Abstract
Replacement of mRNA 5′ UTR sequences by short sequences trans-spliced from specialized, noncoding, spliced leader (SL) RNAs is an enigmatic phenomenon, occurring in a set of distantly related animal groups including urochordates, nematodes, flatworms, and hydra, as well as in Euglenozoa and dinoflagellates. Whether SL trans-splicing has a common evolutionary origin and biological function among different organisms remains unclear. We have undertaken a systematic identification of SL exons in cDNA sequence data sets from non-bilaterian metazoan species and their closest unicellular relatives. SL exons were identified in ctenophores and in hydrozoan cnidarians, but not in other cnidarians, placozoans, or sponges, or in animal unicellular relatives. Mapping of SL absence/presence obtained from this and previous studies onto current phylogenetic trees favors an evolutionary scenario involving multiple origins for SLs during eumetazoan evolution rather than loss from a common ancestor. In both ctenophore and hydrozoan species, multiple SL sequences were identified, showing high sequence diversity. Detailed analysis of a large data set generated for the hydrozoan Clytia hemisphaerica revealed trans-splicing of given mRNAs by multiple alternative SLs. No evidence was found for a common identity of trans-spliced mRNAs between different hydrozoans. One feature found specifically to characterize SL-spliced mRNAs in hydrozoans, however, was a marked adenosine enrichment immediately 3′ of the SL acceptor splice site. Our findings of high sequence divergence and apparently indiscriminate use of SLs in hydrozoans, along with recent findings in other taxa, indicate that SL genes have evolved rapidly in parallel in diverse animal groups, with constraint on SL exon sequence evolution being apparently rare.
Keywords: trans-splicing, spliced leader, evolution, Clytia, Pleurobrachia
INTRODUCTION
The process of spliced leader (SL) trans-splicing joins the short exon (15–50 nucleotides [nt]) from specialized noncoding nuclear SL RNAs to the 5′ ends of assorted pre-mRNAs during their nuclear processing (for reviews, see Davis 1996; Hastings 2005). This unusual form of RNA maturation was first described in trypanosomes, in which all mRNAs are trans-spliced, using a single SL RNA (Sutton and Boothroyd 1986). SL trans-splicing has subsequently been demonstrated to occur in Euglenozoas (Miller et al. 1986; Tessier et al. 1991), in dinoflagellates (Zhang et al. 2007; Zhang and Lin 2009), and in several metazoan lineages: all tested nematodes (Krause and Hirsh 1987; Guiliano and Blaxter 2006), urochordates (Vandenberghe et al. 2001; Ganot et al. 2004), flatworms (Davis 1997), the two tested chaetognath species (Marletaz et al. 2008), a Bdelloid rotifer (Pouchkina-Stantcheva and Tunnacliffe 2005), an Acoel (Marletaz et al. 2008), and the hydrozoan (Cnidaria) Hydra vulgaris (Stover and Steele 2001). In contrast, SL trans-splicing was not detected in vertebrates, echinoderms, arthropods, mollusks, or annelids, or in most nonmetazoan groups including fungi and plants. Furthermore, there are wide differences in SL usage within and between different animal groups, such as in the proportion of trans-spliced mRNA and the number of distinct SL genes (for reviews, see Hastings 2005; Marletaz et al. 2008).
SL trans-splicing is enigmatic not only because of its scattered phylogenetic distribution and variable frequency, but because of the lack of a clear conserved function. In all cases in trypanosomes, and in some nematodes, urochordates, chaetognaths, and possibly flatworms, SL trans-splicing has an important function in the resolution of polycistronic pre-mRNAs into individual transcripts (Davis and Hodgson 1997; Blumenthal and Gleason 2003; Satou et al. 2006; Marletaz et al. 2008). In kinetoplastids (i.e., trypanosomes and Euglenozoas) and nematodes (Lall et al. 2004), SL trans-splicing affects stability and translation by providing a particular 5′ cap structure, the kinetoplastid case being extreme in that the new cap is required for association with ribosomes (Zeiner et al. 2003). It has also been pointed out that the shortening of 5′ UTRs that results from SL trans-splicing could potentially act to “sanitize” overlong pre-mRNAs by removing unwanted sequences potentially affecting translation or stability from the original “outron” (i.e., the part of the original 5′ UTR replaced by the SL sequence), thereby facilitating the use of more distant transcription initiation sites (Davis 1996; Hastings 2005).
Concerning the evolution of SL genes, available evidence has not allowed discrimination between hypotheses of a common origin with multiple losses and independent acquisition in multiple phylogenetic lineages (Lawrence 1999; Nilsen 2001; Stover and Steele 2001; Hastings 2005; Roy and Irimia 2009). Arguments put forward to favor the former hypothesis include the existence of (partially) shared functions, such as polycistronic transcript resolution, between some distantly related taxa, the stereotypic secondary structure of the SL RNA “intron” region, which features three stem–loops and a binding site for the Sm proteins (essential components of the eukaryotic cis-splicing machinery), and the frequent localization of SL genes within 5S rRNA gene clusters. However, given the phylogenetic distribution of species known to employ SL trans-splicing among metazoans, convergent acquisitions of structural and functional features following multiple independent origins cannot be excluded. In order to shed light on SL evolution in metazoans, and, in particular, on their presence or absence in the common metazoan ancestor, we have undertaken a systematic search for SL exons among expressed sequence tag (EST) data sets of early diverging metazoans (cnidarians, ctenophores, placozoa, and sponges) and in four species of unicellular organisms that group together with the metazoans in the clade Holozoa (Fig. 1). We then performed a detailed comparison of SL use in two hydrozoan species, Clytia hemisphaerica and Hydra magnipapillata, since short-range comparisons can be very informative about conserved features and sequence divergence rate, as shown by studies in the nematodes and chaetognaths (Guiliano and Blaxter 2006; Marletaz et al. 2008).
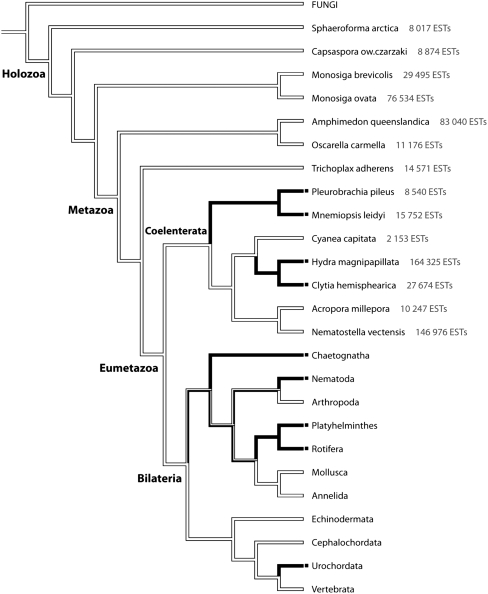
FIGURE 1.
SL usage in holozoan species. The character “presence/absence” of SL genes, deduced from this study (with numbers of ESTs analyzed per species) and from published work, mapped using Mesquite parsimony reconstruction (Maddison and Maddison 2009) onto the phylogeny of Holozoa. This phylogeny was adapted from Dunn et al. (2008) for intrabilaterian relationships and from Philippe et al. (2009) for all other relationships. All SL-positive lineages found in the Metazoa have been included with the exception of the Acoels, whose phylogenetic position remains unclear (Dunn et al. 2008; Hejnol et al. 2009; Schierwater et al. 2009). (Black lines) Presence of SL genes, (white lines) their absence, (mixed lines) unresolved state. The rotifers have been coded as “unresolved” because current evidence indicates SL usage in bdelloid but not monogonont rotifer species (Pouchkina-Stantcheva and Tunnacliffe 2005; Suga et al. 2007). While this study was under review, SL sequences were also discovered in a Hexactinellid sponge and a third ctenophore species (Douris et al. 2009), not included here.
RESULTS
Survey of SL trans-splicing in holozoans from EST data sets
The presence of an identical sequence at the 5′ end of trans-spliced mRNAs means that SL trans-splicing can be readily identified in large EST data sets by looking for identical sequences in the 5′ UTRs of assembled cDNAs. Large EST collections were generated for the ctenophore Pleurobrachia pileus and the hydrozoan cnidarian Clytia hemisphaerica at the Genoscope (Evry, France) using mixed-stage cDNA libraries (Philippe et al. 2009). EST sets for 12 other species were downloaded from NCBI: four additional cnidarians (the Hydrozoan Hydra magnipapillata, the Anthozoans Nematostella vectensis and Acropora millepora, and the Scyphozoan Cyanea capilata), the ctenophore Mnemiopsis leidyi, the sponges Oscarella carmela (Homoscleromorpha) and Amphimedon queenslandica (Demospongia), the placozoan Trichoplax adherens, the choanoflagellates Monosiga brevicollis and Monosiga ovata, the ichthyosporean Sphaeroforma arctica, and the related ameboid protist Capsaspora owczarzaki. For each data set, cDNA sequences were assembled from the ESTs, and identical or nearly identical 5′ leader sequences common to multiple transcripts were identified following alignment (see Materials and Methods for details).
Using this exon detection approach we could clearly identify 5′ SL exon sequences in EST assemblies from the two ctenophore species, P. pileus and M. leidyi, and in the two hydrozoans, C. hemisphaerica and H. magnipapillata (Fig. 1). We were also able to identify several SL sequences from the small EST data set of a third hydrozoan species, Podocoryne carnea (3905 ESTs, downloaded from The National Center for Biotechnology Information [NCBI]; http://www.ncbi.nlm.nih.gov/guide/) (data not shown). In contrast, no conserved 5′ elements were detected in the EST data set from any of the other species tested, strongly suggesting that SL trans-splicing is absent in these species (although we cannot completely rule out the possibility of unprecedentedly low SL trans-splicing frequencies). In each data set from which putative SL exons were identified, many sequence variants were found, defined by their presence in at least five distinct cDNA assemblies (see below for detailed analyses). We confirmed that each variant identified in H. magnipapillata corresponded to a distinct SL gene by BLAST analysis against the draft genome of this species (http://hydrazome.metazome.net); all variants were present in the genome sequence with the exception of one SLB variant.
To see whether our new data could help clarify the picture of SL evolution, we mapped the presence or absence of SL genes obtained from our analyses onto a phylogenetic tree combining recent topologies obtained by phylogenomics approaches focusing on basal metazoan branching (Philippe et al. 2009) and on intrabilaterian relationships (Dunn et al. 2008). SL sequences appear to be restricted to a small number of lineages among Eumetazoa: Ctenophora, Hydrozoa, Urochordata, and several protostome lineages. Parsimony optimization of SL evolution fails to resolve the ancestral state of Protostomia but clearly supports absence of SL trans-splicing in both metazoan and eumetazoan ancestors, followed by multiple independent origins (Fig. 1).
Multiple spliced leaders in ctenophores
Our results provide the first evidence for SL trans-splicing in ctenophores, suggesting that SL usage within the Coelenterate lineage has been acquired (or lost) at least twice (Fig. 1). As in the hydrozoans (Stover and Steele 2001 and see below), multiple SL sequences were recovered from P. pileus. SL exons were detected in 40% of the ∼9000 distinct assembled cDNA sequences. These sequences could be grouped into two distinct but related groups, Ppi_SLA and Ppi_SLB, with a maximum length of 37 nt recovered in both groups (Table 1). Ppi_SLA showed nine distinct variants (probably representing nine distinct genes, see above) and was detected in 77% of trans-spliced cDNAs, while Ppi_SLB had two variants. The Ppi_SL exon sequences obtained were mostly incomplete in their 5′ ends, but the longest variants of each groups exhibited a common 5′ terminal motif, AAC(U) nCA. We found clear evidence that given pre-mRNAs can be joined to alternative SLs, with 103 distinct transcripts in the data set found to be trans-spliced either by Ppi_SLA or Ppi_SLB groups. Among these multiple trans-spliced cDNAs, 12 showed different acceptor splice sites for Ppi_SLA and Ppi_SLB, whereas in all others splicing was at exactly the same position in the 5′ UTR (see the examples in Supplemental Data S1).
TABLE 1.
SL sequences from the ctenophores Pleurobrachia pileus and Mnemiopsis leidyi
Multiple SL sequences were also recovered from EST data of another ctenophore species, M. leidyi. Four different SL groups (Table 1, Mle_SLA, Mle_SLB, Mle_SLC, Mle_SLD), each with only one variant, were found in 3% of the 2968 assembled cDNAs. In terms of sequence similarity, all four Mle_SL groups are closely related to each other but different from the Ppi_SL variants. The absence of identical sequences between these two ctenophore species was confirmed by a negative search of Mle_SL sequences in P. pileus ESTs and of Ppi_SL sequences in M. leidyi ESTs.
High diversity of spliced leader groups in hydrozoans
Analysis of data sets for the hydrozoans H. magnipapillata and C. hemisphaerica revealed a high number of SL sequence variants per species. Given the H. magnipapillata genome analysis (see above), we assume that all of these variants correspond to distinct SL genes. It should be emphasized that the diversity in SL sequences is probably underestimated due to selective transcriptome representation and incomplete 5′ termini in the assembled ESTs. In the most complete EST data set, that of C. hemisphaerica, SL exons were detected in 23% of the approximately 19,000 distinct assembled cDNA sequences. SL sequence variety in C. hemisphaerica was even greater than that detected in ctenophore species, with five distinct groups of SL exon sequences (Table 2, Che_SLA to Che_SLE). Each SL sequence group showed several variants (putative genes), with the exception of Che_SLA, which despite being represented in >20% of trans-spliced cDNA assemblies showed only a single variant. Che_SLB group exons were detected most frequently, in over half of trans-spliced cDNA assemblies, while Che_SLC and Che_SLE exons were rare. The trans-splicing frequency for particular SL groups thus did not correlate with the number of member SL genes. We could not detect a common 5′ motif of Che_SL sequences, perhaps because most 5′ termini are incomplete in the assembled ESTs.
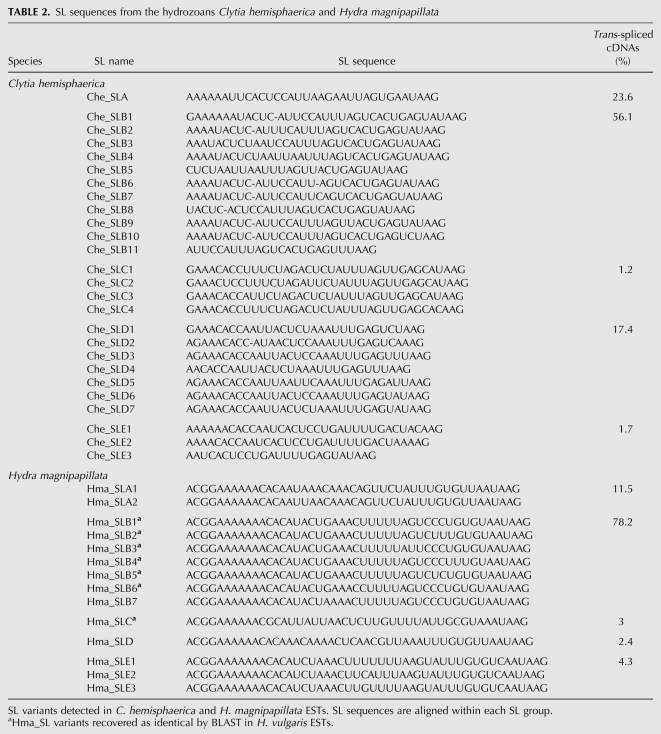
TABLE 2.
SL sequences from the hydrozoans Clytia hemisphaerica and Hydra magnipapillata
The H. magnipapillata EST set showed a similar overall pattern of SL use, with six spliced leader groups and a total of 15 variants detected among ∼3000 of 25,000 assembled cDNA sequences that showed trans-splicing. The Hma_SL groups were again found to be distinct but related, with a common 5′ motif ACGG(A)nC detectable in all six groups (the 5′ end of Hydra SL sequences were completed using genomic data) (Table 2). The relatively low percentage (12%) of trans-spliced sequences detected in the Hydra versus Clytia transcriptome data sets likely reflects in part differences in the origins or qualities of the cDNA libraries used for EST sequencing.
One SL group was detected in nearly 80% of trans-spliced H. magnipapillata cDNAs, and was designated Hma_SLB because of its 100% identical nucleotide sequence with the previously characterized SLB from H. vulgaris (Stover and Steele 2001). No sequence identical to the H. vulgaris SLA exon was detected in the H magnipapillata cDNA data set and genome; however, studies of genomic DNA revealed that H. vulgaris SLA corresponds to a sequence we designated Hma_SLA1, despite the low similarity of the two sequences (see below). Reverse searching of H. vulgaris EST data revealed the presence of most Hma_SLB and Hma_SLC variants, previously unreported, with SLB group exons again detected in the majority, indicating that most of the multiple SL genes are shared between these closely related Hydra species.
The absence of identical SL sequence between Hydra and Clytia was confirmed by negative BLAST searches for C. hemisphaerica SL sequences in the H. magnipapillata draft genome and for Hydra SL sequences in the Clytia ESTs. Although the sequences of SLs from different hydrozoan species (as well as between hydrozoan and ctenophoran SL exons) may well be evolutionarily related, the lack of sequence similarity between them was so great that it precluded phylogenetic analysis to evaluate their evolutionary relationships.
Rapid SL evolution at the genomic level in hydrozoans
The evidence for rapid SL gene evolution obtained from analysis of SL representation in the transcriptome was extended by comparison of two SL gene sequences and the surrounding genomic regions between Hydra species. A previous study in H. vulgaris revealed a spliced leader gene in each of two inter-5S rRNA gene regions amplified by PCR (Stover and Steele 2001). We aligned these with equivalent regions identified by BLAST from H. magnipapillata genome sequences. One of the regions contains the Hma_SLB1 gene in H. magnipapillata and its direct counterpart in H. vulgaris (Fig. 2A). The SLB1 exon is perfectly conserved between the two species, while the intron domain shows one difference per 10 nucleotides (Fig. 2B).
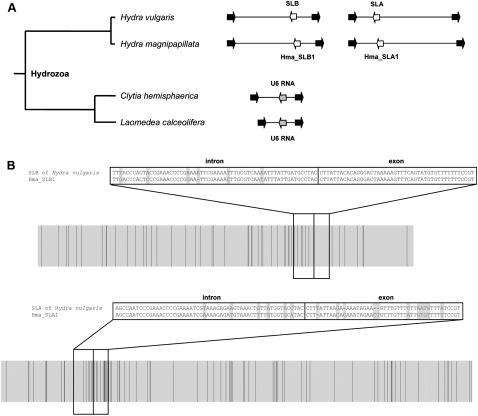
FIGURE 2.
Identification of hydrozoan SL genes. (A) Small RNA genes found in between 5S rRNA genes (black arrows) in hydrozoans. (B) Alignment of H. vulgaris and H. magnipapillata regions. (Light gray) Conserved positions, (darker bars) mutations (each indel was treated as one mutation, independent of length). Sequences of orthologous SL genes (intron and exon) encoded by this genomic regions are shown.
In contrast, the other inter-5S region provided evidence of clear SL gene diversification between the two Hydra species, containing the sequences defined as Hma_SLA1 in H. magnipapillata and as SLA in H. vulgaris. The identical genomic position and orientation of these clearly distinct SL sequences within this 5S rRNA gene cluster suggests a common origin from an ancestral SLA gene. The exon of this SLA gene has clearly evolved faster than the intron domain (2.2 differences per 10 nucleotides and 1.1 differences per 10 nucleotides, for exon and intron domains, respectively); generally, the intron domain shows more differences than the exon domain (as observed for the SLB gene). It is also noteworthy that the SL genes evolved at least as fast as the surrounding 5S intergenic regions (1.6 differences per 10 nucleotides, 0.6 differences per 10 nucleotides, and 0.5 differences per 10 nucleotides, for the SLA gene, SLB gene, and 5S intergenic regions without SL genes, respectively). Although limited, these genomic sequence comparisons provide evidence for rapid SL gene evolution in Hydra, with the exon domains showing different degrees of divergence.
PCR amplification of inter-5S gene regions from genomic DNA of C. hemisphaerica and another hydrozoan belonging to the same family (Campanulariidae), Laomedea calceolifera, yielded a single equivalent genomic fragment flanked by 5S genes (Fig. 2A), but no Clytia SL sequences were identified. In both species, the recovered inter-5S region housed a highly conserved U6 RNA gene in the same orientation (Fig. 2A; Supplemental Data S2). In contrast, in analysis of the H. magnipapillata genome, the multiple copies of U6 RNA genes identified by BLAST did not show an association with 5S rRNA genes, being positioned in different assembled scaffolds. It is clearly not possible to draw strong conclusions about SL evolution at the genomic level from the limited examples examined here; however, our findings are consistent with the marked plasticity in the linkage between SL and 5S ribosomal genes well demonstrated in the nematodes (Drouin and de Sa 1995).
Indiscriminate SL trans-splicing in hydrozoans
The availability of a large transcriptome data set for C. hemisphaerica and H. magnipapillata allowed us to address whether particular SL exons showed any qualitative preferences in trans-splicing within and/or between species. We first assigned each trans-spliced cDNA sequence from C. hemisphaerica and H. magnipapillata to putative cnidarian “orthology clusters” (OCs) (see Materials and Methods), each of them representing a single common gene. As previously noted for HTK32 and Syk genes in H. vulgaris (Stover and Steele 2001), many OCs in our data set showed trans-splicing by different SL groups, with 35% of C. hemisphaerica OCs being trans-spliced by more than one SL group (Fig. 3A; Supplemental Data S3). Trans-splicing of given mRNAs to alternative SLs was also detectable in the H. magnipapillata data set, albeit in only 1% of OCs (Fig. 3A).
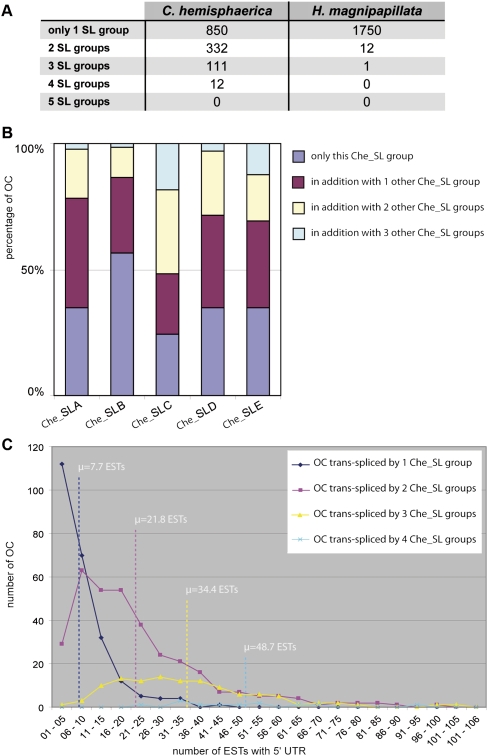
FIGURE 3.
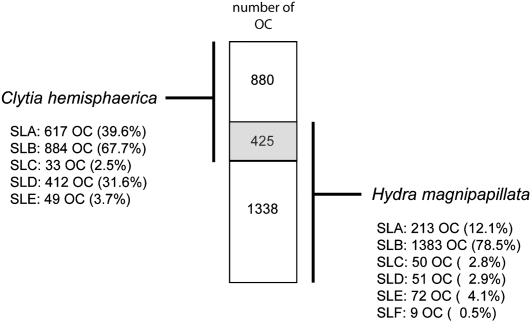
Indiscriminate SL trans-splicing in Hydrozoa. (A) Number of orthologous clusters (OCs) trans-spliced by one, two, three, four, and five SL groups identified in the C. hemisphaerica and H. magnipapillata OC data set. (B) Percentage of OCs in which a given Che_SL group is found solely with or in addition to other Che_SL groups among C. hemisphaerica cDNAs. The numbers of OCs analyzed here are 617, 884, 33, 412, and 49 for Che_SLA, Che_SLB, Che_SLC, Che_SLD, and Che_SLE, respectively. (C) Number of ESTs with a 5′ UTR in OCs trans-spliced in C. hemisphaerica cDNAs by one, two, three, and four SL groups. For each OC subset, the mean number of ESTs with a 5′ UTR is given (μ, dashed line).
The data set of 2643 trans-spliced OCs used in this analysis was assembled from 2967 cDNA sequences from C. hemisphaerica and 3054 from H. magnipapillata. Of these OCs, 880 contained C. hemisphaerica but not H. magnipapillata sequences, 1338 only H. magnipapillata sequences, and 425 sequences from both species (Fig. 4). The representation of each SL group in this 2643 OC data set corresponded to that in the original cDNA sequence collection. Among the 425 OCs trans-spliced in both species there was no preference for particular Che_SL groups and Hma_SL groups to associate with common transcripts, or for particular Che_SLs to do so among the 455 OCs showing trans-splicing to more than one SL. In both cases, the SL groups were found proportionally represented among trans-spliced OCs (data not shown). Moreover, examples were recovered of individual OCs trans-spliced by one of any of the five Che_SL groups and one, two, or three of the other groups (Fig. 3B). Taken together, these observations strongly argue in favor of indiscriminate use of all SL groups in hydrozoans.
FIGURE 4.
Overlap between Clytia and Hydra trans-spliced transcripts. Schematic representation of the orthologous clusters (OCs) data set constructed in this study: Among the 2643 OCs, 425 display trans-spliced cDNAs from both species. For each hydrozoan species, percentages of SL groups observed in this OC data set are indicated.
We further showed that the number of Che_SL groups found spliced to a given mRNA was proportional to the corresponding representation of ESTs in the data set. Thus, OCs trans-spliced by one, two, three, and four Che_SL groups had an average of seven, 21, 34, and 49 corresponding 5′ ESTs, respectively (Fig. 3C). Incidentally, these analyses imply that the proportions of mRNAs able to be trans-spliced by more than one SL calculated from our OC data set, as well as the number of distinct SL exons obtained from the whole EST data sets, are almost certainly underestimates.
Finally, as in Pleurobrachia, we uncovered examples of transcripts trans-spliced at alternative sites. These alternative SL trans-splicing acceptor sites were found in 9% of OCs trans-spliced by at least two SL groups, with the splice acceptor sites always positioned closely together (2–20 nt apart) (Supplemental Data S3).
Spliced leader trans-splicing is favored in adenosine-rich 5′ UTRs in hydrozoans
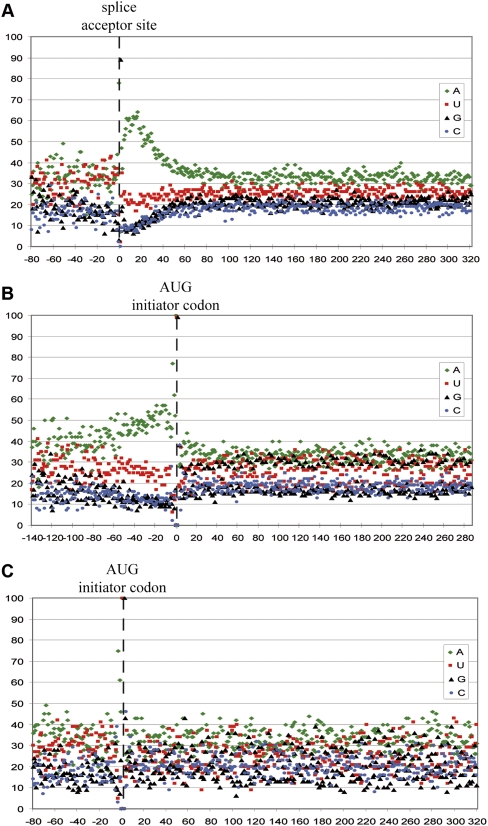
Studies of C. elegans polycistronic mRNAs have demonstrated the presence of particular nucleotide contexts favoring SL trans-splicing (Graber et al. 2007). In an attempt to detect a similar environment in hydrozoan trans-spliced cDNAs, 78 C. hemisphaerica cDNA sequences containing outrons (i.e., non-SL 5′ termini) were identified by comparing non-trans-spliced cDNA sequences with the trans-spliced OC data set. We added to this collection 400 further trans-spliced cDNAs chosen at random, from which SL sequences were removed and aligned with respect to their splice acceptor site (Fig. 5A). Nucleoside proportions per site along this alignment reveal a marked local enrichment of adenosine just downstream of the splice site (reaching a peak of 65% ∼15 bases from the splice acceptor site), whereas no particular enrichment was detected in the outron. An identical enrichment of adenosine was observed using H. magnipapillata trans-spliced cDNAs (400 trans-spliced cDNAs chosen at random and aligned with respect to their splice acceptor site; data not shown), indicating that this feature is common to these two hydrozoan species. When the Clytia cDNA sequence data set was aligned with respect to the AUG translation initiation codon (predicted by GENSCAN), the adenosine enrichment was again detected but was spread more broadly along the 60 bases upstream of the AUG initiation codon (Fig. 5B), indicating that the A-rich area is more likely to be linked to SL trans-splicing than to translation initiation. In contrast, no particular nucleoside enrichment was detected in the 5′ UTRs of 88 non-trans-spliced cDNAs of C. hemisphaerica chosen from conserved and well studied proteins, e.g., ribosomal proteins (Philippe et al. 2009) and developmental regulator genes (Chevalier et al. 2006; Momose et al. 2008; Amiel et al. 2009), except in the Kozac environment (first 3 base pairs [bp] upstream of the AUG initiator codon) (Fig. 5B,C). The random nucleoside distribution in the 5′ UTR of non-trans-spliced cDNAs argues in favor of a functional link between the SL trans-splicing and the A-rich environment and suggests that certain A-rich 5′ UTR compositions are favored for SL trans-splicing.
FIGURE 5.
Analysis of trans-spliced and non-trans-spliced C. hemisphaerica 5′ UTRs. (A) Distribution of nucleotide percentages per position along 478 trans-spliced cDNAs of C. hemisphaerica aligned on the splice acceptor site. (B) Distribution of nucleotide percentages per position along the same data set of trans-spliced cDNAs of C. hemisphaerica aligned on the predicted ATG initiator codon. (C) Distribution of nucleotide percentages per position along 88 non-trans-spliced cDNAs of C. hemisphaerica aligned on the predicted ATG initiator codon. In order to avoid stochastic variations of nucleotide percentages in the 5′ end of incomplete 5′ termini in the assembled ESTs, the positions localized at the 5′ end were removed until the presence of at least 50 nt per position is shown in these three graphics.
A-rich regions in the 5′ UTRs of SL trans-spliced transcripts were not detected outside hydrozoans: No particular nucleotide enrichment was detected downstream of the splice site in SL-trans-spliced cDNAs identified from the ctenophore P. pileus (all 3651 trans-spliced cDNAs) and the nematode Caenorhabditis elegans (300 cDNAs trans-spliced by SL1 plus 300 cDNAs trans-spliced by SL2, randomly chosen), or in trans-spliced cDNAs identified from ESTs of the urochordate Oikopleura dioica (Ganot et al. 2004). It remains possible that more subtle features of the nucleoside context are associated with SL trans-splicing in certain mRNAs from these species.
DISCUSSION
In this study we exploited the availability of large EST data sets to undertake a survey of SL usage in the basal metazoan phyla and related unicellular organisms, and a detailed comparative analysis of SL usage between the hydrozoans C. hemisphaerica and H. magnipapillata. Our results have shed new light on the origin and evolution of SL trans-splicing. They strongly reinforce a scenario in which SL genes have had multiple origins and rapid modification during animal evolution, although of course the possibility of loss of SL usage in one or more evolutionary lineages cannot be ruled out.
Concerning the phylogenetic occurrence of SL usage, our character optimization (Fig. 1) supports its multiple independent origins in ctenophores, hydrozoans, and various bilaterian lineages. This convergence hypothesis is consistent with the high diversity of SL genes in terms of sequences within Eumetazoa and their multiple proposed functions (discussed below). Note that convergence also almost certainly occurred at the wider scale of eukaryotes as unambiguously indicated by the occurrence of SL genes in distantly related eukaryote groups, such as euglenozoans and dinoflagellates. Equivalent conclusions were reached in a parallel study using a very similar approach on EST from a wide range of metazoan species, published while our work was under review (Douris et al. 2009).
An attractive hypothesis to explain multiple evolutionary origins for the SL genes is that they have derived repeatedly from U-rich small nuclear RNAs (snRNAs) of the Sm-class involved in the nuclear spliceosome machinery (for reviews, see Nilsen 2001; Hastings 2005). Like the SL RNAs, these snRNAs (U1, U2, U4, and U5) are characterized by a TMG cap and a U-rich Sm-protein-binding site, and are present in multiple copies per genome. The most likely candidate SL ancestor is the U1snRNA, which is the only snRNA absent from the trans-splicing splicosome, and which can acquire trans-splicing ability with just a few nucleotide changes (Bruzik et al. 1988; Bruzik and Steitz 1990; Hannon et al. 1992). Duplications of the U1 snRNA gene followed by just a few mutations would be sufficient to lead to the acquisition of trans-splicing, since SL pre-RNAs introduced into cells of non-trans-splicing species have been shown to undergo trans-splicing (Bruzik and Maniatis 1992).
Like the SL genes, snRNA genes occupy inter-5S gene regions in some species (Drouin and de Sa 1995; Ebel et al. 1999; Ganot et al. 2004; Lidie and van Dolah 2007), encouraging hypotheses of an evolutionary relationship. Furthermore, the presence of pseudogenes in 5S rRNA clusters (Jacq et al. 1977) is indicative of frequent gene duplications in this region of the genome. The majority of SL-positive species examined, however, including Hydra (this study), do not show this association (Aksoy et al. 1992; Keller et al. 1992; Liu et al. 1996; Zhang et al. 2009), while the U6 genes positioned between 5S rRNA genes Clytia and Laomedea are clearly unlikely to have a recent evolutionary relationship with the Sm-binding U-rich snRNA group or the SL genes, being transcribed by a different RNA polymerase (Pol III) and lacking their TMG cap. Co-accummulation of SL, snRNA, and 5S rRNA genes may have been favored during evolution in some species, either independently or as a result of co-regulation (Lidie and van Dolah 2007). Further analysis of gene content in 5S rRNA gene clusters, especially in non-SL trans-splicing species, may help to resolve this issue (see Drouin and De Sa 1995). Recent analysis in a selection of metazoans uncovered an association of U1 snRNA and SL genes in the amphipod crustacean Parhyale, but not in the ctenophore Mnemiopsis, the chaetognath Spadella, or the bdelloid rotifer Adineta, with no 5S rRNA association detected in any of these species (Douris et al. 2009).
Our comparison of SL trans-splicing in two hydrozoan species provides a striking demonstration of very rapid SL evolution. Both C. hemisphaerica and H. magnipapillata exhibit a high number of different SL exon sequences, divided into five and six SL groups, respectively. In the case of Hydra we were able to identify all but one of the identified variants as a distinct gene from genome sequence data, indicating that the SL groups represent gene families. The SL groups have clearly diversified rapidly, as shown by a lack of clear sequence conservation between these two distant hydrozoan species, and by the marked divergence from a common ancestral gene of H. vulgaris SLA and H. magnipapillata SLA1. Since the rate of SL sequence evolution in these species seems similar to that of the intergenic 5S region, and SL exon vs. intron substitution rates are not significantly different, SL sequences appear essentially unconstrained.
The high diversity and rapid evolution of SL in hydrozoans is very similar to the situation reported in the nematode Trichinella spirallis (Pettitt et al. 2008) and in the Chaetognaths (Marletaz et al. 2008), as well as in ctenophores (this study), where multiple SL groups from P. pileus and M. leidyi EST data were found to be closely related but distinct. In contrast, urochordates (Ganot et al. 2004; Satou et al. 2006) and SL-positive unicellular eukaryotes (Gibson et al. 2000; Zhang et al. 2007), show only one SL exon per species (or one SL group), while in Rhabditida nematodes SL1 and SL2 RNAs show functional dichotomy associated with an atypically high level of conservation of the SL1 sequence between species (Blumenthal 2005; Guiliano and Blaxter 2006). The lower level of SL group diversification in these latter groups may be explained in part by stabilization of SL usage following the acquisition of a function in polycistronic transcript resolution in Rhabditida nematodes and urochordates, or changes in the translational machinery to accommodate the altered 5′ cap structure in trypanosomes. More detailed analysis of the diversity and evolution of SL sequences in other lineages such as flatworms, acoels, and Bdeloid rotifers may help to confirm whether rapid and high diversification of SL genes is a general rule, and to uncover any potential link between evolution rates and SL function. Flatworms may provide an interesting case, as the AUG initiator codons of some trans-spliced mRNAs are provided by the 3′ end of SL sequences, which might be predicted to constrain evolution of the SL sequences (Cheng et al. 2006).
In line with the hypothesis that the acquisition (or retention) of specific functions for SL trans-splicing in certain lineages might constrain SL gene evolution, we were unable to find any evidence for distinct functions for the diversified hydrozoan SL groups. Firstly, there was no evidence for preferential trans-splicing of a particular set of orthologous mRNAs in both Clytia and Hydra. This tendency was confirmed for particular proteins or protein classes. For instance, no common trans-spliced ribosomal protein mRNAs could be identified in the C. hemisphaerica and H. magnipapillata EST data sets: rpl7a, rpl10, rps27a, and rps35 are trans-spliced in C. hemisphaerica ESTs, whereas rpl9, rps2, rps6, and rps19 are trans-spliced in H. magnipapillata ESTs (among 46 and 73 ribosomal transcripts containing 5′ UTRs, respectively). Second, the correspondence in C. hemisphaerica between SL group diversity for a given mRNA and the number of 5′ ESTs in the data set suggests that any of the SL groups can be added to a given mRNA being processed. Furthermore, no preference of particular SLs to splice given mRNAs within a species was found. On the other hand, the different SL variants were not represented proportionally in the hydrozoan EST data set. This suggests that there may be a preference for certain SL genes to be used for trans-splicing, although we cannot completely rule out sampling artifacts. Further analysis of SL genes in hydrozoan genomes should clarify this point. Overall, the apparently indiscriminate use of SL sequences indicates that distinct SL groups have no functional specialization, but may share common function(s) such as 5′ UTR sanitization.
While we found no evidence for a functional specialization of SL usage in hydrozoans, we did uncover a marked tendency for SL trans-spliced transcripts to contain an unexpected local enrichment of adenosine in the 5′ UTR prior to splicing, suggesting that a particular nucleoside environment favors SL trans-splicing. In the two hydrozoan data sets, SL addition was found to occur ∼15 bp upstream of an A-rich region. No such regions were detected in non-trans-spliced mRNAs, indicating that pre-mRNAs may be selected for trans-splicing on the basis of 5′ UTR composition rather than the coded protein. We were not able to detect any nucleoside enrichment in relation to the splice site in other species, although more subtle motifs may be present. Thus, the correlation between SL trans-splicing and an enrichment of adenosine in the 5′ UTR appears to be a hydrozoan-specific feature of SL trans-splicing.
The overall picture of SL evolution emerging from our analyses together with published studies is of frequent SL gene acquisitions, perhaps following mutations in U-rich snRNAs of the spliceosome, followed by rapid sequence evolution and frequent gene duplications. In some cases, modification of the 5′ UTR in some or all mRNAs by SL trans-splicing may have had consequences for the machinery regulating translation, stability, or operon processing, and thus have constrained subsequent SL sequence evolution (Hastings 2005). As more EST data become available from a range of phylogenetic lineages, this hypothesis can be further examined. The Hydrozoa, a broad and diverse monophyletic group comprising about 3700 described species (Collins et al. 2006), is a promising group in which to study SL trans-splicing. Hydra and now C. hemisphaerica (Houliston et al. 2010) are well-established experimental organisms, allowing experimental manipulation. Moreover, as demonstrated by 5S RNA cluster analysis, the close evolutionary scale represented by the Hydra genus offers an appropriate framework to catch SL sequence divergence. Finally, the newly available genome sequence of H. magnipapillata, soon to be joined by that of C. hemisphaerica (sequencing ongoing at the Genoscope), will allow large-scale comparative genomic analysis between these two distant hydrozoan species, for instance to investigate the distribution and evolution of operons (Guiliano and Blaxter 2006; Satou et al. 2006).
MATERIALS AND METHODS
Clytia and Pleurobrachia ESTs
EST sequencing for P. pileus (∼30,000) and C. hemisphaerica (∼90,000) was performed at the Genoscope from a mixture of normalized and nonnormalized cDNA libraries, constructed by Open Biosystems (through BioCat) and Express Genomics from microgram quantities of total RNA extracted from adult (Pp) or mixed embryonic, larval, and adult (Ch) stages (Chevalier et al. 2006; Philippe et al. 2009). All starting material was obtained from Villefranche-sur-Mer, with the C. hemisphaerica derived uniquely from three cultured strains (X, Y, and Z). These EST sequences are available in dbEST/GenBank (http://www.ncbi.nlm.nih.gov/dbEST/).
Detection of SL sequences in EST data sets
To recover SL exon sequences, the following steps were performed independently on each EST data set. The method was validated by recovering known SL exons from C. elegans ESTs and from H. vulgaris ESTs. Cleaned ESTs with vector sequences removed were assembled into contigs using Phrap software. The assemblies were then searched for common sequences of 12 nt at the termini, and all elements present at least three times were aligned manually to reconstruct putative SL sequences. The putative SL sequences were then used to search by nucleotide BLAST the original assembled cDNA data set, and those found identically in at least five 5′ ends of contigs were defined as a SL variants. SL variants displaying three or less nucleotide changes then were considered to form a SL group.
Amplification of 5S rRNA gene repeats in hydrozoans
Genomic DNA was extracted from C. hemisphaerica male medusae by standard methods. L. calceolifera genomic DNA was provided by Peter Schuchert (Muséum d'histoire naturelle de Genève) and Lucas Leclère (Sars Institute). Amplification of inter-5S rRNA regions was performed using primers and PCR cycles as defined in Stover and Steele (2001), and inserted into pGEMt plansmid for sequencing.
Construction of hydrozoan trans-spliced orthologous clusters
Hydrozoan orthology clusters were compiled using the N. vectensis proteome as reference (Putnam et al. 2007), downloaded from the JGI website (http://www.jgi.doe.gov/). Each N. vectensis protein defines one OC. SL trans-spliced assembled cDNAs from C. hemisphaerica and H. magnipapillata ESTs were compared by BLAST against these protein sequences and were assigned to the OC corresponding to their best hit using a threshold value of 1e-10. For all OCs showing multiple SLs and for a selection of 250 among those with single SLs, we also retrieved from the original ESTs all orthologous non-SL trans-spliced transcripts for analysis of splice context. For each OC, cDNAs were aligned using MUSCLE (Edgar 2004), independently for each hydrozoan species, and alignments obtained were carefully checked by eye in order to detect the presence of different transcripts. Rare cases (<5%) in which at least two distinct transcripts were assigned to a single OC for one or both species were discarded to avoid complex multigenic families. The orthology of trans-spliced cDNAs from Clytia and Hydra was confirmed by successful phylogenetic analysis for 20 OCs chosen randomly among the 425 OCs displaying trans-spliced cDNAs from both species (data not shown).
Accession numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accession numbers for the 5S rRNA gene sequences identified in this study are GU126459 (C. hemisphaerica) and GU126460 (L. calceolifera).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Peter Schuchert and Lucas Leclère for providing L. calceolifera genomic DNA, and Sandra Chevalier for technical assistance. We thank our research colleagues and Philippe Ganot (Nice) for useful comments on the study, Mark Carrington (Cambridge) for critical reading of the manuscript, and Rob Steele (Irvine) for much useful input into this study. This work was supported by a grant from the GIS “Institut de la Génomique Marine” – ANR programme blanc NT_NV_52 Genocnidaire and ANR-09-BLAN-0236-01. EST sequencing was performed by the Consortium National de Recherche en Genomique at the Génoscope (Evry, France).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1975210.
REFERENCES
- Aksoy S, Shay GL, Villanueva MS, Beard CB, Richards FF. Spliced leader RNA sequences of Trypanosoma rangeli are organized within the 5S rRNA-encoding genes. Gene. 1992;113:239–243. doi: 10.1016/0378-1119(92)90401-a. [DOI] [PubMed] [Google Scholar]
- Amiel A, Leclere L, Robert L, Chevalier S, Houliston E. Conserved functions for Mos in eumetazoan oocyte maturation revealed by studies in a cnidarian. Curr Biol. 2009;19:305–311. doi: 10.1016/j.cub.2008.12.054. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. The C. elegans Research Community. WormBook. 2005. Trans-splicing and operons. http://www.wormbook.org. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Gleason K. Caenorhabditis elegans operons: Form and function. Nat Rev Genet. 2003;4:112–120. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Maniatis T. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992;360:692–695. doi: 10.1038/360692a0. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Steitz JA. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell. 1990;62:889–899. doi: 10.1016/0092-8674(90)90264-f. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Van Doren K, Hirsh D, Steitz JA. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- Cheng G, Cohen L, Ndegwa D, Davis RE. The flatworm spliced leader 3′-terminal AUG as a translation initiator methionine. J Biol Chem. 2006;281:733–743. doi: 10.1074/jbc.M506963200. [DOI] [PubMed] [Google Scholar]
- Chevalier S, Martin A, Leclere L, Amiel A, Houliston E. Polarised expression of FoxB and FoxQ2 genes during development of the hydrozoan Clytia hemisphaerica. Dev Genes Evol. 2006;216:709–720. doi: 10.1007/s00427-006-0103-6. [DOI] [PubMed] [Google Scholar]
- Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, Schierwater B. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol. 2006;55:97–115. doi: 10.1080/10635150500433615. [DOI] [PubMed] [Google Scholar]
- Davis RE. Spliced leader RNA trans-splicing in metazoa. Parasitol Today. 1996;12:33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- Davis RE. Surprising diversity and distribution of spliced leader RNAs in flatworms. Mol Biochem Parasitol. 1997;87:29–48. doi: 10.1016/s0166-6851(97)00040-6. [DOI] [PubMed] [Google Scholar]
- Davis R, Hodgson S. Gene linkage and steady state RNAs suggest trans-splicing may be associated with a polycistronic transcript in Schistosoma mansoni. Mol Biochem Parasitol. 1997;89:25–39. doi: 10.1016/s0166-6851(97)00097-2. [DOI] [PubMed] [Google Scholar]
- Douris V, Telford MJ, Averof M. Evidence for multiple independent origins of trans-splicing in Metazoa. Mol Biol Evol. 2009 doi: 10.1093/molbev/msp286. [DOI] [PubMed] [Google Scholar]
- Drouin G, de Sa M. The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol Biol Evol. 1995;12:481–493. doi: 10.1093/oxfordjournals.molbev.a040223. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Ebel C, Frantz C, Paulus F, Imbault P. Trans-splicing and cis-splicing in the colorless Euglenoid, Entosiphon sulcatum. Curr Genet. 1999;35:542–550. doi: 10.1007/s002940050451. [DOI] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Kallesoe T, Reinhardt R, Chourrout D, Thompson EM. Spliced-leader RNA trans splicing in a chordate, Oikopleura dioica, with a compact genome. Mol Cell Biol. 2004;24:7795–7805. doi: 10.1128/MCB.24.17.7795-7805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W, Bingle L, Blendeman W, Brown J, Wood J, Stevens J. Structure and sequence variation of the trypanosome spliced leader transcript. Mol Biochem Parasitol. 2000;107:269–277. doi: 10.1016/s0166-6851(00)00193-6. [DOI] [PubMed] [Google Scholar]
- Graber JH, Salisbury J, Hutchins LN, Blumenthal T. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA. 2007;13:1409–1426. doi: 10.1261/rna.596707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiliano D, Blaxter M. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. PLoS Genet. 2006;2:e198. doi: 10.1371/journal.pgen.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Yu YT, Hannon GE, Nilsen TW. Interaction of U6 snRNA with a sequence required for function of the nematode SL RNA in trans-splicing. Science. 1992;258:1775–1780. doi: 10.1126/science.1465612. [DOI] [PubMed] [Google Scholar]
- Hastings KE. SL trans-splicing: Easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Houliston E, Momose T, Manuel M. Clytia hemispherica: A jellyfish cousin joins the laboratory. Trends Genet. 2010 doi: 10.1016/j.tig.2010.01.008. (in press). [DOI] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguñà J, Bailly X, Jondelius U, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq C, Miller JR, Brownlee GG. A pseudogene structure in 5S DNA of Xenopus laevis. Cell. 1977;12:109–120. doi: 10.1016/0092-8674(77)90189-1. [DOI] [PubMed] [Google Scholar]
- Keller M, Tessier LH, Chan RL, Weil JH, Imbault P. In Euglena, spliced-leader RNA (SL-RNA) and 5S rRNA genes are tandemly repeated. Nucleic Acids Res. 1992;20:1711–1715. doi: 10.1093/nar/20.7.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Friedman CC, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE. Contribution of trans-splicing, 5′-leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J Biol Chem. 2004;279:45573–45585. doi: 10.1074/jbc.M407475200. [DOI] [PubMed] [Google Scholar]
- Lawrence J. Selfish operons: The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- Lidie KB, van Dolah FM. Spliced leader RNA-mediated trans-splicing in a dinoflagellate, Karenia brevis. J Eukaryot Microbiol. 2007;54:427–435. doi: 10.1111/j.1550-7408.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- Liu LX, Blaxter ML, Shi A. The 5S ribosomal RNA intergenic region of parasitic nematodes: Variation in size and presence of SL1 RNA. Mol Biochem Parasitol. 1996;83:235–239. doi: 10.1016/s0166-6851(96)02753-3. [DOI] [PubMed] [Google Scholar]
- Maddison W, Maddison D. Mesquite: A modular system for evolutionary analysis. Version 2.71. 2009. http://mesquiteproject.org.
- Marletaz F, Gilles A, Caubit X, Perez Y, Dossat C, Samain S, Gyapay G, Wincker P, Le Parco Y. Chaetognath transcriptome reveals ancestral and unique features among bilaterians. Genome Biol. 2008;9:R94. doi: 10.1186/gb-2008-9-6-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Landfear SM, Wirth DF. Cloning and characterization of a Leishmania gene encoding a RNA spliced leader sequence. Nucleic Acids Res. 1986;14:7341–7360. doi: 10.1093/nar/14.18.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T, Derelle R, Houliston E. A maternally localised Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica. Development. 2008;135:2105–2113. doi: 10.1242/dev.021543. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. Evolutionary origin of SL-addition trans-splicing: Still an enigma. Trends Genet. 2001;17:678–680. doi: 10.1016/s0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Muller B, Stansfield I, Connolly B. Spliced leader trans-splicing in the nematode Trichinella spiralis uses highly polymorphic, noncanonical spliced leaders. RNA. 2008;14:760–770. doi: 10.1261/rna.948008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Pouchkina-Stantcheva N, Tunnacliffe A. Spliced leader RNA-mediated trans-splicing in phylum Rotifera. Mol Biol Evol. 2005;22:1482–1489. doi: 10.1093/molbev/msi139. [DOI] [PubMed] [Google Scholar]
- Putnam N, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov V, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Splicing in the eukaryotic ancestor: Form, function, and dysfunction. Trends Ecol Evol. 2009;24:447–455. doi: 10.1016/j.tree.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Satou Y, Hamaguchi M, Takeuchi K, Hastings K, Satoh N. Genomic overview of mRNA 5′-leader trans-splicing in the ascidian Ciona intestinalis. Nucleic Acids Res. 2006;34:3378–3388. doi: 10.1093/nar/gkl418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R. Concatenated analysis sheds light on early metazoan evolution and fuels a modern ‘urmetazoon’ hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover N, Steele R. Trans-spliced leader addition to mRNAs in a cnidarian. Proc Natl Acad Sci. 2001;98:5693–5698. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga K, Mark Welch D, Tanaka Y, Sakakura Y, Hagiwara A. Analysis of expressed sequence tags of the cyclically parthenogenetic rotifer Brachionus plicatilis. PLoS One. 2007;2:e671. doi: 10.1371/journal.pone.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier LH, Keller M, Chan RL, Fournier R, Weil JH, Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe AE, Meedel TH, Hastings KE. mRNA 5′-leader trans-splicing in the chordates. Genes & Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner GM, Sturm NR, Campbell DA. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J Biol Chem. 2003;278:38269–38275. doi: 10.1074/jbc.M304295200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lin S. Retrieval of missing spliced leader in dinoflagellates. PLoS One. 2009;4:e4129. doi: 10.1371/journal.pone.0004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Campbell DA, Sturm NR, Lin S. Dinoflagellate spliced leader RNA genes display a variety of sequences and genomic arrangements. Mol Biol Evol. 2009;26:1757–1771. doi: 10.1093/molbev/msp083. [DOI] [PMC free article] [PubMed] [Google Scholar]