Abstract
A longitudinal study was conducted to determine the epidemiology of Cryptosporidium in 1,636 children in Nigeria. Oocyst prevalence ranged from 15.6% to 19.6% over one year. Cryptosporidium hominis (34), C. parvum (25), C. parvum/C. hominis (4), C. meleagridis (5), Cryptosporidium rabbit genotype (5), Cryptosporidium cervine genotype (3), and C. canis (1) were identified by polymerase chain reaction–restriction fragment length polymorphism analysis. Glycoprotein 60 subgenotyping showed that 28 amplifiable C. hominis isolates consisted of 12 subtypes that belonged to 5 subtype families (Ia, Ib, Id, Ie, and 1 novel subtype family, Ih) and 23 amplifiable C. parvum isolates consisted of 6 subtypes that belonged to 4 subtype families (IIa, IIc, Iii, and IIm). Three C. meleagridis isolates sub-genotyped by sequence analysis of the small subunit ribosomal RNA gene fragment were type 1. This study is the first one to genetically characterize Cryptosporidium species and subtypes in Nigeria and highlights the presence of a high Cryptosporidium diversity in this pediatric population.
Introduction
The apicomplexan protozoan parasite Cryptosporidium is the causative agent of the diarrheal disease cryptosporidiosis and infection can lead to severe dehydration and death in immunocompromised patients.1 Children are a high risk group for infection, those less than five years of age are most susceptible,2 and cryptosporidiosis may have long-term negative effects on their growth and cognitive development.3,4 Studies indicate that prevalence varies geographically, with increased prevalence in developing countries, and temporally, with higher rates reported during the rainy season in many tropical countries.4–7
A range of Cryptosporidium species, genotypes, and subtypes infect humans, and each may have different sources of infection, transmission routes, and pathogenicity.8–10 Thus, identifying the species present in a population is crucial for identifying risk factors for transmission and implementing control programs to limit exposure to infectious oocysts.
Currently, there are 20 described species of Cryptosporidium of which 8 species (C. hominis, C. parvum, C. meleagridis, C. felis, C. canis, C. suis, C. muris, and C. andersoni) and 6 unnamed species (Cryptosporidium cervine, monkey, skunk, rabbit, horse, and chipmunk genotypes) infect immunocompetent and immunocompromised humans.11–13 Cryptosporidium hominis and C. parvum are the most frequently detected, and C. hominis infections are more common in developing countries.5,7,14–18 Both species have different host ranges. Although C. hominis is confined mostly to humans, C. parvum infects humans and ruminants. However, subgenotyping C. hominis and C. parvum into subtype families by sequencing a locus on the 60-kD glycoprotein (GP60) gene19 has provided a clearer understanding of the transmission dynamics and host specificity of these species. It appears that not all human C. parvum infections are a result of zoonotic transmission as some C. parvum subtypes seem to circulate only in humans.20,21 Although cryptosporidiosis is prevalent in developing countries, genetic characterization of species is lacking, especially in Africa, where only four subgenotyping studies have been conducted in Uganda, Malawi, Kenya, and South Africa.7,15,22,23
Cryptosporidium meleagridis, although not found as commonly as C. hominis and C. parvum, is the third most common infection in humans.4,5,14,18,24,25 Two subtypes of C. meleagridis have been identified at the small subunit (SSU) ribosomal RNA (rRNA) gene locus, and six 6 subtypes have been identified at the GP60 gene locus,26 which indicate possible heterogeneity in host range, and therefore routes of transmission, for C. meleagridis.
The present study determines the prevalence, temporal variability, and molecular epidemiology of Cryptosporidium in a pediatric population in Osun State, Nigeria.
Materials and Methods
Study population and sample collection.
This study was carried out in four semi-urban villages in Ile-Ife, Osun State, Nigeria. These villages (Ipetumodu, Akinlalu, Edunabon, and Moro) are located within 15 km of Ile-Ife town.27 The study was part of a parallel project set up in May 2006 that investigated the interactions between Ascaris and malaria infections.27 A total of 2,332 children (age range = 6 months–6 years) were enrolled into the study during May and September 2006 after informed consent was obtained. Temporary clinics were set up in the center of each village, and mothers from the surrounding area were asked to bring their children for screening of malaria and intestinal worms. Once enrolled, each child was assigned an identification number. To assess the temporal variability of infection, clinics were open at four time points over a one-year period: September 2006, February 2007, May 2007, and August 2007. These time points included the rainy (May–October) and dry (November–April) seasons. Mothers were supplied with 50-mL plastic containers in which to collect their children's feces, and samples were returned and refrigerated at 4°C.
Of the 2,332 children enrolled, 1,636 children submitted fecal samples on at least one of the four time points. A number of children were lost to follow-up; 349 children submitted samples at all four time points. Ethical clearance was provided by the Ethics and Research Committee, Obafemi Awolowo University Teaching Hospital Complex, Ile-Ife, Nigeria.
Stool analysis.
Stool consistency was evaluated by visual examination and classified as formed, unformed, or liquid. A pea-sized amount (200 μL if liquid) of feces from each child was concentrated by using a modified formol-ether technique.28 Concentrates were air-dried onto glass microscope slides (one slide per child), stained with auramine-phenol,29 and examined for the presence of oocysts by using a fluorescent microscope (blue filter block; excitation 490 nm, emission 510 nm). Each slide was scanned at 200× magnification and oocysts were confirmed under 400× magnification. Positive and negative samples were recorded along with the intensity of infection. Intensity was determined as follows: 0 = no oocysts detected in the sample; 1+ = 1–10 oocysts per field of view; 2+ = 10–50 oocysts per field of view, and 3+ = > 50 oocysts per field of view.
DNA extraction.
Three criteria were used for selecting samples for molecular analysis: 1) samples with a high oocyst intensity, 2) positive samples in the month of August because a risk factor analysis was carried out at this time, and 3) a random selection of positive samples from the three remaining time points, resulting in a total of 302 samples. Samples were first purified by using a modified water-ether concentration method.30 For solid and semi-solid stools, a pea-sized stool sample was diluted in 100 µL of reverse osmosis (grade 1) water and emulsified in a 1.5-mL microcentrifuge tube. For liquid samples, 200 µL were transferred to a 1.5-mL microcentrifuge tube by using clean plastic pipettes.
The supernatant was aspirated to a voiume of 100 µL and stored at -20°C prior to DNA extraction. DNA was extracted using 15 cycles of freeze-thawing followed by digestion with proteinase K.29 After digestion (3 hours at 55°C) with proteinase K, samples were centrifuged at 14,000 × g for 10 seconds, and the supernatant was treated with a mixture of polyvinylpolypyrrolidone (PVPP) (catalog no. P6755; Sigma, St. Louis, MO) and Chelex 100 (catalog no. 142-1253; Bio-Rad Laboratories, Hercules, CA) slurry to minimize polymerase chain reaction (PCR) inhibitors. Pre-prepared aliquots of the mixture, each containing 50 µL 10% PVPP and 50 µL of 10% Chelex suspensions in DNase/RNase free water were pipetted into 0.5-mL screw top microcentrifuge tubes, and the slurry was sedimented overnight at 4°C by gravity. When required, the supernatant of an aliquot of PVPP/Chelex 100 slurry was carefully aspirated with a pipette without disturbing the sedimented slurry and replaced with 50 µL of DNA lysate. The tube was vortexed, boiled for 10 minutes, and centrifuged (14,000 × g for 1 minute), and the supernatant was transferred to a clean flip-top tube and frozen at –20°C until used as a template for PCR.
PCR–restriction fragment length polymorphism DNA sequencing analysis.
Cryptosporidium species were determined by nested PCR–restriction fragment length polymorphism (RFLP) analysis and/or PCR sequencing at two SSU rRNA gene loci.31,32 For RFLP analysis, positive secondary DIAG PCR products were digested simultaneously with Ase I and Dra I,31 and the secondary XIAO PCR products were digested separately with Ase I and Ssp I.32 Fragments were separated by electrophoresis on a 2% agarose gel, stained with ethidium bromide and viewed under ultraviolet transillumination.
GP60 sequence analysis.
Cryptosporidium parvum and C. hominis isolates were subgenotyped by GP60 sequence analysis,33 and a previously described subtype nomenclature system was used to differentiate subtypes within each subtype family of C. hominis and C. parvum.19 When the assay of Glaberman and others33 failed to produce sufficient amplicons for sequencing, the assay of Sulaiman and others19 (which produces a 400-basepair product compared with the 800-basepair product of the assay of Glaberman and others33) was used.
Cryptosporidium meleagridis SSU rRNA sequence analysis.
Cryptosporidium meleagridis was subgenotyped by sequence analysis of the SSU rRNA gene as described by Glaberman and others.26
DNA sequencing and analysis.
Amplicons for sequencing were treated enzymatically with ExoSAP-IT (GE Healthcare, Piscataway, NJ) to remove excess dNTPs and primers according to the supplier's instructions. Bi-directional sequencing was performed in an ABI model 3730 sequencer (Applied Biosystems, Foster City, CA) by using Big-Dye version 3.1 chemistry and automated capillary DNA sequencer at the Sequencing Service, Dundee University, Dundee, Scotland (http://www.dnaseq.co.uk/services.html). Bi-directional sequences were aligned using European Molecular Biology Laboratory (EMBL) website tools to obtain a consensus that was manually edited according to the sequence chromatogram. The consensus sequence was used to search the GenBank database for similarities using the CBI Blastn tool. ClustalW alignments using EMBL site was used to compare sequences and Phylogenetic trees were constructed using MEGA4 software (http://www.megasoftware.net/).
Statistical analysis.
Of the 1,636 children assessed in the study, 349 submitted samples at all 4 time points. A generalized linear latent and mixed model analysis was performed in STATA Version10 (Stata Corp., College Station, TX)34 on these 349 children. The model was used to test for associations between month, age, sex, and village of residence, and infection status of the children. This model incorporated random and fixed variables accounting for repeated measures in the longitudinal data.
Chi-squared analysis was performed to test for associations between infection with C. parvum and C. hominis, and demographic features of the children, and associations between oocyst presence and stool consistency.
Results
Prevalence and temporal variability.
A total of 3,840 samples from 1,636 children were examined over the one-year period. For those children where sex data were available, 825 (50.4%) were male and 790 (48.3%) female. Children ranged in age from 6 to 80 months, with a median of 39 months. A total of 266 children (16.3%) were from the village of Akinlalu, 714 (43.6%) from Ipetumodu, 253 (15.5%) from Moro, and 403 (24.6%) from Edunabon.
Cryptosporidium oocysts were detected in 684 samples (17.8%, 95% confidence interval [CI] = 16.61–19.06%). There was no statistical association between the presence of oocysts and stool consistency. The prevalence of infection ranged from 15.6% (95% CI = 13.17–18.16%) in September 2006 to 19.6% (95% CI = 17.31–22.18%) in May 2007 (Figure 1A). Most samples (652 of 684) had an oocyst intensity of 1+. A total of 21 children had an oocyst intensity level of 3+. Of these children, 11 occurred in August, 1 in September, 0 in February, and 9 in May.
Figure 1.
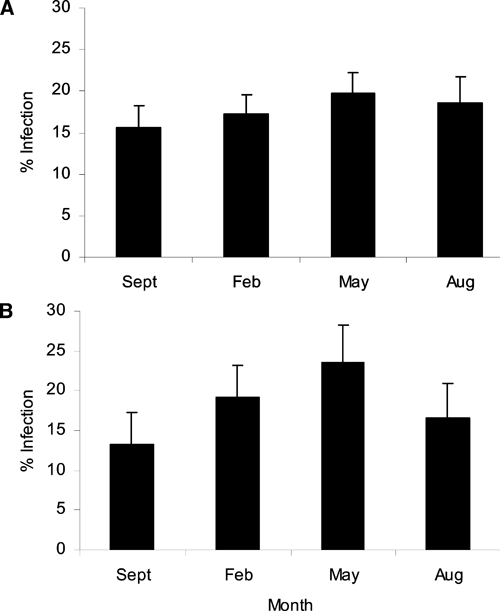
Prevalence (±95% confidence interval) of Cryptosporidium infection in children in Nigeria at four time points over a one-year period. A, All samples collected. B, Samples from 349 children that were tested at each of the four time points.
Of the 1,636 children that submitted fecal sample for analysis, 349 children submitted samples at each of the four time points. Of these children, 180 (51.6%) were male and 165 (47.3%) female. Children ranged in age from 11 to 77 months, with a median of 39 months. Ninety-five children (27.22%) were from the village of Akinlalu, 119 (34.1%) from Ipetumodu, 56 (16.1%) from Moro, and 79 (22.6%) from Edunabon. Prevalence of Cryptosporidium infection ranged from 13.2% (95% CI = 9.81–17.19%) in September to 23.5% (95% CI = 19.15–28.30%) in May 2007 (Figure 1B). Most samples (238 of 252) had intensity levels of 1+. A total of 10 children had an oocyst intensity level of 3+. Of the samples with a 3+ intensity, a higher number occurred in August (8) than in September (0), February (0), and May (2).
Using generalized linear latent and mixed model analysis,34 we determined that month was significantly associated with infection, with increased risk in May (odds ratio [OR] = 1.11, 95% CI = 1.049–1.178, P < 0.0001) and February (OR = 1.06, 95% CI = 1.003–1.127, P = 0.034) in a model adjusted for age, sex, and village of residence. There was no statistically significant association between infection status and age, sex, or village of residence of the children (Table 1).
Table 1.
Results of generalized linear latent and mixed model analysis testing for association between infection status and month, age, sex, and village of residence of 349 children in Nigeria that submitted samples at four time points
| Characteristic | Coefficient | SE | z | P > |z| | 95% confidence interval |
|---|---|---|---|---|---|
| Month | |||||
| Sept | Reference | ||||
| Feb | 0.0621 | 0.029 | 2.12 | 0.034 | 0.005–0.120 |
| May | 0.106 | 0.030 | 3.59 | 0.000 | 0.048–0.164 |
| Aug | 0.040 | 0.030 | 1.34 | 0.179 | −0.018 to 0.010 |
| Age | −0.0002 | 0.001 | −0.36 | 0.719 | −0.002 to 0.001 |
| Sex | |||||
| Male | Reference | ||||
| Female | −0.004 | 0.21 | −0.21 | 0.834 | −0.045 to 0.036 |
| Village | |||||
| Moro | Reference | ||||
| Akinlalu | −0.031 | 0.033 | −0.96 | 0.337 | −0.096 to 0.033 |
| Ipetumodu | −0.043 | 0.032 | −1.37 | 0.172 | −0.106 to 0.019 |
| Edunabon | 0.015 | 0.035 | 0.43 | 0.668 | −0.053 to 0.083 |
Cryptosporidium species, genotypes, and subtypes.
A total of 302 stool samples were analyzed by using two nested PCR-RFLP procedures and/or direct sequencing of PCR products. Of these samples, 77 produced sufficient product for RFLP determination of species. Where required, DNA sequencing was used to confirm Cryptosporidium species/genotypes. Cryptosporidium hominis was detected in 34 samples (44.2%), C. parvum in 25 (32.5%), a mixture of C. parvum and C. hominis in 4 (5.2%), C. meleagridis in 5 (6.5%), Cryptosporidium rabbit genotype in 5 (6.5%), Cryptosporidium cervine genotype in 3 (3.9%), and C. canis in 1 (1.3%) (Table 2).
Table 2.
Species and subtypes of Cryptosporidium identified in 77 samples from children in rural Nigeria
| Species/genotype, subtype family, subtype | No. of children infected |
|---|---|
| C. hominis | 34 (28 subtyped) |
| Ia | 10 |
| IaA18R2 | 3 |
| IaA22R2 | 1 |
| IaA24R2 | 2 |
| IaA25R2 | 2 |
| IaA28R2 | 1 |
| IaA21R1 | 1 |
| Ib | 10 |
| IbA10G2 | 3 |
| IbA13G3 | 7 |
| Id | 4 |
| IdA11 | 2 |
| IdA17 | 2 |
| Ie | 3 |
| IeA11G3T3 | 3 |
| Ih (Novel subtype) | 1 |
| IhA14G1 | 1 |
| C. parvum | 25 (23 subtyped) |
| IIa | 2 |
| IIaA15G2R1 | 1 |
| IIaA16G1R1 | 1 |
| IIc | 17 |
| IIcA5G3a | 9 |
| IIcA5G3b | 8 |
| IIi | 2 |
| IIiA11 | 2 |
| IIm | 2 |
| IImA14G1 | 2 |
| C. hominis/C. parvum | 4 |
| C. meleagridis | 5 (3 subtyped) |
| Type 1 | 3 |
| Cryptosporidium rabbit genotype | 5 |
| Cryptosporidium cervine genotype | 3 |
| C. canis | 1 |
Cryptosporidium hominis was the most common species isolated, followed by C. parvum. There was no statistically significant association between sex, age (< 3 years versus ≥ 3 years), and village of residence and infection with C. hominis or C. parvum.
Further GP60 subgenotyping was conducted successfully on 28 of 33 C. hominis and 23 of 25 C. parvum isolates. Five subtypes of C. hominis (Ia, Ib, Id, Ie, and one novel subtype) and four subtypes of C. parvum (IIa, IIc, Iii, and one unnamed subtype) were identified (Table 2 and Figure 2). Equal numbers of C. hominis subtype families Ia and Ib were detected (10 isolates), and Ia was the most genetically diverse type, consisting of six subtypes. Subtype family Ib and Id (four isolates) each consisted of two genetically distinct subtypes. All three isolates of the subtype family Ie consisted of the subtype IeA11G3T3. One isolate was identified, which has not been previously described (Table 2). This sequence was deposited in the GenBank database (accession no. FJ971716), and according to existing nomenclature is ascribed IhA14G1.
Figure 2.
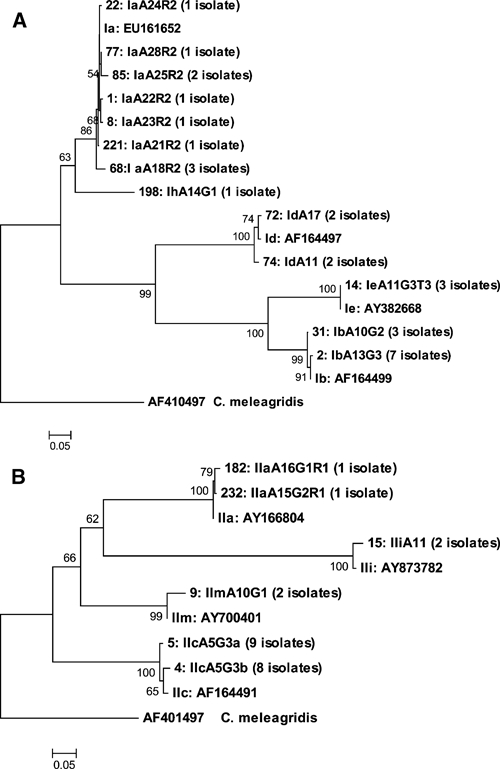
Phylogenetic analysis of A, Cryptosporidium hominis and B, C. parvum subtypes from children in Nigeria and sequences (with accession numbers) previously deposited in GenBank by using neighbor-joining analysis of the glycoprotein (GP60) gene. Values on branches are percentage bootstraps values using 1,000 replicates. Bootstrap values greater than 50% are shown.
Four subtype families of C. parvum were identified. IIc was the most common (17 isolates), and IIc and IIa each had 2 subtypes. Both isolates of the subtype family IIi consisted of the subtype IIiA11, and two isolates were identified that had 99% similarity to sequences deposited in GenBank (accession no. AY700401 (Table 2). The sequence, deposited by Hira KG and others in 2004, has not been published elsewhere. It has been proposed that the subtype family will be named IIm (Ward H, unpublished data).
In addition, three isolates of C. meleagridis were subgenotyped by sequence analysis of the SSU rRNA gene fragment. All three isolates belonged to the subtype, type 1, as previously described26,35 (Table 2).
Discussion
Our findings show that the overall prevalence of Cryptosporidium from a sample of children in Nigeria ranged from 15.6% to 19.6% over a one-year period. Of the few studies conducted in Nigeria, prevalence has ranged from 0% in patients positive and negative for human immunodeficiency virus with diarrhea in Enugu State to 39% in primary school children with diarrhea in the same state.36,37 In the present study, prevalence varied significantly throughout the year. However, rainfall did not appear to be associated with infection rates. It is likely that variation in prevalence is associated with practices or behaviors in the community that exhibit temporal variation. In contrast, other studies in the tropics have indicated that prevalence of infection is highest in months with the greatest rainfall.4–7 However, prevalence was highest during the dry season in Kenya.16 In Venezuela no seasonal variation was detected.38 The overall intensity of infection in this pediatric population was low and few oocysts were observed in the samples. However, in August, higher intensity levels were noted. Data on intensity of infection from previous studies are lacking, particularly at a population level.
This is the first study to genetically characterize Cryptosporidium species in Nigeria, the most populous country in Africa. Isolation of six Cryptosporidium species/genotypes highlights the heterogeneity of Cryptosporidium infections in the children in Nigeria.
In Africa, molecular identification of Cryptosporidium species is generally lacking. Species data have been collected in only five countries; Kenya, Malawi, Uganda, Equatorial Guinea, and South Africa and include C. hominis, C. parvum, C. canis, C. felis, C. meleagridis, and C. muris.5,16,17,22,23,39 However, only in Kenya have all six species been reported.16
Cryptosporidium hominis was the dominant species identified in our study. High levels of C. hominis are consistent with studies from other developing countries such as Peru,18 Kenya,16 India,14 Malawi,5,7 and Uganda,4,15 which indicate that anthroponotic transmission may play a major role in the epidemiology of Cryptosporidium in these areas. A relatively high level of C. parvum was also found in the current study and this finding is in contrast to those of studies from other developing regions where C. parvum infections are generally much lower than that of C. hominis.4,7–9,14,15,18,24,40,41 Higher levels of C. parvum are consistent with results of studies from Kuwait, Equatorial Guinea, and developed countries such as France, Portugal, and the United Kingdom.19,20,25,42 Variations in the distribution of Cryptosporidium species in humans are considered an indication of differences in infection sources.19
We identified C. hominis and C. parvum in similar numbers in males and females and in children < 3 years of age and ≥ 3 years of age. This finding is analogous to findings in southern India, in which no significant differences in age and sex between C. hominis-infected children and those infected with other species.14
Prior to the development of subgenotyping techniques, the presence of high levels of C. parvum would have suggested that zoonotic transmission was as important as anthroponotic transmission in this region in Nigeria. However, results from the current subgenotyping analysis indicate that subtype IIc is dominant in this population. Because IIc is primarily limited to human infections20,21 this finding would indicate that the source of infection in these children in Nigeria is anthroponotic, although this was not determined epidemiologically.
In addition to C. parvum and C. hominis, we also identified C. meleagridis, C. canis, Cryptosporidium cervine genotype, and Cryptosporidium rabbit genotype. Cryptosporidium meleagridis is the third most common Cryptosporidium infection in humans and accounts for 10–20% of cryptosporidiosis cases in Peru and Thailand.10 Few studies have subgenotyped C. meleagridis.26,35 Thus, further analysis of avian and human isolates from various populations and geographic locations is required to address the epidemiologic significance of C. meleagridis subgenotyping.
Our study is the first one to isolate the cervine genotype from humans in Africa. Cryptosporidium canis was isolated on only one occasion previously in an African population.16 Furthermore, our study is the first occasion in which the rabbit genotype was isolated from humans outside the United Lingdom.11,43,44 This genotype was first isolated in an immunocompetent woman in England in 200844 and has since been responsible for a waterborne outbreak in the United Kingdom.43 The rabbit genotype is similar but not identical to C. hominis, with RFLP patterns at the Cryptpsporidium oocyst wall (COWP) locus sharing 99.2% similarity at the SSU rRNA locus and 99.7% at the heat-shock protein (HSP) locus.45 Thus, it is not surprising that human infections have been identified. Two of the four children infected with the rabbit genotype in the current study were twins, indicating that oocysts may have been transmitted anthroponotically between the siblings. Alternatively, it is possible that the children contracted the infection from the same source.
A range of GP60 subtypes of C. hominis and C. parvum were isolated in the current study, including a novel subtype (IhA14G1). Of the six C. hominis subtype families described to date, four (Ia, Ib, Id and Ie) were isolated from these children in Nigeria. These common subtype families are found in humans worldwide10 and were identified in three countries in Africa (Uganda, Malawi, and South Africa).7,15,23 However, subtypes IaA18R2, IaA21R2, IaA24R2, IaA25R2, IaA28R2, and IdA11 have not previously been deposited in GenBank and may indicate the occurrence of new subtypes in the C. hominis families Ia and Id.
Of the 11 C. parvum subtype families described to date, we report the presence of IIa, IIc, IIe, and IIi in addition to an unnamed subtype. The unnamed subtype was previously isolated from children in Bangladesh and the sequence was deposited in GenBank, but the work is unpublished; thus, the GP60 subtype is not yet named. It has been proposed that the subtype will be named IIm (Ward H, unpublished data). The subtype family IIa has been isolated from ruminants and humans, Coupled with IIc, IIa is the most common C. parvum subtype family found to infect humans. Subtype family IIi is less common, being found on only one occasion previously in children in Uganda.15 Of previous studies carried out in Africa, in Malawi, C. parvum subtype families IIc and IIe were present in Malawi,7 and IIc, IIg, IIh, and IIi were identified in Uganda.15 Thus, our study is the first one to report isolation of the C. parvum IIa and IIm subtype families from humans in Africa.
In the present study we identified a high diversity of Cryptosporidium species, genotypes, and subtypes in this pediatric population in Nigeria, but further epidemiologic investigations are required before we can identify anthroponotic, zoonotic, and/or environmental transmission routes of public health significance. Our data indicate that children can be the source of numerous Cryptosporidium species, genotypes, and subtypes, and that their behavior supports both direct and indirect transmission routes. Inadequate treatment of drinking water and indiscriminate outdoor defecation are well recognized risk factors for cryptosporidiosis. Over half (53.9%) of the population in this area do not treat their drinking water, and most children studied defecate indiscriminately outdoors (Molloy S, unpublished data). Both questionnaire- and molecular-based tools will be required to determine the importance of anthroponotic versus other routes of transmission, and we are currently undertaking risk factor analysis that should add to our understanding of the risk factors associated with Cryptosporidium transmission in these tropical environments.
Acknowledgments
We thank all mothers and children who participated in the study, all Obas, fieldworkers, doctors, and nurses for their excellent work, and Cathal Walsh and Andrew Jackson for statistical consultation.
Footnotes
Financial support: Síle Molloy was a recipient of a PhD scholarship from the Irish Research Council for Science, Engineering and Technology. The fieldwork costs were supported by a Health Research Board grant (GHRA/2006/7).
Authors' addresses: Síle Molloy, Patrick Kirwan, and Celia V. Holland, Zoology Department, School of Natural Sciences, Trinity College Dublin, Dublin 2, Ireland, E-mails: molloysi@tcd.ie, kirwanpa@tcd.ie, and cholland@mail.tcd.ie. Huw V. Smith, Rosely A. B. Nichols, and Lisa Connelly, Scottish Parasite Diagnostic Laboratory, Stobhill Hospital, 133 Balornock Road, Glasgow G21 3UW, United Kingdom, E-mails: Huw.Smith@northglasgow.scot.nhs.uk, rosely.nichols@ggc.scot.nhs.uk, and lisa.connelly2@ggc.scot.nhs.uk. Samuel O. Asaolu, Zoology Department, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria, E-mail: sasaolu2002@yahoo.co.uk.
References
- 1.Caccio SM, Thompson RC, McLauchlin J, Smith HV. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Huang DB, Chappell C, Okhuysen PC. Cryptosporidiosis in children. Semin Pediatr Infect Dis. 2004;15:253–259. doi: 10.1053/j.spid.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Guerrant DI, Moore SR, Lima AAM, Patrick P, Schorling JB, Guerrant RL. Association of early childhood diarrhoea and cryptosporidiosis with impaired physical fitness and cognitive function 4–7 years later in a poor urban community in Northeast Brazil. Am J Trop Med Hyg. 1999;61:707–713. doi: 10.4269/ajtmh.1999.61.707. [DOI] [PubMed] [Google Scholar]
- 4.Tumwine JK, Kekitiinwa A, Nabukeera N, Akiyoshi DE, Rich SM, Widmer G, Feng XC, Tzipori S. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am J Trop Med Hyg. 2003;68:710–715. [PubMed] [Google Scholar]
- 5.Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, Smith HV. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect. 2007;135:1307–1315. doi: 10.1017/S0950268806007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman RD, Sears CL, Moore SR, Nataro JP, Wuhib T, Agnew DA, Guerrant RL, Lima AAM. Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J Infect Dis. 1999;180:167–175. doi: 10.1086/314820. [DOI] [PubMed] [Google Scholar]
- 7.Peng MM, Meshnick SR, Cunliffe NA, Thindwa BDM, Hart CA, Broadhead RL, Xiao L. Molecular epidemiology of cryptosporidiosis in children in Malawi. J Eukaryot Microbiol. 2003;50:557–559. doi: 10.1111/j.1550-7408.2003.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 8.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis. 2008;14:1567–1574. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, Vivar A, Ticona E, Navincopa M, Williamson J, Ortega Y, Gilman RH, Bern C, Xiao L. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 10.Xiao L, Ryan UM. In: Cryptosporidium and Cryptosporidiosis. Fayer R, Xiao L, editors. Boca Raton, FL: CRC Press; 2008. pp. 119–171. (Molecular epidemiology). [Google Scholar]
- 11.Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Xiao L, Ryan U, Modha D, Mallaghan C. Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerg Infect Dis. 2009;15:829–830. doi: 10.3201/eid1505.081419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jirků M, Valigurová A, Koudela B, Krízek J, Modrý D, Slapeta J. New species of Cryptosporidium tyzzer, 1907 (Apicomplexa) from amphibian host: morphology, biology and phylogeny. Folia Parasitol (Praha) 2008;55:81–94. [PubMed] [Google Scholar]
- 13.Smith HV, Nichols RA. Cryptosporidium: detection in water and food. Exp Parasitol. 2009 doi: 10.1016/j.exppara.2009.05.014. Jun 6 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Ajjampur SS, Gladstone BP, Selvapandian D, Muliyil JP, Ward H, Kang G. Molecular and spatial epidemiology of cryptosporidiosis in children in a semiurban community in South India. J Clin Microbiol. 2007;45:915–920. doi: 10.1128/JCM.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyoshi DE, Tumwine JK, Bakeera-Kitaka S, Tzipori S. Subtype analysis of Cryptosporidium isolates from children in Uganda. J Parasitol. 2006;92:1097–1100. doi: 10.1645/GE-843R.1. [DOI] [PubMed] [Google Scholar]
- 16.Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, Gatika SM, Kamwati SK, Revathi G, Hart CA. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg. 2006;75:78–82. [PubMed] [Google Scholar]
- 17.Morgan U, Weber R, Xiao LH, Sulaiman I, Thompson RCA, Ndiritu W, Lal A, Moore A, Deplazes P. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38:1180–1183. doi: 10.1128/jcm.38.3.1180-1183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman RH, Lal AA. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis. 2001;183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 19.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao LH. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–2747. doi: 10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallon ME, MacLeod A, Wastling JM, Smith H, Tait A. Multilocus genotyping of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect Genet Evol. 2003;3:207–218. doi: 10.1016/s1567-1348(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 22.Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, Bern C, Sulaiman IM, Glaberman S, Lal AA, Xiao L. A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol. 2001;((Suppl)):28S–31S. doi: 10.1111/j.1550-7408.2001.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 23.Leav BA, Mackay MR, Anyanwu A, RM OC, Cevallos AM, Kindra G, Rollins NC, Bennish ML, Nelson RG, Ward HD. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect Immun. 2002;70:3881–3890. doi: 10.1128/IAI.70.7.3881-3890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cama VA, Bern C, Sulaiman IM, Gilman RH, Ticona E, Vivar A, Kawai V, Vargas D, Zhou L, Xiao L. Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J Eukaryot Microbiol. 2003;50:S531–S533. doi: 10.1111/j.1550-7408.2003.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin J, Amar C, Pedraza-Diaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 faecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–3990. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaberman S, Sulaiman IM, Bern C, Levior J, Peng MM, Morgan U, Gilman R, Altaf A, Xiao L. A multilocus genotypic analysis of Cryptosporidium meleagridis. J Eukaryot Microbiol. 2001;48:S19–S22. doi: 10.1111/j.1550-7408.2001.tb00439.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirwan P, Asaolu SO, Molloy SF, Abiona TC, Jackson AL, Holland CV. Patterns of soil-transmitted helminth infection and impact of four-monthly albendazole treatments in preschool children from semi-urban communities in Nigeria: a double-blind placebo-controlled randomised trial. BMC Infect Dis. 2009;9:20. doi: 10.1186/1471-2334-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casemore DP, Armstrong M, Sands RL. Laboratory diagnosis of cryptosporidiosis. J Clin Microbiol. 1985;38:1337–1341. doi: 10.1136/jcp.38.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols G, Thom BT. Screening for Cryptosporidium in stools. Lancet. 1984;1:734–735. [PubMed] [Google Scholar]
- 30.Nichols RA, Moore JE, Smith HV. A rapid method for extracting oocyst DNA from Cryptosporidium-positive human faeces for outbreak investigations. J Microbiol Methods. 2006;65:512–524. doi: 10.1016/j.mimet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Nichols RA, Campbell BM, Smith HV. Identification of Cryptosporidium spp. oocysts in United Kingdom noncarbonated natural mineral waters and drinking waters by using a modified nested PCR-restriction fragment length polymorphism assay. Appl Environ Microbiol. 2003;69:4183–4189. doi: 10.1128/AEM.69.7.4183-4189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glaberman S, Moore JE, Lowery CJ, Chalmers RM, Sulaiman I, Elwin K, Rooney PJ, Millar BC, Dooley JS, Lal AA, Xiao L. Three drinking-water-associated cryptosporidiosis outbreaks, northern Ireland. Emerg Infect Dis. 2002;8:631–633. doi: 10.3201/eid0806.010368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabe-Hesketh S, Skrondal A, Pickles A. Reliable estimation of generalized linear mixed models using adaptive quadrature. The Stata Journal. 2002;2:1–21. [Google Scholar]
- 35.Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RCA, Fayer R, Lal AA. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nwokediuko SC, Bojuwoye BJ, Onyenekwe B. Apparent rarity of cryptosporidiosis in human immunodeficiency virus (HIV)-related diarrhoea in Enugu, south-eastern, Nigeria. Niger Postgrad Med J. 2002;9:70–72. [PubMed] [Google Scholar]
- 37.Okafor JI, Okunji PO. Prevalence of Cryptosporidium oocysts in faecal samples of some school children in Enugu State, Nigeria. J Commun Dis. 1996;28:49–55. [PubMed] [Google Scholar]
- 38.Chacin-Bonilla L, Bonilla MC, Soto-Torres L, Rios-Candida Y, Sardina M, Enmanuels C, Parra AM, Sanchez-Chavez Y. Cryptosporidium parvum in children with diarrhea in Zulia State, Venezuela. Am J Trop Med Hyg. 1997;56:365–369. doi: 10.4269/ajtmh.1997.56.365. [DOI] [PubMed] [Google Scholar]
- 39.Blanco MA, Iborra A, Vargas A, Nsie E, Mba L, Fuentes I. Molecular characterization of Cryptosporidium isolates from humans in Equatorial Guinea. Trans R Soc Trop Med Hyg. 2009;103:1282–1284. doi: 10.1016/j.trstmh.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 40.Das P, Roy SS, MitraDhar K, Dutta P, Bhattacharya MK, Sen A, Ganguly S, Bhattacharya SK, Lal AA, Xiao L. Molecular characterization of Cryptosporidium spp. from children in Kolkata, India. J Clin Microbiol. 2006;44:4246–4249. doi: 10.1128/JCM.00091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, Akiyoshi DE, Tzipori S. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. 2005;73:921–925. [PubMed] [Google Scholar]
- 42.Guyot K, Follet-Dumoulin A, Lelievre E, Sarfati C, Rabodonirina M, Nevez G, Cailliez JC, Camus D, Dei-Cas E. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39:3472–3480. doi: 10.1128/JCM.39.10.3472-3480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalmers RM, Elwin K, Thomas AL, Guy EC, Mason B. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 2009;14:1–9. doi: 10.2807/ese.14.02.19086-en. [DOI] [PubMed] [Google Scholar]
- 44.Robinson G, Elwin K, Chalmers RM. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg Infect Dis. 2008;14:1800–1802. doi: 10.3201/eid1411.080239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan U, Xiao L, Read C, Zhou L, Lal AA, Pavlasek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl Environ Microbiol. 2003;69:4302–4307. doi: 10.1128/AEM.69.7.4302-4307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


