Abstract
Competition between mammalian RNAi-related gene silencing pathways is well documented. It is therefore important to identify all classes of small RNAs to determine their relationship with RNAi and how they affect each other functionally. Here, we identify two types of 5′-phosphate, 3′-hydroxylated human tRNA-derived small RNAs (tsRNAs). tsRNAs differ from microRNAs in being essentially restricted to the cytoplasm and in associating with Argonaute proteins, but not MOV10. The first type belongs to a previously predicted Dicer-dependent class of small RNAs that we find can modestly down-regulate target genes in trans. The 5′ end of type II tsRNA was generated by RNaseZ cleavage downstream from a tRNA gene, while the 3′ end resulted from transcription termination by RNA polymerase III. Consistent with their preferential association with the nonslicing Argonautes 3 and 4, canonical gene silencing activity was not observed for type II tsRNAs. The addition, however, of an oligonucleotide that was sense to the reporter gene, but antisense to an overexpressed version of the type II tsRNA, triggered robust, >80% gene silencing. This correlated with the redirection of the thus reconstituted fully duplexed double-stranded RNA into Argonaute 2, whereas Argonautes 3 and 4 were skewed toward less structured small RNAs, particularly single-strand RNAs. We observed that the modulation of tsRNA levels had minor effects on the abundance of microRNAs, but more pronounced changes in the silencing activities of both microRNAs and siRNAs. These findings support that tsRNAs are involved in the global control of small RNA silencing through differential Argonaute association, suggesting that small RNA-mediated gene regulation may be even more finely regulated than previously realized.
Keywords: Argonaute, RNA interference, microRNAs
INTRODUCTION
RNAi-related small RNAs have emerged early during evolution, and have subsequently been adapted as guide RNAs in a wide range of genome and gene regulatory pathways (for review, see Zamore and Haley 2005; Ghildiyal and Zamore 2009). Different classes of small RNAs can be distinguished by their size and structure, biogenesis, and often coupled to this, function (for review, see Kim et al. 2009). Many of them are 21–22 nucleotides (nt) long 5′-phosphorylated, 2′–3′-hydroxylated small RNAs, a consequence of the Dicer processing of bimolecular or intramolecular hairpin double-stranded RNAs (dsRNAs). Size and end modifications are also consistent with how they are bound by Argonaute family proteins, which are at the core of small RNA effector complexes (Lingel et al. 2003; Wang et al. 2008). Some small RNAs, such as microRNAs in plants (Yu et al. 2005) and Piwi-associated RNAs (piRNAs) in mammals (Kirino and Mourelatos 2007), are further 2′-O-methylated at the 3′ end, which may stabilize them. Secondary siRNAs in RNAi of Caenorhabditis elegans are 5′-triphosphorylated (Pak and Fire 2007)—apparently, the result of short transcripts produced by an RNA-dependent RNA polymerase (RdRP)—and are loaded into specialized members of the Argonaute family (Aoki et al. 2007).
Small RNA populations can also be classified based on their biogenesis (for review, see Kim et al. 2009), particularly according to their requirement for processing by the RNase III-type endonucleases Drosha, as part of the Microprocessor complex together with DGCR8, and Dicer. This also allows for distinguishing between three major classes of small RNAs: microRNAs, siRNAs, and piRNAs. While in many organisms microRNAs are generated through the sequential cropping by Drosha and Dicer from precursors containing an imperfect hairpin RNA, Drosha is dispensable for the processing of siRNAs from typically long dsRNAs. piRNAs are generated by a pathway that is less well defined, but that is most likely Dicer and Drosha independent, and in the case of primary piRNAs may not involve a dsRNA intermediate at all. In vertebrates, a combination of deep sequencing and bioinformatics has brought to light a number of additional small RNA populations that either somewhat blur the boundaries between the main classes or may even constitute entirely separate classes of small RNAs (for review, see Kim et al. 2009). Mirtrons, for example, appear to be microRNAs in which pre-mRNA intron splicing substitutes for Drosha processing of pri-miRNAs (Berezikov et al. 2007). Similarly, Babiarz et al. (2008) deep sequenced small RNAs from mouse embryonic stem cells and found Drosha/Microprocessor-independent small RNAs that were apparently derived from precursor microRNA-like hairpins termed endo-shRNAs. The same study also identified tRNAs as a source of Microprocessor-independent, Dicer-dependent small RNAs in mice. While the bioinformatic evidence that these were bona fide RNAi-related small RNAs was strong and supported by RNA secondary structure predictions, a more-detailed molecular analysis is needed to better understand the structure, biogenesis, and potential activity of these novel small RNAs. It is also typical of these small RNA sequencing projects that many, often 10%–20% of the small RNAs sequenced, cannot be matched to the genome (e.g., Azuma-Mukai et al. 2008). This raises the possibility that new small RNA populations might have been missed.
Common to all these RNAi-related small RNA pathways is the central role of Argonaute proteins (for review, see Farazi et al. 2008; Hock and Meister 2008). Small RNAs get loaded into Argonautes and guide them to their target RNAs. In humans, there are eight Argonaute proteins: four of the AGO clade, which are ubiquitously expressed, and four of the PIWI clade, which are restricted to the germline and function in the piRNA pathway (for review, see Seto et al. 2007). While the reported Argonaute-associated microRNA profiles do not immediately suggest functional specialization of the four AGOs in mammals (Azuma-Mukai et al. 2008; Ender et al. 2008), Argonaute 2 knockout in mice is embryonic lethal (Liu et al. 2004). Further genetic support of nonredundancy of Argonaute 2 comes from the observation that Argonaute 2 knockout hematopoietic stem cells have defects in hematopoiesis (O'Carroll et al. 2007). Although the most obvious difference between Argonaute 2 and the other Argonautes is in its Slicer activity (Liu et al. 2004), the hematopoiesis phenotype could be rescued with a Slicer-deficient Argonaute 2 mutant, which correlated with restored pre-microRNA processing (O'Carroll et al. 2007). Only little is known about the functions of human Argonautes 1, 3, and 4. Argonaute 1 has been described to be involved in transcriptional gene silencing in humans (Janowski et al. 2006; Kim et al. 2006), and the siRNA knockdown of Argonaute 4 impaired HDV replication (Haussecker et al. 2008).
Small RNA sorting into the different Argonautes in mammals could be due to coupling of small RNA biogenesis with loading and/or preferences of the Argonaute loading complexes for the particular structures of small RNAs or their precursors. Examples for the latter are found in plants where some Argonautes have preferences for certain 5′ bases over others (Mi et al. 2008), in C. elegans where one class of Argonautes specializes in binding triphosphorylated RNAs (Aoki et al. 2007), and in D. melanogaster where the fate of the small RNA is determined by the perfect or imperfect double strandedness of the precursor Dicer substrate (Tomari et al. 2007). It will be of interest to determine whether there are additional small RNA populations that are sorted according to and tightly coupled to their biogenesis. This may be especially likely for single-strand RNA-derived small RNAs that lack pronounced precursor RNA secondary structures.
Considering these complex relationships between small RNAs and the RNAi silencing apparatus, it is important to identify and characterize all RNAi-related small RNAs. In addition to potentially uncovering new classes of small RNAs and their functions, novel modes of gene regulation based on the functional interaction between classes of small RNAs are also of interest.
The existence of such functional interactions in humans is suggested by the demonstrated limited RNAi silencing capacity of mammalian cells, which is subject to a number of autoregulatory feedback regulations (Forman et al. 2008; Han et al. 2009). As a result, RNAi-related competition has been observed between experimentally introduced si/shRNAs and endogenous microRNAs (Grimm et al. 2006), and also between Argonautes for a given small RNA and/or their targets (Diederichs et al. 2008). Based on our studies, we provide a model for how competition between classes of small RNAs may account for the global control of microRNAs, as have been observed in cancer cells (Lu et al. 2005) and in response to changes in cell densities (Hwang et al. 2009).
RESULTS
Small RNA screen uncovers 5′-phosphate, 3′-hydroxyl tRNA-derived small RNAs
In a previous study, we reported on the discovery of two HDV small RNAs, 20–25 nt in length and with mRNA-like cap structures (Haussecker et al. 2008). While the antigenomic small RNA was only seen by 5′-phosphate-dependent semideep sequencing in an RNA preparation enriched for 5′-capped RNAs consistent with biochemical analyses, a corresponding small RNA of genomic polarity was found with and without prior 5′-cap enrichment. Based on the hypothesis that this might be a reflection of the function of this particular class of small RNAs, we set out to discover cellular counterparts of the HDV small RNAs through a Northern blot screen with probes targeting particularly those sequences that occurred in both the 5′-cap-enriched and nonenriched samples (Fig. 1A; Supplemental Table 1). Further selection criteria were a high sequencing frequency to facilitate their subsequent analysis, and not being annotated as obviously deriving from known and abundant noncoding RNAs. The selected candidates included a number of sequences without a perfect match to the genome (nuclear or mitochondrial), as has also been observed, but largely excluded from further analysis in previous small RNA sequencing studies (e.g., Azuma-Mukai et al. 2008).
FIGURE 1.
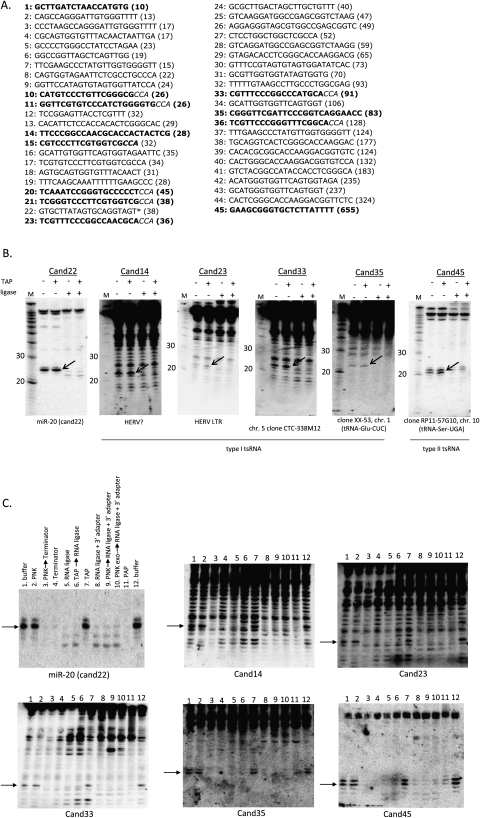
Small RNA Northern blot screen reveals a population of tRNA-derived 21–22-nt small RNAs that are 5′-phosphorylated and 3′-hydroxylated. (A) Northern blot screen candidate sequences. T4 RNA ligase-sensitive small RNAs in bold, except for known microRNA miR-20/cand22, which is indicated by an asterisk (*); tRNA 3′ “CCA” motif in italics; number of sequence hits in parentheses (out of 8554). (B) Northern blot screen examples of ∼21–22-nt T4 RNA ligase-sensitive small RNAs (293 cell RNA). Ligase-sensitive small RNAs evidenced by either disappearance and/or band shift (arrows). Cand22: 5′-phosphorylated, 3′-hydroxylated miR-20 (positive control); Cand14, Cand23, Cand33, and Cand35: type I tsRNA examples; Cand45: type II tsRNA. Initial genomic annotation of the small RNAs shown below the blots; subsequent manual blast revealed Cand35 and Cand45 to be derived from predicted tRNAs (in parentheses); still no perfect match could be identified for Cand14, Cand23, and Cand33. TAP: tobacco acid pyrophosphatase; ligase: T4 RNA ligase; −: untreated; +: treated; M: Decade (Ambion) RNA size marker. (C) tsRNAs are 5′-phosphorylated and 3′-hydroxylated (Northern blot of diagnostic enzyme treatments). Two hundred ninety-three cell RNA was treated with the following enzymes (potential activities described in parentheses), and enzyme susceptibility of the 21–22-nt small RNAs of interest deduced by their shift in gel mobility and/or disappearance in the Northern blot: (1) buffer; (2) T4 polynucleotide kinase (PNK) +ATP (5′ phosphorylation of 5′-OH and 3′ dephosphorylation); (3) T4 PNK, then Terminator (degrades 5′ monophosphorylated, unstructured RNAs; PNK-dependent RNA removal would indicate 5′-OH RNAs); (4) Terminator; (5) T4 RNA ligase +ATP (for 5′P-3′OH RNAs: intramolecular circularization; trans-ligation of RNAs containing either of these modifications); (6) TAP (hydrolyzes phosphoric acid anhydride bonds in triphosphorylated and capped RNAs, leaving 5′ monophosphate), then T4 RNA ligase (TAP-dependent T4 RNA ligation would indicate 5′ cap or 5′ triphosphate); (7) TAP; (8) T4 RNA ligase, no ATP + activated 3′-adapter oligo (adapter ligation would indicate 3′OH); (9) T4 PNK, then T4 RNA ligase, no ATP + activated 3′-adapter oligo (PNK-dependent adapter ligation would indicate either 3′-P, or 2′–3′ cyclic phosphate); (10) 3′-phosphatase-negative T4 PNK, then T4 RNA ligase, no ATP + activated 3′-adapter oligo (would confirm that a reaction in treatment “9” was dependent on 3′ dephosphorylation by T4 PNK); (11) polyA polymerase (PAP; adds polyA to 3′-hydroxyl RNAs); (12) buffer (same as 1). Blots were stripped and rehybridized with the indicated probes. Arrows indicate 21–22-nt RNAs of interest; HERV: human endogenous retroviral element.
To screen for novel small RNAs based on their end modifications, RNA from the human embryonic kidney cell line HEK 293 was treated with the decapping enzyme Tobacco Acid Phosphatase (TAP) and/or T4 RNA ligase. While TAP removes 5′ caps, T4 RNA ligase can circularize 5′-phosphorylated, 3′-hydroxylated RNAs or ligate RNAs that contain either of these end structures to each other in trans. RNAs that have participated in these reactions are marked by either a shift in their gel mobility or their disappearance. In 44 of the 45 probes used in the screen, including those directed at sequences for which no perfect genomic match had been initially identified, RNAs with an apparent size of 70–150 nt were readily detected (Supplemental Fig. 1). We note that probes with random sequences do not recognize such RNAs under the experimental conditions applied (data not shown). Based on their size and apparent abundance, we expected that the detected RNAs might include noncoding RNAs, such as members of the tRNA, snoRNA, and snRNA families. We speculate that the lack of perfect matches to the human genome for many of the sequences could be due to post-transcriptional modifications (well characterized for numerous noncoding RNAs) and consequent nucleotide changes as a result of misincorporations during the reverse transcription step of cDNA library preparation (Kawaji et al. 2008). A recent analysis of apparent RNA sequencing errors strongly supports this notion (Ebhardt et al. 2009).
None of the probes detected 20–25-nt small RNAs that shifted upon TAP treatment, indicating a lack of prominent capped small RNAs and that sequencing alone is not sufficient to conclusively deduce end modifications. Instead, we noticed a number of T4 RNA ligase-sensitive RNAs (Fig. 1A, the 12 candidates highlighted in bold; Supplemental Fig. 1, highlighted in red) that, strikingly, were in the 20–22-nt size range, the typical length of small silencing RNAs. Many of the same probes also detected a number of larger and smaller RNAs around the 20–22-nt size range, which, however, were largely insensitive to T4 RNA ligase treatment (Fig. 1B, for salient examples, see cand14, cand33) and varied in intensity from experiment to experiment, consistent with these being T4 RNA ligase-insensitive degradation products. The T4 RNA ligase-sensitive small RNA detected with the probe directed at candidate 45 (cand45) appeared to be distinct from the other T4 RNA ligase-sensitive small RNAs in that its intensity was comparable to that of its larger, ∼110-nt counterpart (Fig. 1B). Further enzymatic analysis confirmed that these small RNAs, in notable contrast to the T4 RNA ligase-insensitive RNAs, were indeed 5′-phosphorylated and 3′-hydroxylated (Fig. 1C). Accordingly, similar to a control 5′-phosphorylated, 3′-hydroxylated microRNA (miR-20/cand22), treatment of all five accordingly investigated candidates (cand14, cand23, cand33, cand35, cand45) with the Terminator nuclease (Fig. 1C, lanes 4), an exonuclease that degrades (unstructured) 5′-phosphorylated RNAs, selectively removed the T4 RNA ligase-sensitive RNAs; 3′-adapter ligation with an activated 3′-adapter and in the absence of ATP led to the disappearance of the T4 RNA ligase-sensitive small RNAs (Fig. 1C, lanes 8), as did treatment with polyA polymerase (Fig. 1C, lanes 11), both of which are indicative of 3′-hydroxyl ends.
Closer inspection of the T4 RNA ligase-sensitive sequences (Fig. 1A, in bold) strongly suggested that most, if not all of these, were indeed tRNA-derived. First, eight out of the 12 T4 RNA ligase-sensitive small RNA sequences contained a tRNA-like “CCA” motif at their 3′ ends. Some of the reported Dicer-dependent tsRNAs had similarly been reported to be CCA-ylated at their 3′ ends (Babiarz et al. 2008). Second, when blasted manually, perfect genomic matches could now be identified for candidates 11, 15, and 35 and were found to be derived from predicted tRNAs. Cand22 was found to correspond to miR-20 and served as a positive control in subsequent analyses. Moreover, in the absence of a perfect genomic match, candidates 14 and 23 had initially been tentatively annotated as being derived from human endogenous retroviral elements (HERV). This is reminiscent of the human tRNA-derived RNAs reported by Kawaji and colleagues, some of which appeared to be misannotated as endogenous retroviral elements due to the role that tRNAs play in the replicative priming of retroviruses and nucleotide misincorporations opposite of modified bases during the reverse transcription step of cDNA library preparation (Kawaji et al. 2008).
Two types of tRNA-derived small RNAs
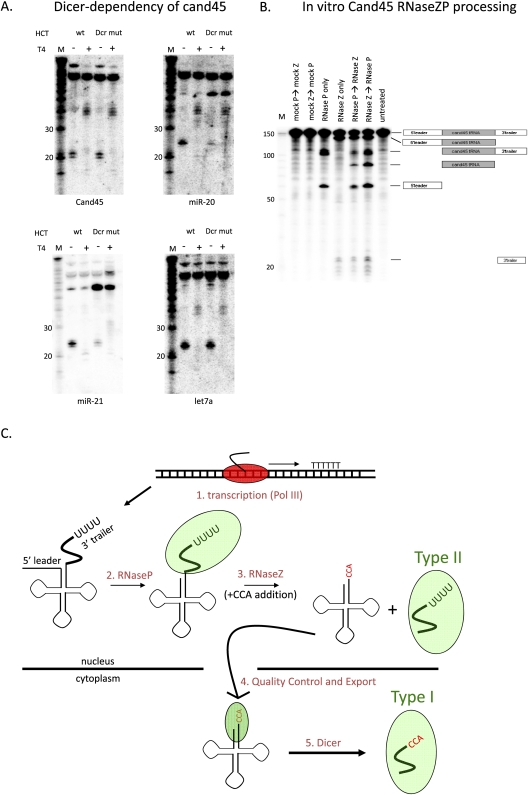
The absence of RNAs detected with probes directed toward the antisense strand of the T4 RNA ligase-sensitive small RNAs (data not shown) is consistent with the previous suggestion (Babiarz et al. 2008) that imperfectly base-paired tRNA stem structures, rather than paired sense-antisense transcripts, were the substrates for Dicer processing. We note that while Dicer-dependent tsRNAs had been identified in mice, based on sequencing and bioinformatic prediction, their direct detection by alternative methods, as well as further structural and functional analyses had been lacking. Manual BLAST analysis of candidate 45 mapped its 5′ end to directly downstream from the discriminator base of a predicted serine-tRNA, and ending in a short stretch of uracils. This immediately suggested a Dicer-independent mode of biogenesis in which the 5′ and 3′ ends are determined by the tRNA processing enzyme RNaseZ, an endonuclease leaving a 3′-hydroxyl and 5′-phosphate at the cleavage site (Mayer et al. 2000), and transcription termination by RNA polymerase III, respectively. Accordingly, we failed to detect a decrease in cand45 abundance either when 293 cells were treated with Dicer siRNAs (although insufficient Dicer knockdown could not be ruled out) (data not shown), or in a HCT116-derived human colorectal cancer cell line in which the Dicer helicase domain had been mutated, leading to a decrease in most, albeit not all, microRNAs (Fig. 2A; Cummins et al. 2006). In contrast, treatment of an in vitro transcribed cand45 tRNA precursor with recombinant RNaseZ yielded the predicted 3′ trailer small RNA (Fig. 2B). Interestingly, at least in this assay, the efficiency of 3′ processing was independent of whether the precursor tRNA had been pretreated with RNaseP or not. RNaseP is the enzyme that cleaves off the 5′ leaders of precursor tRNAs and may be required for the functioning of RNaseZ for at least some tRNAs (Nashimoto et al. 1999; Dubrovsky et al. 2004). We will refer to this type of tsRNA as type II tsRNA, as opposed to the Dicer-dependent type I tsRNAs (see the model illustrated in Fig. 2C).
FIGURE 2.
RNaseZ-dependent, Dicer-independent cand45 biogenesis. (A) Cand45 expression is unchanged in a HCT116-derived cell line that contains a mutation in the Dicer helicase domain. Most (e.g., miR-20 and miR-21), but not all (e.g., let-7a) microRNAs are down-regulated in this cell line. wt: Parental HCT Dicer wild-type cell line; Dcr mut: HCT-derived Dicer helicase mutant cell line; T4: test for T4 RNA ligase sensitivity (−: untreated; +: treated). (B) In vitro RNaseZ/P processing of cand45 tRNA. A radioactively labeled, in vitro transcribed cand45 precursor tRNA was treated with buffer alone (“mock”), recombinant human RNaseZ and/or purified human RNaseP. Arrows indicate that RNA was treated sequentially with stated conditions. Reaction products (schematic for predicted fragments shown on the right) were visualized on a polyacrylamide gel. M: Decade (Ambion) RNA size marker. (C) Model for tsRNA biogenesis: RNA polymerase III (Pol III) generates a precursor tRNA (1). The 5′ leader and 3′ trailers are removed by RNaseP (2) and Z (3), respectively. The mature tRNA is then exported into the cytoplasm (4). There, Dicer recognizes some, potentially misfolded tRNAs to produce Type I tsRNAs (5). The small RNA produced by nuclear RNaseZ cleavage and Pol III termination is a Type II tsRNA. Based on the near-exclusive cytoplasmic localization of type II tsRNAs, it is possible that a cytoplasmic pool of RNaseZ is responsible for the processing into type II tsRNAs of immature tRNAs have evaded nuclear quality control (data not shown).
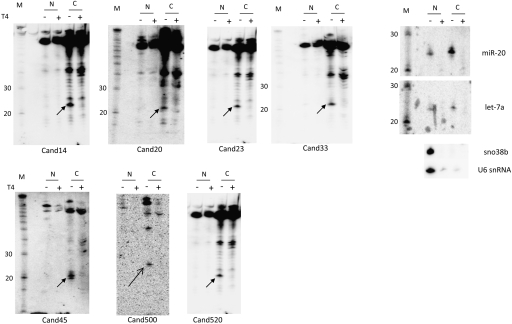
Removal of the 3′ trailers of nuclear encoded pre-tRNAs by RNaseZ is thought to take place in the nucleus (Lund and Dahlberg 1998). It is therefore notable that following nuclear-cytoplasmic fractionation, essentially all detectable cand45 tsRNA was recovered in the cytoplasmic fraction (Fig. 3). We confirmed our fractionation results by examining the distribution of known RNA markers. In particular, both positive controls for nuclear RNAs tested, snoRNA 38b and U6 snRNA, were almost entirely restricted to the nuclear fraction. This suggests that cand45 is either rapidly exported following RNaseZ cleavage or that a population of cytosolic RNaseZ that has been described to function as the effector endonuclease in a new type of gene silencing guided by 5′ half-tRNAs (Elbarbary et al. 2009), may be responsible for RNaseZ-dependent cand45 biogenesis in the cytoplasm. Like cand45, the four type I tsRNAs that we investigated (cand14, cand20, cand23, cand33), which could be identified by size and T4 RNA ligase-sensitivity were each almost exclusively detected in the cytoplasmic fraction and is consistent with cleavage by cytoplasmic Dicer. Of note, while tsRNAs were absent from the nuclear fraction, a portion of microRNAs was always detected in the nucleus (Fig. 3, miR-20, let-7a), albeit at generally lesser intensity than their cytoplasmic counterparts. This is consistent with other observations that microRNAs can be readily detected in the nuclei of mammalian cells (Hwang et al. 2007). More importantly, however, the distinct fractionation patterns of microRNAs and tsRNAs indicate that their intracellular distributions differ.
FIGURE 3.
tsRNAs localize to the cytoplasm (Northern blot analysis of nuclear-cytoplasmic RNA fractionation). Sno38b and U6 snRNA serve as nuclear markers. Equal amounts of nuclear and cytoplasmic RNA were loaded; blots were stripped and rehybridized. N: nuclear RNA fraction; C: cytoplasmic RNA fraction; arrow: T4 RNA ligase (T4)-sensitive RNA of interest (−: untreated; +: treated); M: Decade (Ambion) RNA size marker.
tRNA-derived small RNAs have relative preference for Argonaute 3–4 association
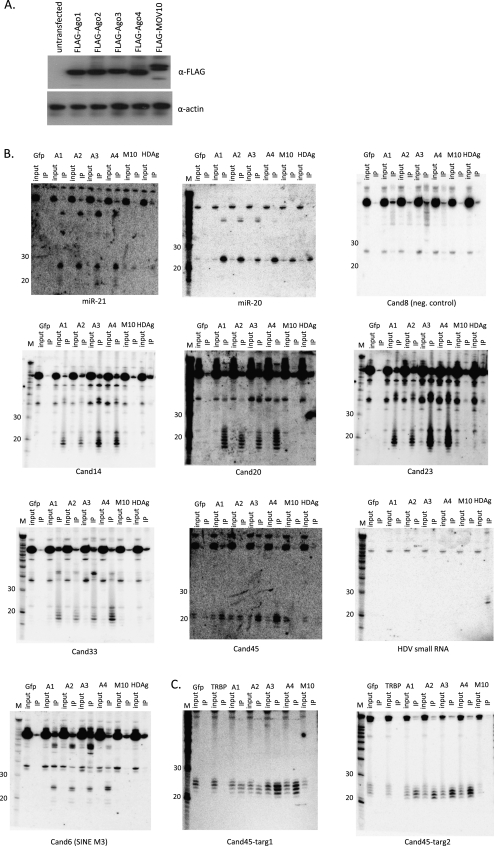
The interpretation of tsRNAs forming a distinct population of small RNAs was also consistent with our analysis of the interactions of tsRNAs and microRNAs with the small RNA effector proteins Argonautes 1–4 and the microRNA factor Mov10. To facilitate this analysis, FLAG-tagged versions of the various proteins were expressed with comparable efficiencies in 293 cells (Fig. 4A) so that associated small RNAs could be immunoprecipitated with the same monoclonal FLAG antibody across all cell lysates. The use of FLAG-tagged Argonautes in elucidating bona fide Argonaute function and small RNA association has been well validated in previous studies. Accordingly, such epitope-tagged Argonautes cofractionate with their endogenously expressed counterparts (Hock et al. 2007), no obvious changes in the small RNA profiles were observed following transient overexpression of a FLAG-tagged Argonaute 2 (Zhang et al. 2009), and no gross differences were noted in small RNA immunoprecipitations with antibodies against endogenous Argonautes when compared with earlier studies immunoprecipitating FLAG-tagged Argonautes (Azuma-Mukai et al. 2008; Ender et al. 2008). On the other hand, overexpression experiments with similar Argonaute constructs have been shown to result in competition with endogenously expressed Argonaute function (Diederichs et al. 2008). A conservative interpretation of such Argonaute co-IP experiments would be, therefore, that they illustrate the relative abilities of Argonautes to load various small RNAs under conditions when they are not limiting. FLAG-Gfp and cand8 served as negative controls for nonspecific FLAG-protein interactions and for non-T4 RNA ligase-sensitive small RNAs (i.e., small RNAs presumably unrelated to RNAi), respectively.
FIGURE 4.
tsRNA Argonaute coimmunoprecipitations. (A) FLAG-Argonautes and FLAG-MOV10 were expressed at similar levels in 293 cells (Western blot); actin was used as a loading control. (B,C) Northern blot analysis of RNA coimmunoprecipitations with FLAG epitope-tagged Gfp (negative control), human Argonautes 1–4 (A1–A4), Mov10 (M10), and either HDAg (B), or TRBP (C). Blots were stripped and rehybridized. (B) tsRNAs associate with human Argonautes 1–4, but not MOV10. (C) Small RNAs that are generated from a cand45 tRNA expression system in which cand45 had been replaced with the (arbitrary) sequences “targ1” and “targ2” preferentially associate with FLAG-Argonaute 3 and 4. Input: RNA isolated from 10% lysate used per immunoprecipitation; IP: immunoprecipitated RNA; M: Decade (Ambion) RNA size marker.
All type I tsRNAs tested (cand14, cand20, cand23, cand33) were readily immunoprecipitated with FLAG-Argonautes 1–4, but could not be detected following FLAG-Mov10 immunoprecipitation (Fig. 4B; Supplemental Fig. 2). This is in contrast to the investigated microRNAs, miR-20, miR-21, and let-7a, which coimmunoprecipitated also with Mov10 (Fig. 4B; Supplemental Fig. 2), albeit to a lesser extent than with the Argonautes. Interestingly, only in the case of type I tsRNAs, there were, in addition to the 21–22-nt species identified in the original screen smaller, T4 RNA ligase-sensitive (data not shown), 18–20-nt RNAs detected with the same oligonucleotide probes that were even more efficiently enriched in the Argonaute immunoprecipitates. The type II tsRNA cand45 similarly coimmunoprecipitated with all Argonautes and could not be detected following Mov10 immunoprecipitation. In this case, however, no <20-nt species was enriched in the immunoprecipitates. Moreover, unlike microRNAs, which appeared to coimmunoprecipitate equally well with all the Argonautes, cand45 associated more efficiently with Argonautes 3 and 4 than with Argonautes 1 and 2 (Fig. 4B; Supplemental Fig. 2). The same Argonaute 3–4 over Argonaute 1–2 preference was observed when the cand45 sequence downstream from the predicted RNaseZ site was replaced with two arbitrary sequences and expressed in 293 cells (Fig. 4C, Cand45-targ1, Cand45-targ2). This suggests that the particular pathway/biogenesis, not the sequence per se determines the Argonaute association pattern (see Supplemental Fig. 3 for a graphical representation of the FLAG-protein association patterns). We also tested tsRNA association with a FLAG-tagged version of the hepatitis delta virus antigen (HDAg) which we had reported to be associated with both Mov10 and HDV small RNAs (Haussecker et al. 2008). Only very little tsRNAs were immunoprecipitated with FLAG-HDAg (<5% of Ago4 immunoprecipitation) suggesting that the tsRNA-associated Argonaute pool was also distinct from the HDV-related small RNA pathway. The FLAG-HDAg coimmunoprecipitation of microRNAs 20 and 21 could reflect its association with Mov10. We also note that small RNAs of ∼22 nt corresponding to cand6, which detects a SINE-derived RNA, were significantly enriched by Argonaute immunoprecipitation. Repetitive elements are a known source of endo-siRNAs in mammalian cells (Tam et al. 2008; Watanabe et al. 2008), and may reflect yet another population of small RNAs associated with Argonautes, but not Mov10. In summary, in addition to the nuclear-cytoplasmic fractionation patterns, tsRNAs are further differentiated from microRNAs by their apparent lack of Mov10 association. Moreover, while our overexpression studies may not quantitatively recapitulate the normal distribution patterns of tsRNAs between the Argonautes, they at the very least demonstrate a relative propensity of the type II tsRNA pathway for Argonaute 3–4 utilization.
Abundance and size of type II tsRNA-like small RNAs are sensitive to Argonaute dosage
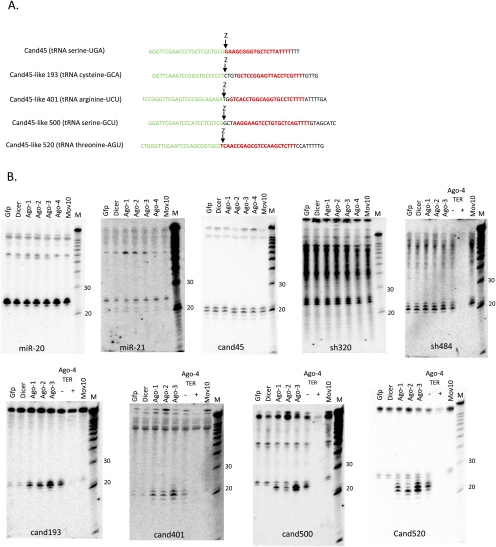
To search for additional cand45-like type II tsRNAs that may have been generated via RNaseZ cleavage and RNA polymerase III termination, we examined four candidates in our small RNA database (cand193, cand401, cand500, and cand520) that met the criteria of mapping to the 3′ ends of predicted tRNAs and ending in a stretch of uracils (Fig. 5A). However, unlike cand45, none of the 5′ ends for these candidates coincided precisely with the predicted major RNaseZ cleavage sites, with those of cand193, cand401, and cand500 mapping slightly upstream of, and that of cand520 downstream from, the RNaseZ site. Northern blot analysis confirmed the existence of corresponding small RNAs (Fig. 5B). Cand500 and cand520 were identified as Terminator-sensitive and therefore 5′-phosphorylated small RNAs of 24–28 nt, while cand193 and cand401 yielded 5′-phosphorylated 20–21-nt RNAs. Like type I and II tsRNAs, cand45-like small RNAs were predominantly localized to the cytoplasm (see Fig. 3 for examples of cand500, cand520). As RNAi knockdown of RNAi components to levels that affected the steady-state abundance of microRNAs proved challenging in 293 cells (data not shown), we sought to investigate the relationship between RNAi and tsRNA pathways by testing the effect of overexpressing Dicer, Ago1–Ago4, and Mov10 on small RNA abundance. Interestingly, all four cand45-like small RNAs were affected when overexpressing Argonautes 1–4 (Fig. 5B). Accordingly, Argonaute overexpression robustly increased the abundances of the 20–21-nt long cand193 and cand401 RNAs, by ∼20-fold in the case of Argonaute 3 overexpression, which showed the strongest effect. In contrast, while the abundances of the 24–28-nt cand500 and cand520 RNAs were essentially unchanged, Argonaute overexpression triggered the appearance of 21–23-nt RNAs that were readily detected by the same probes and were of higher abundance than the uninduced longer counterparts. One possible explanation may be that the proximity of Argonautes with tRNA 3′ processing allowed Argonautes, acting as molecular rulers (Wang et al. 2008), to selectively capture and stabilize the tsRNAs. This is consistent with the observation that although both the induced shorter and noninduced longer cand45-like small RNAs coimmunoprecipitated with Argonautes (but not Mov10), the process was more efficient for the induced species (Supplemental Fig. 2, cand520, arrow b; data not shown). Alternatively, the induced species may be the result of a catalytic activity of Argonautes upstream of target cleavage in the RNAi pathway, as has been suggested for Argonaute 2 (Diederichs and Haber 2007; O'Carroll et al. 2007). Importantly, the abundance of cand45 itself was not affected by Argonaute overexpressions, as was the case for microRNAs 20 and 21 and the bioinformatically predicted endo-shRNAs 320 and 484 (Babiarz et al. 2008), arguing that these are already efficiently incorporated into Argonautes at physiological expression levels (for a graphical representation of the relative changes in small RNA abundances following RNAi-related protein overexpression see Supplemental Fig. 4). It will be interesting to test whether the differences in Argonaute responsiveness is determined by whether or not the 5′ end of the tRNA trailer-derived small RNAs precisely coincides with the predicted major RNaseZ cleavage site.
FIGURE 5.
Investigation of Cand45-like small RNAs. (A) Selected Cand45-like candidate sequences (in red) with predicted RNaseZ cleavage sites (“Z”). Shown are the sequences of the tRNA 3′ ends and the RNA polymerase III termination region. (B) Cand45-like small RNA candidate expression analysis (Northern blot). Cand45-like small RNAs can be detected as discrete 21–28-nt small RNAs and are modulated by Argonaute overexpression. Gfp (negative control), Dicer, Ago1–Ago4, Mov10: transfected expression plasmids. Terminator treatment (“TER”; −: untreated; +: treated) was used to determine the 5′-phosphorylation status of cand45-like small RNA candidates. In these blots, the amount of “+/−” Terminator-treated Argonaute 4-associated RNA loaded was half that of the other samples. M: Decade (Ambion) RNA size marker.
Differential trans-silencing capacity of type I and type II tsRNAs
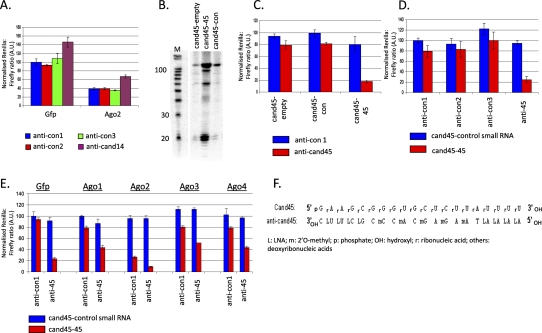
The post-transcriptional trans-silencing capacity of tsRNAs was tested using standard reporter assays (Fig. 6). Specifically, target sites complementary to the type I and II tsRNA cand14 and cand45, respectively, were inserted into the 3′ UTR of a Renilla luciferase reporter gene. These constructs were then transfected into tissue culture cells expressing the tsRNAs. Renilla luciferase activity was normalized for the luminescence of a firefly luciferase reporter gene on the same plasmid, as well as for the Renilla-Firefly ratio of a second plasmid that lacked a tsRNA target site at the corresponding position of the Renilla 3′ UTR. Modest cand14 trans-silencing capacity was deduced based on the ∼30%–40% increase in Renilla activity when cells were treated with a cand14 antisense oligonucleotide, compared to all four negative control (antisense) oligonucleotides tested (P < 0.01) (Fig. 6A). It is possible that the trans-silencing capacity of cand14 is inherently modest, perhaps reflecting preferential Ago3–Ago4 over Ago1–Ago2 interaction (Fig. 4B). It is, however, also possible that the extent of the silencing was limited either due to the specific context of the reporter or as the target site might not have been entirely complementary to cand14, based on the absence of a perfect match in the genome for cand14. Similarly, since simple antisense inhibition of small RNAs, however, may not necessarily be the most efficient way of antagonizing their activities (see Fig. 8B, below, for an example of let-7a), we cannot be certain that the antisense-dependent up-regulation reflects the full degree of cand14-mediated trans silencing. In summary, cand14 exhibits canonical microRNA-/siRNA-like trans-silencing capacity.
FIGURE 6.
Trans-silencing capacity of tsRNAs (dual luciferase assay). (A) Cand14-mediated trans-silencing. Dual luciferase assay with the Renilla luciferase reporter gene carrying a fully cand14-complementary target site in its 3′ UTR (psi-cand14). Addition of a cand14 antisense molecule increased Renilla luciferase expression, as expected, if cand14 had RNAi-like trans-silencing capacity. Likewise, overexpression of Ago2 enhanced cand14-mediated trans-silencing. The specificity of the de-repression with anti-cand14 was confirmed with three (antisense) control oligonucleotides (anti-con1, anti-con2, anti-con3). (B) Cand45 overexpression from a plasmid into which the genomic sequence of cand45 had been cloned (cand45-45; by Northern blot). Cand45-empty: cand45 cloning plasmid with only a cloning site between the cand45 RNaseZ cleavage site and the RNA PolIII terminator; cand45-con: cand45-derived expression plasmid in which the cand45 sequence in cand45-45 was replaced with an arbitrary control sequence; M: Decade (Ambion) RNA size marker. (C) Cand45 overexpression (cand45-45)-dependent, anti-cand45 oligo-induced trans-silencing in HCT116 cells. Dual luciferase assay with reporter gene carrying a fully cand45-complementary target site in the Renilla luciferase 3′ UTR (psi-cand45). Cand45-empty, cand45-con, cand45-45 as in C. (D) Confirmation of the specificity of the anti-cand45 induced trans-silencing effect in panel D by the use of three additional (antisense) control oligonucleotides (anti-con1, anti-con2, anti-con3). (E) Anti-cand45 induced trans-silencing is RNAi-related. Cand45 overexpression (cand45-45)-dependent, anti-cand45 (“anti-45”)-induced trans-silencing is enhanced by overexpression of slicing-competent Argonaute 2, but mitigated by overexpression of the nonslicing Argonautes 1, 3, and 4. (F) Predicted cand45:anti-cand45 duplex; A.U.: arbitrary units; error bars indicate standard deviation from n = 3 transfections.
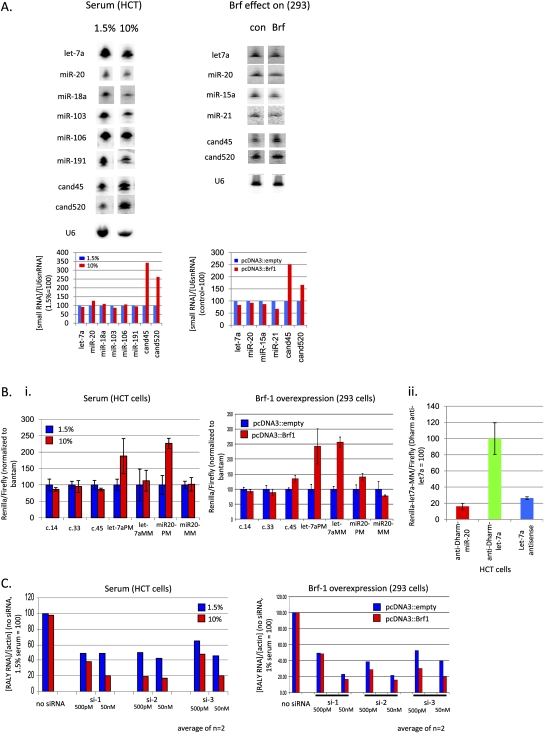
FIGURE 8.
Increased tsRNA abundance correlates with reduction and increase in microRNA and siRNA efficacies, respectively. (A) tsRNA abundance can be modulated by varying serum concentrations or overexpressing the tRNA transcription factor Brf1 (Northern blot; corresponding U6snRNA-normalized phosphorimage quantitations shown below). Blots were stripped and rehybridized. Total RNA from 293 cells was harvested on day 4 after Brf1 transfections on days 0 and 2. For the serum experiments, HCT116 cells cultured for 5 d under 1.5% or 10% serum were chosen. (B, i) MicroRNA silencing capacity is reduced in the presence of increased tsRNA abundance (dual luciferase assay). The target sites of the Renilla luciferase reporters are indicated with “c.14” (cand14) on the X-axis; PM/MM: perfect match/translational reporters. Results are normalized to the Renilla/Firefly ratios of reporter plasmid with no predicted small RNA target site (“bantam”), with 1.5% serum and control pcDNA3∷empty set at 100 for each reporter. (B, ii) Silencing efficiency of translational psi-let-7aMM reporter as indicated by let-7a inhibition (dual luciferase assay). anti-Dharm miR-20: control microRNA hairpin inhibitor; anti-Dharm let7a: let7a hairpin inhibitor; let-7a antisense inhibitor. “anti-Dharm let-7a” where most apparent let-7a inhibition was observed was set = 100. (C) SiRNA silencing is improved in the presence of increased tsRNA abundance (real-time qRT-PCR). Three different siRNAs (si-1–si-3) targeting endogenously expressed RALY RNA were transfected at two concentrations, 500 pM and 50 nM and remaining RALY RNA levels normalized to actin measured.
In contrast to the type I tsRNA cand14, cand45 did not exhibit apparent trans-silencing activity (Fig. 6C). Even when Argonautes were overexpressed (Fig. 6E), there was no strong cand45 antisense-reversible trans-silencing, although Argonaute 2 overexpression caused a small 20%–30% reduction in reporter gene activity in some data sets, which was reversed by an oligonucleotide antisense to cand45 (e.g., Supplemental Fig. 5). We reasoned that by overexpressing cand45 we would be able to achieve cand45-dependent gene silencing. For this, a cloned version of the cand45 tRNA was transfected into HCT116 cells and cand45 overexpression confirmed by Northern blot (Fig. 6B). Nevertheless, the cotransfected Renilla luciferase reporter was still not silenced (Fig. 6C). Unexpectedly, when a cand45 antisense oligonucleotide was added (100 nM), originally intended to relieve any cand45-mediated trans-silencing, there was a robust induction of cand45-Renilla luciferase silencing that was further responsive to the number of target sites in the reporter (an over 80% decrease with two target sites, and a 65% decrease with one target site; Supplemental Fig. 6). The gene knockdown did not depend on the exact antisense modification chemistry as a fully 2′-O-methylated oligonucleotide worked at least as efficiently as the original chemistry which contained unmodified as well as a mix of LNA and 2′-O-methyl modified bases (Fig. 6F; Supplemental Fig. 7). In the case of the fully 2′-O-methylated cand45 antisense, a slight, ∼25%, reduction in target reporter gene activity was already observed in the absence of cand45 overexpression. Similar results, including the observation that sense DNA oligos do not induce such silencing, were obtained with mouse embryonic fibroblast cells, thereby excluding this to be a human-specific phenomenon or a peculiarity of the 293 cell line (Supplemental Fig. 8). This response, in which an oligonucleotide sense to the target gene induces gene knockdown, was quite distinct from our own experience (data not shown) and that of many others in the literature where antisense oligos against RNAi-related small RNAs, such as microRNAs, relieve target gene repression. We therefore refer to this phenomenon as “sense-induced trans-silencing” (SITS). The specificity of SITS was confirmed by the use of three additional control oligonucleotides (Fig. 6D). Cand45 antisense-mediated gene knockdown was moreover RNAi-related as it could be modulated by Argonaute overexpression (Fig. 6E) similar to how Argonaute overexpressions affect the silencing of perfectly complementary small RNA target genes (Diederichs et al. 2008): Argonaute 2 enhanced the anti-45 knockdown effect (to >90% silencing), while the nonslicing Argonautes 1, 3, and 4 each relieved it (to ∼50%–55% silencing), probably by competition with Argonaute 2 for either the target site and/or small RNA.
Argonaute 2 uniquely selects for perfectly complementary dsRNA of ∼21 base pairs
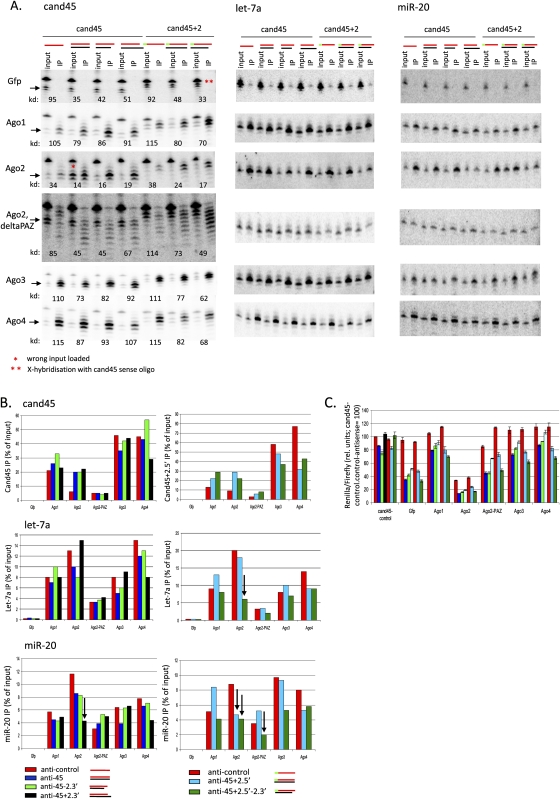
To elucidate the mechanism of SITS, we considered potential changes in Argonaute loading following sense oligo addition. Both the original cand45 tsRNA and a cand45 version extended by 2 nt at its 5′ end were tested in combination with various complementary sense oligonucleotides, differing slightly in the double-strand RNA structure that would be reconstituted (Fig. 7; Supplemental Fig. 9). Strikingly, and in agreement with the notion that Ago2-loaded small RNAs are the main effectors of the trans silencing of perfect complementary target genes, the addition of all complementary sense oligonucleotides enhanced Ago2 loading by two- to fourfold. By contrast, Ago3 and Ago4 loading, which was very efficient for the single-stranded cand45 (∼50% on input was immunoprecipitated) was not further enhanced by sense oligo addition, and in some cases appeared to be slightly impaired by it. Ago1 was somewhat intermediate with no, or slightly enhanced loading following sense oligonucleotide addition. Of note, the sense oligonucleotides that enhanced Ago2 loading in this experiment were fully 2′-O-methylated. As such a modification pattern would be predicted to interfere with passenger strand cleavage by Ago2 (Leuschner et al. 2006), our results suggest that the relative ability of Ago2 for loading fully duplexed small RNAs is independent of its Slicer activity. These findings are consistent with the relative Ago-association patterns observed for microRNAs and tsRNAs and suggest a rule whereby the degree of complementarity of the small RNA loading substrate determines the efficiency with which it is loaded onto the various Argonautes: extensively duplexed small RNAs into preferentially Ago2, and somewhat Ago1, and less stably duplexed and single-stranded RNAs into Ago3 and Ago4 (see Table 2 below). While double-strandedness appeared to be the main determinant for Ago2 loading and SITS efficiency, a more extensive screen of guide strand-sense oligo combinations revealed some differences in SITS efficiencies depending on the exact duplex structure (Supplemental Fig. 9). Although the nature of 5′ and 3′ overhangs could significantly impact SITS, SITS efficiency only poorly correlated with siRNA design rules, such as 2-nt 3′ overhangs. The length of the guide strand, however, had a more obvious impact on SITS efficiency with 20- and 22-nt guides functioning better than 18- and 24-nt guides. Interestingly, Argonaute 2 appeared to discriminate against guide RNAs of unusual length through its PAZ domain, as deleting the PAZ domain of Ago2 conferred onto it the ability to immunoprecipitate cand45-derived small RNAs of various sizes (Fig. 7A, Ago2, delta PAZ). This further correlated with impaired Ago loading, in the case of the original 20-nt cand45 loss of sense-enhanced Ago loading, and ultimately SITS efficiency. To exclude that the heterogeneous small RNAs isolated with the Ago2 PAZ mutant was an experimental artifact as a result of the fact that the PAZ domain binds the 3′ end of the guide RNA (Lingel et al. 2003) and the absence of the PAZ domain might render a bound small RNA susceptible to RNase degradation, we rehybridized the blot for endogenous microRNAs. This showed that such an artifact was unlikely since only single microRNA bands were observed, while microRNA association was similarly diminished in the PAZ deletion mutant. The Argonaute distributions of microRNAs were largely unaffected in the presence of SITS. Interestingly, however, in a number of instances there was a noticeable decrease in Ago2 association, potentially the result of competition between the SITS guide RNA and microRNAs for Ago2 (Fig. 7B, down arrows).
FIGURE 7.
Sense-induced trans-silencing due to preferential loading by Argonaute 2, but not by Argonautes 3 and 4 of the reconstituted double-stranded RNA. (A) Cand45-Argonaute coimmunoprecipitation before and after addition of cand45-complementary sense oligonucleotides (Northern blot; HCT116 cells). FLAG-Gfp and FLAG-Agos used in IPs indicated for each row; “Ago2, deltaPAZ” is a PAZ-deletion mutant of Ago2. Predicted structures of overexpressed cand45 and (2′-O-methyl) complementary oligonucleotides indicated with red (cand45) and black lines (sense); the green line marks the 2-nt 5′ extension of a cand45-derivative (“cand45 + 2”). Arrows indicated cand45 of interest, a double asterisk (**) indicates the results of cross-hybridization of the probe with the transfected sense oligonucleotides; and asterisk (*) marks an input that was incorrectly loaded (correct input requantitated based on separate experiment). For each input/IP pair, the knockdown efficiency is indicated below (“kd,” 100 = no knockdown; summary shown in Fig. 2C) showing correlation between Ago2 IP and silencing efficiencies. Blots were stripped and rehybridized with microRNAs let-7a and miR-20; input: RNA isolated from 10% lysate used per immunoprecipitation. (B) Cand45-AgoIP efficiencies (phosphorimage quantitations of A). Down arrows indicate instances where microRNA-Ago2 associations are reduced under conditions in which cand45-Ago2 associations are increased. (C) Summary of sense-induced trans-silencing results corresponding to the cand45-Ago IP experiment shown in Figure 2A,B (dual luciferase assay with reporter “psi-cand45wt 2x”). Color scheme as in B.
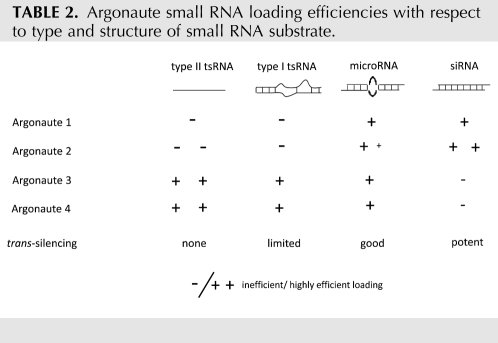
TABLE 2.
Argonaute small RNA loading efficiencies with respect to type and structure of small RNA substrate.
tsRNA levels correlate with microRNA and siRNA silencing activities
To further test whether tsRNAs could affect the function of other classes of small RNAs, we sought to evaluate microRNA and siRNA silencing activity following the modulation of tsRNA levels. To find conditions under which tsRNA levels may be changed, we took two approaches. One was based on changing the serum concentration in the cell growth media since tRNA transcription rates had been linked cell proliferation (for review, see Marshall and White 2008). Alternatively, we overexpressed the RNA polymerase III transcription factor Brf 1 that had been found to specifically up-regulate tRNA transcription in mouse cells (Marshall et al. 2008). Transient Brf 1 overexpression in 293 cells led to a 1.5–2.5-fold increase of tsRNAs cand45 and cand520 (Fig. 8A). At the same time, there was a tendency for microRNAs to be down-regulated. Increasing the serum concentration from 1.5% to 10% had a similar effect on relative small RNA levels: tsRNA cand45 and cand520 were increased at high serum concentrations, while microRNAs were largely unchanged (Fig. 8A), the latter finding being consistent with what had been reported for microRNA levels of subconfluent, serum-starved cells (Hwang et al. 2009). We note that the serum experiments were performed in HCT cells as these proved to be more resistant to outwardly adverse effects of low serum conditions. In both cases, changes in the levels of the type I tsRNA cand14 and cand33 were not consistently observed, although it is possible that some type I tsRNAs not tested for were elevated (data not shown).
We next assessed whether small RNA silencing activity was changed with Brf 1 overexpression or changes in serum concentrations. Depending on the type of competition between tsRNAs and other small RNAs, we would expect the outcome of such experiments to be quite complex. For example, increased tsRNAs would primarily compete with other small RNAs for Ago3 and Ago4 incorporation. The Argonaute distributions of these small RNAs may therefore be shifted toward Ago1 and Ago2, the degree of which, however, would depend on the relative Argonaute affinities and abundances of each particular small RNA. Since tsRNAs will also compete with other small RNAs for Ago1 and Ago2, the net effect of such a redistribution may be muted. In this way, Ago3 and Ago4 may serve as buffers ensuring, e.g., relatively constant microRNA occupancies of Ago1 and Ago2, which may be particularly important for their function. It is also possible that tsRNAs interact with the RNAi machinery upstream of Argonautes, which in turn may make them more available for small RNAs that enter RNAi at the Argonaute stage, e.g., synthetic siRNAs. As a final example of the potentially complex functional outcomes of modulating small RNA levels, although the absolute abundance of a given small RNA may be increased, if it has a higher affinity for the less efficiently silencing Argonautes 3 and 4 and there was competition for target sites, then overall silencing may also be inhibited (both cleavage and translational silencing pathways).
Brf 1 was overexpressed in 293 cells and this was followed by the introduction of various Renilla luciferase reporter genes that only differed in their small RNA target sites located in the 3′ UTRs. Firefly luciferase on the same plasmid and a Renilla luciferase construct containing mock target sites were used for normalization. There was no or little effect of Brf 1 overexpression on the type I cand14, type I cand33, and type II cand45 reporters, consistent with no changes and/or preferential Ago3 and Ago4 incorporation for these tsRNAs. For the microRNA reporters we chose let-7a and miR-20. For each microRNA, two reporters were constructed: one version with three tandem perfect complementary target sites (“PM”) for assessing slicing activity, and a corresponding translational reporter version with complementarity for the microRNA seeds, but mismatched downstream thereof, including at positions 10 and 11 which is predicted to abrogate Ago2 slicing (“MM”). Unlike the tsRNA reporters, both let-7 reporters were de-repressed by ∼2.5-fold in the presence of increased tRNA transcription factor Brf 1 (Fig. 8B), while let-7a steady-state levels were not significantly changed (Fig. 8A). This result is consistent with tsRNAs modulating let-7 silencing activity. The miR-20 reporters were slightly affected by Brf-1 overexpression with a ∼50% increase for miR-20PM, and none for miR-20MM. As discussed above, such differences between reporters for different microRNAs could be due to various factors, such as possible differences in loading efficiencies, absolute abundances and dose sensitivities of the reporter genes. Increased tsRNA levels in the presence of higher serum concentrations were accompanied by ∼twofold de-repressions of both let-7a and miR-20 perfect match reporters, whereas the corresponding translational reporters that differed from the perfect match reporters by only a few nucleotides were unaffected (Fig. 8B). To exclude that inefficient silencing of the translational microRNA reporters in HCT cells was responsible for lack of de-repression in 10% serum, we cotransfected a let-7a antisense inhibitor and observed a ∼75% up-regulation of the reporter gene activity. Interestingly, this simple antisense-mediated inhibition of let-7a underestimated the true extent of let-7a translational repression of the reporter, as the addition of a type of microRNA inhibitor with a region complementary to the microRNA flanked on both sides by small hairpins and that had been reported to be more efficient than simple antisense for microRNA inhibition (Vermeulen et al. 2007), increased reporter gene activity by over fivefold compared to a control inhibitor directed against miR-20 (Fig. 8B, ii). We conclude that the lack of up-regulation of the translational microRNA reporters was not due to their inefficient response to microRNAs. We speculate that given the little changed steady-state level of let-7a at 1.5% versus 10% serum, the difference between the PM and MM reporters is due to a redistribution of let-7a between the Argonautes (more in Ago2 at 1% than 10%, which is predicted to mainly affect PM, and not MM reporters).
To test the effect of Brf-1 overexpression on siRNA efficacy, we transfected three different siRNAs that were directed against the endogenously expressed gene RALY both at a low (500 pM) and a high (50 nM) concentration, and then measured RALY mRNA levels by qRT-PCR (Fig. 8C). “No siRNA” and actin mRNA served for control and normalization purposes, respectively. As expected and in support of the sensitivity of the assay, in each case increased siRNA concentrations more effectively silenced RALY under standard tissue culture conditions of 10% serum. Interestingly, Brf1 overexpression in 293 cells increased siRNA efficacy. In the case of RALY siRNA 3, the fold knockdown was increased from about two- to fourfold at the low, and from about 2.5- to fivefold at the high siRNA concentration. It is unlikely that this is due to Brf 1 increasing transfection efficiency, because (1) plasmid DNA transfection was not affected by Brf1 overexpression based on no significant changes in nontargeted firefly luciferase expression (data not shown) and (2) silencing at 500 pM of siRNA 3 with Brf 1 overexpression exceeded its performance at the 100-fold higher concentration of 50 nM in the absence of Brf 1 overexpression. Instead, we speculate that Brf 1 overexpression further shifts incorporation of the perfectly duplexed siRNA into Ago2 relative to the other Argonautes resulting in reduced competition for the target mRNA. The results are also consistent with tsRNAs competing for components of small RNA silencing upstream of Argonaute loading, thus increasing the availability of Argonautes for the siRNAs. Essentially the same increase in siRNA silencing efficiency was observed in HCT cells when raising serum concentrations from 1% to 10% (Fig. 8B), strengthening the notion that these effects were mediated by tsRNAs and not other changes taking place in the presence of Brf1 overexpression or variations in serum concentrations.
DISCUSSION
Our discovery that there exists in mammals a class of tRNA-derived small RNAs that interacts with the RNAi machinery prompted investigations that contribute to our nascent understanding of the differential properties of the four human Argonautes. Since tsRNAs are not well conserved on a sequence level, yet are observed from yeast to Man (Girard et al. 2006; Babiarz et al. 2008; Buhler et al. 2008), we introduce the concept of how the function of one class of naturally occurring small RNAs may be used to regulate the global activity of another class of small RNAs. Global suppression of microRNA abundance and function has been noted for proliferative diseases such as cancer (Lu et al. 2005) and in response to changes in cell densities (Hwang et al. 2009). It will be of interest to test whether these phenomena converge on signaling pathways regulating the expression of tsRNAs. A general, but modest decrease of microRNA abundance, as seen in cancer (Lu et al. 2005), may be explained by increased turnover of microRNAs that cannot be loaded when increased levels of tsRNAs are present. Indeed, increased tRNA transcription rates and steady-state levels have been linked to cancer (Marshall and White 2008; Pavon-Eternod et al. 2009). Given the critical roles of microRNAs in cancer (Iorio and Croce 2009), it is tempting to speculate that the outcome of such competition between tsRNAs and microRNAs may at least partly explain the link between tRNA overexpression and cancer. In fact, modest changes in steady-state levels of microRNAs could actually mask still more significant functional differences given that competition would involve four, also biochemically distinct, Argonaute proteins in humans. This concept of small RNA class competition, corroborated by our own studies on microRNA and synthetic siRNA efficacies under conditions of varying tsRNA concentrations, is also based on the well-documented limited capacity of RNA silencing, which is subject to autoregulatory feedback and in which Argonautes compete with each other for small RNAs and targets (Grimm et al. 2006; Diederichs et al. 2008; Forman et al. 2008; Han et al. 2009). Clearly, for this reason alone, it is important to identify and characterize all RNAs that interface with the RNA silencing pathways, particularly many of the small RNAs that had previously been sequenced, but have largely been ignored as degradation products of abundant RNA of no particular functional consequence, even when found to coimmunoprecipitate with Argonaute and Piwi proteins. The inability to detect many of them as distinct 21–22-nt small RNAs in conventional Northern blots and to match a large number of them to the genome, recently found to be largely the result of RNA modifications common to highly abundant RNA (Ebhardt et al. 2009), may have contributed to this neglect. Careful bioinformatic analyses, however, have started to reveal that some of the small RNAs derived from abundant noncoding RNAs, particularly tRNAs, are the product of specific processing by endonucleases such as Dicer (Babiarz et al. 2008; Cole et al. 2009).
Additional evidence for competition between tsRNAs and other classes of small RNAs not only in humans comes from the observation that the loss of DGCR8 and Dicer in mouse embryonic stem cells was accompanied by increased tsRNA levels (Babiarz et al. 2008). In fission yeast, deletion of a component of an RNA turnover complex was associated with an increase in tRNA- and rRNA-derived small RNAs that were bound to Argonaute (Buhler et al. 2008). The fact that only little tsRNA was bound to Argonaute in fission yeast may reflect the existence of only one Argonaute protein in this organism. It therefore lacks other Argonautes that could buffer the major functional Argonaute from unwanted small RNAs. That a similar interaction between exosome RNA turnover and RNAi may also exist in mammals is suggested by the observation that elevated beta-globin gene cluster intergenic transcription following Dicer knockdown was particularly noted in cells treated with trichostatin A, a histone deacetylase inhibitor (Haussecker and Proudfoot 2005). As the global increase in histone acetylation is predicted to globally increase intergenic transcription, this may overwhelm other RNA turnover mechanisms and make these transcripts increasingly accessible for RNAi-related turnover.
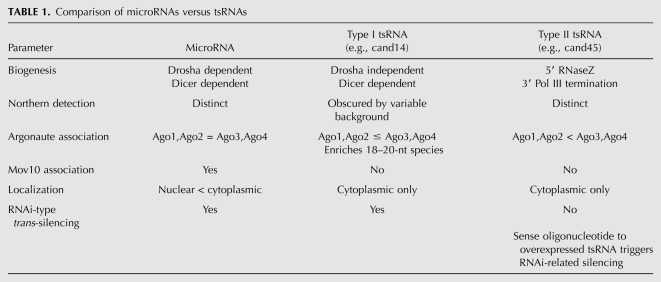
The functional consequence of such competition is predicted to be quite complex, particularly in organisms with multiple Argonautes each with slightly different expression patterns and relative specificities for the different classes of small RNAs and even for different small RNAs within a class. Moreover, as shown here, competition between classes of small RNAs may also be a dynamic property depending on the physiologic state of the cell. tsRNAs differ in a number of respects from microRNAs and can be grouped into two subclasses based on differences in biogenesis and biological activity (Table 1). We demonstrate here that the Microprocessor-independent, Dicer-dependent type I tsRNAs, when appropriately pretreated, can in fact be observed as distinct 5′-phosphorylated, 3′-hydroxylated small RNAs that are incorporated into Argonautes and have trans-silencing capacity. Unlike microRNAs, however, neither tsRNA subclass associated with Mov10 and tsRNAs were essentially restricted to the cytoplasm. Type II tsRNAs, as exemplified by cand45, are generated by RNaseZ cleavage at the discriminator base of tRNAs to generate the phosphorylated 5′ end and by RNA polymerase III termination leaving a stretch of uracils at the 3′ end. As these processes are thought to occur in the nucleus and quality control mechanisms exist to ensure that only properly processed, mature tRNAs are exported (Lund and Dahlberg 1998), this suggests type II tsRNAs to be efficiently exported following synthesis. Since some Argonautes, like microRNAs (Hwang et al. 2007), have been demonstrated in the nuclei of mammalian cells (e.g., Rudel et al. 2008) and are able to shuttle between the nucleus and cytoplasm (Guang et al. 2008; Weinmann et al. 2009), Argonautes themselves may be responsible for the export and cytoplasmic localization of type II tsRNAs, possibly through their association with the tsRNAs soon after RNA polymerase III transcription termination. Alternatively, type II precursor tRNAs may escape nuclear quality control and are processed by the less well-defined pool of cytoplasmic RNaseZ (Elbarbary et al. 2009).
TABLE 1.
Comparison of microRNAs versus tsRNAs
Overlap between various noncoding RNA biogenesis pathways is not uncommon. In this regard, the type II tsRNAs are reminiscent of the RNaseZ-mediated separation of dicistronic RNAs into upstream tRNAs and downstream snoRNAs in plants (Kruszka et al. 2003). RNaseZ has also been recently reported to act on the nascent long noncoding human MALAT1 precursor RNA (Wilusz et al. 2008), thereby generating the mature 3′ end of MALAT1 RNA and liberating a downstream cytoplasmic tRNA-like small RNA. The only relatively recently discovered RNaseZ may therefore function in a much wider array of biological pathways than previously anticipated. A link between snoRNA and microRNA biogenesis was established by Ender and colleagues who described a human gene originally thought to function only as a snoRNA, but that was then found to be also processed in a Microprocessor-independent, Dicer-dependent manner into an Argonaute-associated silencing small RNA (Ender et al. 2008). Overall, the evolution of small RNA biology appears to be highly experimental and flexible in that various mechanisms that can generate hairpins and/or 5′-phosphorylated small RNAs may all enter into RNAi-related pathways.
Although it remains to be elucidated why exactly Argonaute 2 is genetically the most important Argonaute for mammalian cell viability, its importance may also be reflected in the relatively high molecular selectivity of guide RNA loading (Table 2). Both the Argonaute distribution of microRNAs and tsRNA and the mechanism of action of the sense-induced trans-silencing phenomenon show that Argonaute 2 has a preference for 20–22 base-pair (bp), fully duplexed dsRNAs relative to the other Argonautes. Deletion studies further indicated that the PAZ domain contributes to this size and structural selectivity. By contrast, particularly Argonautes 3 and 4, which lack slicing capacity and have intrinsically less translational silencing capacity (Su et al. 2009), may act as buffers soaking up unstructured, especially small single-stranded RNAs. They may thus serve to protect the cells from adventitious degradation products, therefore preventing them from having widespread impact on cellular gene expression through guiding off-targeting. This property of Argonautes 3 and 4 is also of interest for the application of single-stranded RNAs to induce RNAi (ssRNAi), since as the cand45 example shows, the mere presence of a small 5′-phosphorylated RNA is not sufficient for effective trans-silencing. Because RNAi can be elicited by the transfection of single-stranded RNAs (Martinez et al. 2002), small RNA biogenesis may also play a role in determining the small RNA Argonaute distribution pattern. The germline-restricted (primary) piRNAs are another example of a small RNA population that is apparently generated from single-stranded precursor RNAs with no obvious secondary structures via an unknown mechanism that, however, also does not seem to involve Dicer (Vagin et al. 2006), yet loads into Argonaute family proteins. While piRNAs in lower eukaryotes serve to control transposon activity by a so-called ping-pong mechanism, their function and molecular mechanism of action is less well understood in mammals (for review, see Aravin et al. 2007). Of note, ∼6% of piRNA complexes isolated from mouse testes contained tRNA-derived small RNAs, yet were essentially entirely depleted of ribosomal and micro-RNAs (Girard et al. 2006).
Given that RNAi efficiency is determined by siRNA-guided slicing of target mRNA by Argonaute 2 (Liu et al. 2004), our results further highlight the importance of achieving comparatively efficient Argonaute 2 loading. Differential duplex end stabilities and consequently biased passenger-guide strand loading is thought to be the single most important determinant for siRNA efficacy (Khvorova et al. 2003; Schwarz et al. 2003). This is surprising, however, since absolute Argonaute 2 occupancy of a passenger strand derived from a highly abundant siRNA should still be higher than that of the preferred guide strand from a much less abundant siRNA, yet silencing efficiencies are not correlated with absolute Argonaute 2 occupancy. We therefore speculate that the improved performance of asymmetric siRNAs is due to their relative efficient loading into Ago2 rather than their absolute ability to be loaded into Argonautes in general. It is then the ability of Ago2 to rapidly cleave the passenger strand (Matranga et al. 2005; Rand et al. 2005) of asymmetric siRNAs that is ultimately responsible for the observed strand bias. Passenger strands of siRNAs that are equally well recognized by all Argonautes, including nonslicing Ago1, Ago3, and Ago4 should be more stable and such siRNAs should therefore exhibit less apparent strand bias. Poor siRNA performance may thus result from competition for the target with the nonslicing Argonautes. This suggests relative Ago2 loading efficiency to be an important consideration for siRNA design.
An unanticipated finding from our studies were that despite the apparent lack of classical trans-silencing activity by cand45, there was robust, >80% down-regulation of a cand45 reporter gene in cells overexpressing cand45 upon the addition of an oligonucleotide antisense to cand45, i.e., sense to the target gene. This is a quite unusual response to oligonucleotides that are antisense to small guide RNAs as this usually relieves, but does not induce gene repression. We show that reconstitution of a fully duplexed siRNA that is now preferentially loaded into Ago2 is responsible for this phenomenon. In addition for exploiting this system to learn about Argonaute substrate specificities, equally exciting is the prospect of harnessing this mechanism as a new type of RNA silencing tool in which genes can be silenced by the addition of an oligonucleotide with sense polarity to the target gene. This could not only be useful for studying gene function in vitro, but it may be particularly valuable for knocking down genes in vivo for target validation and therapeutic purposes by combining the relative ease and simplicity of delivering unformulated single-stranded oligonucleotides to organs like the liver and spleen to, in a temporally regulated manner, tap into the inherently more potent RNAi pathway once inside cells. The ability to use at least two different modification chemistries raises hopes that the pharmacological requirements for in vivo applications will not be limiting.
MATERIALS AND METHODS
Tissue culture
Human 293 and HCT116 cells (wild-type and derived Dicer helicase mutant cells were a kind gift from B. Vogelstein, John Hopkins University) were maintained in standard 10% FCS DMEM medium. For the RNA immunoprecipitation experiment in Figure 4B, the 293-derived FLAG-HDAg expressing cell line and the induction of HDV replication by plasmid transfection have been described before (Haussecker et al. 2008). Cells were transfected with Lipofectamine 2000 (L2K, Invitrogen) for plasmid DNA and RNAiMax (Invitrogen) for siRNA according to the manufacturer's instructions. For the serum starvation experiments, HCT116 cells were cultured at the indicated serum concentrations by daily media change and split such that they were growing at comparable cell densities. For the Brf1 overexpression experiments, 293 cells were transfected with a Brf1 overexpression plasmid on days 0 and 2 with 2 μg of plasmid DNA per 6 cm dish. For testing steady-state small RNA levels as a function of serum concentrations/Brf1 overexpression, RNA was isolated for Northern blot analysis on day 5; for testing small RNA trans-silencing capacity under these conditions, luciferase reporter constructs were introduced on day 4 of serum starvation/Brf1 overexpression and dual luciferase assays performed the next day.
Plasmids
For RNA analyses, 2 μg expression plasmids were used per six wells of 293 cells and RNA analyzed 48 h after transfection. FLAG-Argonautes 1 and 2 and FLAG-EGFP are as described by Meister et al. (2004). To obtain similar expression levels for FLAG-Argonautes 1–4, FLAG-Argonaute 3 and 4 described by Meister et al. 2004 were codon optimized by GENEART AG. Codon optimization did not change amino acids, only expression levels (detailed sequence available upon request). The FLAG-Ago2 PAZ deletion mutant in Figure 7 was generated according to Gu et al. (2009). FLAG-Mov10 is as described in Meister et al. (2005), FLAG-TRBP as described by Kok et al. (2007); and pCMV-Dicer was a gift of Ian G. Macara (University of Virginia). A cand45 cloning vector (“cand45-empty”) genomic sequence was amplified with primers hsCand45gen-F cttaaAAGCTTaagcttCTCTCGCAGAAATGCCAAAT and hsCand45gen-R cttaatctagaAAAAAAAtgGTCTTCAGTGAAGCGAAGACgcaggg TTCGAACCTGCGCGGGGAGAC, and cloned into the HindIII-XbaI sites of pCRII-TOPO (Invitrogen). For cand45-45, cand45-targ1 (referred to as “cand45-con” in Fig. 6C), cand45-targ2, cand45-+2.45, cand45-+4.45, and cand45-2.45 overexpression, the cloning vector was digested with BbsI, de-phosphorylated, and the following phosphorylated and annealed oligos inserted:
Cand45-45: ccctGCTCGCTGCGGAAGCGGGTGCTCTTA and aaaaTAAGAGCACCCGCTTCCGCAGCGAGC;
Cand45-targ1: ccctGCTCGCTGCGttcagcccgtcctctaggc and AAAAgcctagaggacgggctgaaCGCAGCGAGC;
Cand45-targ2: ccctGCTCGCTGCGctcctcgagcgtcagacgc and AAAAgcgtctgacgctcgaggagCGCAGCGAGC;
Cand45-+2.45: ccctGCTCGCTGCGGAGAAGCGGGTGCTCTTA and aaaaTAAGAGCACCCGCTTCTCCGCAGCGAGC;
Cand45-+4.45: ccctGCTCGCTGCGGAGAGAAGCGGGTGCTCTTA and aaaaTAAGAGCACCCGCTTCTCTCCGCAGCGAGC; and
Cand45-2.45: ccctGCTCGCTGCGAGCGGGTGCTCTTA and aaaaTAAGAGCACCCGCTCGCAGCGAGC.
In the cotransfection studies in Figure 4C, 2 μg of cand45 and 2 μg of FLAG-protein expression vectors were cotransfected into 6-cm dishes of 293 cells. The Brf1 overexpression vector was created by cloning the Brf1 ORF from pTRE2-Brf1 (Marshall et al. 2008) into pcDNA3 just downstream from an N-terminal FLAG epitope (Cao et al. 2009), while in the negative control vector the Brf1 ORF had been replaced with the truncated ORF of a replication-deficient early nonsense mutant version of HDAg (Haussecker et al. 2008).
Northern blot
For small RNA Northern blotting, RNA was separated by 20% urea-polyacrylamide gel electrophoresis, transferred onto Hybond-N (Amersham) nitrocellulose by semidry transfer, and hybridized to T4 PNK end-labeled oligonucleotide probes overnight at 32°C with PerfectHyb Plus (Sigma). Blots were washed three times with 6X SSC, 0.2% SDS (32°C, 34°C, 36°C), and then once with 0.5X SSC, 0.1% SDS (42°C) for 10 min each. Images were obtained by a PhosphorImager. Ambion's “Decade” was used as a size marker. Northern probe oligos were the following:
Cand1: gCACATGGTTAGATCAAGC;
Cand2: gAAAACCCACAATCCCTGGCTG;
Cand3: gAAAACCCACAATCCCTGGCTTA;
Cand4: gTCAATTAGTTGTAAACACCACTG;
Cand5: gTTCTAGGATAGGCCCAGGGGC;
Cand6: gCCAACTGAGCTAACCGGCC;
Cand7: gAACCCCACCAACATAGGGCTTCG;
Cand8: GGGCAGGCGAGAATTCTACCAC;
Cand9: GGATAACCACTACACTATGGAA;
Cand10: gTGGCGCCCGAACAGGGACA;
Cand11: GGCACCCCAGATGGGACACGA;
Cand12: gAAACGAGGTAACTCCGGA;
Cand13: GTGCCCGAGTGTGGTGGAGAATG;
Cand14: GAGTAGTGGTGCGTTGGCCGG;
Cand15: gTGGCGACCACGAAGGGACG;
Cand16: GAATTCTACCACTGAACCACAAT;
Cand17: GGCGACCACGAAGGGACACGA;
Cand18: GTTGTAAACACCACTGCACT;
Cand19: GGGCTTCAAAAAATTTGCTTGA;
Cand20: GGAGGGGGCACCCGGATTTGA;
Cand21: GGCGACCACGAAGGGACCCGA;
Cand22 (also known as miR-20): gACTACCTGCACTATAAGCAC;
Cand23: GGTGCGTTGGCCGGGAAACGA;
Cand24: gAAACAGCAAGCTAGTCAAGC;
Cand25: gCTTAGACCGCTCGGCCATCCTT;
Cand26: GACCGCTCGGCCACGCTACCCTC;
Cand27: gTGGCGAGCCAGCCAGGAG;
Cand28: gCCTTAGACCGCTCGGCCATCCT;
Cand29: GTCCTTGGTGCCCGAGGTGTCTA;
Cand30: GTGATATCCACTACACTACGGA;
Cand31: gCACCACTATACCACCAACGC;
Cand32: gCTCGCCAGGGCAAGGCTTACAA;
Cand33: GGTGCATGGGCCGGGAAACG;
Cand34: gACCACTGAACCACCAATGC;
Cand35: GGTTCCTGACCGGGAATCGAAC;
Cand36: GGTGCCGAAACCCGGGAACGA;
Cand37: gAACCCCACCAACATAGGGCTT;
Cand38: GTCCTTGGTGCCCGAGTGACCT;
Cand39: GACACCGTCCTTGGTGCCGCGT;
Cand40: GGACACCGTCCTTGGTGCCCAG;
Cand41: GCCCGAGGTGGTATGGCCGTAG;
Cand42: gTCTACCACTGAACCACCCATG;
Cand43: gACCACTGAACCACCCATGC;
Cand44: GAGAACCGTCCTTGGTGCCCGA;
Cand45: gAAAATAAGAGCACCCGCTTC;
Cand193: gCGAGGTAACTCCGGAGC;
Cand401: GAGGCACCTGCCAGGTGAC;
Cand500: gCTGAGCACAGGACTTCCTT;
Cand520: GAGCTTGGACGCTCGGTTGA;
sh320: gTCGCCCTCTCAACCCAGCTTTT;
sh484: gATCGGGAGGGGACTGAGCCTGA;
MALAT mascRNA: gtcctggaaaccaggagtgc;
let-7a: AACTATACAACCTACTACCTCA;
miR-15a: acaaaccattatgtgctgcta;
miR-18a: Gctatctgcactagatgcacct;
mir-21: Gtcaacatcagtctgataagc;
mir-103: Gtcatagccctgtacaatgctg;
mir-106a: Gctacctgcactgtaagcacttt;
mir-191: Gcagctgcttttgggattccgtt;
U6 snRNA: gccatgctaatcttctctgtatc;
sno38b: AGAACTGGACAAAGTTTTCATCAC;
Cand45-targ1: gcctagaggacgggctgaa;
Cand45-targ2: gcgtctgacgctcgaggag; and
HDV small RNA: ggcggcagtcctcagtactctta.
For the Northern blot screen in Figure 1 (Supplemental Fig. 1) and the Dicer helicase mutant analysis in Figure 2A, 4 μg mirVana (Ambion) low-molecular weight RNA was sequentially treated with TAP and/or T4 RNA ligase as described in the following section).
Analysis of small RNA 3′ and 5′ ends
Enzyme treatments were performed by denaturing 4 μg mirVana (Ambion) RNA per sample at 65°C for 5 min, chilled on ice for 2 min, and followed by the addition of enzyme buffer, rRNasin (Promega; except for Terminator Exonuclease treatments), and, finally, enzyme. Fifteen-microliter reactions were incubated with the indicated enzymes at 37°C (Terminator Exonuclease: 30°C) for 60 min, acid phenol/chloroform extracted, ethanol precipitated, and resuspended for the second round of enzyme treatments, which was again followed by acid phenol/chloroform extraction, ethanol precipitation, and resuspension in PAGE loading buffer for Northern blot. Buffer indicates that no enzyme was added. Amounts of enzymes used: 15 units (U) of T4 PNK, 3′ phophatase ± (NEB M0201/m0236); 8 U of Tobacco Acid Pyrophosphatase (Epicentre Biotechnologies); 3 U of Terminator Exonuclease (Epicentre Biotechnologies); 4 U polyA polymerase (PAP; Ambion); and 15 U T4 RNA ligase (NEB). For 3′-adapter ligation with T4 RNA ligase, a noncommercial buffer without ATP was made up and 1 μg of the following activated 3′-adapter added: 5′-AppCTGTAGGCACCATCAAT–NH2-3′ (NEB 1315).
RNA Immunoprecipitation
To test for the distribution of tsRNAs, cell lysate corresponding to a 6 cm dish of confluent 293 cells were used for each immunoprecipitation. Lysates were prepared by washing cells 48 h after FLAG-protein transfection twice with ice-cold PBS and then lysing them with 0.6-mL/10-cm dish M-PER lysis buffer (Pierce) containing protease inhibitor cocktail (EDTA, Roche). Lysates were diluted by the addition of 3X volumes IP buffer (20 mM Tris-HCl [pH 8.0], 50 mM KCl, 0.2 mM EDTA, 10% glycerol). One-tenth of the volume lysate per immunoprecipitation was removed and RNA isolated with TRIzol (Invitrogen) for the input RNA control. Per sample, 20 uL anti-Flag M2 agarose beads (Sigma A2220) were added to the lysate for incubation with rotation overnight at 4°C. The next day, immunoprecipitates were washed extensively with IP buffer and RNA isolated by the addition of TRIzol to the beads. To delineate the mechanism of sense-induced trans-silencing, HCT116 cells were cotransfected in 6 cm dishes with 2 μg of FLAG-bait, 2 μg of the cand45 overexpression vector, and 0.1 μg of the luciferase reporter, 2 h after which 100 nM of the sense oligonucleotide were added. Cells were split the next day into one 6 cm dish and three 24 wells each, with lysates harvested and processed for RNA immunoprecipitation (6 cm dish) or luciferase assay (24 wells) 48 h after the first transfection.
RNaseZP processing assay
RNaseZP processing assays were as previously described (Wilusz et al. 2008). Purified HeLa RNaseP and recombinant His-tagged tRNaseZL (delta30) were generously provided by Sidney Altman (Yale University) and Masayuki Nashimoto (Niigata University), respectively. After the first enzyme treatments, RNA was acid phenol/chloroform extracted and ethanol immunoprecipitated, and resuspended for performing the second enzyme reaction. Mock treatment was buffer without enzyme addition. An internally alpha-P32 UTP-labeled, gel-purified cand45 precursor tRNA substrate was prepared by SP6 in vitro transcription of XbaI-linearized plasmid SP6-cand45. SP6-cand45 was obtained by cloning into the HindIII-XbaI sites of pCRII-TOPO (Invitrogen) a genomic cand45 tRNA PCR fragment using primers SP6-cand45-F (cttaaAAGCTTACTAAAGTGTCTCCGCCTG) and SP6-cand45-R (ttaatctagaAAATAAGAGCACCCGCTTCCGCAGCGAGCAGGGTTCGAACCTGCGCGGGG; starting 34 nt upstream of the predicted RNaseP cleavage site and ending with the RNA polymerase termination oligo-dT stretch).
Nuclear-cytoplasmic RNA fractionation
Nuclear-cytoplasmic fractionation was performed as previously described (Haussecker et al. 2008). Two micrograms of each fraction were used for Northern blot analysis.
Dual luciferase assay
The Dual-Luciferase Reporter (DLR) Assay (Promega) was performed according to the manufacturer's instructions, with the following modifications. Cand14 and Cand45 reporter vectors were derived by inserting the following phosphorylated and annealed oligos into the XhoI-SpeI sites of Renilla luciferase in psi-check2 (Promega):
psi-cand14: TCGAcgagtagtggtgcgttggccgggaaAAAAAcgagtagtggtgcgttggccgggaa and CTAGttcccggccaacgcaccactactcgTTTTTttcccggccaacgcaccactactcg;
psi-cand45 wt (also known as “psi-cand45wt 2x” in Supplemental Fig. 6): tcgaAAAATAAGAGCACCCGCTTCaaaaAAAATAAGAGCACCCGCTTC and ctagGAAGCGGGTGCTCTTATTTTttttGAAGCGGGTGCTCTTATTTT;
psi-cand45wt 1x: TCGAAAAATAAGAGCACCCGCTTC and CTAGGAAGCGGGTGCTCTTATTTT;
psi-cand45wt_extended (Fig. 7; Supplemental Fig. 9): tcgaAAAAAAAATAAGAGCACCCGCTTCTCTCaaaaAAAAAAAATAAGAGCACCCGCTTCTCTC and ctagGAGAGAAGCGGGTGCTCTTATTTTTTTTttttGAGAGAAGCGGGTGCTCTTATTTTTTTT;
psi-let-7aPM: TCGAaactatacaacctactacctcAaaaaaaactatacaacctactacctcAaaaaaaaactatacaacctactacctcA and ctagTgaggtagtaggttgtatagttttttttTgaggtagtaggttgtatagtttttttTgaggtagtaggttgtatagtt;
psi-let-7aMM : TCGAaTcTAtTGaaGGAactacctcAaaaaaaaaTcTAtTGaaGGAactacctcAaaaaaaaaTcTAtTGaaGGAactacctcA and ctagTgaggtagtTCCttCAaTAgAttttttttTgaggtagtTCCttCAaTAgAttttttttTgaggtagtTCCttCAaTAgAt;
psi-miR-20PM: TCGActacctgcactataagcactttaaaaaaactacctgcactataagcactttaaaaaaactacctgcactataagcacttta and ctagtaaagtgcttatagtgcaggtagtttttttaaagtgcttatagtgcaggtagtttttttaaagtgcttatagtgcaggtag;
psi-miR-20MM: TCGActTGctGCaGtaTAagcactttaaaaaaactTGctGCaGtaTAagcactttaaaaaaactTGctGCaGtaTAagcacttta and ctagtaaagtgctTAtaCtGCagCAagtttttttaaagtgctTAtaCtGCagCAagtttttttaaagtgctTAtaCtGCagCAag; and
psi-bantam: tcgaTAGTTTTCACAATGATCTCGGTAGTTTTCACAATGATCTCGGTAGTTTTCACAATGATCTCGGTAGTTTTCACAATGATCTCGG and ctagCCGAGATCATTGTGAAAACTACCGAGATCATTGTGAAAACTACCGAGATCATTGTGAAAACTACCGAGATCATTGTGAAAACTA.
Each transfection was performed in triplicate 24 wells by cotransfecting the following amounts of nucleic acids with L2K into 293 (cand14 assays) or HCT116 (cand45 assays) cells: Fifty-nanogram psi-check reporter plasmid, and either 225ng+225ng FLAG-protein expression+cand45-45/-targ.1/-targ.2 or 450 ng FLAG-protein expression vector. A number of antisense oligonucleotides (100 nM) were applied 2 h after plasmid transfection, lysates harvested for luciferase assays 48 h after the tranfections. With the exception of the experiment shown in Supplemental Fig. 7, the Renilla:Firefly luciferase ratios of the target vectors were further normalized to the Renilla:Firely luciferase ratio of a psi-check2-derived reporter in which the tsRNA target sequences were replaced by bantam microRNA target sequences (not expressed in human cells). Antisense oligonucleotides were the following (where “+” denotes the number of nucleotides extended at the 5′ end):
Anti-14: 5′-+T+A+G+T+GGmUGmCGmUTmG+G+C+C+GG-3′;
Anti-cand45/anti-45 LNA/methyl: 5′-+A+A+A+ATmAAmGAmGCmACmCC+G+C+T+TC-3′;
Anti-cand45-methyl: 5′-mAmAmAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmC-3′;
Anti-cand45-LNA: 5′-+A+A+A+A+U+A+A+G+A+G+C+A+C+C+C+G+C+U+U+C-3′;
Anti-con1: ATGGCCTCGAGCCTCCTCAATTCACAACCTG;
Anti-con2: mAmGmGmCmGmGmCmAmGmUmCmCmUmCmAmGmUmAmCmUmCmUmUmA; and
Anti-con3: mUmAmAmGmAmGmUmAmCmUmGmAmGmGmAmCmUmGmCmCmGmCmCmU.
Anti-cand45 oligonucleotides for testing structure-function of sense-induced trans-silencing (Fig. 7; Supplemental Fig. 9) were the following:
Anti-cand45 (2-nt matching cand45 5′end removed): 5′-mAmAmAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmU-3′;
Anti-cand45 (with an additional 2-nt matching cand45-derivative cand45-+2.45): 5′-mAmAmAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmCmUmC-3′;
Anti-cand45 (with an additional 4-nt matching cand45-derivative cand45-+4.45): 5′-mAmAmAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmCmUmCmUmC-3′;
Anti-cand45 (with an additional 2 nt that would be able to match a cand45-derivative with extended oligo-U 3′ tail): 5′-mAmAmAmAmAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmC-3′;
Anti-cand45 (resulting in 2-nt 3′ overhang of cand45 with 4Us at the 3′ end): 5′-mAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmC-3′;
Anti-cand45 (with an additional 4 nt matching cand45-derivative cand45-+4.45 and resulting in 2-nt 3′ overhang of cand45 with 4Us at the 3′ end): 5′-mAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmCmUmCmUmC-3′;
Anti-cand45 (with a 3′ extension that would extend complementarily toward cand45, 2 nt upstream of the predicted RNaseZ cleavage site): 5′-mAmAmAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmCmCmG-3′;
Anti-cand45 (with a 3′ extension that would extend complementarily toward cand45, 2 nt upstream of the predicted RNaseZ cleavage site, and resulting in a 2-nt 3′ overhang of cand45 with 4Us at the 3′ end): 5′-mAmAmUmAmAmGmAmGmCmAmCmCmCmGmCmUmUmCmUmC-3′; and
Let-7a antisense: mAmAmCmUmAmUmAmCmAmAmCmCmUmAmCmUmAmCmCmUmCmA (miRIDIAN miR-20 Hairpin Inhibitor: Dharmacon Cat. No. IH-300491-05: miRIDIAN let-7a Hairpin Inhibitor: Dharmacon Cat. No. IH-300473-07; +: LNA; m: 2′-O-methyl; others: DNA bases).
RNAi
Two hundred ninety-three (Brf1) and HCT116 (serum) cells were cultured for 4 d under the indicated conditions and then transfected with siRNAs targeting the endogenously expressed RALY gene using RNAiMax (Invitrogen). RNA was harvested the next day with TRIzol (Invitrogen) and quantitative reverse transcription realtime PCR (qRT-PCR) performed according to Haussecker and Proudfoot (2005); actin was used for normalization. Two-nucleotide dTdT 3′ overhang siRNAs of the following sequences (sense/passenger) were obtained from Dharmacon/Thermo Fischer:
si-1: GAUCAAGUCCAAUAUCGAUdtdt;
si-2: GCGUGUCAAAACUAACGUAdtdt;
si-3: AGACGACGGCGAUGAGGAAdtdt;
RT-PCR primers:
Actin RT: cttaatgtcacgcacgatttcc;
Actin forward: aaatctggcaccacaccttc;
Actin reverse: agaggcgtacagggatagca;
RALY RT: tcttcctcgctgtgtgtcag;
RALY forward: ttctgtgcacaagggctatg; and
RALY reverse: atggcagatgctgctctctt.
Western blot
Western blot was performed according to standard protocols. Two micrograms of protein from 293 cells transfected with the indicated expression plasmids were run on 4%–20% polyacrylamide gradient gels, blotted, and probed with the following antibodies: mAb FLAG M2 (Sigma, A8592); mAb anti-actin (Sigma, A5316).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH, NIAID 71068, and DK 078424 to M.A.K. and by GM 37706 to A.Z.F. We thank Masayuki Nashimoto (Niigata University) and Sidney Altman (Yale University) for recombinant RNaseZ and purified human RNaseP, respectively.
NOTE ADDED IN PROOF
Since submitting our manuscript, Lee et al. (2009) published on tRNA-derived small RNAs in human cells. That study identified three populations of 17–26 nt tRNA-derived small RNAs, one corresponding to the 5′ end of mature tRNAs, the other two to the class I and II tsRNAs reported here. Lee et al. (2009) also picked cand45 (referred to as tRF-1001 in Lee et al. [2009]) for more detailed analyses and found its expression to correlate with cell proliferation, to have almost no trans-silencing capacity, and to be the result of processing by an isoform of RNaseZ/ELAC possibly in the cytoplasm. Beyond that, their studies suggest cand45 to be necessary for cell proliferation. Where the two studies overlap, the conclusions are consistent with one another.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2000810.
REFERENCES
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;26:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA. 2009;15:1971–1979. doi: 10.1261/rna.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNaseZ processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 2004;9:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewski N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:84–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Joplin CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Proudfoot NJ. Dicer-dependent turnover of intergenic transcripts from the human β-globin gene cluster. Mol Cell Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol. 2008;15:714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210.1–210.8. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Cell–cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena S. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- Kruszka K, Barneche F, Guyot R, Ailhas J, Meneau I, Schiffer S, Marchfelder A, Echeverria M. Plant dicistronic tRNA-snoRNA genes: A new mode of expression of the small nucleolar RNAs processed by RNaseZ. EMBO J. 2003;22:621–632. doi: 10.1093/emboj/cdg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- Marshall L, White RJ. Noncoding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8:911–914. doi: 10.1038/nrc2539. [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNAiMet synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luehrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Mayer M, Schiffer S, Marchfelder A. tRNA 3′ processing in plants: Nuclear and mitochondrial activities differ. Biochemistry. 2000;39:2096–2105. doi: 10.1021/bi992253e. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;23:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel Argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M, Wesemann DR, Geary S, Tamura M, Kaspar RL. Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 1999;27:2770–2776. doi: 10.1093/nar/27.13.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes & Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore P. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes & Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidnandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutz J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets Argonaute proteins to distinct mRNAs. Cell. 2009;136:1–12. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang X, Graves PR, Zeng Y. Stable Argonaute2 overexpression differentially regulates microRNA productions. Biochim Biophys Acta. 2009;1789:153–159. doi: 10.1016/j.bbagrm.2008.11.004. [DOI] [PubMed] [Google Scholar]