Abstract
Seven new arginine binding motifs have been selected from a heterogeneous RNA pool containing 17, 25, and 50mer randomized tracts, yielding 131 independently derived binding sites that are multiply isolated. The shortest 17mer random region is sufficient to build varied arginine binding sites using five different conserved motifs (motifs 1a, 1b, 1c, 2, and 4). Dissociation constants are in the fractional millimolar to millimolar range. Binding sites are amino acid side-chain specific and discriminate moderately between L- and D-stereoisomers of arginine, suggesting a molecular focus on side-chain guanidinium. An arginine coding triplet (codon/anticodon) is highly conserved within the largest family of Arg sites (72% of all sequences), as has also been found in minimal, most prevalent RNA binding sites for Ile, His, and Trp.
Keywords: amino acid, triplet, genetic code, stereochemical, origin
INTRODUCTION
Among the canonical amino acids, arginine may be bound by RNA in the greatest variety of ways. Binding sites for free amino acids are known on the Tetrahymena self-splicing rRNA introns (Yarus 1988), and five independent populations of such sites have been selected in vitro (Connell et al. 1993; Connell and Yarus 1994; Famulok 1994; Geiger et al. 1996; Tao and Frankel 1996).
This multiplicity of sites can be explained because arginine offers two different polar centers with which RNA can strongly associate: at α-carbon groups and the side-chain guanidinium (Yarus et al. 2009). The strongest RNA binding sites should therefore be double ended, directed at both ends of free arginine. However, selection of smaller sites, as in the present experiments, will force simplification of the RNA site. Simple sites tend toward interaction with one end or the other of an arginine ligand. Because only sites that are side-chain specific are accepted after our selections, the result is that our selection of sites both simple and specific emphasizes interaction with guanidinium (as indeed did some sites selected without explicit constraints). This is observed here, for example, in the tendency to only moderate stereoselectivity for D- and L-arginine in the present sites.
Simpler, single-ended binding sites emphasizing arginine guanidinium are also of interest because they are crucial in many cases of regulatory peptide–RNA interaction, where α-carbon groups sequestered in the peptide backbone are necessarily less accessible to RNA. For example, messenger RNAs of HIV-1 form a 5′-terminal bulged hairpin, called transactivation responsive (TAR), that binds the viral Tat protein. Binding focuses on a single arginine within a nine-residue basic stretch of Tat (Calnan et al. 1991). Free arginine also binds specifically to the TAR hairpin and in a guanidine-dependent manner similar to arginine in the Tat peptide (Tao and Frankel 1992). The HIV–Rev interaction employs multiple specific arginine–base interactions (Battiste et al. 1996). The HTLV-1 Rex peptide makes multiple specific arginine guanidinium contacts with paired G bases and the RNA backbone (Jiang et al. 1999). The Arg–RNA regulatory interaction is phylogenetically broad, also being the crux of the phage λ Box B (Legault et al. 1998) and phage P22 Box B (Cai et al. 1998) antiterminator interactions.
As might be expected from this robust and widespread association, arginine side-chain–RNA interactions are also among the most frequent contacts observed within the entire population of structurally characterized RNA–peptide interfaces (Allers and Shamoo 2001; Treger and Westhof 2001; Morozova et al. 2006; Ellis et al. 2007).
Arginine binding sites with apparent double- and single-ended specificities are therefore well known. However, there are several unusual observations associated with recently selected free L-arginine sites. No arginine site for free amino acid has been re-isolated in a later selection. Therefore arginine, perhaps because it interacts with RNA so easily, seemed potentially different from other amino acids, whose most prevalent (simplest) biding sites (for isoleucine, tryptophan, and histidine) have been shown to be prevalent by repetitive re-isolation (Majerfeld and Yarus 1998; Lozupone et al. 2003; Majerfeld and Yarus 2005; Majerfeld et al. 2005; M Illangasekare and M Yarus, pers. comm.).
This in turn becomes a crucial point with regard to a stereochemical origin for the genetic code. It has been argued that the simplest RNA binding sites for amino acids contain essential cognate coding triplets unexpectedly frequently (Yarus 1988), (Yarus et al. 2005); (Yarus et al. 2009). Arginine has been a crucial part, in fact, of the argument for such a stereochemical code from the beginning (Yarus and Christian 1989). However, although the disparate populations of binding sites from varied independent arginine selections also have this property of triplet concentration (Yarus et al. 2005), the relationship of this observation to our other results with easily isolated RNA binding sites was unclear. Therefore, we have re-examined arginine by our usual selection methods to see if simple RNA binding sites can be repetitively isolated, and to determine whether these show unusual sequence frequencies related to coding.
Here we describe seven new arginine binding motifs, selected from RNA pools with 17, 25 and 50mer randomized tracts. Arginine shows small, simple recurring site sequences, as do other amino acids, which again exhibit an improbable concentration of arginine triplets.
RESULTS
In vitro selection
We used affinity chromatography and selection-amplification (SELEX) to isolate aptamers eluted by free L-arginine. The initial selection pool of 2.4 × 1015 independent DNA templates was transcribed to give RNAs with 17, 25, and 50mer randomized regions. Calculated representation using Poisson estimation for zero copies (Ciesiolka et al. 1996) suggests 17,400 copies of each 17mer, one copy of each 25mer, and for 50mers, only 10−15 of all possible sequences. Therefore, all possible 17mer sequences are probably represented among RNAs entering the first cycle of the selection.
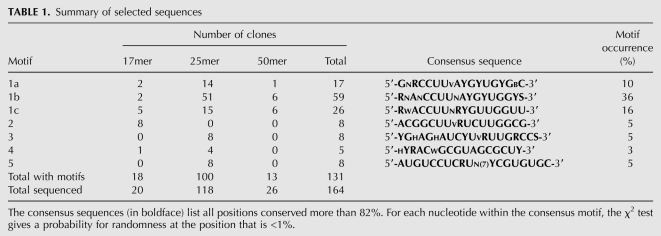
Arginine-specific aptamers were isolated from selection cycles 5 and 6. At these rounds of selection, 8.4% and 23% of the RNA applied, respectively, were eluted by L-arginine at pH 7.0. Sequencing of 164 clones from arginine-eluted RNAs revealed 160 sequences, all apparently of independent origin; that is, from parental molecules of different sequences. Table 1 summarizes these selected sequences. A complete sequence list is available in Supplemental Table 1S. The final population was dominated by 25mer RNA (118 clones, 72%); in addition 20 clones were 17mer RNAs (12%), and 26 clones were 50mers (16%).
TABLE 1.
Summary of selected sequences
Sequence analysis (CLUSTALW) suggested seven different groups of conserved sequences (motifs 1a, 1b, 1c, 2, 3, 4, and 5) together accounting for 80% of all sequenced molecules (see Table 1). The remaining 20% not represented in Table 1 do not have conserved nucleotides, and are likely less frequent kinds of arginine sites. These sequences were not further tested for arginine binding.
As might be expected, all prevalent active structures have apparently simple, repetitively isolated structures. In 20 sequenced 17mers, 18 (90%) contained conserved sequences. Among 50mers, 50% of all sequenced clones contained conserved motifs. The most abundant 25mer group contained conserved motifs in 100 isolates (85%) (Table 1).
Six motifs (1a, 1b, 1c, 2, 3, and 4) are composed of single modules (contiguous nucleotides) comprising 17–20 nucleotides (nt). Motif 5 consists of two conserved sequence modules (10- and 8-nt long) separated by seven nonconserved nucleotides (see Table 1).
Conserved motifs 1a, 1b, and 1c were found within all three sizes of randomized tracts, but were most abundant among 25mers. Motifs 1a, 1b, and 1c share two highly conserved core segments: CCUU and RYGYUG, but 3′- and 5′-flanking regions around these are distinctly different, having different recurring forms in different groups of conserved sequences. Therefore 1a, 1b, and 1c sites are distinguished in the discussion below.
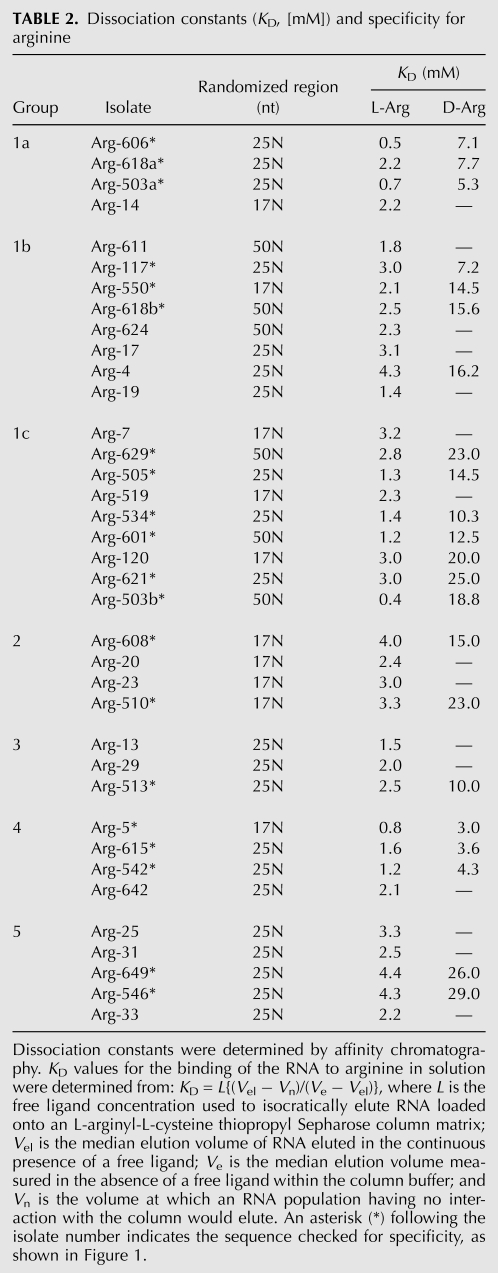
Binding affinity and selectivity
Dissociation constants (KD) for free L- and D-arginine were determined by combining data from two affinity columns: buffer and isocratic L-arginine elution from L-dipeptide-Thiopropyl Sepharose. Table 2 contains these KD's, as well as specificity measured for individual aptamers. KD's are approximately millimolar, similar to previous KD's for selected arginine binding sites (Connell et al. 1993; Connell and Yarus 1994; Tao and Frankel 1996). Observed discrimination between L- and D-stereoisomers was four- to sevenfold for motif 1a, three- to sevenfold for motif 1b, seven- to 12-fold for motif 1c, four- to sevenfold for motif 2, fourfold for motif 3, two- to fourfold for motif 4, and six- to sevenfold for motif 5. Thus, a significant, but moderate, stereoselectivity characterizes all these arginine binding sites, consistent with the idea that prevalent selected (simple) sites emphasize side-chain guanidinium (Yarus et al. 2009).
TABLE 2.
Dissociation constants (KD, [mM]) and specificity for arginine
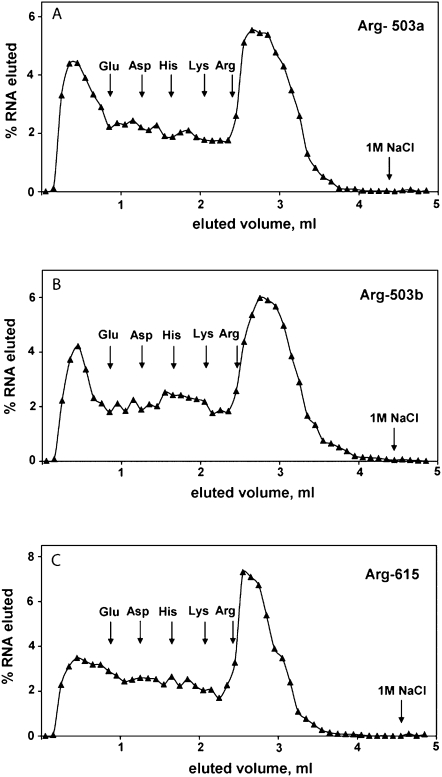
Figure 1 illustrates a test of the side-chain specificity for arginine aptamers with conserved motifs 1a, 1c, and 4. Elution with consecutive blocks of 10-mM L-glutamic acid, L-aspartic acid, L-histidine, L-lysine, and L-arginine are shown for RNA Arg-503a (Fig. 1A), Arg-503b (Fig. 1B), and Arg-615 (Fig. 1C). Binding appears specific for arginine; other basic or acidic (and therefore multiply charged) amino acids do not observably elute affinity column-bound RNA. Wherever asterisks occur in Table 2, similar specificity was observed.
FIGURE 1.
Affinity chromatography and specificity of L-arginine aptamers. Eluants: L-Glu, L-Asp, L-His, L-Lys, and L-Arg were employed at 10 mM in selection buffer S. (A) RNA Arg-503a carrying motif 1a; (B) RNA Arg-503b carrying motif 1c; and (C) RNA Arg-615 carrying motif 4.
Secondary structures
The secondary structure of the arginine aptamers was examined by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (Wilkinson et al. 2006). The 2′-hydroxyl-selective electrophile, N-methylisatoic anhydride (NMIA), modifies RNA ribose. NMIA forms stable 2′-O-adducts more rapidly at conformationally unconstrained RNA nucleotides. Modifications are identified as reverse transcriptase primer extension stops on phosphorimaged sequencing gels.
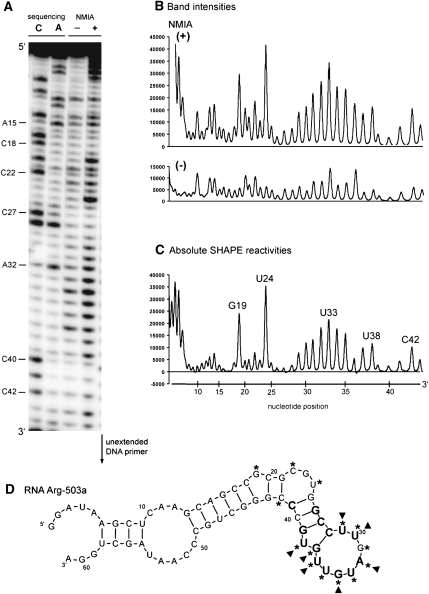
Figure 2A shows typical data for (summarized below) +NMIA, −NMIA, and sequencing reactions for RNA Arg-503a (motif 1a) resolved in a 10% denaturing polyacrylamide gel. In Figure 2B, band intensities quantified by a PhosphorImager are shown for −NMIA controls and +NMIA reactions. By subtracting control intensities from the +NMIA lanes, relative NMIA reactivities are assessed at nucleotide resolution (Fig. 2C). Superposition of relative NMIA reactivities on a secondary structure predicted by BayesFold (Knight et al. 2004) for RNA Arg-503a identifies bulge loop G19, C22, and U24, U29–U36 in the hairpin loop and bulged U38 as NMIA reactive; these outcomes agree with the thermodynamic and Bayesian predictions, thereby supporting the bulged-hairpin-and-stem fold shown for RNA Arg-503a (Fig. 2D).
FIGURE 2.
SHAPE analysis for RNA Arg-503a: a Group 1a aptamer. (A) Bands in the (−) and (+) NMIA lanes are the cDNA fragments of nonreacted and reacted with NMIA. U, G, and C are sequencing reactions performed using adenine, cytosine, and guanosine dideoxy nucleotides. These marker lanes are 1 nt longer than the corresponding NMIA lanes. (B) PhosphorImager profile of reverse transcription analysis for the RNA Arg-503a treated (+) and untreated (−) with NMIA. (C) Absolute SHAPE reactivities as a function of nucleotide position. Band intensities were calculated by subtracting (−) NMIA data from (+) NMIA data. (D) Superposition of absolute NMIA reactivities on a secondary structure of RNA Arg-503a predicted by BayesFold. Asterisks, SHAPE data; triangles, NMIA modification–interference.
The motifs
Motif 1a
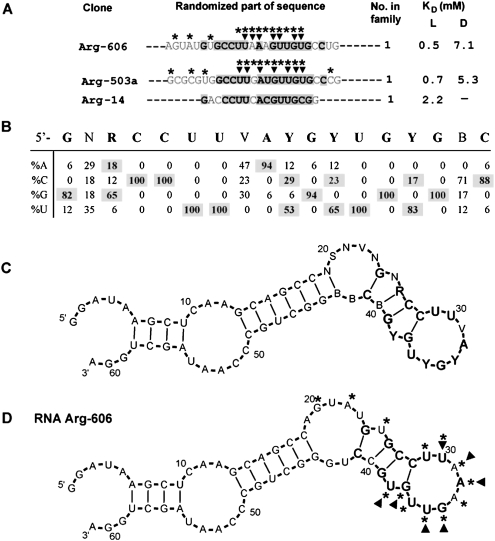
Motifs 1a, 1b, and 1c can be considered members of a single structural family with a common core sequence and a varied supporting periphery. Arginine binding motif 1a consists of 17 independent isolates with 17, 25, and 50mer initially randomized regions. In Figure 3A, randomized parts of representative clones are shown (for the full list of group 1a, see Supplemental Table1S). Shaded nucleotides comprise the conserved segments. Sequence conservations (Fig. 3B) were calculated within a 17-nt span for the 17 independent aligned clones.
FIGURE 3.
Summary of motif 1a. (A) Representative sequences from group 1a. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks or triangles were identified by SHAPE or NMIA modification–interference experiment, respectively. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) and NMIA modification–interference (triangles) data on computed secondary structure of RNA Arg-606.
Folding of 11 aligned 25mer sequences suggests a bulged hairpin structure within the conserved G24–C41 (Fig. 3C, region marked in boldface). The four nucleotide pairs of the G–C-rich stem show highly conserved bulges at N25 and are entirely conserved at Y38. The invariant sequences CCUU and UGYG form the stem–loop boundary. Seven out of eight hairpin loop nucleotides are ≥82% conserved.
SHAPE confirms the predicted secondary structure for the aligned sequences. Results for representative RNA Arg-606 are shown in Figure 3D (asterisks). SHAPE confirms an unconstrained backbone at G20 and A22 in the nonconserved region, nonconserved U25, and conserved pyrimidine U38 in the loop–adjacent bulge. All hairpin loop nucleotides (U29–U36) appear free in conformation. Thus, the predicted motif 1a secondary structure is supported by SHAPE modification data.
NMIA modification–interference was used to identify nucleotides essential for arginine binding. Nucleotides with 2′ modifications at positions essential for the binding site are enriched in affinity column void and early eluting fractions. Modifications at crucial nucleotides are depleted in more strongly bound and arginine-eluted fractions. Ribose in hairpin loops U30, A31, A32, G34, and U35 and nucleotides G37 and U38 in the conserved stem bulge must be unmodified for arginine binding by RNA Arg-606 (Fig. 3D, triangles).
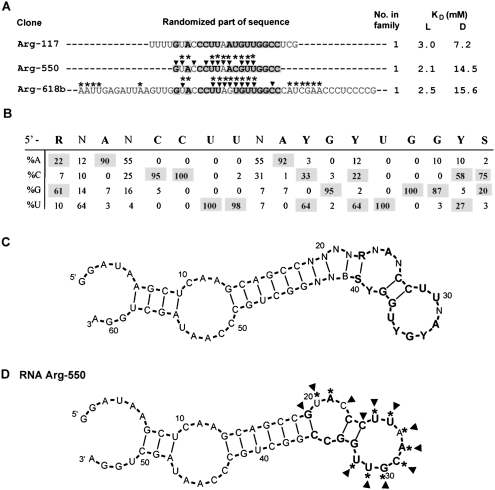
Motif 1b
The second member of the 1a, 1b, and 1c family, and the most abundant arginine site (motif 1b), was found in 59 independent isolates (36% of total sequences). Figure 4A shows randomized parts of representative clones with 17, 25, and 50mer randomized positions (the full sequence list for group 1b is in Supplemental Table 1S). In Figure 4A, shaded nucleotides comprise the conserved segments. Sequence variation for motif 1b was calculated based on the shortest 18mer examples. The observed 18mers apparently are insertions of 1 nt in the initial random 17mer. Figure 4B lists the frequencies of the nucleotides within this 18-nt motif. Conserved nucleotides within the consensus are in boldface (≥83% are taken as nonrandom because χ2 testing gives a probability of nonrandomness of <1%).
FIGURE 4.
Summary of motif 1b. (A) Representative sequences from group 1b. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks or triangles were identified by SHAPE or NMIA modification–interference experiment, respectively. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) and NMIA modification–interference (triangles) data on computed secondary structure of RNA Arg-550.
The calculated most probable fold for 22 aligned 25mer isolates is shown in Figure 4C. In the secondary structure prediction, conserved nucleotides R23–S40 (in boldface) form an asymmetrical internal loop and a terminal loop connected by two G–C base pairs. This same secondary structure was predicted for another group of 12 aligned 25mer sequences, for two aligned 18mer sequences, and for two separate 50mer sequences (data not shown). This strongly supports the structure proposed in Figure 4C for the conserved motif. Highly conserved CCUU and GYUGG form the stem–terminal-loop junction. Seven of eight terminal-loop nucleotides are also ≥86% conserved.
SHAPE was used to test the predicted secondary structure for the aligned sequences. Results for RNA Arg-550 (a representative 18mer clone) are shown in Figure 4D. SHAPE suggests less constrained ribose at U20 and A21 in the internal loop (Fig. 4C, corresponding to N24 and A25, respectively) and for all nucleotides in the terminal loop (U25–U32) (Fig. 4C, corresponding to U29–U36). The predicted secondary structure therefore agrees with the motif 1b NMIA data.
NMIA modification–interference for RNA Arg-550 points to G19 and C24 in the stem base pairs, U20 and C22 in the stem internal loop, and every nucleotide in the terminal loops (U25–U32), as crucial nucleotides for arginine binding (Fig. 4D, triangles).
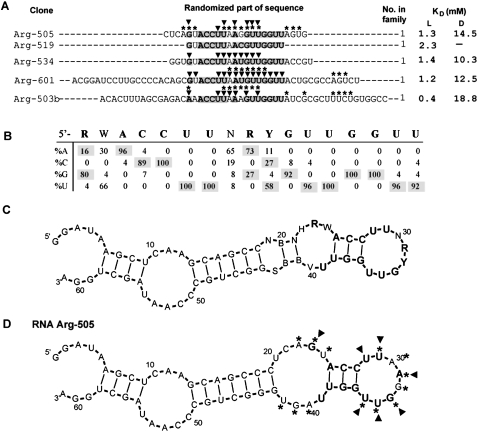
Motif 1c
The final variation in the 1a, 1b, and 1c family is arginine binding motif 1c, derived from 26 independent isolates with 17, 25, 50mer initially randomized regions. In Figure 5A, randomized parts of representative sequences are shown (for the full sequence list for group 1c, see Supplemental Table 1S). Shaded nucleotides comprise conserved segments. Nucleotide variation through the 17-nt-long segment was calculated and the data are presented in Figure 5B. Nucleotides in the consensus are >85% conserved.
FIGURE 5.
Summary of motif 1c. (A) Representative sequences from group 1c. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks or triangles were identified by SHAPE or NMIA modification–interference experiment, respectively. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) and NMIA modification–interference (triangles) data on computed secondary structure of RNA Arg-505.
Figure 5C presents a BayesFold-generated secondary structure for seven aligned 25mer sequences. Two conserved segments, A25–U29 and R31–U38, form a 3 base-pair (bp) stem and terminal loop. The remaining two conserved nucleotides (R23 and U39) are within the internal loop adjacent to the stem. Folding of five aligned 17mer sequences predicts conserved segments forming a 4-bp stem adjacent to a hairpin bulged by the W24 nonconserved nucleotide (data not shown).
SHAPE was used to test the predicted secondary structure for the aligned 25mer sequences. Results for representative RNA Arg-505 are shown in Fig. 5D. SHAPE picks out internal loop nucleotides A22, G23, U24, A40, G41, and U42, and nucleotides U29–U35 in the terminal loop as unconstrained. Thus, the BayesFold-predicted structure for aligned sequences is in agreement with the modification data.
An NMIA modification–interference experiment suggested that ribose of conserved G23 in the internal loop, and U28, U29, A31, G33, U34, and U35 in the terminal loop are crucial for motif 1c arginine affinity (Fig. 5D, triangles).
Motifs 1a, 1b, and 1c (in aggregate, 78% of all recurring motifs) share the consensus sequence 5′-CCUU(N)RYGYUG-3′ that contains a highly conserved arginine triplet CCU (see below and Table 3). Conserved CCU always resides in a predicted stem–loop junction, confirmed by SHAPE for representative sequences 1a (Fig. 3D), 1b (Fig. 4D), and 1c (Fig. 5D). Terminal loop nucleotides have been implicated in NMIA modification–interference experiments; therefore also appearing essential for arginine binding. Strikingly, structures adjacent to the conserved stem–loop are quite varied. They include a bulged stem (Fig. 3D, motif 1a), an asymmetrical bulge (Fig. 4D, motif 1b), and a bulge and 3-bp helix (Fig. 5D, motif 1c).
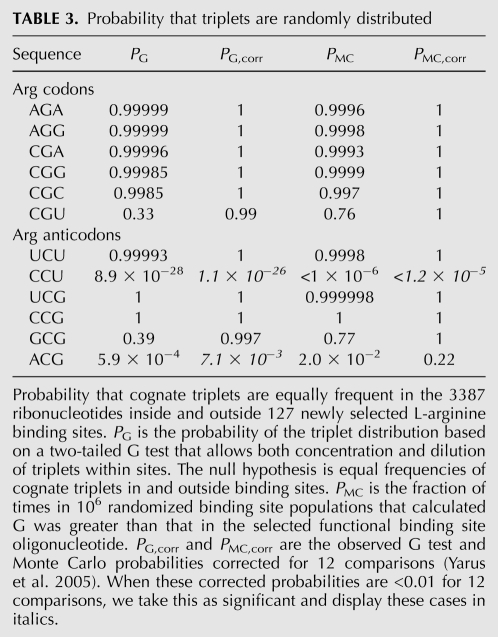
TABLE 3.
Probability that triplets are randomly distributed
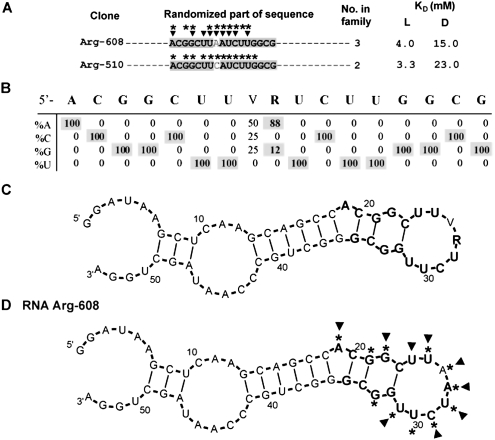
Motif 2
Arginine binding motif 2 was detected only among 17mers. A group of eight clones was derived from four independent origins. Random regions for representative clones are shown in Figure 6A (for the full sequence list for motif 2, see Supplemental Table 1S). Conserved nucleotides are shaded in Figure 6A. Nucleotide variation was calculated from eight isolates and is listed in Figure 6B. Within the 17-nt initially randomized sequence, 16 nt were completely conserved.
FIGURE 6.
Summary of motif 2. (A) Representative sequences from group 2. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks or triangles were identified by SHAPE or NMIA modification–interference experiment, respectively. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) and NMIA modification–interference (triangles) data on computed secondary structure of RNA Arg-608.
Simultaneous folding of eight aligned sequences predicts a conserved bulged hairpin (Fig. 6C). The three G–C base-pair stem of the hairpin is bulged by conserved A19, G22, and G33.
This BayesFold-predicted structure was confirmed by SHAPE performed with the motif 2 RNA Arg-608. Bulged A19 and looped G22, G33, as well as U25–U31 in the terminal loop, appeared reactive (Fig. 6D, asterisks).
In an NMIA modification–interference experiment, ribose of bulged A19 and G22, and all ribose in the terminal loop except for U30, appear crucial for arginine binding to motif 2 (Fig. 6D, triangles).
Motif 2 appears to be a variation of the 1a, 1b, and 1c sites described above. Here, two mutations alter the underlined nucleotides in 5′-GCUU(V)RUCUUG-3′ (Table 1). These mutations decrease arginine affinity twofold in comparison with motifs 1a, 1b, and 1c, providing an additional item of evidence that the terminal loop and the stem are the binding site for arginine.
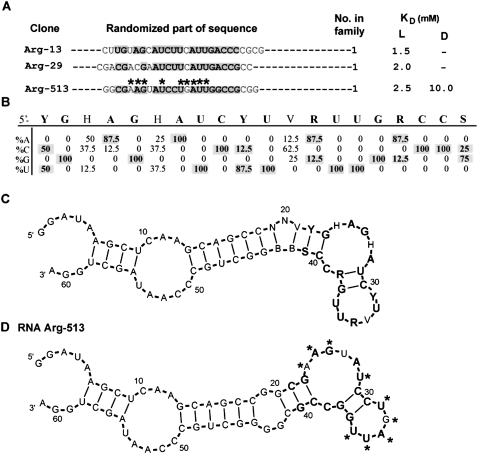
Motif 3
Arginine binding motif 3 was only detected in eight independently derived 25mers. Random regions for representative clones are shown in Figure 7A (for the full sequence list of group 3, see Supplemental Table 1S). Conserved nucleotides are shaded in Figure 6A. Nucleotide variation was calculated and is listed in Figure 7B. The 20-nt-long motif contains 16 absolutely conserved nucleotides, one nucleotide that is 87.5% conserved, and three unconserved nucleotides.
FIGURE 7.
Summary of motif 3. (A) Representative sequences from group 3. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks were identified by SHAPE. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) data on computed secondary structure of RNA Arg-513.
The BayesFold-predicted (Knight et al. 2004) secondary structure, consistent with six aligned sequences, forms an asymmetrical internal loop and a 2-bp stem adjacent to a terminal pentaloop (Fig. 7C).
The predicted structure was confirmed by SHAPE on RNA Arg-513. Nucleotide ribose at A23, A24, G25, and U28 in the internal loop, and all terminal loop nucleotides (U31–U35) appeared less constrained (Fig. 7D, asterisks).
Motif 3 contains 17 highly (more than 88%) conserved nucleotides and is a sequence related to the most prevalent site family—it shares sequence conservation and a potential cognate coding triplet with motifs 1a, 1b, 1c, and 2 within the seven similar nucleotides: 5′-CYU(V)RUUG-3′ (Table 1).
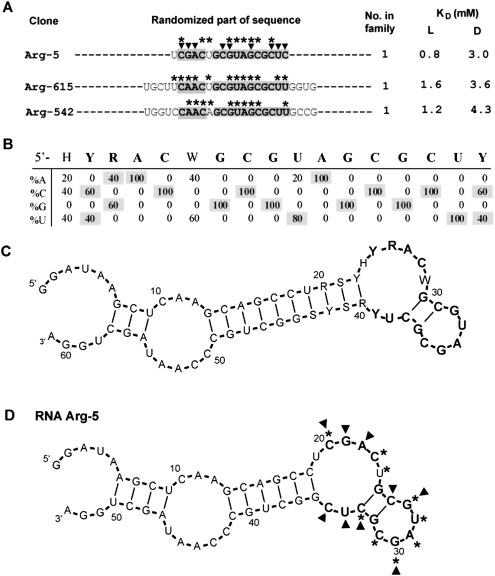
Motif 4
Arginine binding motif 4 was derived from five independent 17 and 25mer sequences. In Figure 8A, randomized parts of representative clones are shown (for the full list of group 4 sequences, see Supplemental Table 1S). Shaded nucleotides comprise the conserved segments. Nucleotide variation spanning 16 nt was calculated (Fig. 7B). Fourteen of 16 nucleotides in this tract are completely conserved.
FIGURE 8.
Summary of motif 4. (A) Representative sequences from group 4. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks or triangles were identified by SHAPE or NMIA modification–interference experiment, respectively. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) and NMIA modification–interference (triangles) data on computed secondary structure of RNA Arg-5.
The calculated stable secondary structure for three aligned 25mer sequences and for an individual 17mer isolates predicts an asymmetrical internal loop connected to the terminal loop by a conserved stem of two G–C pairs (Fig. 8C).
SHAPE for group 4 RNA Arg-5 is shown in Figure 8D (asterisks). SHAPE confirms unconstrained C20, C23, and U24 in the asymmetrical loop (Fig. 8C, corresponding to Y24, C27, and W28, respectively), and an unconstrained terminal loop G27–C31 (Fig. 8C, corresponding to G31–C35).
NMIA modification–interference picks out the ribose backbone at internal loops C20–A22 and U34–C35, stems C26 and C33, and G27 and G30 in the terminal loop as crucial for arginine binding to motif 4 (Fig. 8D, triangles).
Motif 4 is unrelated to motifs 1a, 1b, 1c, and 2, but also forms a binding site within 17mer sequences (Table 1). Most conserved nucleotides within the loop, stem, and bulge are important for arginine binding. The conserved site contains evident arginine coding triplets, concentrated within the conserved stem–pentaloop structure (Fig. 8C).
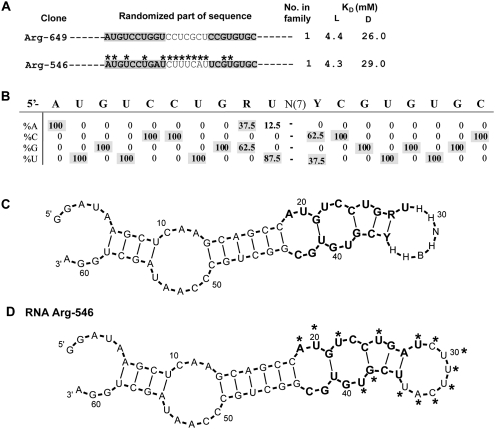
Motif 5
Arginine binding motif 5 appeared in eight independently derived 25mers only. Random regions for representative clones are shown in Figure 9A (for the full sequence list of group 5, see Supplemental Table 1S). Conserved nucleotides are shaded in Figure 9A. Nucleotide variation was calculated from eight isolates (Fig. 9B). The consensus contains two modules, both completely conserved.
FIGURE 9.
Summary of motif 5. (A) Representative sequences from group 5. Only randomized regions are shown. Shaded nucleotides comprise the conserved motif. KD's for free L- and D-arginine are listed if available. Nucleotides marked with asterisks were identified by SHAPE. (B) Sequence variation for motif 1a. (C) BayesFold secondary structure prediction for the alignment of group 1a sequences. (D) Superposition of SHAPE (asterisks) data on computed secondary structure of RNA Arg-546.
A BayesFold-computed secondary structure for eight aligned 25mer sequences predicts three consecutive bulges adjacent to a nonconserved terminal loop (Fig. 9C).
This predicted stable structure was supported by SHAPE on RNA Arg-546. Bulged nucleotides A19, U20, U22, U25, and U39 and all terminal loop nucleotides U28–U35 were reactive (Fig. 9D, asterisks).
Motif 5 consists of two absolutely conserved modules: 5′-AUGUCCUGR-3′ (module 1) and 5′-UCGUGUGC-3′ (module 2). Unlike the other motifs above, this terminal loop, located between conserved stem sequences, consists mostly of nonconserved nucleotides (except for one uracil) (Fig. 9C). Two absolutely conserved arginine triplets CCU and CGU are conjoined within the complex bulged stem, which is the arginine binding site (Fig. 9C).
DISCUSSION
Small matters
An RNA world (White 1976; Gilbert 1986) anticipates that selection of functional RNAs from randomized mostly nonfunctional sequences was an essential step for biological evolution (there being no plausible alternative origin). This has the important implication that active RNAs with the fewest essential nucleotides will usually have been selected early on because the smallest sites should have been more abundant among initially arbitrary sequences (for example, Knight et al. 2005).
Here we describe the first example of selection for an arginine aptamer that seeks the simplest site(s). That is, we seek sites that appear reproducibly within three successively smaller (50, 25 and 17mer) random regions under simultaneous squeezed selections. This experiment has previously been carried out for isoleucine binding (Lozupone et al. 2003), tryptophan binding (Majerfeld and Yarus 2005), and histidine binding (Majerfeld et al. 2005; M Illangasekare and M Yarus, in prep.).
In the present experiments, even 17-nt random regions yielded arginine sites. Therefore, these arginine experiments lacked the earlier demonstration that selection fails when the randomized region is too small. Consequently, here it is not shown experimentally that the smallest possible sites have been observed. To the contrary, the ready occurrence of small sites in a space that was unproductive or less productive for Ile, His, and Trp suggests that Arg provides an easier target for RNA affinity. In any case, the recurrence of specific site sequences in the shortest randomized regions argues that Arg sites among the simplest are being observed.
The smallest arginine binding sites
Five new possibilities for the simplest specific arginine-binding sites have been isolated here: motifs 1a, 1b, 1c, 2, and 4. Every motif is composed of a single continuous tract of highly conserved nucleotides (see Table 1) that fold into a structured stem adjacent to a terminal loop. These are previously uncharacterized sequences that form sites using 17 randomized nucleotides, apparently the smallest binding sites selected for the arginine so far. The larger motif 3 and the more complex motif 5 were recovered only within 25mer sequences.
Distribution of arginine triplets
We have assessed the overall significance of the cognate triplet content of these arginine binding sites in two ways. First, with the G statistic (Sokal and Rohlf 1995) for significant differences between the proportion of triplets inside and outside the site nucleotides, tested against the null hypothesis of equal triplet frequencies. The G statistic resembles χ2, but is more robust. PG in Table 3 is the probability of a triplet concentration equal to or greater than that actually observed, on the basis of the G distribution. Second, observed RNA sequences were randomized 1 million times, and the number of times that the G statistic computed for a randomized sequence, with a binding site in the same position, exceeded that for the initially selected sites was recorded. This comparison of site populations with the same composition, but shuffled sequences, is independent of any a priori assumption about distributions or frequencies. It empirically determines the probability that the selected binding site sequence is as biased as observed. The observed fraction of higher computed G statistics (more biased triplet compositions) in randomized libraries is PMC, the Monte Carlo probability listed on the right in Table 3. Because very improbable initial triplet biases will not be exceeded even among the 106 computed randomizations of the site oligomers, the lowest observable PMC is listed as <1 × 10−6, or PMC,corr < 1.2 × 10−5 after adjustment for multiple comparisons.
As can be seen in Table 3, no arginine codon triplet recurs significantly in this new population of arginine binding sites. The arginine anticodon ACG is partially conserved within the functional hairpin loops of binding sites in motifs 1a, 1b, 1c, and 2. ACG recurrence in the complete population of sites is barely significant by the G test (with our customary criterion of P < 0.01 corrected for 12 comparisons), but not significant in the distribution-free Monte Carlo simulation. For the moment, we do not know whether ACG represents a significant result.
However, the arginine anticodon CCU is found in a highly conserved core sequence whose surroundings vary in independent RNAs to yield binding motifs 1a, 1b, 1c, 3, and 5. The triplet occurs in CCUUNRYGYUG in motifs 1a, 1b, and 1c, for example. The CCU (anticodon of AGG) is therefore very decisively concentrated within selected amino acid binding sites (Table 3). In fact, it recurs in 72% of all sequenced RNAs in this selection, so its abundance would be striking to the eye even without statistical analysis. Thus, the simplest L-arginine sites do have recurring structures, which share a property previously noted for the simplest RNA sites for histidine, tryptophan, and isoleucine (Yarus et al. 2005). That is, cognate coding triplets perform an indispensable function within the most accessible RNA binding sites. This property has now been reproducibly observed in all four cases where the simplest sites have been sought via a squeezed simultaneous selection (that is, with differently sized randomized regions analyzed together). Of course, triplets may be concentrated in simple sites in other cases where the squeezed selection has not been applied.
This newly isolated arginine site population may be more impressive for coding studies than previous similar outcomes, however, because simple, frequent arginine sites have surrounding structural elements that can take multiple forms, as well as invariant core sequence elements conserved in most independent isolations (Figs. 3–9). The CCU cognate anticodon triplet is in the latter class. This work thereby adds to other evidence (Yarus et al. 2009) that suggests the current genetic code is seen with unexpectedly high frequency among the RNA–amino acid interactions that appear most readily after selection. In fact, this type of squeezed selection experiment, which concentrates on a small subset of binding sites (the smallest and simplest), and yet repeatedly finds coding triplets, is among the most persuasive individual arguments in favor of a chemical relation between genetic code triplets and bound amino acids.
MATERIALS AND METHODS
Chemicals were analytical-reagent grade whenever available. Thiopropyl Sepharose 6B freeze-dried powder was obtained from Pharmacia. Lyophilized peptide (N-Arg-Cys-COOH; MW 277.34; purity >98.38%) was synthesized by Bio Basic It was stored in the dark at −70°C. Fresh solutions of dipeptide were prepared before each use. NMIA (“high purity,” MW 177.16) was obtained from Invitrogen Molecular Probes. dimethyl sulfoxide (DMSO) (>99.9%) was obtained from Sigma Chemical.
Methods
Covalent attachment of L-arginyl-L-cysteine to thiopropyl Sepharose 6B
Thiopropyl Sepharose 6B contains reactive 2-thiopyridyl disulfides. ArgCys dipeptide was coupled to sepharose via its cysteine thiol. All buffers and water were de-gassed to avoid oxidation of free thiols. Usually, 0.5 g of dried sepharose powder was suspended in distilled water; after swelling this yielded about 2 mL gel. Gel was washed with 50 volumes of water and then equilibrated with 30 volumes of selection buffer (S; 250 mM NaCl, 50 mM Hepes [pH 7.0], 5 mM MgCl2, 5 mM CaCl2, 0.1 mM EDTA) before coupling. Coupling used 75% settled medium to 25% of buffer S containing L-cysteine (85% of total Sepharose thiol) and L-dipeptide (85% of Sepharose thiol). The suspension was gently rotated at 4°C for 12 h Then the gel was washed with 40 volumes of buffer S. Substitution was calculated by measuring absorbance at 343 nm—released 2-thiopiridone has molar absorption = 8.08 × 103 M−1 cm−1 (Stuckbury et al. 1975). The total initial activated thiol was 27 mM. The final concentration of column L-arginine was 4 mM.
Selection procedure
The initial synthetic single-stranded template DNAs were: (50/25/17N) taatacgactcactatatccagctattgggcagcc N(50/25/17) ggctgcttgagcttatcc, where underlined nucleotides indicate the T7 promoter. Three DNA libraries with 17N, 25N, and 50N were prepared individually by PCR amplification of DNAs. Selections from 50, 25, and 17N templates were started from 9 × 1014, 1.2 × 1015, and 3 × 1014 DNA sequences, respectively, individually amplified by PCR fourfold. An initial RNA pool was generated by ∼14-fold transcription by T7 RNA polymerase for each DNA template. 32P-labeled RNA (7.5 nmol in the first selection round, decreasing in subsequent rounds) was heated at 65°C for 5 min in water, 5X buffer S was added, and heated at room temperature for 10 min to allow for RNA folding. Folded RNA was applied to a 0.25 mL L-dipeptide-thiopropyl Sepharose column equilibrated with buffer S at room temperature. After washing with 10 column volumes of buffer S, RNA was eluted with S + 10 mM L-arginine. Eluted RNA was precipitated, reverse transcribed, and PCR amplified, and the resulting DNA was transcribed for the next selection cycle. After the first cycle, selection was preceded by counterselection on an L-cysteine-thiopropyl sepharose column and the initial ∼85% of void RNA was used for subsequent selection.
RNA secondary structure probing
SHAPE chemistry was performed following the method of Merino et al. (2005). RNA (100 pmol) in 72 μL of water was denatured by heating at 65°C for 5 min, then 18 μL of 5X folding buffer (250 mM Hepes [pH 7.6], 1.25M NaCl, 25 mM MgCl2, 25 mM CaCl2, 0.5 mM EDTA) were added and incubated (24°C) for 20 min. Folded RNA in a single reaction was separated into (+) and (−) NMIA reactions, 45 μL volume of each. NMIA (5 μL at 130 mM in anhydrous DMSO) was added to the RNA solution and allowed to react with RNA for 155 min (∼5 half-lives) (Merino et al. 2005, Wilkinson et al. 2006) at 24°C. Control reactions contained DMSO only. Modified RNA was ethanol precipitated and redissolved in water at 1 μM.
5′-32P-labeled primer (5′-tccagctattgggca-3′) 1 μL at 0.5 μM was added to the (+)/(−) NMIA modified RNA (5 pmol in 5 μL). An RNA-oligoDNA mixture (6 μL) was heated at 65°C for 5 min, and then immediately placed on ice for 5 min for primer annealing. Hybridized RNA-oligoDNA solution and SuperScript III enzyme mix were prewarmed to 48°C in a thermal cycler for 2 min. An enzyme mix (4 μL) was aliquoted, the temperature was raised to 50°C, and the DNA primers were reverse transcribed to sites of modification in the presence of dNTPs. The reaction was terminated at 85°C for 5 min and then chilled on ice. The RNA was degraded by addition of 1 μL of 2 M NaOH. cDNA fragments were subsequently resolved on a 10% (w/v) polyacrylamide gel.
NMIA modification–interference experimental procedure
32P-labeled RNA (150 pmol) was folded and modified by NMIA as above. The modification buffer was exchanged out by passage through a Micro-Bio-Spin P6 column equilibrated with buffer S. Modified RNA was fractionated by passage through an L-arginine affinity column into unbound and specifically eluted fractions, and then ethanol precipitated. Modified nucleotides were detected after primer extension with SuperScript III reverse transcriptase (as above).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by USPHS research Grant GM 48080. We thank laboratory members for their comments on the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1979410.
REFERENCES
- Allers J, Shamoo Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J Mol Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. α Helix-RNA major groove recognition in an HIV-1 rev peptide–RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- Cai Z, Gorin A, Frederick R, Ye X, Hu W, Majumdar A, Kettani A, Patel DJ. Solution structure of P22 transcriptional antitermination N peptide–boxB RNA complex. Nat Struct Biol. 1998;5:203–212. doi: 10.1038/nsb0398-203. [DOI] [PubMed] [Google Scholar]
- Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: The arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Ciesiolka J, Illangasekare M, Majerfeld I, Nickles T, Welch M, Yarus M, Zinnen S. Affinity selection-amplification from randomized ribo-oligonucleotide pools. Methods Enzymol. 1996;267:315–335. doi: 10.1016/s0076-6879(96)67021-9. [DOI] [PubMed] [Google Scholar]
- Connell GJ, Yarus M. RNAs with dual specificity and dual RNAs with similar specificity. Science. 1994;264:1137–1141. doi: 10.1126/science.7513905. [DOI] [PubMed] [Google Scholar]
- Connell GJ, Illangsekare M, Yarus M. Three small ribooligonucleotides with specific arginine sites. Biochemistry. 1993;32:5497–5502. doi: 10.1021/bi00072a002. [DOI] [PubMed] [Google Scholar]
- Ellis JJ, Broom M, Jones S. Protein–RNA interactions: Structural analysis and functional classes. Proteins. 2007;66:903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- Famulok M. Molecular recognition of amino acids by RNA-aptamers: An L-citrulline binding RNA motif and its evolution into an L-arginine binder. J Am Chem Soc. 1994;116:1698–1706. [Google Scholar]
- Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M. RNA aptamers that bind L-arginine with submicromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Origin of life:The RNA world. Nature. 1986;319:618. doi: 10.1038/319618a0. [DOI] [Google Scholar]
- Jiang F, Gorin A, Hu W, Majumdar A, Baskerville S, Xu W, Ellington A, Patel DJ. Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples. Structure. 1999;7:1461–1472. doi: 10.1016/s0969-2126(00)88337-9. [DOI] [PubMed] [Google Scholar]
- Knight R, Birmingham A, Yarus M. BayesFold: Rational secondary folds that combine thermodynamic, covariation, and chemical data for aligned RNA sequences. RNA. 2004;10:1323–1336. doi: 10.1261/rna.5168504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, De Sterck H, Markel R, Smit S, Oshmyansky A, Yarus M. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005;33:5924–5935. doi: 10.1093/nar/gki886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Changayil S, Majerfeld I, Yarus M. Selection of the simplest RNA that binds isoleucine. RNA. 2003;9:1315–1322. doi: 10.1261/rna.5114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerfeld I, Yarus M. Isoleucine: RNA sites with essential coding sequences. RNA. 1998;4:471–478. [PMC free article] [PubMed] [Google Scholar]
- Majerfeld I, Yarus M. A diminutive and specific RNA binding site for L-tryptophan. Nucleic Acids Res. 2005;33:5482–5493. doi: 10.1093/nar/gki861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerfeld I, Puthenvedu D, Yarus M. RNA affinity for molecular L-histidine; Genetic code origins. J Mol Evol. 2005;61:226–235. doi: 10.1007/s00239-004-0360-9. [DOI] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Morozova N, Allers J, Myers J, Shamoo Y. Protein–RNA interactions: Exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics. 2006;22:2746–2752. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- Sokal R, Rohlf F. Biometry: The principles and practice of statistics in biological research. Freeman; New York: 1995. [Google Scholar]
- Stuckbury T, Shipton M, Normis R, Malthouse JPG, Brocklehurst K, Herbert JAL, Suschitzky H. A reporter group delivery system with both absolute and selective specificity for thiol groups and an improved fluorescent probe containing the 7-nitrobenzo-2-oxa-1,3-diazole moiety. Biochem J. 1975;151:417–432. doi: 10.1042/bj1510417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Frankel AD. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Frankel AD. Arginine-binding RNAs resembling TAR identified by in vitro selection. Biochemistry. 1996;35:2229–2238. doi: 10.1021/bi951844b. [DOI] [PubMed] [Google Scholar]
- Treger M, Westhof E. Statistical analysis of atomic contacts at RNA–protein interfaces. J Mol Recognit. 2001;14:199–214. doi: 10.1002/jmr.534. [DOI] [PubMed] [Google Scholar]
- White HB., III Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Yarus M. A specific amino acid binding site composed of RNA. Science. 1988;240:1751–1758. doi: 10.1126/science.3381099. [DOI] [PubMed] [Google Scholar]
- Yarus M, Christian EL. Genetic code origins. Nature. 1989;342:349–350. doi: 10.1038/342349b0. [DOI] [PubMed] [Google Scholar]
- Yarus M, Caporaso JG, Knight R. Origins of the genetic code: The escaped triplet theory. Annu Rev Biochem. 2005;74:179–198. doi: 10.1146/annurev.biochem.74.082803.133119. [DOI] [PubMed] [Google Scholar]
- Yarus M, Widmann JJ, Knight R. RNA–amino acid binding: A stereochemical era for the genetic code. J Mol Evol. 2009;69:406–429. doi: 10.1007/s00239-009-9270-1. [DOI] [PubMed] [Google Scholar]