Abstract
Upstream open reading frames (uORFs) are protein coding elements in the 5′ leader of messenger RNAs. uORFs generally inhibit translation of the main ORF because ribosomes that perform translation elongation suffer either permanent or conditional loss of reinitiation competence. After conditional loss, reinitiation competence may be regained by, at the minimum, reacquisition of a fresh methionyl-tRNA. The conserved h subunit of Arabidopsis eukaryotic initiation factor 3 (eIF3) mitigates the inhibitory effects of certain uORFs. Here, we define more precisely how this occurs, by combining gene expression data from mutated 5′ leaders of Arabidopsis AtbZip11 (At4g34590) and yeast GCN4 with a computational model of translation initiation in wild-type and eif3h mutant plants. Of the four phylogenetically conserved uORFs in AtbZip11, three are inhibitory to translation, while one is anti-inhibitory. The mutation in eIF3h has no major effect on uORF start codon recognition. Instead, eIF3h supports efficient reinitiation after uORF translation. Modeling suggested that the permanent loss of reinitiation competence during uORF translation occurs at a faster rate in the mutant than in the wild type. Thus, eIF3h ensures that a fraction of uORF-translating ribosomes retain their competence to resume scanning. Experiments using the yeast GCN4 leader provided no evidence that eIF3h fosters tRNA reaquisition. Together, these results attribute a specific molecular function in translation initiation to an individual eIF3 subunit in a multicellular eukaryote.
Keywords: translation regulation, uORF, reinitiation, eIF3, modeling
INTRODUCTION
Upstream open reading frames (uORFs) are short protein coding regions located in the 5′ leader of many eukaryotic mRNAs. Upstream ORFs are found in about one-third of experimentally supported 5′ leaders in Arabidopsis and occur at even higher frequencies among transcription factors and protein kinases (Kim et al. 2007). According to the scanning model of translation initiation, uORFs are expected to generally suppress efficient initiation at the start codon of the main ORF. However, when the translational repression by the uORF is compensated in response to specific signals, translation becomes regulated (Hanfrey et al. 2005). For example, in Arabidopsis, an uORF-encoded peptide in the mRNA for the basic leucine zipper transcription factor, AtbZip11, mediates translational inhibition by sucrose (Rook et al. 1998; Wiese et al. 2004; Rahmani et al. 2009). In yeast, the uORFs in the 5′ leader of the bZip transcription factor GCN4 cause a translational derepression in response to amino acid starvation (Hinnebusch 2005). A mechanistically similar regulatory module exists in mammalian cells, where phosphorylation of eIF2α by one of a number of different eIF2 kinases depresses general translation, all the while boosting translation of mRNAs such as ATF4 with suitably balanced uORF patterns (Harding et al. 2000). Upstream ORFs also function in complex regulatory modules dedicated to internal ribosome entry (Yaman et al. 2003) as well as translation reinitiation or shunting (Park et al. 2001, 2004; Ryabova et al. 2004).
The great majority of uORFs are not conserved at the peptide level (Hayden and Jorgensen 2007). However, occasionally, the peptide sequence encoded by the uORF determines the fate of the mRNA (Hanfrey et al. 2005; Rahmani et al. 2009), in particular if rare codons are present, if the peptide blocks elongation while in the ribosome exit channel (Wang et al. 1998), or if translation termination is stalled (Gaba et al. 2001, 2005). According to the scanning model of translation initiation, the 40S ribosomal subunit in association with eIF3 and the ternary complex consisting of eIF2-GTP-methionyl-tRNAMet scans along the mRNA and is poised to recognize the first AUG start codon it encounters (Kozak 2002). The start codon is recognized primarily through codon–anticodon pairing with tRNAMet, although the accuracy of AUG recognition is enhanced by eIF1 and eIF1A (Pestova et al. 1998; Maag et al. 2006; Fekete et al. 2007) and eIF3 (Nielsen et al. 2004). At this point, the 60S large ribosomal subunit joins the complex and translation elongation commences. A uORF poses a barrier to the scanning 40S ribosome because, upon recognition of the uORF start codon, the uORF peptide must be translated and terminated. A ribosome whose 40S subunit disassociates from the mRNA after termination can be regarded as having suffered a permanent loss of reinitiation competence. Conversely, a 40S ribosome that resumes scanning downstream from the uORF displays a conditional loss of reinitiation competence because it lacks a ternary complex. The regaining of full reinitiation competence requires, at the least, that the 40S reacquire a fresh ternary complex, in order to successfully recognize the start codon of the main ORF further downstream (Kozak 1987, 2001). The molecular factors responsible for resumption of scanning and acquisition of a fresh ternary complex are largely unknown. The efficiency with which scanning resumes is affected by a number of factors, including the length of the uORF, the time it took to translate it (Kozak 2001; Rajkowitsch et al. 2004), as well as the translation initiation factors involved in the initial initiation event (Pöyry et al. 2004). These data point to a form of molecular memory, possibly in the form of initiation factors clinging to the 80S ribosome for a limited period of time during translation elongation on a uORF. Because eIF2 loses its tRNA and GTP cofactor in the process of start codon recognition, and because eIF1 and 1A bind at the 40S–60S interface and must surely be displaced before subunit joining can take place (Lomakin et al. 2003; Maag and Lorsch 2003), the most likely known eIF to maintain a physical or functional association with the ribosome during the initial phase of elongation is eIF3. This notion gained additional credibility when cryoelectron microscopy of the 40S-eIF3 complex located eIF3 on the back side of the 40S ribosome, near the mRNA exit channel (Siridechadilok et al. 2005). Reinitiation requires, first, that the post-termination 40S ribosome has not permanently lost its competence to resume the scanning mode, and second that the 40S overcome its conditional loss of reinitiation competence by regaining a fresh ternary complex. Studies in yeast have implicated specific eIF3 subunits in this process (Valasek et al. 2002; Nielsen et al. 2004; Szamecz et al. 2008), extending earlier biochemical data demonstrating that eIF3 bolsters ternary complex loading onto the 40S (Benne and Hershey 1978).
The eIF3 complex is the largest initiation factor. While functions have been assigned in yeast to the five subunits shared between yeast and other eukaryotes, the individual contributions of the remaining seven eIF3 subunits, which are not recognizably conserved in budding yeast, are largely unknown (Hinnebusch 2006). The h subunit of eIF3 is a 38-kDa MPN (Mpr1-Pad1-N-terminus)-domain protein conserved among fission yeast, plants, and metazoans. Plants mutated in eif3h show defects in translation of mRNAs harboring uORFs (Kim et al. 2004, 2007). To identify the role of eIF3 more precisely, we performed a detailed mutational dissection of the Arabidopsis AtbZip11 leader, which harbors a cluster of phylogenetically conserved uORFs. The major contribution of eIF3h is not for start codon recognition, but to facilitate reinitiation. Specifically, our data suggest that eIF3h helps to prevent the permanent loss of reinitiation competence, so that scanning can resume after termination. These conclusions were informed by, compatible with, and extended using computational modeling of translation initiation in wild-type and eif3h mutant plants.
RESULTS
Mutant eIF3h protein does not associate tightly with 43S complexes
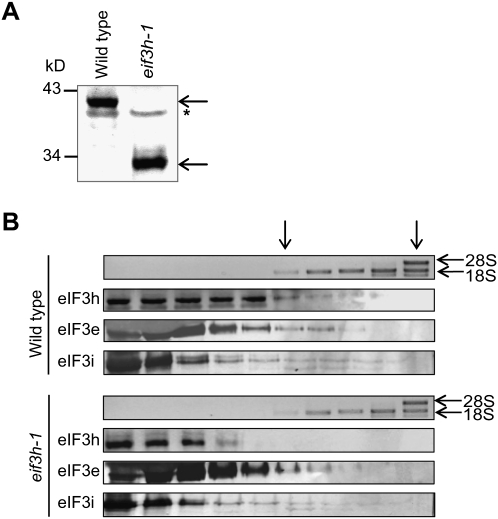
Plants harboring T-DNA insertions in eIF3h continue to express carboxyl-terminally truncated eIF3h protein (Fig. 1A; Kim et al. 2004). We examined whether the truncated eIF3h-1 protein is associated with multifactor complexes harboring eIF3, such as the 43S complex (Fig. 1B). In wild-type plants, a small portion of eIF3e, eIF3i, and wild-type eIF3h were detected in sucrose gradient fractions containing fast sedimenting complexes (fractions 5–9), including 40S ribosomes marked by the 18S rRNA. In eif3h-1 mutant plants, eIF3e and eIF3i were found in similar fractions, while the truncated eIF3h-1 protein was found primarily at the top of the gradient. We conclude that the eIF3h-1 mutant protein has a reduced tendency to associate with other eIF3 subunits into multifactor complexes, and may be lacking from 43S and 48S preinitiation complexes.
FIGURE 1.
In eif3h mutant plants, 43S complexes lack eIF3h. (A) Wild-type eIF3h and truncated eIF3h expressed in eif3h-1 mutant plants were visualized by immunoblotting with anti-eIF3h antibody. (*) Crossreacting protein. (B) Sucrose gradients from wild-type and eif3h mutant plants were examined for 18S ribosomal RNAs to identify 40S subunits using electrophoresis and ethidium bromide staining. Corresponding protein extracts were examined for eIF3 subunits by SDS-PAGE and immunoblotting. (Arrows) Topmost fractions containing 40S and 60S subunits.
eIF3h supports translation of the uORF-containing AtbZip11 mRNA
To elucidate the molecular mechanism of the h subunit of eIF3 in a living biological context, we defined the deficiencies in expression that can be observed on a panel of mutant 5′ leader sequences derived from our model system, the 5′ leader of Arabidopsis AtbZip11, which is dependent on wild-type eIF3h for optimal expression (Kim et al. 2004).
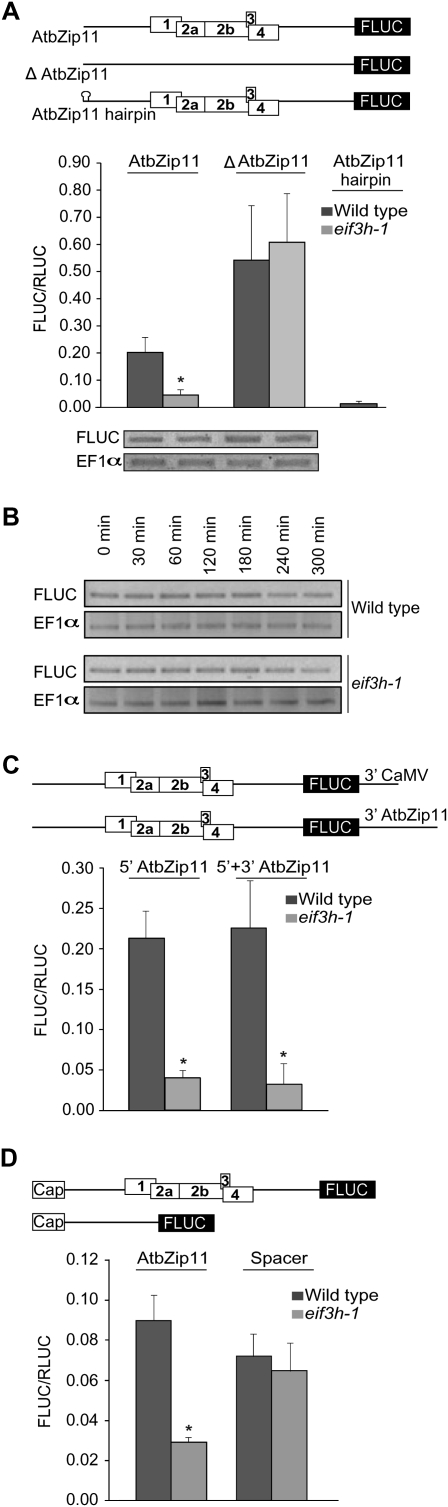
In transient expression assays using firefly luciferase (FLUC) as a reporter, the cluster of four uORFs in the AtbZip11 leader repressed expression by about twofold in the wild type but by about 10-fold in eif3h mutant seedlings. The dependence on eIF3h was attributed primarily to direct translational inhibition, rather than reduced abundance or decreased stability of the AtbZip11-FLUC mRNA (Fig. 2A,B). Notably, translation of the AtbZip11 leader in the eif3h mutant was no longer impeded once all five upstream AUG start codons (uAUGs) were removed by site-directed mutagenesis. Introducing a stable hairpin (ΔG = −42.8 kcal/mol) (Kozak 1986) near the 5′ end dramatically reduced expression, thus ruling out that translation occurs by internal ribosome entry or from a truncated transcript (Fig. 2A). Repression by uORFs is sometimes controlled by elements in the 3′ UTR (Mehta et al. 2006). However, the translational inhibition was not rescued by including the native 3′ untranslated region (UTR) from AtbZip11 instead of the heterologous 3′ UTR from cauliflower mosaic virus (Fig. 2C). Similar results were obtained using transformation of Arabidopsis seedlings with expression plasmids (Fig. 2A) and transformation of Arabidopsis protoplasts with capped in vitro transcripts (Fig. 2D), which rules out that expression in the wild type is high because uORFs are spliced out. In summary, the data indicate that initiation on the AtbZip11 leader occurs by ribosome scanning from the 5′ cap. Moreover, the wild-type ribosome must traverse the hurdle imposed by the uORF cluster, either by leaky scanning across the uAUGs or by uORF translation followed by reinitiation.
FIGURE 2.
eIF3h-dependent translational regulation of the AtbZip11 5′ leader. (A) (Top) Schematic of the FLUC reporter coding region fused to original and mutated 5′ leaders of AtbZip11 (590 nt long). Only the four uORFs are drawn to scale (white boxes; 18, 42, 5, and 19 amino acids, respectively). (Middle) Transient expression data from 10-d-old wild-type and eif3h mutant seedlings. The efficiency of translation was calculated as the activity ratio of FLUC versus Renilla luciferase (RLUC) as a co-transformed reference gene. Bars, standard error. (*) P < 0.002 for wild-type (WT) versus eif3h in a two-sided Students t-test. (Bottom) Respective transcript levels of FLUC reporter and translation elongation factor 1α (EF1α) as a control were compared by RT-PCR. (B) Turnover of AtbZip11-FLUC mRNA expressed in stable transgenic plants (Kim et al. 2007) was measured by RT-PCR after blocking transcription with 0.1 mg/mL cordycepin. EF1α served as a control. Neither mRNA was labile over this time course. (C) Translation of the AtbZip11-FLUC reporter construct with the default 3′ UTR from cauliflower mosaic virus was similar to that with the native AtbZip11 3′ UTR. (*) P < 0.002 (n = 5). (D) Undertranslation of the AtbZip11 5′ leader in the eif3h mutant in Arabidopsis protoplasts transformed with in vitro transcripts. “Spacer” is an example for a uORF-less leader that is unaffected by eif3h.
Which uORFs are most inhibitory in the eif3h mutant?
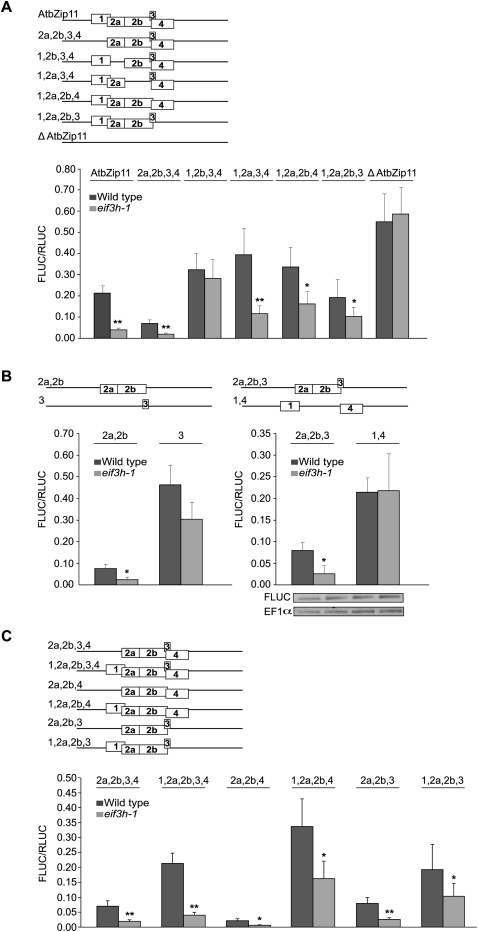
To determine whether the translational defect in the eif3h mutant could be attributed to one specific uORF, each uAUG in AtbZip11 was mutated systematically (Fig. 3A). The only uORF that was clearly essential for eIF3h-dependent translation was uORF 2a. However, individual removal of uORFs 2b, 3, or 4 also caused derepression, especially in eif3h. The presence of uORF 1, which should impede recognition of uORF 2 by virtue of overlapping with it, had an anti-inhibitory effect (Fig. 3A,C). The simplest uORF arrangement with the clearest eIF3h dependence consisted of uORFs 2 and 3 (Fig. 3B). Taken together, all uORFs affect expression and contribute to the requirement for eIF3h, either directly or indirectly, with uORF2 playing a predominant role.
FIGURE 3.
Contribution of individual uAUGs to translational repression and eIF3h-dependent translation of AtbZip11-FLUC. (A) Single uAUGs were eliminated by site-directed mutagenesis. Note the anti-inhibitory effect of uORF 1 (also see C). Error bars represent standard error (n = 8–11). Data on the original and uORF-less leaders are included from Figure 2A for comparison. (*) Statistical significance at P < 0.01, (**) P < 0.002. (B) The combination of uORFs 2 and 3 caused a robust dependence on eIF3h, in contrast to uORFs 1 and 4 together (n = 8–12). eIF3h dependence of uORF2 alone was also significant but that of uORF3 alone was not. (*) Statistical significance at P < 0.01. (Right panel) AtbZip-FLUC and endogenous EF1α mRNA levels were estimated by RT-PCR. (C) A pairwise comparison demonstrating the anti-inhibitory effect of uORF1 in the AtbZip11 leader. Note that leaders harboring uORF1 are still eIF3h-dependent. Bars, standard error (n = 9–22). (*) Statistical significance at P < 0.04, (**) P < 0.01.
eIF3h and uAUG start codon recognition
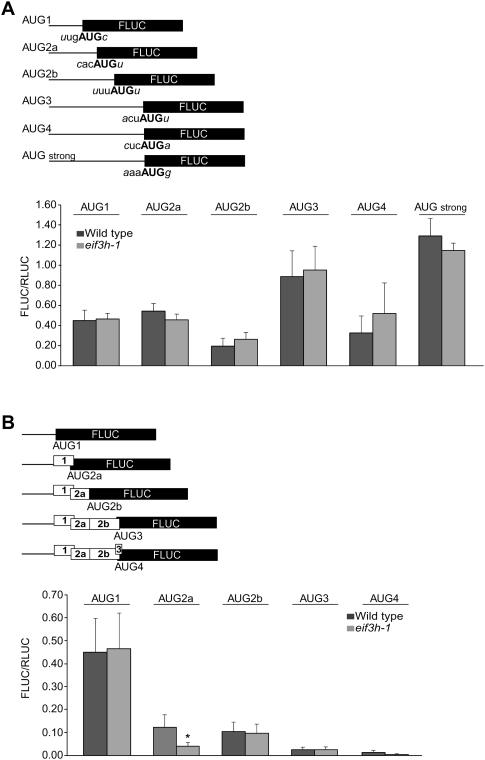
One possible role of eIF3h that would be consistent with the observations is that eIF3h helps wild-type plants to disregard certain uAUG start codons, in particular uAUGs in a weak sequence context (Kozak 1986, 1989). To examine this, each uAUG start codon in the AtbZip11 leader was coupled directly to the FLUC coding sequence. Because FLUC expression was indistinguishable between wild-type and mutant seedlings, we concluded that eIF3h does not affect uAUG start codon recognition in AUG contexts considered weak (uAUG1, 2a, 2b, 4), medium (uAUG3), or strong (modified uAUG4; Fig. 4A).
FIGURE 4.
uAUG recognition. (A) The uAUGs of the AtbZip11 uORFs were fused directly to FLUC while retaining the start codon context up to the +4 position as well as their distance from the transcription start site as shown. Note that all uAUGs appear to be recognized in an eIF3h-independent fashion (n = 7–14). Experimental conditions are as in Figure 2. (B) uAUG-FLUC fusion constructs to examine the (re)initiation potential at individual uAUGs downstream from uORF1. Bars, standard error (n = 5–6). (*) Statistical significance at P < 0.02.
Leaky scanning and reinitiation at individual uAUGs were further examined using the uORF arrangements shown in Figure 4B. Compared with the number of successful initiation events at uAUG1 (construct AUG1), gradually fewer ribosomes initiated at the downstream AUGs. We note that construct AUG2b, which reports on initiation at either 2a or 2b, did not show eIF3h dependence, and neither did AUG3. Initiation at uAUGs 3 and 4 was much lower than at the main ORF (cf. Fig. 2A and Fig. 4B), which argues for poor reinitiation immediately downstream from uORF1 in these constructs. The apparent eIF3h dependence of initiation at uAUG2a was unexpected given the prior data. We considered whether eIF3h-dependent backward scanning from the Stop of uORF1 toward uAUG2a might occur. However, an additional construct in which uAUG2a was moved further 5′, which should eliminate backward scanning, continued to express the reporter gene, but in an eIF3h-independent fashion (data not shown). This result favors leaky forward scanning over backward scanning as the primary route for initiation at uAUG2a.
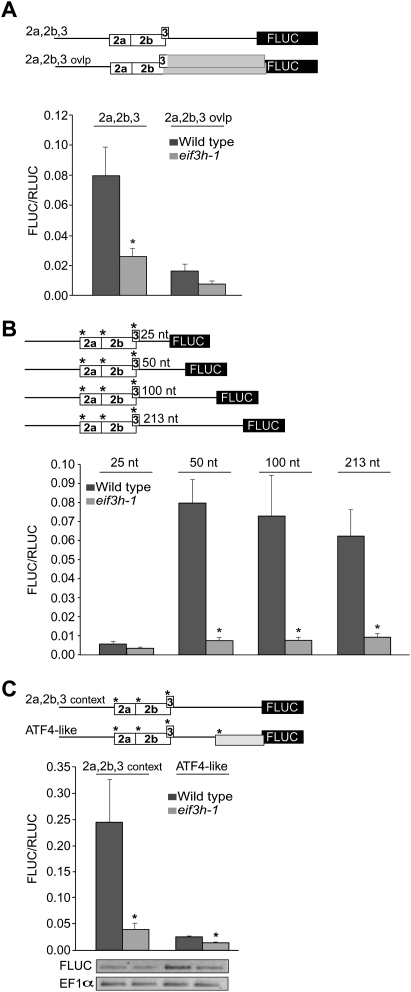
To examine leaky scanning and reinitiation around uAUGs 2a, 2b, and 3, uORFs 2 and 3 were extended so as to overlap the FLUC start codon by 44 and 45 codons, respectively (Fig. 5A, 2a,2b,3 ovlp), an overlap sufficient to suppress any hypothetical backward scanning (Jackson et al. 2007). In the case of rampant leaky scanning, the 2a,2b,3 ovlp transcript should be expressed at the same level as 2a,2b,3. If the uORF cluster was recognized every time, FLUC expression would be zero. Because expression was reduced dramatically, translation of FLUC downstream from the AtbZip11 leader must require a reinitiation event. If one considers that uORF3 is masked by uORF2 and is marginally eIF3h-dependent on its own (Fig. 3B), it becomes clear that the eIF3h-dependent reinitiation event must occur downstream from uORF2. Because the ribosomes got trapped by the overlap uORFs, one can also conclude that ribosomes do not simply bypass the entire uORF cluster by shunting (discontinuous scanning).
FIGURE 5.
Requirement for eIF3h for efficient reinitiation. (A) To delineate leaky scanning across uORFs 2 and 3, all in-frame stop codons were eliminated by point mutations, thus causing them to overlap the main FLUC ORF by 44 and 45 amino acids, respectively (2a,2b,3 ovlp). Residual expression of FLUC is attributed to ribosomes that have leaky-scanned uORFs 2a, 2b, and 3. (B) The effect of eIF3h on reinitiation downstream from uORF 2a,2b,3 was tested by shortening the intercistronic spacer length in the AtbZip11 leader by deletion from the 3′ end of the spacer. uORFs 2a, 2b, and 3 were placed in a strong Kozak context (−3aaaAUGg+4) to minimize the effect of leaky scanning. Error bars, standard error (n = 5–8). (*) Statistical significance at P < 0.002. (C) Inhibition of translation by an ATF4-like (Harding et al. 2000) overlap-uORF downstream from uORF2 demonstrates consistent reinitiation in wild type but marginal reinitiation in eif3h. A uORF (gray box) in a strong Kozak context (A−3AAAUGG+4) was created starting 60 nt upstream of the FLUC ORF to overlap the main ORF. (*) AUGs in a strong context. Bars, standard error (n = 8–12). AtbZip-FLUC and endogenous EF1α mRNA levels were estimated by RT-PCR.
eIF3h controls the efficiency of certain reinitiation events
To directly test whether eIF3h is responsible for reinitiation after uORF translation, the cluster of uORFs 2a, 2b, and 3 was first altered to commence with start codons in a strong Kozak context (Fig. 5B, “*”) in an effort to minimize leaky scanning. Reinitiation is generally poor if the spacer between the uORF stop codon and the main ORF's start codon is too short, an effect generally attributed to Met-tRNAMet acquisition being a rate-limiting step (Kozak 1987; Abastado et al. 1991a,b). We clearly demonstrated such a relationship with four leader constructs in which the spacer length was varied between 25 and 213 nucleotides (nt). Reinitiation in the wild type had reached a plateau with a spacer length of 50 nt. For comparison, the eif3h mutant showed only marginal expression, regardless of the spacer length (Fig. 5B). This result indicates that eIF3h is specifically required for reinitiation downstream from uORF 2 or 3.
Placing a uORF that overlaps the main ORF downstream from the uORF2,3 cluster greatly diminished the residual expression, especially in the wild type (Fig. 5C). This result confirms that ribosomes resume scanning after uORF2,3 and do acquire a fresh ternary complex over the time needed to scan across the ∼150-nt spacer between the uORFs.
Computational modeling of translation defects in eif3h mutant plants
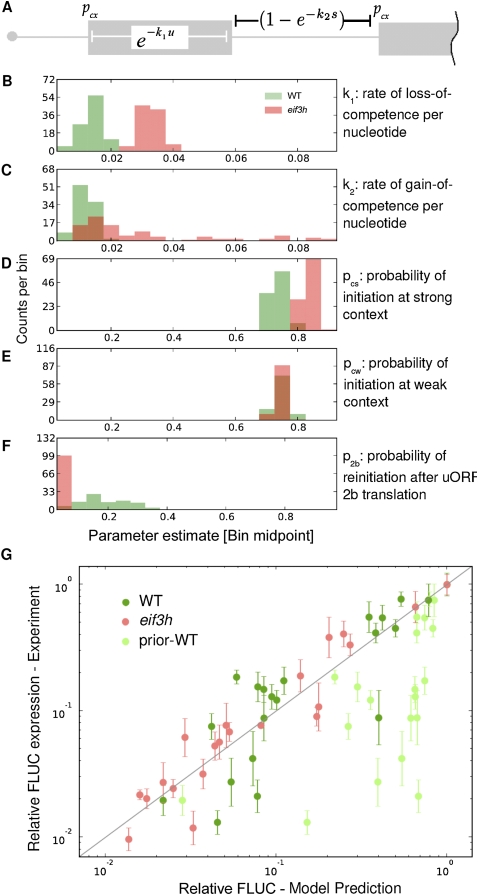
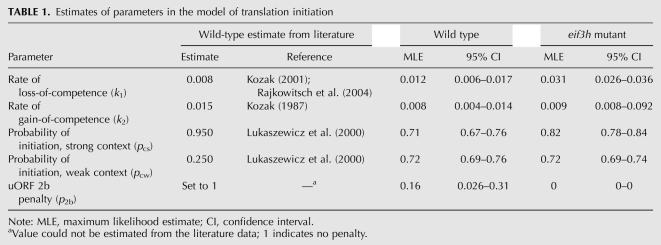
With the experimental work described above we hoped to define the molecular defects that distinguish eif3h mutant plants from wild-type plants. To facilitate quantitatively rigorous conclusions, we constructed a computational model of translation initiation on the uORF-containing AtbZip11 leader. The five variables of the model correspond to canonical events underlying uORF translation and reinitiation (Fig. 6A). Variables pcs and pcw represent the probability that the 40S ribosome recognizes an AUG in strong or weak sequence context, respectively (Kozak 1986; Lukaszewicz et al. 2000). Variable k1 describes the rate per nucleotide at which the ribosome loses its reinitiation competence, permanently, during uORF translation (Kozak 1987, 2001; Rajkowitsch et al. 2004). The remaining ribosomes, said to have suffered only conditional loss of competence due to loss of ternary complex, will terminate translation and resume scanning. Variable k2 describes the rate per nucleotide scanned at which the ribosome regains its full reinitiation competence (Kozak 1987), in part by acquiring a ternary complex. Finally, certain uORFs trigger permanent loss of reinitiation competence in a fashion dependent on the peptide sequence, independent of their length. Variable p2b represents the probability of escape from the attenuation caused by translating the peptide of uORF2 of AtbZip11 (Rahmani et al. 2009), where p2b = 0 indicates dissociation of the ribosome from the mRNA every time, i.e., no escape. Parameters culled from the literature (Table 1) yielded a poor fit between model and wild-type experimental data (Fig. 6G). We therefore adopted an evolutionary algorithm to generate estimates for each parameter for both wild-type and eif3h mutant plants, using as a fitness criterion the least sum-squares fit between model and experimental data (see Materials and Methods). To estimate the uncertainty with regard to experimental variation, we generated 95% confidence intervals (CIs) for these parameters by bootstrapping from the experimental data (Fig. 6B–F). Significantly, the maximum likelihood estimate (MLE) for k1, the rate at which reinitiation competence is lost during uORF translation, was lower in wild-type plants than in eif3h mutant plants (Table 1). Thus, in the wild type, 50% loss of competence was estimated at 58 nt of uORF translated, whereas in the mutant 50% loss of competence was more rapid, at 22 nt.
FIGURE 6.
Computer simulation of the translational defect in eif3h mutant plants. (A) Model parameters. (Gray boxes) ORFs. Spanning bars indicate the range over which a given term applies. (B–F) Distributions of parameter estimates. x-axis length reflects the manually set boundaries of possible parameter estimates. y-axis counts indicate the number of times out of 100 trials that a parameter fell into one of 20 x-axis bins. (B) Loss rate of reinitiation competence (k1) as a function of uORF length (u [nt]). (C) Regaining of reinitiation competence (k2) as a function of intercistronic spacer length (s [nt]). (D) Probability of AUG recognition in a strong context (pcs). (E) AUG recognition in a weak context (pcw). (F) Escape from attenuation upon translation of uORF2b peptide (p2b). (G) Scatter plot illustrating the match between model output using MLEs (x-axis) and experimental data (y-axis, ± one standard error) for all 21 AtbZip11 5′ leader constructs in wild-type (dark green) and eif3h plants (red). (Light green symbols) Predicted expression values using model parameters culled from the literature (see Table 1).
TABLE 1.
Estimates of parameters in the model of translation initiation
In contrast, the parameter estimates for AUG recognition (pcx) were affected little by eIF3h. Weak context was the same between the two genotypes. Strong context differed by a small, yet statistically significant, margin (Table 1).
The escape from attenuation at uORF2 [MLE(p2b) = 0.16 for wild type] suggests that translation of uORF2b allows only one out of six wild-type ribosomes to retain reinitiation competence. In contrast, in eif3h p2b was effectively nil (Fig. 6F), indicating complete loss. The low MLEs for p2b suggest that uORF2 strips ribosomes of their reinitiation competence more effectively than suggested by its length. It stands to reason that the mechanisms driving k1 and p2b are intimately related. Loss of competence (k1) and uORF2 dissociation penalty (p2b) were negatively correlated in the wild type; i.e., modeling trials in which loss of competence was mild tended to assign a very strong dissociation penalty to uORF2 and vice versa (data not shown).
The k2 parameter for regain of initiation competence during scanning was smaller in wild type than in eif3h; i.e., slower recovery in the wild type. However, k2 had a weak effect on fit, and hence was poorly constrained, in the eif3h background (Fig. 6C; Table 1). In summary, the new parameters generally improved the fit between model and experimental data (Fig. 6G). The modeling work supports two conclusions: (1) translation initiation on the AtbZip11 leader can be explained to a large degree by translation of inhibitory uORFs, reinitiation, and leaky scanning; and (2) the most evident molecular function of eIF3h is the retention of reinitiation competence during uORF translation.
eIF3h and the AtbZip11 attenuator peptide
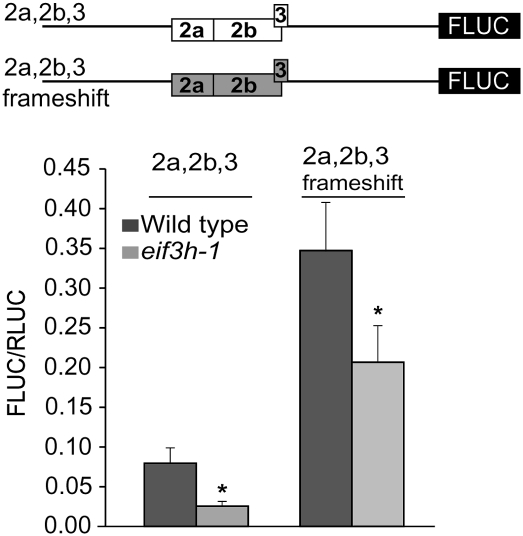
The 2b portion of uORF2 is an inhibitory attenuator peptide that steps into action when AtbZip11 translation is repressed by sucrose (Rahmani et al. 2009). The computer model incorporated an equivalent uORF2 penalty. Although we worked at a comparatively low sucrose concentration of 1%, changing the peptide sequences of uORF2 and 3 via compensatory frameshift mutations resulted in translational derepression, the extent of which was more pronounced in eif3h than in wild type. Despite alteration of the uORFs' peptide sequences, translation remained dependent on eIF3h, albeit at a diminished level (Fig. 7). Several versions of the frameshifted uORF with slightly different coding sequences gave similar results (data not shown). In conclusion, the role of eIF3h in reinitiation is not restricted to the uORF2 peptide. Instead, as suggested by the model, eIF3h helps to retain reinitiation competence in peptide sequence-dependent and sequence-independent ways.
FIGURE 7.
Stimulation of translation across uORFs 2 and 3 by eIF3h is not dependent on the uORF peptide sequences. Two independent frameshift mutations were introduced into uORF2a/2b to change the sequence of the encoded peptide while retaining its native length and also the flanking sequences. For uORF3, point mutations were introduced to change three out of its five amino acids. The frameshift data (wild type versus eif3h) were statistically significant by paired t-test (P < 0.002).
Evidence that eIF3h supports the resumption of scanning, not tRNA acquisition
The modeled data did not provide a clear answer as to whether eIF3h might affect the regaining of reinitiation competence after the ribosome has resumed scanning; i.e., ternary complex/tRNA reacquisition.
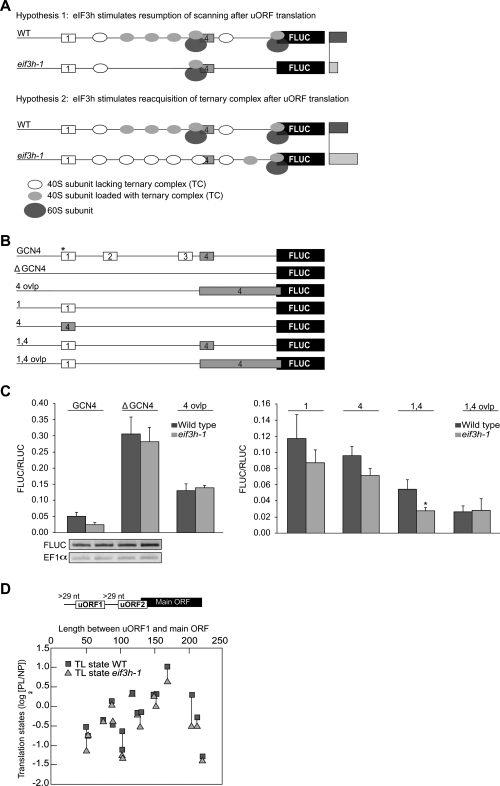
In yeast, the GCN4 leader reports on ternary complex reacquisition by ribosomes that have resumed scanning after translating uORF1. A delay in ternary complex reacquisition, as occurs in yeast general control derepressed (gcd) mutants, results in skipping of the inhibitory uORF4, which in turn permits some translation initiation at the main ORF (Hinnebusch 2005). We asked whether eif3h shows a gcd-like delay in tRNA acquisition on the GCN4 leader (Fig. 8A). Individually, uORF1 and 4 were both inhibitory when compared with the uORF-less leader (Fig. 8B,C). uORF4 alone was not as strongly inhibitory as it is in yeast, however, and neither uORF 1 nor 4 caused significant eIF3h dependence in Arabidopsis, as expected because these uORFs are short. Defects in resumption of scanning and tRNA reaquisition make opposite predictions about the expression from mRNAs containing both uORF1 and uORF4 (Fig. 8A). Experimentally, FLUC expression downstream from uORF 1 plus 4 (1,4) was reduced at least as strongly in eif3h as in wild type, a result more consistent with a defect in resumption of scanning than a delay in tRNA acquisition (Fig. 8C). The residual expression from 1,4 in the eif3h mutant is due to leaky scanning past uAUG4, which is in a medium context, as judged from the 4 ovlp and 1,4 ovlp constructs. The near-background expression with 1,4 ovlp also suggests that ribosomes reinitiating after the strong uORF1 reacquire a fresh tRNA, soon enough to be trapped efficiently by uAUG4 (Fig. 8C). These data are consistent with the scenario that the wild-type initiation machinery can traverse the GCN4 leader with two reinitiation events, whereas in the eif3h mutant, resumption of scanning downstream from the uORFs is reduced.
FIGURE 8.
Translation initiation on the 5′ leader from yeast GCN4 in Arabidopsis, and microarray meta-analysis. (A) The schematic illustrates the predictions of two contrasting hypotheses concerning the molecular function of eIF3h (after Hinnebusch 2005). uORF4 is shaded (gray) for emphasis. If eIF3h supported resumption of scanning, few ribosomes would scan the intercistronic spacer in the eif3h mutant, but those that do would easily acquire a fresh ternary complex (TC) and initiate at the inhibitory uORF4. Hence, adding uORF4 to uORF1 will reduce FLUC translation to similar degrees in wild type and eif3h (bars on the right symbolize predicted FLUC expression levels). In contrast, if the eif3h mutation delayed TC acquisition, some mutant ribosomes would leaky-scan past uORF4 and thus reach the main FLUC ORF. Meanwhile, wild-type ribosomes, having acquired their TC early, will be preferentially intercepted by uORF4, resulting in a reversal of the eIF3h dependence (bars on right). (B) GCN4 5′ leader sequences tested. The version with four uORFs is the original. (*) uAUGs in a strong context. (C) Expression data for the constructs shown in panel B. Error bars, standard error (n > 4). (*) Statistical significance at P < 0.05. Where shown, GCN4-FLUC and endogenous EF1α mRNA levels were estimated by RT-PCR. (D) The translation state of Arabidopsis mRNAs corresponding to the scheme outlined at the top was mined from polysome microarray data (Kim et al. 2007) and displayed for wild type and eif3h mutant. Translation state is defined as the log(2) of the ratio of polysomal (PL) to nonpolysomal (NP) mRNA signal. Note that the translation state is equal or lower in eif3h than in wild type, as predicted by the resumption-of-scanning hypothesis.
For an independent test of the tRNA acquisition hypothesis, we examined the polysome loading in wild-type and eif3h mutant seedlings for native Arabidopsis mRNAs with a uORF pattern reminiscent of ATF4, a mammalian mRNA that is regulated similarly to GCN4 (Harding et al. 2000). If tRNA acquisition was reduced in the eif3h mutant, such ATF4-type leaders would show higher polysome loading in the eif3h mutant than in the wild type. However, this was not the case (Fig. 8D). In fact, the polysome loading defect was about the same for ATF4-type leaders as for other leaders with the same number of uORFs (data not shown). We also checked whether the cohort of mRNAs that shows higher polysome loading in the eif3h mutant than in the wild type (Kim et al. 2007) contains leaders resembling ATF4; again, the results came out negative. These data favor the notion that resumption of scanning rather than tRNA acquisition is defective in eif3h.
DISCUSSION
The role of AtbZip11 in amino acid biosynthesis and its translational regulation by nutrient levels, i.e., sucrose, are reminiscent of other translationally regulated bZips, such as GCN4 in yeast and ATF4 in mammalian cells (Hanson et al. 2008). All available evidence indicates that AtbZip11 is translated according to the scanning model, involving at least one reinitiation event, but no significant ribosome shunting or internal ribosome entry.
Transit routes of ribosomes on the AtbZip11 leader
Our experimental data and modeling results lead to the following scenario for ribosome transit routes on the wild-type AtbZip11 leader. The majority of ribosomes translate uORF1, notwithstanding its weak AUG context. In light of the length of uORF1 (18 codons) and the different values of k1 (Table 1), reinitiation after uORF1 is predicted to be moderately eIF3h-dependent. Most of the FLUC expression seen with the wild-type AtbZip11 leader in both the wild type and eif3h mutant is likely attributable to this transit route.
Of the ribosomes that skip uAUG1, most initiate at uAUG2a or 2b. Translation of uORF2 makes initiation at uAUGs 3 or 4 unlikely, because their start codons lie upstream of the uORF2 stop codon. Reinitiation downstream from uORF2 at the main FLUC start codon is overall inefficient (cf. Fig. 4A, wt expression “AUG 2a” and Fig. 5B, wt expression “213 nt”). It is also eIF3h-dependent, first because uORF2 is long enough to make reinitiation highly inefficient in the mutant, and second because uORF2 encodes an attenuator peptide (Rahmani et al. 2009) that appears to be more inhibitory in the mutant (Figs. 6F, 7).
Estimating ribosome initiation behavior
The computational model is founded on the notion that four types of variables drive initiation efficiency at the main start codon: the context of uORF start codons (Kozak 1986; Lukaszewicz et al. 2000), the length of the previously translated uORF (Kozak 2001; Rajkowitsch et al. 2004), any attenuation caused by the nature of the uORF peptide (Rahmani et al. 2009), and the spacer length between a uORF stop codon and the next AUG start codon (Kozak 1987). Granted that eIF3h stimulates reinitiation, is it not possible that eIF3h simply increases the affinity between the 40S subunit and eIF3, such that a post-termination 40S subunit can effectively recruit a fresh eIF3 complex from the soluble cytosolic pool? Speaking against this is that eIF3h's effect is conditional on attributes of the uORF, and in two distinct ways. First, eIF3h suppresses the permanent loss of reinitiation competence during uORF translation. Specifically, it reduces the rate parameter, k1, by about twofold (Fig. 6B). Second, eIF3h reduces the additional loss of reinitiation competence caused by uORF2 (p2b) (Figs. 6F, 7). The effect of eIF3h on parameters k1 and p2b may well be due to one and the same molecular activity of eIF3h. For example, eIF3h may facilitate post-initiation retention of eIF3 on the ribosome, and ribosomes that have retained eIF3 may be more likely to resume scanning and reinitiate than ribosomes that have not.
Discrepancies between model and experiment were dispersed over the entire data set and were generally within two standard errors of the experimental mean (Fig. 6G). The set of variables was deliberately kept to the minimum that is well supported by prior knowledge. Therefore, some data are not fitted well by the model, which is evidence of mechanistic events that have yet to be modeled. For example, uAUG2a is very inhibitory, especially in the eif3h mutant (Fig. 3A), even though it is in a weak context and masked by uORF1 (Fig. 4A). This might point to ribosome–ribosome interactions. Let us consider that the uORF2 attenuator peptide slows the progression of elongating or terminating 80S ribosomes (Rahmani et al. 2009), consistent with the uORF2 penalty in both wild type and mutant (Fig. 6F, 7). We now postulate that ribosome occupancy by uORF2 affects the trajectory of upstream ribosomes that are poised to reinitiate after uORF1. The block on uORF2 would block 40S ribosomes that are scanning downstream from uORF1 and this might cause them to dissociate from the mRNA, possibly in an eIF3h-dependent way. Another plausible mechanism is that stacking of initiation-competent 40S ribosomes may foster AUG recognition at uAUG2a or 2b, which would exacerbate exponentially the eIF3h-dependent inhibition of expression. AUG recognition by 40S ribosomes can also be enhanced when the ribosome is blocked in its track by RNA secondary structure (Kozak and Shatkin 1978; Kozak 1990; Dinesh-Kumar and Miller 1993; Doohan and Samuel 1993). Stacking on top of uAUG2a might arise from a block of eif3h mutant ribosomes upon termination of uORF1, a block of 60S subunit joining on uAUG2b, a block in elongation over a triplet of rare arginine codons present in uORF2b, or a combination of these. These possibilities remain to be tested. At pcw = 0.78, our parameter estimate for initiation at AUG in a weak context was fairly high, but not unprecedented, compared with published values (Lukaszewicz et al. 2000; David-Assael et al. 2005). The postulated ribosome stacking effect may be the reason underlying the high AUG recognition and may explain why leaders harboring uORF1 are generally eIF3h-dependent, but in a fashion dependent on uAUG2a (Fig. 3).
The h subunit of eIF3 is a reinitiation factor
Reinitiation after translation of a short ORF is not uncommon, but the molecular events needed for successful reinitiation are still not well understood. Reinitiation after translation of long ORFs is exceedingly rare but involves eIF3 as well (Park et al. 2001; Pöyry et al. 2007). Budding yeast has a poor reinitiation capacity on all but short uORFs, and lacks eIF3h. However, several yeast eIF3 mutants have defects in reinitiation. While few yeast eif3 mutants display the general control derepressed (gcd) phenotype that is suggestive of defects in tRNA acquisition (Nielsen et al. 2004), a point mutant of eIF3b has a decrease in scanning during reinitiation on the GCN4 leader in (Nielsen et al. 2004). And, eIF3a helps to recognize the specific sequence context around uORF1 of GCN4 that drives efficient reinitiation (Szamecz et al. 2008).
For Arabidopsis eIF3h, there are two canonical biochemical activities of eIF3 that may underlie its role in reinitiation: effective recruitment of a fresh ternary complex to the 40S subunit, which our data did not favor; and separation of the 40S and 60S subunits (Benne and Hershey 1978; Pisarev et al. 2007). Our data indicate that eIF3h assists the ribosome to retain competence for reinitiation during translation of a uORF; i.e., more efficient resumption of scanning. This function of eIF3h might be related to eIF3's role in ribosomal subunit separation, but it need not be. Which ribosomal subunits or proteins might partner with eIF3h? One might expect that the cognate partner of eIF3h in the ribosome displays a similar mutant phenotype as eif3h. Arabidopsis STV1 encodes ribosomal protein RPL24B, mutation of which interferes with translation of some uORF-containing mRNAs (Nishimura et al. 2005). One other plant eIF3 subunit, eIF3g, has been linked to reinitiation because it interacts with the cauliflower mosaic virus reinitiation factor, TAV (Park et al. 2001, 2004). Furthermore, eIF3 and TAV also interact with RPL24 (Park et al. 2001) and with a recently discovered cellular protein that is auxiliary to reinitiation (Thiebeauld et al. 2009). Taken together, eIF3h appears to be part of a larger functional module that is responsible for efficient reinitiation after uORF translation and that involves cooperation between eIF3 and the large ribosomal subunit.
MATERIALS AND METHODS
Molecular cloning and generation of AtbZip11 5′ leader mutants
The AtbZip11-leader/FLUC reporter plasmid for testing translation efficiency in transient expression assays (Kim et al. 2004) served as the template to generate AtbZip11 5′ leader mutants by site-directed mutagenesis, which were confirmed by sequencing. Primer sequences are available upon request. To introduce multiple uAUG mutations, single mutants were used as PCR templates. For the set of constructs to test initiation at uAUGs, the AtbZip11 5′ leader until +4 position with respect to the uAUG was amplified with XhoI/NcoI sites and fused to FLUC. Frameshift mutations in uORF2 were produced by introducing +1 and −1 mutations in uORFs 2a and 2b separately while maintaining the internal AUG in frame. Spacer lengths were varied by truncating its 3′ end. To study the extent of leaky scanning, uORFs 2 and 3 were extended past the start of the main ORF by mutating all in-frame stop codons. The AtbZip11 3′ UTR was amplified as a BglII/XbaI fragment from Arabidopsis genomic DNA and subcloned so as to replace the cauliflower mosaic virus 35S terminator in the AtbZip11-FLUC construct. The 5′ leaders from the GCN4-LacZ constructs described previously as B180, B976, B235, B227, pM114, B1002, B1010, and B1014 (Abastado et al. 1991a,b) were amplified with XhoI and NcoI sites, subcloned to the 35S:FLUC plasmid, and confirmed by DNA sequencing.
Transient expression and translation assay
Arabidopsis seedlings were grown in MS medium for up to 20 d and were transformed via particle bombardment (PDS-1000/He; Bio-Rad) with plasmid DNAs harboring either AtbZip11 or GCN4 5′ leaders and firefly luciferase (FLUC) coding sequence. The promoter (35S) and 3′ UTR are from cauliflower mosaic virus. A 35S-driven Renilla luciferase construct containing the translational leader of tobacco etch virus served as a reference (Kim et al. 2004). Luciferase activities were measured in protein extracts prepared 15–17 h after bombardment using the Dual Luciferase System (Promega) and a TD 20/20 luminometer (Turnerdesigns). The ratio of FLUC to RLUC activity indicates the translation efficiency. P-values for two-sided t-tests were routinely calculated after log-transformation of the FLUC/RLUC ratios.
For in vitro transcription, expression plasmids harboring a SP6 promoter and a 70base poly(A) tail were linearized by PvuII digestion and column purified (Qiagen). For cap-dependent translation initiation, mRNA was in vitro transcribed and capped using SP6 RNA polymerase and m7G cap analog (New England Biolabs) following the manufacturer's protocols. The 5′ UTR designated as “Spacer” contains a polylinker sequence from pGL3-basic (Promega). mRNA quality and quantity were estimated by agarose gel electrophoresis. Protoplasts were prepared from wild-type or mutant 7-d-old Arabidopsis seedlings (Yoo et al. 2007). Seedlings with roots removed were cut into 0.5 mm slices and digested with digestion buffer (1.5% cellulase R10, 0.3% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 20 mM MES at pH 5.7, 10 mM CaCl2, 5 mM mercaptoethanol, 1% BSA) for 3 h after 30 min vacuum infiltration. Protoplasts were released by gently swirling and were filtered through a 40-μm mesh into a plastic centrifuge tube on ice. After centrifugation in a tabletop centrifuge, the protoplast pellets were washed with 10 mL of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES at pH 5.7) and resuspended in 2 mL of W5 solution. The W5 solution was then substituted with MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES at pH 5.7) before mRNA transformation (Gallie 1993). Eighty micrograms of sheared and denatured salmon sperm DNA were added to 0.1 mL of protoplasts in MMG solution in a 1.7 mL microcentrifuge tube. Two hundred nanograms of mRNA were added immediately before the addition of 0.11 mL of PEG solution (40% PEG, 240 mM mannitol, and 100 mM CaCl2), which was subsequently mixed by gently inverting three to four times. The PEG transformation was terminated by addition of 0.43 mL of W5 and centrifugation. The protoplasts were resuspended in 1 mL of W5 solution and incubated in a 24-well plate for 3 h in the dark at room temperature, then harvested by centrifugation for luciferase assays.
RNA expression analysis
Total RNA was isolated from seedlings transiently expressing AtbZip11 5′ leader constructs by harvesting tissues in liquid nitrogen after 15–17 h and extracting with TRIzol (Sigma). For RT-PCR analysis, 1 μg of total RNA was treated with RNase-free DNase I (Promega), and cDNA was synthesized with a 15-mer oligo(dT) primer using MMLV reverse transcriptase (Promega). Amplification was performed as described before (Kim et al. 2004). Half-life of mRNAs was determined after blocking transcription with cordycepin. Transgenic Arabidopsis seedlings harboring the AtbZip11-FLUC transgene shown in Figure 2A (Kim et al. 2007) were grown for 2 wk in germination medium and were transferred to incubation buffer (1 mM Pipes at pH 6.25, 1 mM sodium citrate, 1 mM KCl, 15 mM sucrose) for 30 min, followed by addition of cordycepin (Sigma) to a final concentration of 100 μg/mL. RNA samples were harvested at specific time points, treated with DNase, and analyzed by RT-PCR for the levels of FLUC and eukaryotic elongation factor 1α. Control experiments demonstrated that the amplifications had not reached saturation and that no DNA contamination was present.
Polysomal protein isolation and eIF3 association
Seedling extract was prepared in 1mL of extraction buffer containing 200 mM Tris-Cl (pH 8.4), 50 mM KCl, 25 mM MgCl2, 50 μg/mL cycloheximide, and 1× protease inhibitor cocktail (Sigma; Park et al. 2001). The extract was layered on a 10-mL 7.5%–30% sucrose gradient. After centrifugation for 5 h at 4°C in a Beckman SW41Ti rotor at 39,000 rpm, 11 fractions were collected manually (Nielsen et al. 2004). Fractions were concentrated using an Amicon centrifugal filter device (100 kDa cutoff, Millipore) at 6500 rpm for 10 min. Concentrated fractions were resolved on 10% acrylamide gels, and immunoblots were performed with anti eIF3h (Kim et al. 2004) and anti eIF3 (Lauer et al. 1985) antibody.
Structure of the computational model
The program used to run the model and to generate parameter distributions is available upon request (InitiationModel.pl). Each 5′ UTR sequence was parsed for uORFs using a custom Perl module (uORF.pm). Initiation efficiency was modeled as the sum probability of a strongly binary tree representing all possible initiation events that a ribosome could experience prior to encountering the start codon of the main ORF (Abastado et al. 1991a,b). The probability of initiation at a given AUG was calculated as the context of that AUG (weak or strong) multiplied by the effects of the previously translated uORF's length (Kozak 2001; Rajkowitsch et al. 2004) and distance from that previously translated uORF's stop (Kozak 1987):
where P(u,s) = probability of initiation; pcx = probability of initiation based on context (strong or weak); k1 = rate of loss-of-competence per nucleotide; k2 = rate of gain-of-competence per nucleotide; u = length of last translated uORF; and s = length from last translated uORF stop to the start of the current ORF (for further description of parameters, see Figure 6 and Results).
AUG contexts were considered strong (pcs, [GA]nnAUGG, [GA]nnAUG[ATC], or nnnAUGG) and weak (pcw, [CT]nnAUG[ATC]), with n being any other nucleotide and brackets indicating alternatives (Kozak 1986; Angenon et al. 1990; Lukaszewicz et al. 2000). The uORF2b penalty (p2b) was applied when uORF2b sequence was translated, which included the translation of uORF2a. The experimental data are from 21 AtbZip11 reporter constructs, the majority of which are shown in Figures 2, 3, and 5.
Parameter estimation
The five parameters in the model were optimized for best fit with the experimental data using an evolutionary strategy (Moles et al. 2003; Ashlock 2006) implemented in Perl with Math∷Evol (http://www.cpan.org/authors/id/P/PJ/PJB/Math-Evol-1.10.tar.gz) and run for 80 cpu sec maximum with default parameters and with a relative and absolute convergence criteria of 10−11 and 10−16, respectively; if fit did not improve by the given criteria within the last 25 generations, then optimization was terminated. In order to correct for differential efficiency of reporters, fit was evaluated based on the following equation:
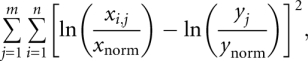 |
where n = the number of experiments per condition; m = the number of conditions; xnorm = experiment median of the uORF-less leader condition; ynorm = model prediction of the uORF-less leader; x = the experimental result; and y = model prediction.
Each of the 100 optimization trials was started with parameter values selected from a uniform distribution delimited by the biologically relevant minima and maxima described below. The sampled range and initial value of the evolvable step-size, in parentheses, for all parameters are as follows: weak (pcw) and strong (pcs) contexts as well as p2b, 0–1 (0.204); k1 and k2, 0–0.10 (0.025). To obtain confidence intervals for our estimates, the reference data set used to evaluate fit was bootstrapped from the original data set for each trial of parameter estimation (Hunt et al. 1998; Efron and Tibshirani 1993). Increasing the trial number further had only a nominal effect on confidence intervals. To assure parameter convergence, 100 trials were run against the set of 21 conditions without bootstrapping. All but one of the resulting parameter estimates varied to within <0.001% of the parameter mean, suggesting that a global minimum (within the bounded region) was consistently found. The one exception was rate of gain-of-competence (k2) in eif3h, which found local minima 12 out of 100 times and, excluding these values, varied to within 1% of the parameter mean. Meta-analysis suggests that this is not a result of rough topology around the MLE but of k2's lack of impact on model fit to the eif3h data once the parameter exceeds 0.009.
ACKNOWLEDGMENTS
We thank Alan Hinnebusch for plasmids harboring 5′ leaders of yeast GCN4, and Karen Browning for an anti-eIF3 antibody. We thank David Brian and Andreas Nebenführ for numerous discussions. This work was supported by grants from the Department of Energy (DE-FG02-96ER20223), the National Science Foundation (DBI-0820047), and US-Israel BARD (IS3901-06C) to A.G.v.A.
Footnotes
Abbreviations: eIF3, eukaryotic translation initiation factor 3; uORF, upstream open reading frame; nt, nucleotides; FLUC, firefly luciferase; RLUC, Renilla luciferase; UTR, untranslated region; RT-PCR, reverse transcriptase-polymerase chain reaction; MLE, maximum likelihood estimate; CI, 95% confidence interval.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2056010.
REFERENCES
- Abastado JP, Miller PF, Hinnebusch AG. A quantitative model for translational control of the GCN4 gene of Saccharomyces cerevisiae. New Biol. 1991a;3:511–524. [PubMed] [Google Scholar]
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991b;11:486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenon G, Van Montagu M, Depicker A. Analysis of the stop codon context in plant nuclear genes. FEBS Lett. 1990;271:144–146. doi: 10.1016/0014-5793(90)80392-v. [DOI] [PubMed] [Google Scholar]
- Ashlock D. Evolutionary computation for modeling and optimization. Springer; New York: 2006. [Google Scholar]
- Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- David-Assael O, Saul H, Saul V, Mizrachy-Dagri T, Berezin I, Brook E, Shaul O. Expression of AtMHX, an Arabidopsis vacuolar metal transporter, is repressed by the 5′ untranslated region of its gene. J Exp Bot. 2005;56:1039–1047. doi: 10.1093/jxb/eri097. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Miller WA. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doohan JP, Samuel CE. Biosynthesis of reovirus-specified polypeptides. Analysis of ribosome pausing during translation of reovirus S1 and S4 mRNAs in virus-infected and vector-transfected cells. J Biol Chem. 1993;268:18313–18320. [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. Chapman & Hall; London: 1993. [Google Scholar]
- Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Gallie DR. Introduction of mRNA to plant protoplasts using polyethylene glycol. Plant Cell Rep. 1993;13:119–122. doi: 10.1007/BF00235304. [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J Biol Chem. 2005;280:39229–39237. doi: 10.1074/jbc.M509340200. [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 2008;53:935–949. doi: 10.1111/j.1365-313X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hayden CA, Jorgensen RA. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 2007;5:32. doi: 10.1186/1741-7007-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hunt CA, Givens GH, Guzy S. Bootstrapping for pharmacokinetic models: Visualization of predictive and parameter uncertainty. Pharm Res. 1998;15:690–697. doi: 10.1023/a:1011958717142. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A, Pöyry TAA. Coupled termination–reinitiation events in mRNA translation. In: Mathews MB, et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 197–223. [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG. Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell. 2004;16:3341–3356. doi: 10.1105/tpc.104.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, von Arnim AG. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8:R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M, Shatkin AJ. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J Biol Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- Lauer SJ, Browning KS, Ravel JM. Characterization of initiation factor 3 from wheat germ. 2. Effects of polyclonal and monoclonal antibodies on activity. Biochemistry. 1985;24:2928–2931. doi: 10.1021/bi00333a017. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz M, Feuermann M, Jerouville B, Stas A, Boutry M. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 2000;154:89–98. doi: 10.1016/s0168-9452(00)00195-3. [DOI] [PubMed] [Google Scholar]
- Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- Mehta A, Trotta CR, Peltz SW. Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells. Genes & Dev. 2006;20:939–953. doi: 10.1101/gad.1388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles CG, Mendes P, Banga JR. Parameter estimation in biochemical pathways: A comparison of global optimization methods. Genome Res. 2003;13:2467–2474. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG. Functions of eIF3 downstream from 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell. 2005;17:2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. A plant viral ‘reinitiation’ factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Park HS, Browning KS, Hohn T, Ryabova LA. Eucaryotic initiation factor 4B controls eIF3-mediated ribosomal entry of viral reinitiation factor. EMBO J. 2004;23:1381–1391. doi: 10.1038/sj.emboj.7600140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes & Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;150:1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JE. Reinitiation and recycling are distinct processes occurring downstream from translation termination in yeast. J Mol Biol. 2004;335:71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998;15:253–263. doi: 10.1046/j.1365-313x.1998.00205.x. [DOI] [PubMed] [Google Scholar]
- Ryabova L, Park HS, Hohn T. Control of translation reinitiation on the cauliflower mosaic virus (CaMV) polycistronic RNA. Biochem Soc Trans. 2004;32:592–596. doi: 10.1042/BST0320592. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valasek L. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes & Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebeauld O, Schepetilnikov M, Park HS, Geldreich A, Kobayashi K, Keller M, Hohn T, Ryabova LA. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 2009;28:3171–3184. doi: 10.1038/emboj.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Hinnebusch AG. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fang P, Sachs MS. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol. 1998;18:7528–7536. doi: 10.1128/mcb.18.12.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell. 2004;16:1717–1729. doi: 10.1105/tpc.019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, et al. The zipper model of translational control. A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]