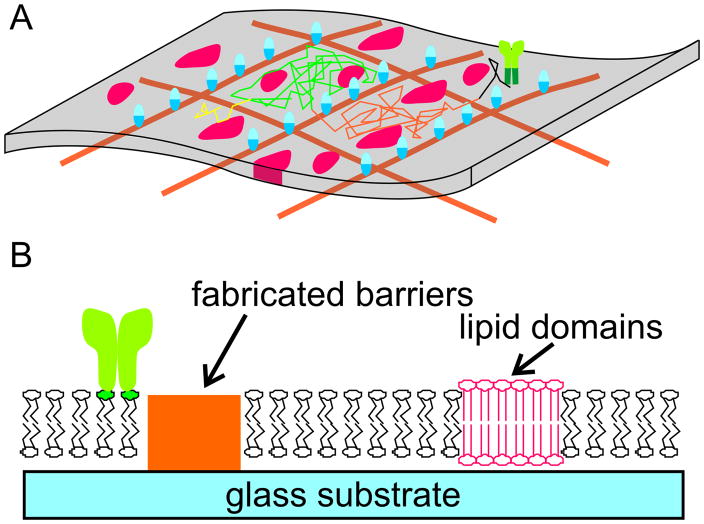
The first is the concept of microdomains, localized aggregates of membrane biomolecules (suggested by the red islands in Figure 1A) that dynamically assemble, reorganize, and dissolve against a background of otherwise fluid-phase membrane. The archetypical model of this concept, the lipid raft, was proposed as a mechanism for trafficking of membrane proteins from synthesis to delivery on the cell surface, mediated by local, liquid-ordered regions rich in sphingolipids and cholesterol (Simons and Ikonen, 1997), interspersed throughout a liquid-disordered matrix. The concept of lipid rafts and microdomains in general as spatial modulators of cell signaling has become a major organizational principle (Dustin, 2002; Edidin, 2003; Gomez-Mouton et al., 2004; Jury et al., 2007; Shaw, 2006; Simons and Toomre, 2000) that is applied across multiple cell types and a wide range of signaling protein pathways. In each situation, important questions include the size and duration of domain formation, domain composition, and inclusion/exclusion of the molecule of interest as well as associated signaling partners into the microdomain.
Figure 1.
Nanoscale organization of cell membranes. (A) Two mechanisms for this organization in living cells are the presence of microdomains (red forms) and cytoskeletal structures (orange grid and blue transmembrane proteins) both of which limit long-range diffusion. The random-walk trajectory of a cell signaling molecule (green) that is excluded from these domains and restricted by the cytoskeletal elements is illustrated in this panel. (B) Strategies for capturing nanoscale membrane complexity in supported lipid bilayers include the patterning of non- compatible materials (orange) onto the substrate, or the use of lipid mixtures that demix to form stable analogs of the native microdomains.
The second source of nanoscale order we will focus on is the cell cytoskeleton that underlies the cell membrane, which modulates molecular diffusion of membrane components (Fujiwara et al., 2002; Iino et al., 2001; Kusumi et al., 1993; Kusumi et al., 1999; Ritchie and Kusumi, 2004; Ritchie et al., 2003). The original observation driving this concept is that membrane proteins exhibit diffusion coefficients that are often much smaller than that predicted from the aggregate properties of the membranes. High-resolution microscopy revealed that this slower diffusion is consistent with a model in which diffusing membrane molecules interact either directly with the cytoskeleton or through intermediate proteins (Figure 1A) tethered to the cytoskeleton; these two complementary models are termed the “skeleton fence” and “picket fence” models, respectively. In both cases, the cytoskeleton forms semi-permeable barriers that delineate corrals of the membrane, leading to a process of diffusion along the surface characterized by a relatively slow “hop diffusion” across multiple corrals (suggested by the multicolor trajectory shown in Figure 1A).
These two complementary sources of membrane organization share several properties. First, both act to reduce diffusion along the cell membrane. Second, both involve structures that are generally believed to be nanoscale. Microdomains are composed of clusters as small as a countable number of lipids or up to several hundred nanometers in extent; the specific size and dynamics vary tremendously as a function of signaling mechanism, physical conditions, and experimental analysis method. The size of corrals estimated to be present in a variety of cells ranges over a similar range, tens to hundreds of nanometers (Kusumi et al., 2005a; Kusumi et al., 2005b). Third, both sources of nanoscale organization are based on phenomena that are difficult to fully verify and demonstrate; the formation and stability of microdomains, as well as the nature of interactions between the cytoskeleton and membrane proteins, are transient and sensitive to the experimental techniques. In particular, the presence of semi-permeable barriers has not been observed concurrently with molecular diffusion. As such, the term “hop diffusion” will be applied to diffusion in the presence of explicit corrals; more generally, the term “hindered diffusion” will be used to expresses the concept of non-Brownian subdiffusion.
Capturing these sources of membrane nanostructure in substrate-supported lipid bilayers would provide a powerful tool for investigating the impact of these mechanisms on cell signaling. The basic structure of the supported membrane system is a single bilayer of lipids closely apposed with but not attached to an underlying substrate. This separation between the membrane and support imparts lateral fluidity to the membrane. Proteins tethered to the membrane are also laterally mobile, forming a system for studying the role of protein mobility on cell signaling. This approach has been highly successful in the context of immune cell function, and is finding application in an increasing range of cellular systems (Baksh et al., 2005; Dori et al., 2000; Groves et al., 2001; Kam and Boxer, 2001; Lenz et al., 2004; Orth et al., 2003; Pautot et al., 2005; Perez et al., 2005; Ratto and Longo, 2003; Sackmann, 1996; Torres et al., 2008). Moreover, the development of approaches to micropatterning these bilayers has set much promise for imposing nanoscale order on these systems. A widely-used method of forming lipid bilayers is through fusion of lipid vesicles (essentially lipid bilayers patches rolled on themselves) onto a support. This fusion process occurs on only a few, select materials, including silicon oxide surfaces such as glass and quartz but also mica and, perhaps surprisingly, oxidized polydimethyl siloxane (Brian and McConnell, 1984; Hafeman et al., 1981; Hovis and Boxer, 2001). Micropatterning of metals, plastics, or even proteins onto these oxide surfaces prior to vesicle exposure serves to restrict, limit, and pattern lipid bilayers formation (Figure 1B) (Boxer, 2000; Groves and Boxer, 2002; Groves et al., 1997; Kam and Boxer, 2000; Kung et al., 2000b; van Oudenaarden and Boxer, 1999), and extensions of these methods into the nanoscale has allowed capture of both microdomains and cytoskeletal barriers (Furukawa et al., 2007; Lenhert et al., 2007; Nabika et al., 2005; Nabika et al., 2009; Shi et al., 2008; Tsai et al., 2008; Werner et al., 2009). While these methods offer a high degree of control over the barrier and substrate layout, they are expensive to use over large areas. An alternative approach, inspired by the microdomain concept, is to form bilayers from binary and ternary mixture of lipids, each with physical/chemical properties that lead to demixing or phase separation in the final structure (Johnston, 2007; Ratto and Longo, 2002; Ratto and Longo, 2003). The resulting structures tend to be less well-defined than fabricated systems, but are highly effective in capturing the concept of microdomains on supported bilayers. In particular, these surfaces have had recent success as a platform for understanding the targeting and segregation of specific classes of biomolecules in to the gel phase domains, which are relatively immobile (Alves et al., 2005; Devanathan et al., 2006; Giocondi et al., 2007; Ira and Johnston, 2008; Saslowsky et al., 2002).
Notably, this review will focus on diffusion of molecules that do not preferentially associate with raft or microdomain lipids. The motion of raft-associated protein are expected to be tied strongly to those lipid structures, a very rich topic that is beyond the scope of this review.
3. Observation and quantification of diffusion in complex environments
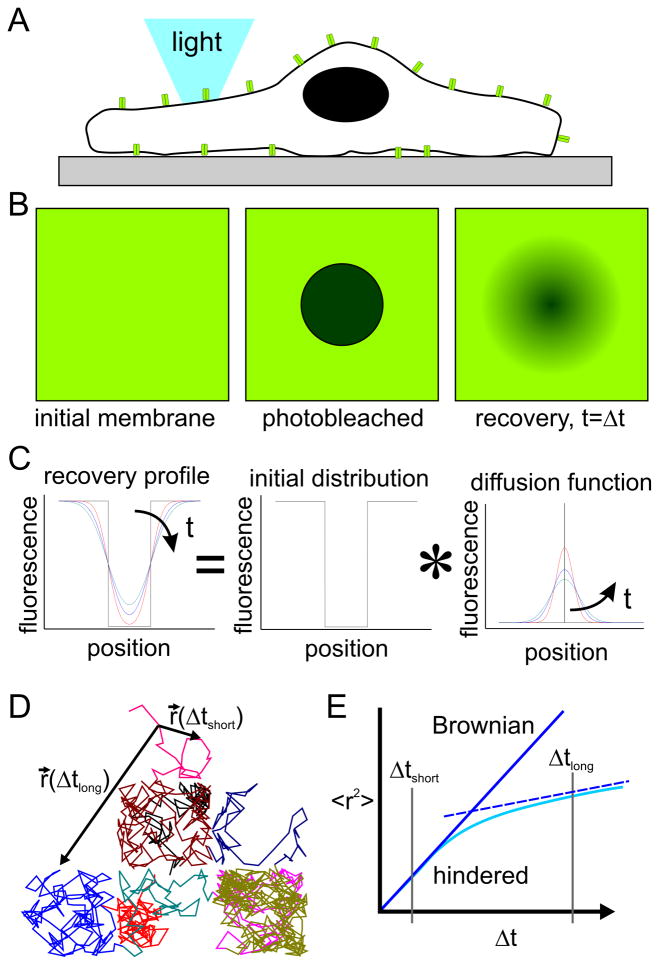
To understand the impact of these local membrane and cytoskeleton structures on molecular diffusion, it is useful to review the experimental methods used to measure this property. This section covers two methods for measuring the diffusion coefficient of a mobile molecule, as this is an important parameter in describing molecular motion and serves as a point of comparison between membranes of living cells and on engineered surfaces. The first technique is Fluorescence Recovery After Photobleaching (FRAP), in which specific membrane components of interest are tagged with a fluorescent marker and imaged using any of a range of microscopy modes, even simple wide-field fluorescence (Figure 2A); based on its relative technical simplicity, wide-field FRAP has a long, established history (Axelrod et al., 1976; Poo and Cone, 1974) and is built into many modern microscopy platforms. In an individual experiment, illustrated in Figure 2B, high-intensity illumination is used to photobleach a localized population of these molecules (middle panel). Images are then recorded over a period of time; owing to the mobility of the target molecule, the photobleached molecules mix locally with their non-bleached counterparts, resulting in a gradual softening and recovery of the photobleached spot. The shape and speed of this recovery are used to derive fundamental diffusive properties of the target molecules, such as diffusion coefficient and mobile fraction (the fraction of molecules that are free to traverse the membrane).
Figure 2.
(A) Live cell imaging allows measurement of molecular diffusion. (B,C) Illustration of the basic FRAP experiment. (D,E) Single particle tracking provides a different, complementary insight into diffusive motion.
An alternative approach (Groves and Boxer, 1995; Kung et al., 2000b; Tsai et al., 2008; Tsay and Jacobson, 1991) used in several laboratories including our own follows a different analysis route and is better-suited for obtaining complex parameters of molecular motion. In this method, illustrated in Figure 2C, the process of diffusion of a population of molecules is treated as the sum of diffusive motion of a large (ideally, infinite) collection of smaller sub-populations, each independently undergoing its own free diffusion. For Brownian motion, the concentration profile of a population that starts as an impulse function is a Gaussian curve of width that increases with time and diffusion coefficient (Figure 2C, right panel). The time evolution of a non-uniform, starting concentration profile can be predicted by convolving the profile with the diffusion function. For 1-dimensional diffusion, this is expressed as: I (x,t) = I (x,t =0))* G(x,t), where G(x,t) = (4πDt)−1/2 exp(x2/(4Dt)), and D is the diffusion coefficient of the molecule being followed. Conversely, the diffusion coefficient can be computed by finding the G(x,t) that provides the best fit between profiles collected at known time intervals. This approach offers several advantages, including the ability to use photobleach profiles of arbitrary, nonuniform shape and the ability to extend this analysis into systems exhibiting anisotropic diffusion (as will be discussed later in this review). From this discussion, it is clear that FRAP-based approaches are tied to the measurement of how a population of molecules spreads over time. Conceptually, this means that FRAP-based methods are sensitive to the area covered by the spreading of an impulse function. Given the conventional optics and cameras that are used in these experiments, FRAP-based measurements are capable of identifying micrometer-scale changes in membrane properties, but essentially average or otherwise combine the effect of smaller-scale phenomena over micrometer-scale areas.
As an alternative method to measuring the diffusive properties of membrane molecules, continual improvement in optical imaging equipment has made it possible to follow individual, tagged molecules over time and with fine precision (Kusumi et al., 2005a; Ritchie and Kusumi, 2003; Sheetz, 1995). This approach, generally termed single particle tracking (SPT), requires a sparse population of labeled molecule in order to be certain that a single entity is being followed, and generates a map of particle position over time as illustrated by the simulation in Figure 2D, left panel. We recognize that there are a number of variations on this basic method, including the use of fluorescent moieties vs. beads to tag proteins (trading off a smaller label that does not crosslink proteins for a more durable tag that may bind to multiple proteins). However, we will wrap these variations into the label of SPT, as the goal of each method, providing a trajectory of protein movement with nanoscale precision over a long duration trajectory, is the same. The simulation of Figure 2D illustrates a single trajectory of a particle diffusing along a surface broken up into a 3 X 3 array of corrals, defined by a series of barriers that are partially permissive to diffusion thus mimicking the presence of membrane-cytoskeleton interactions. To calculate values of diffusion coefficient from this type of data, the displacement of the particle over a specific interval of time, Δt, is calculated and averaged over the total trajectory and multiple particles. For a system exhibiting purely Brownian motion in a 2-dimensional plane, the expected value of the square of this distance is predicted to be linear with the observation time interval, <r(Δt) 2>=4DΔt. Conversely, the diffusion coefficient can be calculated as D=<r(Δt) 2>/(4*Δt). For Brownian diffusion this ratio is expected to be independent of the observation interval, but analysis of systems such as that illustrated in Figure 2D reveals a more complex relation, illustrated in Figure 2E. Interpretation of this behavior will be covered in more detail in the next section.
Finally, Fluorescence Correlation Spectroscopy (FCS) provides a complementary view of molecular diffusion within the membrane. In this method, fluorescent molecules passing through a defined, illuminated volume give rise to a fluorescence signal that fluctuates with time; autocorrelation analysis of this signal, along with an understanding of the dimensions of the illumination volume, provides a measurement of the diffusion coefficient of the labeled molecules. Continued improvements in this method have provided new insights into the organization of membranes, including the effect of large domains and position on a cell surface (Chiantia et al., 2009; Ries and Schwille, 2008; Schwille et al., 1999). However, the dimensions of an illuminated volume are currently on the order of a fraction of the light wavelength, thus on the order of several hundred nanometers. While these methods are a valuable and complementary tool to the bulk FRAP and SPT methods described above, they don’t currently provide the resolution required to examine features that are on the order of tens of nanometers, that of microdomains and the smallest corrals postulated to be present on the cell surface.
4. Impact of nanostructure on molecular diffusion
The application of SPT methods to proteins and lipids revealed a complex pattern of diffusive motion along the surface of living cells(Kusumi et al., 1993), an important step in development of the skeleton- and picket-fence models. In those experiments, membrane molecules exhibited locally random movement associated with Brownian motion, but appeared to be largely confined to patches of the cell surface. This behavior is illustrated in the simulation of Figure 2D, and has a startling impact on the parameter <r(Δt)2>. Specifically, the slope of <r(Δt)2>, which reflects the diffusion coefficient, changed with observation time Δt, as illustrated in Figure 2E. To understand this behavior, two r(Δt) vectors, originating from a specific timestep of this simulation, are shown in Figure 2D. The first of these, r(Δtshort), corresponds to the displacement measured over a small observation time, one in which the particle has not encountered a barrier on the edge of the corral and is essentially undergoing free, Brownian diffusion within that structure. The second, r(Δtlong), represents the displacement associated with a longer time period over which the particle has had time to cross between several barriers and interact with the barriers multiple times. For short observation times associated with diffusion within a corral, the slope of <r(Δt)2> vs. Δt is simply linear, reflecting diffusion in the free membrane. In the presence of barriers, diffusion associated with long observation times, Δtlong, is slowed by interactions with the barrier. As such, the parameter <r(Δtlong)2> will be less than that associated with free diffusion within the corral. In fact, diffusion over a large number of corrals will be represented by a value of r(Δtlong)2 that is linear with time and an apparent diffusion coefficient that is lower than that associated with short-range motion (illustrated by the dotted line in Figure 2E). Formally, motion on the membrane can be described by two different diffusion coefficients: one short-range diffusion coefficient, Dshort, which is associated with motion in a corral and timescales of Δtshort, and a long-range counterpart, Dlong, which described motion between corrals. Moreover, the value of Δt over which the slope changes from Dshort to Dlong gives a measure of the average size of the corrals; this reflects the average time over which diffusing molecules encounter the corral walls.
Importantly, diffusion within the corrals can only be observed as the spatial- and temporal-resolution of SPT becomes finer than that of these patches. This motion only came to light as the resolution of SPT improved to this level; current SPT implementations, which require specialized hardware, revealed that the residence time in corrals from a variety of cells ranges from roughly 1 to 20 ms, in compartments measuring tens to hundreds of nm. This is much finer than that used in traditional FRAP analysis, and not typically available for routine microscopy. The low diffusion coefficients observed on cells result from a mismatch between the observation method and nanoscale structure of the cell membrane/cytoskeleton. A second aspect of this process is that traditional FRAP analysis on cell membranes did appear to be Brownian. In other words, structures below the resolution of the FRAP analysis (below the scale of the impulse spreading function illustrated in Figure 2C) can be manifested into traditional interpretations of molecular diffusion; this hierarchy of diffusional scales can be seen as the analysis of a metamaterial. Conversely, FRAP measurements, with proper caveats on interpretation, can be used to probe the sub-diffraction spatial details of diffusion.
Diffusion of molecules in a fluid matrix populated with immobile microdomains follows overall pattern as that in the presence of the cytoskeletal matrix. That is, diffusion coefficients decrease with increasing observation time, as molecules are able to freely diffuse in spaces between domains, but experience barriers to long range diffusion. The specific relations governing short- vs. long-range diffusion coefficients are more complicated than those predicted for hop diffusion, and are strongly related to the size and shape of microdomains, the overall ratio of fluid to gel phase lipids, and the organization of microdomains with respect to each other (Bussell et al., 1995; Ratto and Longo, 2003; Vaz, 1994). This aspect is discussed in greater detail in the following sections.
5. Capturing cytoskeletal barriers on nanofabricated surfaces
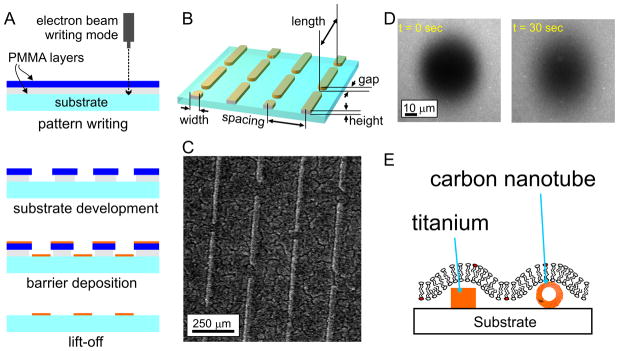
With a framework in place for the design and analysis of nanostructured lipid bilayers, this section focuses on implementation of these systems, beginning with the use of pre-patterned, non-lipid barriers (orange features, Figure 1B) to direct membrane formation and diffusion. Micro-/nano-scale lithography remains one of the predominant ways of patterning these barriers, particularly when high precision and accuracy is needed and when the desired geometry is complex. A typical workflow, in this case a “lift off” process used to pattern a metal on a surface, is shown in Figure 3A. The working substrate (glass, silicon, quartz) is coated with a plastic resist material, which is sensitive to exposure to radiation of specific energy/frequency. A desired pattern is created by controlling the exposure of the resist, either by use of an optical mask (photolithography) or by scanning a beam across the surface (ebeam lithography), inducing local chemical/physical changes that allow the resist to be either removed (as in the case of electron-beam lithography of Figure 3A), or hardened to prevent removal in a subsequent development stage. The substrate is then coated with a barrier material such as a metal, and subsequently developed, lifting off the barrier material from regions previously coated with resist. Numerous variations on this basic process exist; in the strategy of Figure 3A, a dual layer of resist is used, such that development removes more of the underlying layer, providing an undercut that improves the quality of the liftoff process. The barrier material, thickness, and lateral dimensions are all open to design in this process. Given the ability to control exposure over an entire wafer in a single process, photolithography has been the workhorse process for much of this type of fabrication. However, photolithographic processes that are available to the academic community, which typically are highly customized, low run devices, are limited to features with dimensions on the order of several hundred nanometers. To access the smaller dimensions required for mimicking membrane structures, electron beam lithography (Cherniavskaya et al., 2005) is currently required.
Figure 3.
(A) Patterning of barrier materials using lithographic approaches. (B-D) Anisotropic diffusion of lipids on a nanopatterned surface containing chromium barriers. Panel B details the nanoscale layout of barriers used to induce anisotropic, long-range diffusion. Panel C shows a surface patterned with chromium barriers of this geometry. Panel D shows an anisotropic FRAP experiment on these surfaces. The barriers run vertically in this image. (E) Schematic of an alternative bilayer configuration, based on the use of materials that support formation of a continuous supported membrane.
An important demonstration of this process was reported by Mossman and colleagues (Mossman et al., 2005), in which they used electron beam lithography to pattern a gridwork of chromium lines onto a substrate. These surfaces were used to break up a lipid bilayers surface, to which ligands to the T Cell Receptor (TCR) complex and LFA-1 were tethered, into an array of corrals, each measuring on the order of single micrometers on a side. Effectively, this gridwork, as well as other geometries they examined, served as extended, linear barriers to diffusion, mimicking the presence of cytoskeletal interactions. T cells interacting with these surfaces attempted to reorganize the tethered receptors, leading to the formation of complex patterns of ligand engagement; using these surface, Mossman and colleagues demonstrated that maintaining the localization of TCR clusters in the periphery of the cell-substrate interface, an in vitro model of the immune synapse, enhanced downstream signaling within the T cell. The presence of diffusional barriers within this structure modulates cell response. However, these gridwork patterns consisted of continuous, linear barriers. While conceptually similar to a cytoskeletal element, a key feature of the cytoskeleton/membrane interaction is that the barriers occasionally allow lipids and proteins to diffuse across the structure, giving rise to the “hop diffusion” process. This is difficult to embody on a fabricated surface, as the barriers commonly formed by microfabrication normally act by completely disrupting the bilayers continuity.
In an attempt to capture the semi-permeable property proposed for cytoskeleton-membrane interactions, we recently described the use of a pattern of linear barriers which, in contrast to being continuous over extended lengths, were interspersed with periodic gaps, designed to allow a limited amount of communication across the barriers(Tsai et al., 2008). The probability of a lipid or protein to cross these barriers was effectively a function of position rather than time. The dimensions of these patterns are detailed in Figure 3B, with a resultant surface shown in Figure 3C. Using electron beam lithography, we were able to pattern features as narrow as 30 nm in width, with controlled gaps as small as 30 nm in size. The use of chromium as a barrier material, as was done in the Mossman et al. study, provided an effective disruption of the lipid bilayers and control over diffusion. These barriers were aligned into parallel, linear arrays with staggered gaps, taking advantage of the ability of lithography to create patterns of near-arbitrary layout. This layout was chosen to simplify the analysis of diffusion on these surfaces. Specifically, diffusion in a direction parallel to the barriers is unhindered. Long-range diffusion coefficients measured in this direction are thus representative of the short-range diffusion coefficient of membrane molecules. Diffusion across the barriers, by contrast, involves the effect of the barrier on diffusion. The design of this pattern allowed us to use the relatively low-tech approach of FRAP to measure both short-range and long-range diffusion in the presence of barriers. This is manifested as anisotropic diffusion of the FRAP spot, as illustrated in Figure 3D; recovery of fluorescently-labeled lipids following formation of a photobleach spot occurs faster in the direction parallel to the barriers, which is unhindered by the barriers. We adapted our standard FRAP analysis software, which implements the Gaussian spread function analysis illustrated in Figure 2C, to accommodate anisotropic diffusion. With this approach, we demonstrated that the presence of these semi-permeable barriers can be used to controllably modulate the long-range diffusion of membrane molecules, while retaining short-range diffusion in this system. This, to our knowledge, is the first demonstration of controlled, experimental system that captures the phenomenon of hindered diffusion.
One drawback of that study is that the use of interspersed gaps provided control over long-range diffusion, it is unclear how well this approach mimics the actual cell membrane, particularly as the nanoscale barriers completely penetrate the overlying membrane. A system in which the membrane remains continuous, yet diffusion is reduced over an underlying membrane, would be experimentally more attractive. Two recent developments suggest that by proper selection of the barrier material and construction, such systems can be achieved (Figure 3E). When patterning barriers at the scale of micrometers, different materials (such as chromium, titanium, or photoresist) can be used essentially interchangeably for the purpose of limiting bilayer diffusion. However, while the 50-nm wide chromium barriers used in the Tsai et al. study behaved as ideal barriers and completely blocked diffusion, membrane molecules were able to diffuse over the barriers when they were composed of Titanium. The properties of titanium (and most likely other materials, but not chromium) allow a limited amount of interaction with lipid bilayers, stabilizing the structure and allowing membranes to bridge over short expanses; notably, the width of the barriers was similar to that of the lipid vesicles used to form the bilayers. A second material for this purpose was described in 2007 by Zhou and colleagues (Zhou et al., 2007). That study focused on designing hybrid chemical-electrical sensors of membrane proteins based on lipid bilayers deposited over carbon nanotubes (Figure 3E) which served as a gate of a transistor device. That study demonstrated that both leaflets of the membrane are continuous over the nanotube. Moreover, as the radius of the nanotube increased (up to several nm in diameter) membrane proteins and lipids encountered an increasingly strong barrier to diffusion. In comparison to the use of lithographic approaches, carbon nanotubes are highly attractive as they more closely match the dimensions of cytoskeletal elements. However, the ability to accurately place multiple tubes on a single substrate, a strength of nanopatterning, currently limits the use of carbon nanotubes as a well-defined, controllable system. As techniques for improving the accuracy of carbon nanotube placement continue to be developed, new models for capturing the organization of the cell cytoskeleton will undoubtedly emerge.
Such systems, when paired with appropriate cell-based experiments will provide new insight into how nanoscale structure modulates cellular signaling and the underlying molecular processes. As an example of this emerging field, DeMond and colleagues (DeMond et al., 2008) used lithographically patterned lipid bilayers, containing arrays of short, linear barriers, to investigate transport of TCR complexes in the immune synapse. This system allowed observation of the interaction of TCR/pMHC clusters with these barriers in real time, and revealed that the normal motion of these structures from the periphery to the center of the T cell/surface interface can be temporarily impeded by the barriers. Surprisingly, the clusters advanced beyond the barriers by being diverted around the barrier edges, as opposed to jumping over or disassembling on one side of the barrier to reform on the other. This observation provides new insight into the molecular machinery that drives the motion of TCR clusters, and how the nanoscale structure of the immune synapse may alter T cell signaling.
Lastly, we note that the methods presented above all aim at modulating diffusion within an essentially planar substrate, which is not necessarily representative of the cell membrane. Emergins studies use micro- and nano-scale fabrication to capture the topological aspects of membrane, and have demonstrated the ability to direct diffusion through this surface engineering (Goksu et al., 2009; Vinchurkar et al., 2008; Werner et al., 2009).
6. Capturing microdomains and other obstacles on nanofabricated surfaces
While models of cytoskeletal interactions suggest the presence of corrals within which molecular movement is relatively unhindered, the presence of microdomains or other relatively immobilized structures gives rise to a different form of hindered diffusion. Again, this review focuses on the motion of molecules excluded from the microdomains, as the motion of molecules that sequester into these regions is typically small, and under the influence of factors beyond the scope of this discussion. Quantitiative treatment of diffusion in these systems is complex, given that the obstacles are typically assumed to be randomly distributed and of irregular (but generally round) geometry. Of these approaches, Monte Carlo simulations and analytical approaches based on percolation theory and viscosity provide compelling predictions of long-range membrane diffusion (Bussell et al., 1995; Deverall et al., 2005; Saxton, 1994; Saxton, 2001; Vaz, 1994). As a broad generalization, these models predict that long-range diffusion coefficients decrease with increasing fraction of the membrane associated with immobile structures, as well as closer packing and increased irregularity of the domain geometries. However, the rate at which this decrease occurs varies between models.
The development of highly controllable supported lipid bilayer systems provides both a mechanism for testing the validity of these different models as well as the effect of this behavior on signaling by membrane-bound molecules. To this end, systems containing two or more lipid species that, by virtue of different melting points, well-know associations between components (i.e., lipid raft formulations), and/or tethering to an underlying substrate have been the most widely deployed experimentally, given the relative ease of preparation associated with these self-assembling structures and the physiological relevance of the resultant lipid interactions. In particular, Ratto and Longo (Ratto and Longo, 2002) demonstrated through FRAP based experiments that diffusion of a tracer lipid incorporated into the fluid phase of demixed DLPC/DSPC preparations on solid supports decreased with increasing gel phase. Their results demonstrate a particularly sharp decrease in diffusion coefficient as the gel fraction approached 68%, in agreement with predictions from percolation models. However, the use of lipid mixtures to capture the presence of nanoscale domains faces a fundamental challenge; the process of domain formation, particularly on solid supports, is highly dynamic and sensitive to the conditions used to create and process the membrane (Johnston, 2007; McConnell, 1991). Domain formation is governed by thermodynamic processes, tending to form large, circular structures that minimize the interface between different lipid types. For reasons that are not entirely known, the domains formed in artificial membranes are typically larger, up to several micrometers, than those posed to be operating in cell membranes. However, recent reports have demonstrated finer characterization and control over domain formation (Coban et al., 2007; Ratto and Longo, 2002), including the ability to selectively promote micro- vs. nano-scale domains by modulating the surface pressure of a deposited monolayer. Surface topology has also been reported to spatially control the distribution of raft-like lipid domains in supported membranes (Yoon et al., 2006). Against this backdrop of dynamic reordering of lipid structures, the use of nanofabrication to pattern bilayers(DeMond et al., 2008; Mossman et al., 2005; Tsai et al., 2008) may experience a resurgence. For example, the Murakoshi group (Nabika et al., 2005; Nabika et al., 2009) has investigated lipid bilayer function on arrays of metallic nanodots patterned onto surfaces. These studies have focused predominantly on lipid bilayer spreading, demonstrating that the presence and dimensions of these metallic barriers modulates the spreading speed on surfaces. Moreover, different fluorescent dyes, incorporated into the same bilayer and use for visualization, may exhibit different spreading properties, indicating a type of selective sorting of control of lipid motion based on the size of the diffusing entity.
The ability of microdomain structures to hinder diffusion along the plasma membrane is well established; Douglass and Vale (Douglass and Vale, 2005) present an elegant view into how crowded this surface may be. However, the methods discussed above for capturing these effects in substrate supported bilayers have not, to date, been applied significantly to understanding their impact on cell signaling. We envision that experiments examining how cells respond to this nanoscale structure, as has been discussed above for cytoskeletal elements, will be carried out in the near future. Alternatively, we envision that these systems will be used to understand the motion of intracellular, membrane-associated signaling proteins and the role of this in modulating cell response.
7. Conclusions
The role of the nanoscale structure of cell membranes in modulating cell signaling (as the matrix on which many of these reactions occur) is becoming increasing recognized and invoked. As a basic example, short-range diffusion coefficients of signaling molecules modulate the apparent affinity of receptor-ligand pairs, while long-range diffusion affects the cell-level recruitment and dynamics of signaling proteins. The ability to decouple diffusion coefficients at these scales has wide implications in cell signaling(Kusumi et al., 2005a; Kusumi et al., 2005b). Nanopatterned lipid bilayers promise a route towards capturing these aspects of molecular diffusion in a controllable environment, allowing the precise investigation of their impacts on cell signaling. Recent studies have demonstrated the power of this approach, particularly in the study of immune cell function, but more widespread application is currently hampered by the lack of patterning throughput at the appropriate scales. Continued development of high-throughput techniques such as nanoimprint lithography (Guo, 2004; Schvartzman et al., 2008) as well as increased availability of more conventional, optical methods will make these systems available to a wider audience.
References
- Alves ID, Salamon Z, Hruby VJ, Tollin G. Ligand modulation of lateral segregation of a G-protein-coupled receptor into lipid microdomains in sphingomyelin/phosphatidylcholine solid-supported bilayers. Biochemistry. 2005;44:9168–78. doi: 10.1021/bi050207a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophysical Journal. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh MM, Dean C, Pautot S, DeMaria S, Isacoff E, Groves JT. Neuronal Activation by GPI-Linked Neuroligin-1 Displayed in Synthetic Lipid Bilayer Membranes. Langmuir. 2005;21:10693–10698. doi: 10.1021/la051243d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer SG. Molecular transport and organization in supported lipid membranes. Current Opinion in Chemical Biology. 2000;4:704–709. doi: 10.1016/s1367-5931(00)00139-3. [DOI] [PubMed] [Google Scholar]
- Brian AA, McConnell HM. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6159–63. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell SJ, Koch DL, Hammer DA. Effect of hydrodynamic interactions on the diffusion of integral membrane proteins: diffusion in plasma membranes. Biophysical Journal. 1995;68:1836–49. doi: 10.1016/S0006-3495(95)80360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PY, Lawrence MB, Dustin ML, Ferguson LM, Golan DE, Springer TA. Influence of Receptor Lateral Mobility On Adhesion Strengthening Between Membranes Containing Lfa-3 and Cd2. Journal of Cell Biology. 1991;115:245–255. doi: 10.1083/jcb.115.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniavskaya O, Chen CJ, Heller E, Sun E, Provenzano J, Kam L, Hone J, Sheetz MP, Wind SJ. Fabrication and surface chemistry of nanoscale bioarrays designed for the study of cytoskeletal protein binding interactions and their effect on cell motility. Journal of Vacuum Science & Technology B. 2005;23:2972–2978. [Google Scholar]
- Chiantia S, Ries J, Schwille P. Fluorescence correlation spectroscopy in membrane structure elucidation. Biochim Biophys Acta. 2009;1788:225–33. doi: 10.1016/j.bbamem.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Coban O, Popov J, Burger M, Vobornik D, Johnston LJ. Transition from nanodomains to microdomains induced by exposure of lipid monolayers to air. Biophysical Journal. 2007;92:2842–53. doi: 10.1529/biophysj.106.088419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer PS, Groves JT, Kung LA, Boxer SG. Writing and Erasing Barriers to Lateral Mobility into Fluid Phospholipid Bilayers. Langmuir. 1999;15:3893–3896. [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nature Neuroscience. 2003;6:708–16. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophysical Journal. 2008;94:3286–92. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanathan S, Salamon Z, Lindblom G, Grobner G, Tollin G. Effects of sphingomyelin, cholesterol and zinc ions on the binding, insertion and aggregation of the amyloid Abeta(1–40) peptide in solid-supported lipid bilayers. FEBS J. 2006;273:1389–402. doi: 10.1111/j.1742-4658.2006.05162.x. [DOI] [PubMed] [Google Scholar]
- Deverall MA, Gindl E, Sinner EK, Besir H, Ruehe J, Saxton MJ, Naumann CA. Membrane lateral mobility obstructed by polymer-tethered lipids studied at the single molecule level. Biophysical Journal. 2005;88:1875–86. doi: 10.1529/biophysj.104.050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori Y, Bianco-Peled H, Satija SK, Fields GB, McCarthy JB, Tirrell M. Ligand accessibility as means to control cell response to bioactive bilayer membranes. Journal of Biomedical Materials Research. 2000;50:75–81. doi: 10.1002/(sici)1097-4636(200004)50:1<75::aid-jbm11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. Single-Molecule Microscopy Reveals Plasma Membrane Microdomains Created by Protein-Protein Networks that Exclude or Trap Signaling Molecules in T Cells. Cell. 2005;121:937. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML. Membrane domains and the immunological synapse: keeping T cells resting and ready. Journal of Clinical Investigation. 2002;109:155–160. doi: 10.1172/JCI14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin M. THE STATE OF LIPID RAFTS: From Model Membranes to Cells. Annual Review of Biophysics and Biomolecular Structure. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. Journal of Cell Biology. 2002;157:1071–81. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Sumitomo K, Nakashima H, Kashimura Y, Torimitsu K. Supported Lipid Bilayer Self-Spreading on a Nanostructured Silicon Surface. Langmuir. 2007;23:367–371. doi: 10.1021/la062911d. [DOI] [PubMed] [Google Scholar]
- Giocondi MC, Besson F, Dosset P, Milhiet PE, Le Grimellec C. Remodeling of ordered membrane domains by GPI-anchored intestinal alkaline phosphatase. Langmuir. 2007;23:9358–64. doi: 10.1021/la700892z. [DOI] [PubMed] [Google Scholar]
- Goksu EI, Nellis BA, Lin WC, Satcher JH, Groves JT, Risbud SH, Longo ML. Effect of Support Corrugation on Silica Xerogel-Supported Phase-Separated Lipid Bilayers. Langmuir. 2009 doi: 10.1021/la803851b. [DOI] [PubMed] [Google Scholar]
- Gomez-Mouton C, Lacalle RA, Mira E, Jimenez-Baranda S, Barber DF, Carrera AC, Martinez-A C, Manes S. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J Cell Biol. 2004;164:759–768. doi: 10.1083/jcb.200309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Groves JT, Boxer SG. Electric Field-Induced Concentration Gradients in Planar Supported Bilayers. Biophysical Journal. 1995;69:1972–1975. doi: 10.1016/S0006-3495(95)80067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves JT, Boxer SG. Micropattern formation in supported lipid membranes. Accounts of Chemical Research. 2002;35:149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- Groves JT, Dustin ML. Supported planar bilayers in studies on immune cell adhesion and communication. Journal of Immunological Methods. 2003;278:19–32. doi: 10.1016/s0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- Groves JT, Ulman N, Boxer SG. Micropatterning of Fluid Lipid Bilayers on Solid Supports. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- Groves JT, Mahal LK, Bertozzi CR. Control of cell adhesion and growth with micropatterned supported lipid membranes. Langmuir. 2001;17:5129–5133. [Google Scholar]
- Guo LJ. Recent Progress in Nanoimprint Technology and its Applications. Journal of Applied Physics. 2004;D 37:R123–41. [Google Scholar]
- Hafeman DG, von Tscharner V, McConnell HM. Specific antibody-dependent interactions between macrophages and lipid haptens in planar lipid monolayers. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:4552–6. doi: 10.1073/pnas.78.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovis JS, Boxer SG. Patterning and Composition Arrays of Supported Lipid Bilayers by Microcontact Printing. Langmuir. 2001;17:3400–3405. [Google Scholar]
- Iino R, Koyama I, Kusumi A. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophysical Journal. 2001;80:2667–77. doi: 10.1016/S0006-3495(01)76236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, Johnston LJ. Sphingomyelinase generation of ceramide promotes clustering of nanoscale domains in supported bilayer membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2008;1778:185–197. doi: 10.1016/j.bbamem.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Johnston LJ. Nanoscale Imaging of Domains in Supported Lipid Membranes. Langmuir. 2007;23:5886–5895. doi: 10.1021/la070108t. [DOI] [PubMed] [Google Scholar]
- Jury EC, Flores-Borja F, Kabouridis PS. Lipid rafts in T cell signalling and disease. Seminars in Cell & Developmental Biology. 2007;18:608–615. doi: 10.1016/j.semcdb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam L, Boxer SG. Formation of supported lipid bilayer composition arrays by controlled mixing and surface capture. Journal of the American Chemical Society. 2000;122:12901–12902. [Google Scholar]
- Kam L, Boxer SG. Cell adhesion to protein-micropatterned-supported lipid bilayer membranes. Journal of Biomedical Materials Research. 2001;55:487–495. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Kung LA, Groves JT, Ulman N, Boxer SG. Printing via photolithography on micropartitioned fluid lipid membranes. Advanced Materials. 2000a;12:731–734. [Google Scholar]
- Kung LA, Kam L, Hovis JS, Boxer SG. Patterning Hybrid Surfaces of Proteins and Supported Lipid Bilayers. Langmuir. 2000b;16:6773–6776. [Google Scholar]
- Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophysical Journal. 1993;65:2021–40. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Suzuki K, Koyasako K. Mobility and cytoskeletal interactions of cell adhesion receptors. Current Opinion in Cell Biology. 1999;11:582–90. doi: 10.1016/s0955-0674(99)00020-4. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Ike H, Nakada C, Murase K, Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Seminars in Immunology. 2005a;17:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annual Review of Biophysics & Biomolecular Structure. 2005b;34:351–78. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- Lenhert S, Sun P, Wang Y, Fuchs H, Mirkin Chad A. Massively Parallel Dip-Pen Nanolithography of Heterogeneous Supported Phospholipid Multilayer Patterns13. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- Lenz P, Ajo-Franklin CM, Boxer SG. Patterned supported lipid bilayers and monolayers on poly(dimethylsiloxane) Langmuir. 2004;20:11092–9. doi: 10.1021/la048450i. [DOI] [PubMed] [Google Scholar]
- McConnell HM. Structures and Transitions in Lipid Monolayers at the Air-Water Interface. Annual Review of Physical Chemistry. 1991;42:171–195. [Google Scholar]
- Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–3. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- Nabika H, Sasaki A, Takimoto B, Sawai Y, He S, Murakoshi K. Controlling Molecular Diffusion in Self-Spreading Lipid Bilayer Using Periodic Array of Ultra-Small Metallic Architecture on Solid Surface. 2005;127:16786–16787. doi: 10.1021/ja0559597. [DOI] [PubMed] [Google Scholar]
- Nabika H, Iijima N, Takimoto B, Ueno K, Misawa H, Murakoshi K. Segregation of Molecules in Lipid Bilayer Spreading through Metal Nanogates. Analytical Chemistry. 2009;81:699–704. doi: 10.1021/ac802130e. [DOI] [PubMed] [Google Scholar]
- Orth RN, Wu M, Holowka DA, Craighead HG, Baird BA. Mast Cell Activation on Patterned Lipid Bilayers of Subcellular Dimensions. Langmuir. 2003;19:1599–1605. [Google Scholar]
- Pautot S, Lee H, Isacoff EY, Groves JT. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nat Chem Biol. 2005;1:283. doi: 10.1038/nchembio737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez TD, Nelson WJ, Boxer SG, Kam L. E-Cadherin Tethered to Micropatterned Supported Lipid Bilayers as a Model for Cell Adhesion. Langmuir. 2005;21:11963–11968. doi: 10.1021/la052264a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo M-m, Cone RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247:438–441. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- Ratto TV, Longo ML. Obstructed Diffusion in Phase-Separated Supported Lipid Bilayers: A Combined Atomic Force Microscopy and Fluorescence Recovery after Photobleaching Approach. Biophysical Journal. 2002;83:3380–3392. doi: 10.1016/S0006-3495(02)75338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratto TV, Longo ML. Anomalous Subdiffusion in Heterogeneous Lipid Bilayers. Langmuir. 2003;19:1788–1793. [Google Scholar]
- Ries J, Schwille P. New concepts for fluorescence correlation spectroscopy on membranes. Phys Chem Chem Phys. 2008;10:3487–97. doi: 10.1039/b718132a. [DOI] [PubMed] [Google Scholar]
- Ritchie K, Kusumi A. Single-particle tracking image microscopy. Methods Enzymol. 2003;360:618–634. doi: 10.1016/s0076-6879(03)60131-x. [DOI] [PubMed] [Google Scholar]
- Ritchie K, Kusumi A. Role of the membrane skeleton in creation of microdomains. Sub-Cellular Biochemistry. 2004;37:233–45. doi: 10.1007/978-1-4757-5806-1_7. [DOI] [PubMed] [Google Scholar]
- Ritchie K, Iino R, Fujiwara T, Murase K, Kusumi A. The fence and picket structure of the plasma membrane of live cells as revealed by single molecule techniques. Molecular Membrane Biology. 2003;20:13–18. doi: 10.1080/0968768021000055698. [DOI] [PubMed] [Google Scholar]
- Sackmann E. Supported membranes: scientific and practical applications. Science. 1996;271:43–8. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- Saslowsky DE, Lawrence J, Ren X, Brown DA, Henderson RM, Edwardson JM. Placental Alkaline Phosphatase Is Efficiently Targeted to Rafts in Supported Lipid Bilayers. J Biol Chem. 2002;277:26966–26970. doi: 10.1074/jbc.M204669200. [DOI] [PubMed] [Google Scholar]
- Saxton MJ. Anomalous diffusion due to obstacles: a Monte Carlo study. Biophysical Journal. 1994;66:394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Anomalous subdiffusion in fluorescence photobleaching recovery: a Monte Carlo study. Biophysical Journal. 2001;81:2226–40. doi: 10.1016/S0006-3495(01)75870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman M, Mathur A, Kang Y, Jahnes C, Hone J, Wind SJ. Fluorinated diamondlike carbon templates for high resolution nanoimprint lithography. 6. Vol. 26. AVS; 2008. pp. 2394–2398. [Google Scholar]
- Schwille P, Korlach J, Webb WW. Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry. 1999;36:176–82. doi: 10.1002/(sici)1097-0320(19990701)36:3<176::aid-cyto5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Shaw AS. Lipid rafts: now you see them, now you don’t. Nat Immunol. 2006;7:1139–1142. doi: 10.1038/ni1405. [DOI] [PubMed] [Google Scholar]
- Sheetz MP. Cellular plasma membrane domains. Molecular Membrane Biology. 1995;12:89–91. doi: 10.3109/09687689509038501. [DOI] [PubMed] [Google Scholar]
- Shi J, Chen J, Cremer PS. Sub-100 nm patterning of supported bilayers by nanoshaving lithography. J Am Chem Soc. 2008;130:2718–9. doi: 10.1021/ja077730s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–72. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL. The Fluid Mosaic Model of the Structure of Cell Membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- Torres AJ, Vasudevan L, Holowka D, Baird BA. Focal adhesion proteins connect IgE receptors to the cytoskeleton as revealed by micropatterned ligand arrays. Proc Natl Acad Sci U S A. 2008;105:17238–44. doi: 10.1073/pnas.0802138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Sun E, Gao Y, Hone JC, Kam LC. Non-Brownian Diffusion of Membrane Molecules in Nanopatterned Supported Lipid Bilayers. Nano Letters. 2008;8:425–430. doi: 10.1021/nl072304q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay T, Jacobson K. Spatial Fourier analysis of video photobleaching measurements. Principles and optimization. Biophys J. 1991;60:360–368. doi: 10.1016/S0006-3495(91)82061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulman N, Groves JT, Boxer SG. Micropatterning fluid membranes. Advanced Materials. 1997;9:1121–1123. [Google Scholar]
- van Oudenaarden A, Boxer SG. Brownian Ratchets: Molecular Separations in Lipid Bilayers Supported on Pattered Arrays. Science. 1999;285:1046–1048. doi: 10.1126/science.285.5430.1046. [DOI] [PubMed] [Google Scholar]
- Vaz WLC. Diffusion and chemical reactions in phase-separated membranes. Biophysical Chemistry. 1994;50:139–145. doi: 10.1016/0301-4622(94)85026-7. [DOI] [PubMed] [Google Scholar]
- Vinchurkar MS, Bricarello DA, Lagerstedt JO, Buban JP, Stahlberg H, Oda MN, Voss JC, Parikh AN. Bridging Across Length Scales: Multi-Scale Ordering of Supported Lipid Bilayers via Lipoprotein Self-assembly and Surface Patterning. Journal of the American Chemical Society. 2008;130:11164–11169. doi: 10.1021/ja803110v. [DOI] [PubMed] [Google Scholar]
- Werner JH, Montaño GA, Garcia AL, Zurek NA, Akhadov EA, Lopez GP, Shreve AP. Formation and Dynamics of Supported Phospholipid Membranes on a Periodic Nanotextured Substrate. Langmuir. 2009;25:2986–2993. doi: 10.1021/la802249f. [DOI] [PubMed] [Google Scholar]
- Yoon TY, Jeong C, Lee SW, Kim JH, Choi MC, Kim SJ, Kim MW, Lee SD. Topographic control of lipid-raft reconstitution in model membranes. Nat Mater. 2006;5:281–285. doi: 10.1038/nmat1618. [DOI] [PubMed] [Google Scholar]
- Zhou X, Moran-Mirabal JM, Craighead HG, McEuen PL. Supported lipid bilayer-carbon nanotube hybrids. Nat Nano. 2007;2:185–190. doi: 10.1038/nnano.2007.34. [DOI] [PubMed] [Google Scholar]