Abstract
The Mouse Disease Information System (MoDIS) is a data capture system for pathology data from laboratory mice designed to support phenotyping studies. The system integrates the mouse anatomy (MA) and mouse pathology (MPATH) ontologies into a Microsoft Access database facilitating the coding of organ, tissue, and disease process to recognized semantic standards. Grading of disease severity provides scores for all lesions that can then be used for quantitative trait locus (QTL) analyses and haplotype association gene mapping. Direct linkage to the Pathbase on line database provides reference definitions for disease terms and access to photomicrographic images of similar diagnoses in other mutant mice. MoDIS is an open source and freely available program (The Jackson Laboratory, Bar Harbor, ME; http://research.jax.org/faculty/sundberg/index.html).
This provides a valuable tool for setting up a mouse pathology phenotyping program.
Keywords: Access, medical records, pathology training, Pathbase, MPATH, Mouse Anatomy, Mouse Tumor Biology Database, ontology, OBO
Introduction
The relationship between genetic variation and phenotype is at the heart of the model organism approach to the study of human disease. In recent years the mouse has become the model organism of choice for the study of human disease, partly as a consequence of its physiological and genomic similarities, but also because of the developments in mouse genetics, that now provide powerful tools for the manipulation of the mouse genome (Rosenthal and Brown 2007). The last five years have also seen rapid advances in the instrumentation and technology available for detailed phenotyping, and these factors together provide enormous potential for the advancement of our understanding of gene function in health and disease.
The torrent of phenotype data currently being generated from both gene-driven and phenotype-driven experimental approaches to functional genomics will accelerate over the next few years. Together with the accumulation of data now emerging from the large ethyl-nitrosourea (ENU) mutagenesis projects (Auwerx et al. 2004) and the ambitious whole mouse genome mutagenesis projects represented by the International Knockout Mouse Consortium (Collins et al. 2007) there is the risk that this will overwhelm our ability to retain, share and exploit the resulting information. The challenges presented by the collection and analysis of this volume of phenotype data are unprecedented, not only because of the quantity, but also the range and depth of the information. This requires specifically tailored approaches to the capture and representation of radically different types of data, for example craniofacial morphology and blood chemistry (Brown et al. 2006; Gkoutos et al. 2005). The dominant approach to this set of problems is exemplified by that adopted by the EUMORPHIA consortium using EmPRESS (Green et al. 2005), where phenotype is represented by a standard assay, which then defines a set of measurements or descriptions derived from formal description frameworks and ontologies (Mallon et al. 2008). The power of this approach is that it allows for high resolution data to be captured on individual mice for one or more assays and then combined to provide data which can be compared with that from background or control strains. Relating this accumulated variant phenotype data to genetic information is then a matter for new computational tools and resources, many of which are newly available or under development (Chen et al. 2007; Groth et al. 2007; Swertz et al. 2004).
Crucial to the utility of this data is that it is presented in a formalized way to facilitate data sharing, which requires databases using standard data structures and semantics.Currently two databases present raw data for individual mouse strains: the Mouse Phenome Database (Bogue et al. 2007) (http://www.jax.org/phenome) and EuroPhenome Database (http://www.europhenome.org) (Mallon et al. 2008).
Pathology is an essential aspect of phenotyping that requires labour intensive work-up and detailed knowledge of laboratory mouse anatomy, physiology and genetics to be fully effective. There are two major problems with recording this aspect of phenotype: standardization of pathology data, and the availability of pathology expertise to derive and interpret that data.
The latter is a well recognized problem; “The importance of pathology in mouse phenotyping cannot be underestimated. However, the laborious nature of pathology analysis and the dependence on a small cadre of experts continues to represent a significant stumbling block to unraveling the mouse phenome.” (Brown et al. 2006).Such expertise is not easy to find, (Barthold et al. 2007; Cardiff 2007; Valli et al. 2007) and the perils of “DIY pathology” are well illustrated in the paper from Cardiff et al. (Cardiff 2007). The gold standard is represented in the systematic pathology segment of the German Mouse Clinics phenotyping process where there is standardized morphological phenotyping of potential mouse models (Mossbrugger et al. 2007).
The depth of data captured, data structure and description semantics are not yet fully standardized and require not only community agreement on the minimal information needed to record a phenotype but also data capture tools that allow for rapid and accurate recording of data in a form in which it may be uploaded to central databases (The_Mouse_Phenotype_Database_Integration_Consortium 2007). The terminology for lesions in widespread use is a mixture of veterinary and human diagnostic names that do not always correspond, although recent recommendations by the Mouse Models of Human Cancer Consortium (MMHCC) have gone some way towards standardization of nomenclature for neoplastic diseases (Cardiff et al. 2000; Kogan et al. 2002; Nikitin et al. 2004a; Nikitin et al. 2004b; Shappell et al. 2004). Unfortunately, uptake of these recommendations has been slow amongst pathologists working in different environments and traditions. Much needed resources are being developed to provide standard reference vocabularies for mouse anatomy at the gross level (mouse anatomy ontology) and disease processes (mouse pathology ontology) useful at both the gross and microscopiclevels. Integrating these with annotated and labeled line drawings, gross photographs or photomicrographs and literature references provides tools that can be rapidly used for reference and for training the next generation of mouse specialist pathologists. These are adjuncts to, not replacements for traditional training and mentorship approaches (Barthold et al. 2007; Sundberg et al. 2004). Unfortunately these types of resources are spread all over the world at many different institutions and, if online, are often unlinked.
Diagnostic laboratories face record keeping problems that can be overwhelming. Using traditional approaches to diagnostic case record keeping linked to flat files of anatomy (Hayamizu et al. 2005) and pathology ontologies (Schofield et al. 2005) provides one approach to rapidly coding case materials, standardizing the diagnoses and retrieving all case materials. Development of a disease diagnostic field with assessment of the disease severity provides a definitive answer that is also semi-quantitative. These are necessary for quantitative trait locus analysis (QTL mapping) as well as for defining the pathogenesis of a novel disease in a mouse model system.
Web-based systems can now integrate all of these activities to allow a pathologist to review slides from a case, rapidly enter the diagnosis, which is automatically accurately coded, that can be exported in a defined format, e.g. XML, to other databases. More importantly, clicking onto the links to the appropriate web site provides access to photomicrographs of representative cases in other genetically engineered mice or inbred strains to provide the pathologist with reference information, descriptions and original papers on the disease process. This approach provides tools for verification of a diagnosis, training for those not familiar with laboratory mice and a means to improve the quality of the service to the molecular biologist submitting the samples. Furthermore, because panels of veterinary and physician pathologists volunteer to maintain the quality of these databases, it is possible to access expertise not otherwise available, to accurately define diseases and make appropriate comparisons with human diseases.
We describe here a system, MoDIS (Mouse Disease Information System), that integrates all of these tools using a Microsoft Access based database, which is open source and freely available (The Jackson Laboratory, Bar Harbor, ME;http://research.jax.org/faculty/sundberg/index.html). This provides a valuable tool for setting up a mouse pathology phenotyping program.
Materials and Methods
Case materials
Routine disease surveillance cases received by The Jackson Laboratory’s Laboratory Animal Health Disease Surveillance Program from 1987-2000 were used to develop the original medical records database (Sundberg and Sundberg 1990, 2000). This used a traditional free text diagnostic field. This was converted to use the MPATH pathology ontology in 2006 to standardize detailed histopathological phenotyping methods for defining and describing diseases. This conversion was done to expedite large scale phenotyping and haplotype mapping of chronic diseases in the most important inbred strains of mice used today in biomedical research. Complete systematic necropsies (Seymour et al. 2004) were performed on 15 males and 15 females of each of 31 inbred strains designated in the Mouse Phenome Database (http://phenome.jax.org) at 12 and 20 months of age. All tissues were screened by one pathologist (JPS) to standardize the first screen interpretation (Yuan et al. submitted). Additional protocols from the major international research consortiums doing phenotyping can be accessed through a common website (www.interphenome.org) (Consortium et al. 2007).
Databases
MoDIS (mouse disease information system) is implemented on a Microsoft Access database (Microsoft Corp., Redman, WA) platform and is the descendent of earlier versions of our pathology medical records database, which were converted from dBASE III Plus (Ashton-Tate, Torrance, CA) (Sundberg and Sundberg 1990), to FoxPro 2.6 for Windows (Microsoft) (Sundberg and Sundberg 2000), to its current form. Accumulated practical experience of using this database for pathology data capture in a large institutional setting has greatly improved its development. MoDIS stores information from individual mice and cross references to any images captured in an image database, for example Extensis Portfolio (http://www.extensis.com, Portland, Oregon). The database has the facility for recording information associated with husbandry, pedigree, strain, assays carried out, location of material, as well as recording of pathological diagnosis.
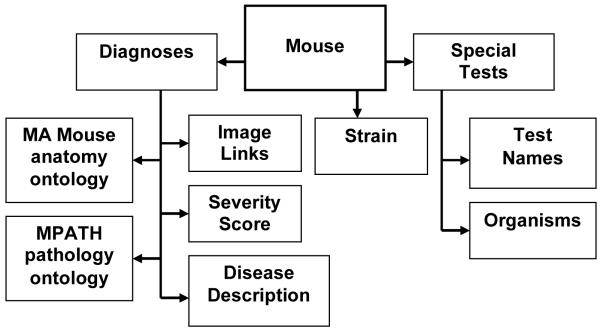
The primary entity in the database is a necropsy case consisting of one mouse. A mouse can have many diagnoses and several special tests, such as immunohistochemistry or microbiology, associated with the record. Each mouse case can have many types of materials associated with it (histology, photographs, frozen tissue, etc.) (Fig. 1).
Figure 1.
Diagram of work and data flow. Investigators/collaborators submit a mouse. The animal is necropsied at which point samples are collected for histopathology and special tests. When all results are received these are added to the case file including diagnoses for all lesions and final diagnoses. The finalized report can then be emailed to the submitter or printed out and signed to create a legal diagnostic medical report.
The diagnostic information includes a “Disease Description” field, which uses an extendable controlled vocabulary containing high level diagnostic terms input by the pathologist, in which summative pathological diagnosis may be recorded. This allows for locally preferred terminology to be recorded and defined. The recording of standardized pathological terminology uses a pair of terms from the MPATH and MA ontologies for each lesion.
It is possible to record several “Disease Description” and several MA/MPATH pairs of terms for each mouse. An additional field grades severity of lesions building on the commonly used adjectives no lesions (0), mild (1), moderate (2), severe (3) and extreme (4). This provides an estimate of the variation of severity within a group of mice of the same strain and genotype or treatment group which provides a semi-quantitative set of parameters for comparison which can be used in quantitative trait locus analyses.
Ontologies and controlled vocabularies
MoDIS uses the MPATH (Schofield et al. 2005) and MA ontologies (Hayamizu et al. 2005) downloaded from the OBO foundry web site (http://www.obofoundry.org/) as flat files. Terms from the two ontologies are used to specify the intersection of anatomy and pathology for each lesion. Strains and genotypes are recorded in compliance with standard nomenclature (http://www.informatics.jax.org/mgihome/nomen/gene.shtml). Special tests, organisms found in testing, submitters and housing locations may also be recorded in free text or locally controlled vocabulary (CV). The facility is available to build a local controlled vocabulary (CV) for high level summative disease diagnoses.
Results
MoDIS is currently designed for local installation but with the facility to output files to other databases and programmes. The local MoDIS database at The Jackson Laboratory now contains nearly 40,000 records and forms the core resource for other databases and resources at The Jackson Laboratory, such as the Mouse Tumor Biology Database (Begley et al. 2007) and elsewhere. These clinical records can be quickly searched for individual cases or case series by disease, organ, strain, etc. to output to comma-separated values (CSV) or a Microsoft Excel file that can be further sorted and analyzed. All case materials are linked by a common accession number code for each animal in other databases of the laboratory. Individual or groups of images representing example sections have been placed online with annotations. Summaries of studies are also being put online (Mouse Phenome Database (MPD), http://phenome.jax.org; (Bogue et al. 2007; Consortium et al. 2007) Mouse Tumor Biology Database (MTB),http://tumor.informatics.jax.org; (Krupke et al. 2008) and Pathbase, http://www.Pathbase.net (Schofield et al. 2004a; Schofield et al. 2004b)) as they are completed and curated. Complete tables of spontaneous background diseases, although currently only published for strain disease surveillance (Mikaelian et al. 2004; Sundberg and Ichiki 2005a), will soon be online with links to the specific photomicrographs of lesions from the strain in question.
Pathologic diagnosis recording is complex and depends to a great extent on the tradition in which the diagnostician was trained. This problem has generated much discussion over the semantic standardization of pathologic diagnoses over recent years. From experience we believe the solution is to use standard defined ontologies for formal recording, but to leave the clinician with the opportunity to make local annotations in other formalisms. This means that eventual export of key data to central public databases such as Europhenome (Mallon et al. 2008) can be semantically and syntactically compatible with accepted standards, and can be achieved automatically. The MPATH ontology is continuously under review by a panel of pathologists at annual meetings of the Pathbase European Mouse Pathology Consortium. The pathologists (both veterinarians and physicians) review new terms, edit and arrive at a consensus on the term and its definition. Similarly MA is under constant revision and refinement and in this way there is an ongoing system of checks and balances on the terminology used as well as a formal means to upgrade the system especially to expand to a higher level of sophistication and utility.
MoDIS as a training resource
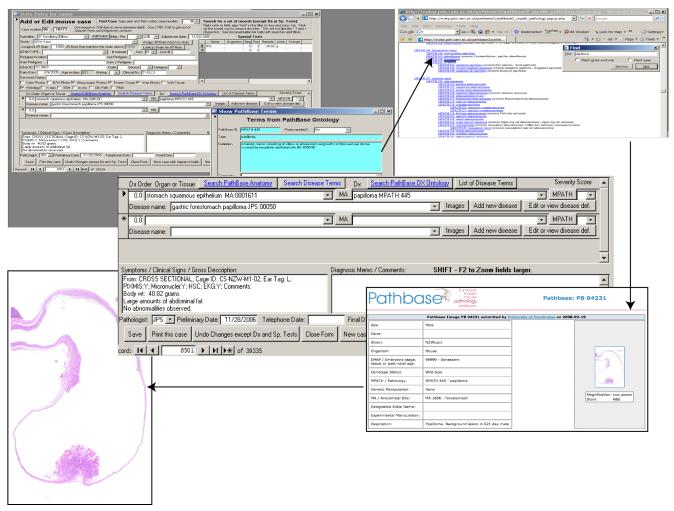
When online and if the ontology flat files used for coding are current and linked to Pathbase (http://www.pathbase.net), it is possible to move from the ontology term to retrieve a formal definition with literature or web references and, where appropriate, a series of images of similar lesions with their annotation posted by pathologists working with mice worldwide on Pathbase becomes available to the viewer (Fig. 2).
Figure 2.
Steps to move from a diagnosis to definitions, references and images. Once a diagnosis is arrived at and MoDIS is linked to Pathbase one can move to the MPATH definition and from there to images of lesions given this diagnosis. In this way one can quickly verify their tentative diagnosis using virtual mentoring.
Discussion
Standardization and integration
The capture of detailed primary data from phenotyping experiments, with appropriate structure, is a sine qua non for high throughput large-scale studies. As more structured descriptions of phenotypes are developed this will allow data to be processed and interpreted in a consistent manner and facilitate the development of new computational software. The capture of primary data from individual mice allows for its re-analysis and re-use in the light of new hypotheses and new information, and maximizes the added value of the studies. Development of tools which address this is a recognized need. For example, the MPHASYS system (Calder et al. 2007) is designed to capture and integrate phenotype information, though not in this case in a format using recognized ontologies and structure. This example emphasizes the importance of semantic and syntactic standardization for the general application of such data capture tools, and the centrality of adherence to community consensus standards, which we have begun to implement in MoDIS. Removal of multiple manual curation steps in data entry reduces the risk of error and the cost of large database curation, which can be substantial. Importantly, then, data capture tools need to be intuitively and readily usable by the “phenotyper”, in this case the pathologist, and consideration of the user’s expertise is an important aspect of their design.
Training and referencing
To meet the need of biomedical researchers world-wide for pathologists to interpret lesions that develop in inbred, genetically engineered or experimentally manipulated mice, small pathology programs are being set up in medical centers and universities world-wide. While ideally these would be peer-driven, mentor-based programs, (Sundberg et al. 2004) in fact, they are usually isolated junior level pathologists or clinicians with marginal training in histopathology. While many books are now available (Bannasch and Goessner 1994; Bannasch and Gossner 1994; Frith and Ward 1988; Kaufman et al. in press; Kaufman 1992; Kaufman and Bard 1999; Maronpot et al. 1999; Mohr 2001; Mohr et al. 1996; Smith et al. 2002; Sundberg 1994; Sundberg and Boggess 2000; Sundberg and Ichiki 2005b; Ward et al. 2000) these are only a partial substitute for a team of pathologists with whom one can consult. Formal national training programs (Barthold et al. 2007; Sundberg et al. 2007), mentored by established senior pathologists or other organ or disease experts can provide support for these junior pathologists. The high volume of case materials many are faced with combined with the fact that senior pathologists with expertise in rodent pathology are not readily available at many institutions results in less than optimal working conditions. We provide here a freely available system that addresses the problem of record keeping and case retrieval, continuing education and confirmation (second opinions) on the cases. We provide a relatively simple system that can be easily integrated into larger databases or the data downloaded into formats for use in larger, more comprehensive databases. This database and full documentation on how to use it are available free online (http://research.jax.org/faculty/sundberg/index.html).
Investigators usually want a summative diagnosis rather than a list of lesions. Pathologists understand that lesions can be independent of each other or linked, and that they can be specific to the strain used, husbandry conditions or be related to the experimental manipulation being done on the animals. For those mapping complex genetic traits, keeping lesions separated by organ system and quantified is critical for these types of analyses. By adding a field for disease severity, accomplished by simply converting adjectives commonly used by pathologists to describe lesions (mild, moderate, severe and extreme) to a graded scale (1, 2, 3 and 4, respectively), one now has a simple semi-quantitative scale for all lesions. If this is done consistently one can immediately run a quantitative trait analysis. We have used this successfully for many years for mapping inflammatory bowel disease severity and resistant genes in the mouse (Bleich et al. 2004; Bristol et al. 2000; Farmer et al. 2001; Mahler et al. 1998; Mahler et al. 2002; Mahler et al. 1999).
Future developments
Whilst the use of small locally instantiated MoDIS databases can be useful for a wide range of users, migration to a server based relational database management system (RDBMS) such as MySQL opens up a range of further possibilities with regard to sophistication of structure, access and interoperability. Live linkage of ontologies to the Ontology Lookup Service (OLS) (http://www.ebi.ac.uk/ontology-lookup/) will maintain currency with the standard ontologies and remove any requirement for manual updating. Standards for reporting of environmental and husbandry conditions as well as other assays are now being developed and inclusion of compatibility with these standards will enhance interoperability with larger databases and computational tools to generate a data capture, coding and uploading resource of wide utility.
Acknowledgements
This work was supported by grants from the Ellison Medical Foundation, the National Institutes of Health (CA34196, CA089713, and AG025707) and Commission of the European Community, Framework Programme 6 contract no. LSH-2005-1.1.3-2.
References
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch B, Goessner W. Pathology of neoplasia and preneoplasia in rodents. vol 2. Schattauer; Stuttgart: 1994. [Google Scholar]
- Bannasch P, Gossner W. Pathology of neoplasia and preneoplasia in rodents. vol 1. Schattauer; Stuttgart: 1994. [Google Scholar]
- Barthold SW, Borowsky AD, Brayton C, Bronson R, Cardiff RD, et al. From whence will they come? - A perspective on the acute shortage of pathologists in biomedical research. J Vet Diagn Invest. 2007;19:455–456. doi: 10.1177/104063870701900425. [DOI] [PubMed] [Google Scholar]
- Begley DA, Krupke DM, Vincent MJ, Sundberg JP, Bult CJ, et al. Mouse Tumor Biology Database (MTB): status update and future directions. Nucleic Acids Res. 2007;35:D638–642. doi: 10.1093/nar/gkl983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich A, Mahler M, Most C, Leiter EH, Lieber-Tenorio E, et al. Refined histopathologic scoring systems improves power to detect colitis QTL in mice. Mammalian Genome. 2004;15:865–871. doi: 10.1007/s00335-004-2392-2. [DOI] [PubMed] [Google Scholar]
- Bogue MA, Grubb SC, Maddatu TP, Bult CJ. Mouse Phenome Database (MPD) Nucleic Acids Res. 2007;35:D643–649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristol IJ, Farmer MA, Cong Y, Zheng XX, Strom TB, et al. Heritable susceptibility for colitis in mice induced by IL-10 deficiency. Inflamm Bowel Dis. 2000;6:290–302. doi: 10.1002/ibd.3780060407. [DOI] [PubMed] [Google Scholar]
- Brown SD, Hancock JM, Gates H. Understanding mammalian genetic systems: the challenge of phenotyping in the mouse. PLos Genetics. 2006;2:e118. doi: 10.1371/journal.pgen.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder RB, Beems RB, vanSteeg H, Mian IS, Lohman PH, et al. MPHASYS: a mouse phenotype analysis system. BMC Bioinformatics. 2007;8:183. doi: 10.1186/1471-2105-8-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD. Pathologists needed to cope with mutant mice. Nature. 2007;447:528–528. doi: 10.1038/447528c. [DOI] [PubMed] [Google Scholar]
- Cardiff RD, Moghanaki D, Jensen RA. Genetically engineered mouse models of mammary intraepithelial neoplasia. J Mammary Gland Biol Neoplasia. 2000;5:421–437. doi: 10.1023/a:1009534129331. [DOI] [PubMed] [Google Scholar]
- Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Consortium MPDI. Hancock JM, Adams NC, Aidinis V, Blake A, et al. Mouse Phenotype Database Integration Consortium: integration [corrected] of mouse phenome data resources. Mamm Genome. 2007;18:157–163. doi: 10.1007/s00335-007-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer MA, Sundberg JP, Bristol IJ, Churchill GA, Li R, et al. A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10-deficient mice. Proc Natl Acad Sci USA. 2001;98:13820–13825. doi: 10.1073/pnas.241258698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith CH, Ward JM. Color atlas of neoplastic and non-neoplastic lesions in aging mice. Elsevier; Amsterdam: 1988. [Google Scholar]
- Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EC, Gkoutos GV, Lad HV, Blake A, Weekes J, et al. EMPReSS: European mouse phenotyping resource for standardized screens. Bioinformatics. 2005;21:2930–2931. doi: 10.1093/bioinformatics/bti441. [DOI] [PubMed] [Google Scholar]
- Groth P, Pavlova N, Kalev I, Tonov S, Georgiev G, et al. PhenomicDB: a new cross-species genotype/phenotype resource. Nucleic Acids Res. 2007;35:D696–699. doi: 10.1093/nar/gkl662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The adult mouse anatomical dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman M, Nikitin A, Sundberg JP. Comparative embryology of the endocrine system. CRC Press; Boca Raton: (in press) [Google Scholar]
- Kaufman MH. The atlas of mouse development. Academic Press; London: 1992. [Google Scholar]
- Kaufman MH, Bard JBL. The anatomical basis of mouse development. Academic Press; San Diego: 1999. [Google Scholar]
- Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- Krupke D, Begley D, Sundberg J, Bult C, Eppig J. The mouse tumor biology database. Nature Rev Cancer. 2008 doi: 10.1038/nrc2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler M, Bristol I, Leiter E, Workman A, Birkenmeier E, et al. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am J Physiol. 1998;274:G544–G551. doi: 10.1152/ajpgi.1998.274.3.G544. [DOI] [PubMed] [Google Scholar]
- Mahler M, Most C, Schmidtke S, Sundberg JP, Li R, et al. Genetics of colitis susceptibility in IL-10-deficient mice: backcross versus F2 results contrasted by principal component analysis. Genomics. 2002;80:274–282. doi: 10.1006/geno.2002.6840. [DOI] [PubMed] [Google Scholar]
- Mahler M, Sundberg JP, Birkenmeier EH, Bristol JJ, Elson CO, et al. Genetic analysis of susceptibility to dextran sulfate sodium-induced colitis in mice. Genomics. 1999;55:147–156. doi: 10.1006/geno.1998.5636. [DOI] [PubMed] [Google Scholar]
- Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic acids research. 2008;36:D715–718. doi: 10.1093/nar/gkm728. (2008) Mallon, A.M., Blake, A. and Hancock, J.M. Nucleic Acids Res. 36, D715-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maronpot RR, Boorman GA, Gaul BW. Pathology of the mouse. Reference and atlas. Cache River Press; Vienna, IL: 1999. [Google Scholar]
- Mikaelian I, Ichiki T, Ward JM, Sundberg JP. Diversity of spontaneous neoplasms in commonly used inbred strains and stocks of laboratory mice. In: Hedrich HJ, editor. The Laboratory Mouse. Academic Press; London: 2004. pp. 345–354. [Google Scholar]
- Mohr U. International classification of rodent tumors: the mouse. Springer Verlag; Berlin: 2001. [Google Scholar]
- Mohr U, Dungworth DL, Capen CC, Carlton WW, Sundberg JP, et al. Pathobiology of the aging mouse. ILSI Press; Washington, D.C.: 1996. [Google Scholar]
- Mossbrugger I, Hoelzlwimmer G, Calzada-Wack J, Quintanilla-Martinez L. Standardized morphological phenotyping of mouse models of human diseases within the German Mouse Clinic. Verh Dtsch Ges Pathol. 2007;91:98–103. [PubMed] [Google Scholar]
- Nikitin A, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004a;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- Nikitin AY, Connolly DC, Hamilton TC. Pathology of ovarian neoplasms in genetically modified mice. Comp Med. 2004b;54:26–28. [PubMed] [Google Scholar]
- Rosenthal N, Brown S. The mouse ascending: perspectives for human-disease models. Nature cell biol. 2007;9:993–999. doi: 10.1038/ncb437. [DOI] [PubMed] [Google Scholar]
- Schofield PN, Bard JBL, Boniver J, Covelli V, Delvenne P, et al. Pathbase: a new reference resource and database for laboratory mouse pathology. Radiation Protection Dosimetry. 2004a;112:525–528. doi: 10.1093/rpd/nch101. [DOI] [PubMed] [Google Scholar]
- Schofield PN, Bard JBL, Booth C, Boniver J, Covelli V, et al. Pathbase: a database of mutant mouse pathology. Nucleic Acids Res. 2004b;32:D512–515. doi: 10.1093/nar/gkh124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PN, Bard JBL, Rozell B, Sundberg JP. Computational pathology: challenges in the informatics of phenotype description in mutant mice. In: Sundberg JP, Ichiki T, editors. Handbook on genetically engineered mice. CRC Press; Boca Raton: 2005. pp. 61–81. [Google Scholar]
- Seymour R, Ichiki T, Mikaelian I, Boggess D, Silva KA, et al. Necropsy methods. In: Hedrich HJ, editor. Laboratory mouse. Academic Press; London: 2004. pp. 495–516. [Google Scholar]
- Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- Smith RS, John SWM, Nashina PM, Sundberg JP. Systematic evaluation of the mouse eye. anatomy, pathology, and biomethods. CRC Press; Boca Raton, FL: 2002. [Google Scholar]
- Sundberg BA, Sundberg JP. A database system for small diagnostic laboratories. Lab. Anim. 1990;19:55–58. [Google Scholar]
- Sundberg BA, Sundberg JP. Medical record keeping for project analysis. In: Sundberg JP, Boggess D, editors. Systematic approach to evaluation of mouse mutations. CRC Press; Boca Raton: 2000. pp. 47–55. [Google Scholar]
- Sundberg JP. Animal Models and Biomedical Tools. CRC Press, Inc; Boca Raton, Florida: 1994. Handbook of Mouse Mutations with Skin and Hair Abnormalities. [Google Scholar]
- Sundberg JP, Boggess D. Systematic characterization of mouse mutations. CRC Press; Boca Raton: 2000. [Google Scholar]
- Sundberg JP, Hackman RC, HogenEsch H, Nikitin AY, Ward JM. Training Mouse Pathologists: Five years of Pathology of Mouse Models of Human Disease Workshops. Toxicol Pathol. 2007;35:447–448. doi: 10.1080/01926230701302566. [DOI] [PubMed] [Google Scholar]
- Sundberg JP, Haschek WM, Hackman RC, HogenEsch H. Developing a comprehensive mouse pathology program. Comp Med. 2004;54:615–619. [PubMed] [Google Scholar]
- Sundberg JP, Ichiki T. Common diseases found in inbred strains of laboratory mice. In: Sundberg JP, Ichiki T, editors. Handbook on genetically engineered mice. CRC Press; Boca Raton: 2005a. pp. 223–229. [Google Scholar]
- Sundberg JP, Ichiki T. Genetically engineered mice handbook. CRC Press; Boca Raton: 2005b. [Google Scholar]
- Swertz MA, DeBrock EO, VanHijum SA, DeJong A, Buist G, et al. Molecular Genetics Information System (MOLGENIS): alternatives in developing local experimental genomics databases. Bioinformatics. 2004;20:2075–2083. doi: 10.1093/bioinformatics/bth206. [DOI] [PubMed] [Google Scholar]
- The_Mouse_Phenotype_Database_Integration_Consortium Mouse Phenotype Database Integration Consortium: integration of mouse phenome data resources. Mamm Genome. 2007;18:157–163. doi: 10.1007/s00335-007-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli T, Barthold SW, Ward JE, Brayton C, Nikitin A, et al. Over 60% of NIH extramural funding involves animal-related researchh. Vet Pathol. 2007;44:962–963. doi: 10.1354/vp.44-6-962. [DOI] [PubMed] [Google Scholar]
- Ward J, Mahler J, Maronpot R, Sundberg JP. Pathology of genetically engineered mice. Iowa State University Press; Ames: 2000. [Google Scholar]
- Yuan R, Tsaih S-T, Petkova S, deEvsikova CM, Xing S, et al. Aging in 31 inbred strains of mice: study design and interim report on median lifespans. Aging Cell. doi: 10.1111/j.1474-9726.2009.00478.x. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]