Abstract
In both prokaryotes and eukaryotes, the expression of a large number of genes is controlled by negative feedback, in some cases operating at the level of translation of the mRNA transcript. Of particular interest are those cases where the proteins concerned have cell-wide function in recognizing a particular codon or RNA sequence. Examples include the bacterial translation termination release factor RF2, initiation factor IF3, and eukaryote poly(A) binding protein. The regulatory loops that control their synthesis establish a negative feedback control mechanism based upon that protein's RNA sequence recognition function in translation (for example, stop codon recognition) without compromising the accurate recognition of that codon, or sequence during general, cell-wide translation. Here, the bacterial release factor RF2 and initiation factor IF3 negative feedback loops are reviewed and compared with similar negative feedback loops that regulate the levels of the eukaryote release factor, eRF1, established artificially by mutation. The control properties of such negative feedback loops are discussed as well as their evolution. The role of negative feedback to control translation factor expression is considered in the context of a growing body of evidence that both IF3 and RF2 can play a role in stimulating stalled ribosomes to abandon translation in response to amino acid starvation. Here, we make the case that negative feedback control serves primarily to limit the overexpression of these translation factors, preventing the loss of fitness resulting from an unregulated increase in the frequency of ribosome drop-off.
Keywords: release factor RF2, initiation factor IF3, negative feedback, eukaryote release factor eRF1, ribosomal frameshifting
INTRODUCTION
Many ribosomal proteins and translation factors have their expression governed at both the transcriptional level and translational levels, matching protein expression levels to cell growth rate (for review, see Kaczanowska and Ryden-Aulin 2007). For example, Escherichia coli ribosomal protein S15 controls translation of its own mRNA through binding of an operator-type element in the mRNA that overlaps the mRNA's ribosome binding site. Overexpression of the protein (relative to rRNA levels) feeds back to limit translation of S15 mRNA (Portier et al. 1990). In a similar way, E. coli threonyl-tRNA synthetase also autoregulates its own expression (Brunel et al. 1995). In these, and other examples, the protein regulated by feedback only regulates translation of its own mRNA (Freedman et al. 1987; Dabeva and Warner 1993; Boni et al. 2001). A more intriguing and complex class of autogenous translational control is defined by examples where the protein whose expression is regulated naturally functions as an mRNA-interacting translation factor, and which thus interacts with all mRNAs in the cell. For example, elegant negative feedback control regulates expression of E. coli translation initiation factor 3 (IF3) and release factor 2 (RF2). These autogenous negative feedback loops operate in an analogous way to the thermostatic control of room temperature. As the temperature rises, negative feedback, via a thermostat with a given set point, switches off heat production. In the case of RF2 and IF3 feedback control, the set point of the system is regulated by the mRNA sequence of the cis-acting control element within the respective mRNAs. However, the IF3 and RF2 negative feedback loops must exhibit one particularly important feature; the normal dynamic range of their control should not interfere with ordinary recognition of the relevant codon (for example, UGA in the case of RF2) during translation of the bulk of other mRNAs in the cell. It is therefore important that translation factor levels fluctuate around some optimal level that maintains accurate translational decoding. In what follows, we review the mechanism underpinning the negative feedback loop, and show how the system set point is established. We discuss how the primary role of these negative feedback loops is to prevent overproduction of RF2 and IF3, which in both cases is likely to compromise translational fidelity and the regulation of the normal responses of the ribosomal population to amino acid starvation stresses.
Translational regulation of RF2 expression
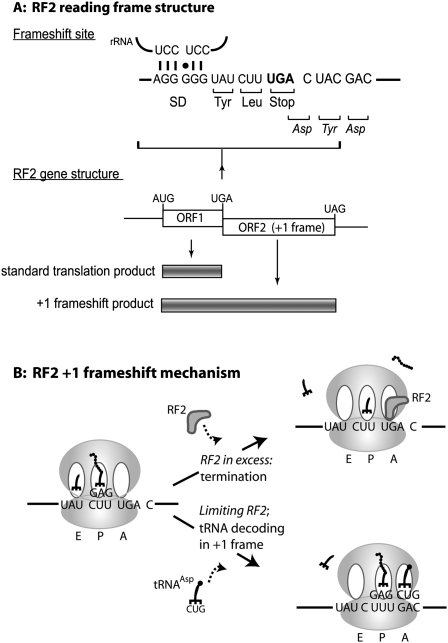
In prokaryotes, stop codons located in the ribosomal A-site during translation will be recognized by one of two release factors with semi-redundant specificity: RF1 recognizes UAA and UAG, whereas RF2 recognizes UAA and UGA. E. coli RF2 is encoded by the prfB gene that comprises two open reading frames (ORFs), a short one just 26 codons long and terminated by a UGA codon (ORF1), followed immediately by a second ORF located in the +1 frame relative to ORF1. For translation of the full-length 365-amino acid protein, a +1 frameshift must therefore occur at the internal UGA codon (Craigen et al. 1985; Craigen and Caskey 1986). Frameshifting at this site occurs with the extremely high frequency of 50% (Craigen and Caskey 1986; Weiss et al. 1988). The internal UGA stop codon can be recognized by RF2, which therefore autoregulates its own production. When RF2 levels drop below the optimal concentration, the ORF1 UGA codon will be recognized slowly. The resulting ribosomal pause triggers a +1 frameshift event with the consequential translation of an ORF1-ORF2 encoded fusion protein and synthesis of full-length RF2 (Fig. 1). Conversely, overabundance of RF2 causes termination at the ORF1 UGA codon rather than frameshifting, and the synthesis of a rapidly turned-over short peptide, assumed to be functionless.
FIGURE 1.
Translation of RF2 expression is autogenously controlled via a negative feedback loop. (A) The nucleotide sequence of the mRNA frameshift site, showing the ORF1 UGA stop codon (bold), with a 3′ C residue, the UAU E-site codon, the internal Shine–Dalgarno (SD) sequence, and its predicted base-pairing to the 16S rRNA. The structure of the RF2 gene shows the second ORF in the +1 frame relative to ORF1. (B) The frameshift mechanism is in part triggered by a leucyl tRNA wobble base paired in the ribosomal P-site. This unstable codon–anticodon interaction is one of the factors stimulating +1 frameshifting, along with a poor context UGA stop codon terminating ORF1, and a SD sequence within ORF1 that stimulates the +1 nucleotide shift.
There are many contributing factors to the efficiency of the frameshift event. The leucine codon (CUU) that precedes the premature ORF1 stop codon is decoded by a tRNA using third base wobble (Craigen et al. 1985; Weiss et al. 1988). Overall, the sequence CUUUGA is particularly prone to frameshifting because a forward slip of 1 nucleotide (nt) allows the CUN-decoding tRNALeu (anticodon GAG) to decode UUU using first and third position G-U wobble base pairing (Curran and Yarus 1988; Weiss et al. 1988). Furthermore, the position 3 nt upstream of the leucine codon marks the end of a Shine–Dalgarno (SD) sequence that is specifically positioned to drive frameshifting (Fig. 1). Mutation either of this SD sequence or the binding helix of the 16S rRNA by only 1 nt decreased the frameshift frequency at this site (Weiss et al. 1988). Specific mRNA–rRNA interactions at this SD sequence appear to encourage the +1 repositioning of the ribosome paused with a P-site CUU leucine codon, thus elevating frameshift efficiency. The cryptic SD site is, however, necessary but not in itself sufficient for frameshifting; a shifty codon context is required in addition to the internal SD sequence (Weiss et al. 1988). A further contributor to frameshifting is the codon preceding the CUU shifty codon, which in a preframeshift ribosome would occupy the ribosomal E-site. In vitro, tRNA occupancy of the E-site appears to repress frameshift efficiency, by stabilizing the P-site codon–anticodon interaction. Conversely, a rapid ejection of E-site tRNA stimulates frameshifting on the shifty CUUUGA sequence (Marquez et al. 2004). These results have been confirmed in vivo, with a demonstration that those E-site codons directing a strong codon–anticodon interaction repress frameshift efficiency at the RF2 frameshift site. Weaker codon–anticodon interactions stimulate frameshifting (Sanders and Curran 2007). In fact, this study provided evidence not only that E-site codon–anticodon interactions are important to modulate A-site decoding fidelity but also that of the E-site codons surveyed, the wild-type frameshift E-site codon UAU was one of the most efficient frameshift stimulators (Sanders and Curran 2007).
Further downstream, the nucleotides 3′ of UGA stop also reduce termination efficiency and thus promote the competing frameshift event. At the frameshift site (CUUUGACUA), the C nucleotide immediately following the stop codon creates a particularly inefficient termination context (Poole et al. 1995) . Varying this “+4” nucleotide immediately following the stop codon and measuring the ratio of termination:frameshift products showed that for both UAA and UGA, the preferred fourth base hierarchy was U>G>A–C. This observation is supported by the fact that the UGAC termination sequence is under-represented in E. coli genes (Brown et al. 1990; Poole et al. 1995) . While the +5 position was inert with respect to termination efficiency, an A at the +6 position also reduced termination efficiency (Major et al. 1996). Therefore the prfB UGA 3′ sequence UGACUA is an extremely poor termination context. Because RF2 frameshifting occurs in competition with termination, inefficient stop codon recognition dictated by a poor 3′ context will promote the efficiency of frameshifting (Adamski et al. 1993).
In summary, there are several important factors that govern the RF2 frameshift frequency and thus the operation of the RF2 negative feedback loop. Overall, these factors combine to govern the “set point” of this homeostatic mechanism and are vital to allow efficient expression of RF2 to balance the requirements for cell growth and to compensate for RF2 protein turnover. However, negative feedback limits overexpression of RF2 by governing the ratio of frameshifting to translation termination at the premature stop codon.
Establishing negative feedback control de novo: Nonsense alleles of eukaryote release factor genes
The identification of an autogenous regulatory mechanism governing RF2 translation suggests that it should be possible to establish such a feedback loop to control the translational expression of eukaryote release factors. In eukaryotes, translation termination is catalyzed by a class I release factor, eRF1, which recognizes all three stop codons (Frolova et al. 1994). eRF1 forms a heterodimeric complex with another protein, eRF3, a class II GTPase release factor (Stansfield et al. 1995; Zhouravleva et al. 1995). Although eRF1 and eRF3 are both encoded by essential genes in yeast, a number of reports describe viable premature termination codon (PTC) mutants in both SUP45 (encoding eRF1) (Stansfield et al. 1996; Moskalenko et al. 2003) and SUP35 (eRF3-encoding) (Chabelskaya et al. 2007). All such PTC release factor mutants exhibit increased frequencies of stop codon read-through and reduced levels of full-length release factor (Stansfield et al. 1996; Chabelskaya et al. 2007). Some of the SUP45 PTC mutants were identified by selecting for an allosuppressor phenotype, an enhancement of the suppressor activity of an ordinarily weak UAA suppressor tRNA SUQ5oc (Stansfield et al. 1996). Many such allosuppressor mutants defined nonsense alleles of the essential SUP45 gene. In the other cases, PTC mutants of SUP45 were isolated in a wild-type tRNA background (Moskalenko et al. 2003). The investigators of this latter study have provided convincing evidence that in fact a suppressor tRNA is probably not necessary for the establishment of a viable eRF1 autogenous feedback loop. It is therefore assumed that viability can be maintained in the PTC mutants by read-through of the premature stop codon by ordinary cellular tRNAs such as 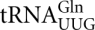 , that can misread UAA via first base wobble (Edelman and Culbertson 1991).
, that can misread UAA via first base wobble (Edelman and Culbertson 1991).
In both SUP45 gene studies, the mutant stop codons were found at a range of codon positions across the breadth of the coding region. Stop codon read-through assays were used to measure UAA read-through of between 25% and 45%. Western blot analysis of eRF1 protein revealed the mutants expressed a mix of truncated eRF1 produced from translation termination at the nonsense codon, and full-length eRF1 produced by read-through of the nonsense codon and subsequent termination at the natural stop codon (Stansfield et al. 1996; Moskalenko et al. 2003). In both cases, the introduction of the nonsense codon establishes a negative feedback loop regulating the expression of full-length eRF1. The nonsense allele directs expression of an inactive N-terminal eRF1 fragment. The consequential deficit in levels of full-length active release factor drives stop codon read-through of the PTC to produce full-length eRF1 at some level. This reduced level of full-length eRF1 is in turn able to inefficiently recognize the premature stop codon. An autogenous negative feedback loop is thus established analogous to the regulation of bacterial RF2 expression.
In summary, therefore, the eRF1 nonsense alleles identified represent examples of an autogenous translational control mechanism, regulated by negative feedback. They demonstrate that the establishment of an RF2-like control mechanism is possible for both prokaryote and eukaryote release factor genes. However, in the bacterial RF2 system, the ribosomal frameshift site represents a cis element of the feedback loop intrinsic to the mRNA, which drives expression of full-length RF2 as a default mechanism; only when the level of RF2 begins to overshoot, does termination limit the production of full-length peptide. Crucially, it is the cis-frameshift site that serves to insulate RF2 expression control from any knock-on effects on general cell translation termination. In contrast, for the eukaryote eRF1 nonsense alleles, there is little or no such insulation; cis effects driving read-through are limited to a slightly leaky termination context in some mutants (0.5%–2% read-through in a wild-type tRNA background) (Kiktev et al. 2009). For this reason, the levels of stop codon read-through needed to generate enough full-length eRF1 for viability must originate through trans effects, namely a suboptimal level of eRF1 (Stansfield et al. 1996; Moskalenko et al. 2003). This allows the competitive advantage for stop codon recognition to shift toward tRNA species, either those cognate for the stop codon (SUQ5) (Stansfield et al. 1996) or those miscognate for the stop codon (e.g., 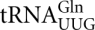 ) (Edelman and Culbertson 1991), and the establishment of the feedback loop.
) (Edelman and Culbertson 1991), and the establishment of the feedback loop.
Initiation factor IF3: An autogenous control system using codon-specific mRNA recognition
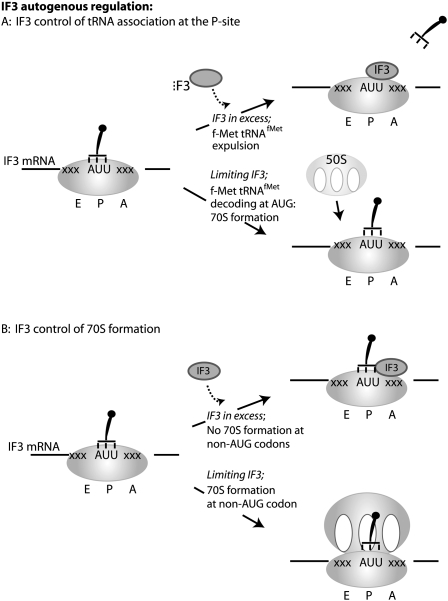
During translation initiation, the bacterial 70S ribosome must be dissociated into 30S and 50S subunits, the fmet-tRNAfmet initiator tRNA must be delivered to the AUG codon, located at the P-site, and accurate AUG recognition achieved. Elements of this process are aided by IF3. The bacterial translation initiation factor IF3 is encoded by the infC gene, unusual because it begins not with an AUG codon, but with AUU. The discovery that this gene could not be overexpressed in E. coli suggested that it is regulated at the level of translation by a negative feedback loop (Fig. 2; Gold et al. 1984; Butler et al. 1986) and that the AUU initiation codon plays a central role in this regulation. In fact, replacing the AUU codon with an AUG abolishes this autogenous regulation and boosts IF3 production 10-fold (Butler et al. 1987). Furthermore, replacing the AUG initiation codon of a neighboring gene thrS with AUU also placed this gene under IF3 autoregulatory control (Sacerdot et al. 1996).
FIGURE 2.
Translation of IF3 is autogenously controlled via a negative feedback loop acting at the level of initiation codon detection. (A) In vitro evidence suggests IF3 catalyzes continuous eviction of tRNA from the ribosomal P-site (Antoun et al. 2006). This ejection mechanism is faster if the initiator tRNA is noncognate for the initiation codon being used. For this reason, in the presence of IF3, initiation on the IF3 mRNA is highly inefficient due to the use of an AUU initiation codon. In the absence of IF3, initiator tRNA is not ejected, and IF3 translation is begun. (B) Some studies indicate a model where IF3 selectively prevents 70S formation if a noncognate P-site interaction is encountered (Grigoriadou et al. 2007). In the absence of IF3, 70S formation is not sensitive to initiator tRNA decoding fidelity, thus initiation at the infC (IF3) AUU codon is permitted.
The precise molecular function of IF3, enabling the ribosome to discriminate against non-AUG codons in the P-site, is the subject of some debate. Mutations in the infC gene encoding IF3 enable translation initiation to take place at non-AUG codons, for instance, in a GUA initiation codon recJ mutation in E. coli (Haggerty and Lovett 1997). Use of mutant initiator tRNAs cognate for non-AUG codons, but nevertheless capable of initiating translation, allowed the conclusion that IF3 was capable of detecting mismatches between the initiator tRNA anticodon and the mRNA (Meinnel et al. 1999). Recent work suggests that the principal role of IF3 is to enhance the natural rate of tRNA expulsion from the P-site, no matter its identity (Antoun et al. 2006); AUG-cognate initiator tRNAs are expelled more slowly, because it is known that a cognate P-site tRNA–mRNA interaction is more effectively clamped by the ribosome (Ringquist et al. 1993). That being the case, the negative feedback loop controlling IF3 synthesis is explained as follows: during translation of the IF3 mRNA when IF3 levels are high, the initiator tRNA, noncognate for the IF3 AUU codon, is rapidly evicted from an initiating ribosome. Conversely, when IF3 is limiting, the initiator mRNA is evicted from the initiating ribosome at the AUU codon much more slowly, providing some opportunity for IF3 translation to begin; autoregulation is thus achieved (Fig. 2; Antoun et al. 2006). Other studies, however, suggest a significantly different role for IF3: as a fidelity checking agent governing formation of a 70S ribosomal complex from a 30S initiation complex (Grigoriadou et al. 2007). In this study, the use of an AUU initiation codon markedly reduced 70S formation in the presence of IF3. IF3 did not exert this 70S gate-keeper function with an AUG initiation codon. Further research will be needed to reconcile these alternative explanations. However, it is nevertheless clear that the infC cis-regulatory element, namely, the AUU codon, insulates the fidelity of cell-wide initiation at AUG codons from regulation of IF3 expression. The AUU codon is thus analogous to the RF2 premature UGA codon in the sense that both codons are either inefficiently recognized (RF2) or poorly discriminated against (IF3) when the abundance of their cognate factor is suboptimal.
Other RNA binding protein examples of autogenous control
In eukaryotic cells the poly(A) tails of protein-encoding mRNAs are important determinants of mRNA stability and translational efficiency. The poly(A) binding protein PABP is a polypeptide with four RNA binding domains that specifically binds the poly(A) tail to mediate stability of that mRNA. There is evidence to suggest that PABP expression is regulated at the translational level by a negative feedback loop. Within the 5′ leader sequence of the PABP mRNA from a wide range of species lies a 50–70-nt, A-rich sequence (de Melo Neto et al. 1995). This is thought to be a weak binding site for PABP, impeding ribosomal scanning during AUG location (de Melo Neto et al. 1995). Addition of PABP to an in vitro translation reaction programmed with a PABP mRNA caused inhibition of PABP translation, and this inhibition was dependent upon the PABP mRNA leader sequence (de Melo Neto et al. 1995; Bag and Wu 1996). There is also evidence that PABP may destabilize its own mRNA through binding to the A-rich 5′ leader sequence (Hornstein et al. 1999).
Why control by negative feedback: Translational fidelity as a driver
The above examples raise the key question of why certain translation factors are regulated at the level of translation in this way. Of importance here is the understanding that while a factor like RF2 regulates its own expression through regulated bypass of a UGA stop codon, in the general bacterial translation system, UGA recognition must be highly efficient. Thus extremely high levels of UGA read-through (via frameshift) occur on the RF2 mRNA, driving RF2 expression, while ordinary translation termination is unaffected and insulated from this autogenous regulation. This insulation effect is achieved using cis factors on the RF2 mRNA to drive RF2 expression, namely, the ribosomal frameshift slippery sequence. The resultant RF2 expression must be sufficient to compensate for depletion of RF2 by protein turnover and the growth of the cell, maintaining RF2 levels at between 5300 and 24,900 molecules per cell, depending upon growth rate (Adamski et al. 1994). We therefore advance the argument that while negative feedback maintains output around a set point, both up-regulating and down-regulating protein expression, the RF2 negative feedback loop has the primary function of preventing overproduction of RF2. Why should this be? It is known that the release factors can recognize a subset of sense codons with significant frequency (Freistroffer et al. 2000). RF2 is at risk of falsely recognizing UGG as a stop codon, while RF1 can recognize UAU in error. It is estimated that UAU and UGG are between them responsible for 50% of false stops in E. coli (Freistroffer et al. 2000). It is also known that overexpression of RF1 or RF2 can increase this type of false stop error. The RF2 negative feedback loop, by limiting overproduction of RF2, will ameliorate the frequency of this type of mistake, which in terms of energy expended in amino acid polymerization, is costly for the cell. Therefore, while some estimates for processivity errors caused by release factor dependent false stops are low (Jorgensen et al. 1993), it seems plausible that the cell would be advantaged by limiting overexpression of a factor that can misrecognize some types of sense codon.
Furthermore, it has become clear recently that ribosomes have a P-site sensitivity to codon–anticodon mispairing—in other words, the ribosome has a fidelity maintenance function that can “atone” for recent decoding mistakes made during A-site decoding (Zaher and Green 2009). If a mispairing event occurs at the A site, and this is fixed into the P-site by a peptidyl-transferase event, using a yet-to-be identified mechanism, there is an increased likelihood that a release factor will misrecognize an A-site sense codon to trigger peptidyl release and premature termination (Zaher and Green 2009). It seems possible that if overexpressed, the RF2 population might erroneously catalyze A-site termination events for some weaker, perhaps wobble base-paired, codon–anticodon interactions at the P-site, triggering premature abandonment of translation of the nascent peptide. This fidelity function seems to have evolved to prevent mistaken incorporation of amino acids into a peptide but is dependent on a release factor misrecognition event at the A-site. Limiting RF2 overexpression using a negative feedback loop could serve to limit inappropriate A-site termination.
Similar arguments can be advanced for IF3, which evidence suggests may play a role in fine-tuning amino acid starvation responses. Overexpression of the IF3 encoding gene, infC, is also predicted to be deleterious to the cell. It is known that ribosome recycling factor (RRF), and elongation factor G (EF-G) cooperate to release small peptidyl tRNAs from ribosomes stalled early in the translation of an open reading frame—this is particularly frequent during the first seven codons translated (Karimi et al. 1998; Heurgue-Hamard et al. 2000; for review, see Buchan and Stansfield 2007). This may help rapid abandonment of translation under conditions where aminoacyl tRNAs are in limiting supply, namely, as an amino acid starvation early warning system. The released peptidyl-tRNAs are hydrolyzed by the essential enzyme peptidyl tRNA hydrolase. Recent evidence suggests IF3 may have a role in this process; infC mutants rescue the phenotype of a peptidyl-tRNA hydrolase mutant, and the same IF3 mutant enhances the temperature-sensitive phenotype of an frrts RRF mutant; IF3 overexpression (from an AUG initiated ORF) suppresses the same frrts mutant (Singh et al. 2005). Finally, in vitro assays show IF3 is necessary to allow RRF/EF-G to catalyze peptidyl-tRNA release (Singh et al. 2005). IF3 may therefore help evict peptidyl tRNA from the P-site of stalled ribosome complexes immediately post-initiation. Other research shows that IF3 antagonizes translation initiation on leaderless mRNAs, some of which are expressed in E. coli (Tedin et al. 1999). Overexpression of IF3 would be expected to have a detrimental effect on this minor class of mRNAs (Tedin et al. 1999) and may more generally compromise fitness by stimulating inappropriate abandonment of early translation events, thereby limiting translation system productivity.
In summary, we suggest that the use of negative feedback loops to regulate translation factor synthesis, both RF2 and IF3, has been selected primarily to prevent overexpression of these essential factors. This control will limit premature and inappropriate abandonment of translation that would otherwise compromise cell fitness.
The evolution of translational feedback control
One important question that arises is why translational control of eRF1 production has apparently not arisen in eukaryotes over the course of evolution, if it (RF2 regulation) is otherwise so wide-spread in bacterial species (70% of species surveyed) (Baranov et al. 2002). This may be because of the different nature of stop codon recognition in eukaryotes and prokaryotes. In prokaryotes, RF1, recognizing UAA and UAG, and RF2, with responsibility for terminating at UAA and UGA, act as the primary stop codon recognizing activities. The GTPase RF3 plays a nonessential role in recycling the RF1 and RF2 off the ribosome once peptidyl-release has been triggered (Zavialov et al. 2001; Gao et al. 2007). However, in eukaryotes, a single release factor, eRF1, recognizes all three stop codons (Frolova et al. 1994), in a complex with eRF3 (Stansfield et al. 1995; Zhouravleva et al. 1995). The requirement for a eukaryote autogenous feedback loop is avoided for two reasons. First, there is evidence that eRF1 recognition of UGA codons (and presumably therefore discrimination against the near cognate UGG) may be particularly aided by the involvement of eRF3 (Fan-Minogue et al. 2008). In vitro, and acting alone, eRF1 can inefficiently recognize UGG with a dissociation constant only 75-fold greater than that for a stop codon (Chavatte et al. 2003). However, in vivo, discrimination against termination at UGG is likely to be much more efficient. Evidence to support the idea that eRF3 may particularly aid UGA recognition comes from the observation that some eRF3 mutants with defective GTP hydrolysis are specifically compromised in their ability in vivo to help eRF1 terminate at UGA stop codons (Salas-Marco and Bedwell 2004). Second, some mutations in the eRF1 TASNIKS stop recognition motif eliminate an eRF3 requirement for UAA and UAG recognition but not for UGA recognition (Fan-Minogue et al. 2008).
In addition, there is almost certainly far less selective pressure for autogenous control loops to evolve in eukaryotes to prevent class I release factor overexpression; it is well established that eRF1 requires eRF3 for termination activity and that overexpression of eRF1 alone does not increase termination efficiency. In order to increase release factor competition at stop codons in yeast, it is necessary to overexpress both eRF1 and eRF3 (Stansfield et al. 1995; Salas-Marco and Bedwell 2004). The requirement for a hetero-dimeric termination complex to recognize stop codons thus provides a natural protection against the deleterious effects of individually overexpressing either one release factor.
In contrast to bacterial release factors, and as far as is currently known, eRF1 does not seem to play a role in starvation responses and emergency translational abandonment at stalled ribosomes. Instead, in eukaryotes this responsibility seems to have been devolved away from the eRF1 termination factor, to paralogs such as Dom34. In yeast, the eRF1-like Dom34 plays a role in releasing stalled ribosome complexes in the process known as “no-go” mRNA decay (Doma and Parker 2006). Taken as a whole, there is therefore no biological imperative to deploy an RF2-like negative feedback loop to prevent eRF1 overexpression.
The control principle of translational negative feedback loops
The negative feedback loops that govern IF3, RF2, and PABP can be considered from the perspective of control engineering with the aim of examining the control structure and properties of these pathways. In engineering, control systems are employed to operate processes robustly (in the face of disturbances) and maintain specific operating conditions. While the effects of particular disturbances can be anticipated and corrected for (feed-forward control), a more robust way of implementing control is through feedback: here corrective action is implemented based on knowledge of the state of the process (and its deviation from the prescribed value, the set point). Negative feedback is often used in control engineering to robustly attenuate the effect of disturbances. Typically the control action involves a negative action on the process, often dependent proportionally on the error (difference between the actual state of the process, as measured by sensors, and the set point), namely, a greater corrective action is taken given a greater difference between measured and set points. In more sophisticated control circuits, the negative corrective action is dependent upon the proportion to the integral; namely, the summed deviation from the set point over some time interval is considered when correcting using feedback. In the case of RF2 and IF3, the equivalent of a proportional feedback control is being used (there is no evidence of a “metabolic record” kept of cumulative deviation from the set point). Typically negative feedback has a system output stabilizing effect. However, it is also known that introducing a time delay in the implementation of negative feedback can introduce oscillatory behavior in the system, for instance in the synthetic “repressillator” circuit comprising a sequence of three elements, each repressing the next element in a cyclical configuration resulting in a negative feedback loop with delay (Elowitz and Leibler 2000). Since in the case of RF2 and IF3, both proteins will be almost immediately functional following their translation and protein folding, there is no intrinsic delay in the control circuit, and oscillatory behavior is not expected.
Both positive and negative feedback are seen at various levels in biological processes. Strong positive feedback can introduce bistabilty, namely, the property of a network to possess multiple stable steady states (Ferrell 2002). On the other hand, negative feedback can attenuate disturbances and noise. Using a synthetic circuit, it was demonstrated that the use of a transcriptional negative feedback loop conferred a clear effect of noise reduction, in comparison to ordinary unregulated gene transcription (Becskei and Serrano 2000). Since transcription of proteins expressed at low level can occur in bursts, with considerable stochastic variation, it is possible that the negative feedback loops discussed here have a noise attenuating function important for smoothing protein expression, and avoiding spikes in IF3 or RF2 production. In yeast, where the noisiness of gene expression has been studied using GFP-tagged proteins on a genome-wide scale, proteins involved in protein synthesis such as translation initiation factors and ribosomal proteins typically exhibited the least noisy expression profiles (Newman et al. 2006). However, the degree to which an autoregulatory translation control mechanism might reduce noise is far from clear, and indeed, some reports indicate that translational regulation may actually increase the noise component of a gene expression system (Komorowski et al. 2009).
The characterization of the RF2, IF3, and eRF1 translational feedback loops described here will undoubtedly throw more light onto the regulatory properties of such control circuits. Mathematical modeling has frequently been employed in this respect to dissect the complex nonlinear effects that govern feedback behavior, including when multiple feedback loops interact to generate nontrivial effects (Brandman et al. 2005). In fact, such interlocking, or nested, feedback is a feature of some unusual mutations isolated in the prfB gene encoding RF2 (Kawakami and Nakamura 1990). In this mutant, in addition to the natural UGA codon located at codon 26, an additional UGA stop codon was introduced by mutation at codon 144, generating a form of nested negative feedback. Such mutations therefore define an unusual case of double feedback that is nevertheless able to maintain viability, although the control properties of these alleles have yet to be investigated.
Modeling approaches are now being employed in our laboratories to examine the behavior of such feedback quantitatively from a dynamical systems and control engineering perspective. This approach will enable a quantitative examination of the termination process, its interplay with the competing tRNA population, and the role of cis factors such as stop codon context and frameshifting (de Silva et al. 2010).
SUMMARY
Overall, this review has described a diverse set of examples of negative feedback loops governing the translation of components of the translation apparatus itself. They represent a fascinating subject for research into the evolution and mechanism of control mechanisms and will in future undoubtedly shed more light onto the molecular functions of proteins such as IF3 and RF2 in canonical translation, their additional roles in mediating the stress responses of the ribosome population, and, finally, the control properties of finely tuned translational regulatory systems.
ACKNOWLEDGMENTS
Work in our laboratories is supported by BBSRC (BB/F019084/1, BB/G010722/1) and EPSCR (EP/E056644/1) (to I.S.), and EPSRC (EP/E057012/1) (to J.K.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1796210.
REFERENCES
- Adamski FM, Donly BC, Tate WP. Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 1993;21:5074–5078. doi: 10.1093/nar/21.22.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamski FM, McCaughan KK, Jorgensen F, Kurland CG, Tate WP. The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J Mol Biol. 1994;238:302–308. doi: 10.1006/jmbi.1994.1293. [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell. 2006;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Bag J, Wu J. Translational control of poly(A)-binding protein expression. Eur J Biochem. 1996;237:143–152. doi: 10.1111/j.1432-1033.1996.0143n.x. [DOI] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Boni IV, Artamonova VS, Tzareva NV, Dreyfus M. Noncanonical mechanism for translational control in bacteria: Synthesis of ribosomal protein S1. EMBO J. 2001;20:4222–4232. doi: 10.1093/emboj/20.15.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel C, Romby P, Sacerdot C, de Smit M, Graffe M, Dondon J, van Duin J, Ehresmann B, Ehresmann C, Springer M. Stabilized secondary structure at a ribosomal binding site enhances translational repression in E. coli. J Mol Biol. 1995;253:277–290. doi: 10.1006/jmbi.1995.0552. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Stansfield I. Halting a cellular production line: Responses to ribosomal pausing during translation. Biol Cell. 2007;99:475–487. doi: 10.1042/BC20070037. [DOI] [PubMed] [Google Scholar]
- Butler JS, Springer M, Dondon J, Graffe M, Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J Mol Biol. 1986;192:767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- Butler JS, Springer M, Grunberg-Manago M. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc Natl Acad Sci. 1987;84:4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabelskaya S, Gryzina V, Moskalenko S, Le Goff C, Zhouravleva G. Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae. BMC Mol Biol. 2007;8:71. doi: 10.1186/1471-2199-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Frolova L, Laugaa P, Kisselev L, Favre A. Stop codons and UGG promote efficient binding of the polypeptide release factor eRF1 to the ribosomal A site. J Mol Biol. 2003;331:745–758. doi: 10.1016/s0022-2836(03)00813-1. [DOI] [PubMed] [Google Scholar]
- Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen WJ, Cook RG, Tate WP, Caskey CT. Bacterial peptide chain release factors: Conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF, Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988;203:75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Dabeva MD, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J Biol Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- de Melo Neto OP, Standart N, Martins de Sa C. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 1995;23:2198–2205. doi: 10.1093/nar/23.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva E, Krishnan J, Betney R, Stansfield I. A mathematical modelling framework for elucidating the role of feedback control in translation termination. J Theor Biol. 2010 doi: 10.1016/j.jtbi.2010.01.015. (in press). [DOI] [PubMed] [Google Scholar]
- Doma M, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman I, Culbertson MR. Exceptional codon recognition by the glutamine tRNAs in Saccharomyces cerevisiae. EMBO J. 1991;10:1481–1491. doi: 10.1002/j.1460-2075.1991.tb07668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- Freedman LP, Zengel JM, Archer RH, Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: Genetic dissection of transcriptional and posttranscriptional regulation. Proc Natl Acad Sci. 1987;84:6516–6520. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Gold L, Stormo G, Saunders R. Escherichia coli translational initiation factor IF3: A unique case of translational regulation. Proc Natl Acad Sci. 1984;81:7061–7065. doi: 10.1073/pnas.81.22.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Pan D, Gualerzi CO, Cooperman BS. The translational fidelity function of IF3 during transition from the 30 S initiation complex to the 70 S initiation complex. J Mol Biol. 2007;373:551–561. doi: 10.1016/j.jmb.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty TJ, Lovett ST. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J Bacteriol. 1997;179:6705–6713. doi: 10.1128/jb.179.21.6705-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Dincbas V, Buckingham RH, Ehrenberg M. Origins of minigene-dependent growth inhibition in bacterial cells. EMBO J. 2000;19:2701–2709. doi: 10.1093/emboj/19.11.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Harel H, Levy G, Meyuhas O. Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett. 1999;457:209–213. doi: 10.1016/s0014-5793(99)01039-x. [DOI] [PubMed] [Google Scholar]
- Jorgensen F, Adamski FM, Tate WP, Kurland CG. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol. 1993;230:41–50. doi: 10.1006/jmbi.1993.1124. [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Heurgue-Hamard V, Buckingham RH, Ehrenberg M. Initiation factors IF1 and IF2 synergistically remove peptidyl-tRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J Mol Biol. 1998;281:241–252. doi: 10.1006/jmbi.1998.1953. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Nakamura Y. Autogenous suppression of an opal mutation in the gene encoding peptide chain release factor 2. Proc Natl Acad Sci. 1990;87:8432–8436. doi: 10.1073/pnas.87.21.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev D, Moskalenko S, Murina O, Baudin-Baillieu A, Rousset JP, Zhouravleva G. The paradox of viable sup45 STOP mutations: A necessary equilibrium between translational readthrough, activity and stability of the protein. Mol Genet Genomics. 2009;282:83–96. doi: 10.1007/s00438-009-0447-5. [DOI] [PubMed] [Google Scholar]
- Komorowski M, Miekisz J, Kierzek AM. Translational repression contributes greater noise to gene expression than transcriptional repression. Biophys J. 2009;96:372–384. doi: 10.1016/j.bpj.2008.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major LL, Poole ES, Dalphin ME, Mannering SA, Tate WP. Is the in-frame termination signal of the Escherichia coli release factor-2 frameshift site weakened by a particularly poor context? Nucleic Acids Res. 1996;24:2673–2678. doi: 10.1093/nar/24.14.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: The influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Sacerdot C, Graffe M, Blanquet S, Springer M. Discrimination by Escherichia coli initiation factor IF3 against initiation on noncanonical codons relies on complementarity rules. J Mol Biol. 1999;290:825–837. doi: 10.1006/jmbi.1999.2881. [DOI] [PubMed] [Google Scholar]
- Moskalenko SE, Chabelskaya SV, Inge-Vechtomov SG, Philippe M, Zhouravleva GA. Viable nonsense mutants for the essential gene SUP45 of Saccharomyces cerevisiae. BMC Mol Biol. 2003;4:2. doi: 10.1186/1471-2199-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Poole ES, Brown CM, Tate WP. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C, Dondon L, Grunberg-Manago M. Translational autocontrol of the Escherichia coli ribosomal protein S15. J Mol Biol. 1990;211:407–414. doi: 10.1016/0022-2836(90)90361-O. [DOI] [PubMed] [Google Scholar]
- Ringquist S, MacDonald M, Gibson T, Gold L. Nature of the ribosomal mRNA track: Analysis of ribosome-binding sites containing different sequences and secondary structures. Biochemistry. 1993;32:10254–10262. doi: 10.1021/bi00089a048. [DOI] [PubMed] [Google Scholar]
- Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NS, Das G, Seshadri A, Sangeetha R, Varshney U. Evidence for a role of initiation factor 3 in recycling of ribosomal complexes stalled on mRNAs in Escherichia coli. Nucleic Acids Res. 2005;33:5591–5601. doi: 10.1093/nar/gki864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Eurwilaichitr L, Akhmaloka, Tuite MF. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol Microbiol. 1996;20:1135–1143. doi: 10.1111/j.1365-2958.1996.tb02634.x. [DOI] [PubMed] [Google Scholar]
- Tedin K, Moll I, Grill S, Resch A, Graschopf A, Gualerzi CO, Bläsi U. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. A post-termination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]