Abstract
Splicing of vertebrate introns involves recognition of three consensus elements at the 3′ end. The branch point (BP) and polypyrimidine tract (PPT) are usually located within 40 nucleotides (nt) of the 3′ splice site (3′ ss), AG, but can be much more distant. A characteristic of the region between distant BPs (dBPs) and the 3′ ss is the absence of intervening AG dinucleotides, leading to its designation as the “AG exclusion zone” (AGEZ). The human HTR4 gene, which encodes serotonin receptor 4 and has been associated with schizophrenia, bipolar disease, and gastrointestinal disorders, has four exons with extensive AGEZs. We have mapped the BPs for HTR4 exons 3, 4, 5, and g generated by in vitro splicing, and validated them by mutagenesis in exon-trapping vectors. All exons used dBPs up to 273 nt upstream of the exon. Strikingly, exons 4 and 5 used combinations of both distant and conventionally located BPs, suggesting that successful splicing of these exons can occur by distinct pathways. Our results emphasize the importance for single nucleotide polymorphism resequencing projects to take account of potential dBPs, as the extended AGEZs are vulnerable to mutations that could affect splicing itself or regulation of alternative splicing.
Keywords: branch point, alternative splicing, HTR4, SNP
INTRODUCTION
Accurate splicing is a prerequisite for mRNA formation by the removal of introns from pre-mRNAs and by the joining together of exons. This reaction is catalyzed by the spliceosome, and several elements within the RNA sequence need to be recognized for functional splicing (Wahl et al. 2009). The upstream exon–intron boundary is defined by the 9 nucleotide (nt) 5′ splice site (5′ ss) consensus sequence with an invariant GU dinucleotide at the most 5′ part of the intron. Recognition of the 3′ splice site (3′ ss) requires three essential elements: the branch point (BP) sequence, the polypyrimidine tract (PPT), and an invariant AG dinucleotide at the 3′ end of the intron. These three elements are normally located within the last 40 nt of the intron and are recognized during the early steps of spliceosome formation. The importance of the consensus splice site elements is illustrated by the observation that at least 10% of human genetic diseases are caused by mutations within them (Cooper et al. 2009).
During the early steps of splicing the BP is one of the first elements to be recognized and is first identified by SF1, the BP-binding protein (Abovich and Rosbash 1997; Berglund et al. 1997). In close vicinity, U2AF65 interacts with the PPT and has a preference for uridine (U)-rich sequences (Singh et al. 1995). Additionally, in so-called AG-dependent introns (Reed 1989), U2AF35 recognizes the 3′ ss AG (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). In the next steps of splicing, U2 snRNP replaces SF1 at the BP, by base-pairing of the U2 snRNA with the BP sequence, after which the U4/5/6 snRNPs are recruited and conformational rearrangements lead to formation of the active spliceosome. The first step of splicing involves a hydrophilic attack by the 2′ OH of the BP adenosine on the 5′ ss, breaking the bond between the 5′ exon and intron. Following the first step of splicing, the spliceosome must locate the 3′ ss AG, where the second step–the joining of exons and the lariat excision—takes place (Wahl et al. 2009). In introns with distant BPs (dBPs) and PPTs, the 3′ ss is located by a scanning process from the BP to the first downstream AG (Smith et al. 1989, 1993; Liu et al. 1997). Similar scanning behavior is observed in bimolecular exon ligation experiments (Chen et al. 2000). Consequently, one prerequisite of the region between distant BPs and the 3′ ss is that it lacks any other AG dinucleotides leading to its designation as the AG-exclusion zone (AGEZ) (Gooding et al. 2006).
The dBP introns are likely to be of interest from a number of standpoints. The dBP arrangement may facilitate particular mechanisms of splicing regulation. A uniform feature of those that have been dissected experimentally is the presence of negative regulatory elements within the AGEZ between the dBP and the exon (Helfman et al. 1990; Gooding et al. 1994; Southby et al. 1999; Wollerton et al. 2004). Moreover, the extended AGEZ is likely to be vulnerable to mutations, the effects of which could range from catastrophically deleterious effects upon splicing, such as the introduction of new AG dinucleotides that would be recognized as 3′ss (Gooding et al. 2006), to more subtle misregulation of alternative splicing. Either type of effect could be associated with pathologies (Cooper et al. 2009).
Based upon the small number of known alternatively spliced dBP exons we previously applied the concept of AGEZs as a basis for a bioinformatics algorithm systematically searching alternative human exons in the AltExtron database for intronic regions at the 3′ ss with extended zones lacking AG dinucleotides. More than 800 AltExtron exons had AGEZs ≥100. One of the most extreme examples, an intron in the tyrosine phosphatase σ (PTPRS) gene has an AGEZ of 1126 nt. We estimated that between 20% and 50% of the identified introns would be associated with dBPs. Consequently, up to 0.6% of all human introns potentially contain dBP and 2%–5% of all human genes have at least one intron with a dBP (Gooding et al. 2006). The serotonin receptor 4 gene (HTR4 or 5-HT4) is a particularly interesting example. Exons 3, 4, 5, and g have extended AGEZs of 149, 291, 221, and 101 nt, respectively, and the large AGEZ arrangement is maintained between several mammalian species, suggesting functional significance, even though the primary sequence in these regions is not highly conserved (Fig. 1B,C; see also the first section in Materials and Methods). Despite the tendency for exons with extended AGEZ to be alternatively spliced (Gooding et al. 2006), only HTR4 exon g was annotated as a cassette exon. HTR4 is a member of the G-protein coupled receptors (GPCRs) family, typically with seven trans-membrane regions. Most of the known 10 isoforms vary in their cytoplasmic—the most C-terminal— part and arise from the use of alternative 3′ terminal exons (Figs. 1C; see also Fig. 6D below). HTR4 expression is prevalent in the brain, specifically in the hippocampus and basal ganglia, and also in intestinal tissue and the heart (Medhurst et al. 2001). Mouse knock-out models suggest a potential involvement in stress-induced eating disorders (Compan et al. 2004). HTR4 agonists also have been shown to improve the performance of animals in learning and memory tasks, but also hold potential for antidepression treatment (Fontana et al. 1997; Letty et al. 1997; Marchetti et al. 2004; Lucas et al. 2007). Several studies have associated HTR4 single nucleotide polymorphisms (SNPs) with disorders such as schizophrenia and mood or attention-deficit disorders (Ohtsuki et al. 2002; Suzuki et al. 2003; Li et al. 2006). At least one SNP was suggested to affect the BP of exon 5 (Suzuki et al. 2003). Hence, HTR4 is not only interesting from a splice regulatory point of view but also for its physiological properties.
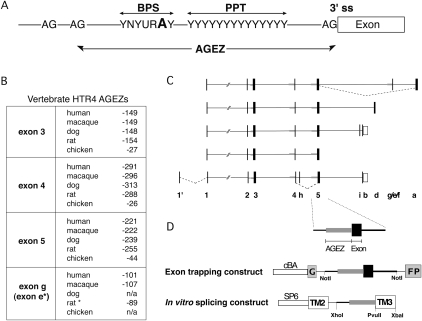
FIGURE 1.
AG dinucleotide exclusion zones and distant branch points. (A) The branch point sequence (BPS), polypyrimidine tract (PPT), and the invariant AG at the 3′ ss are normally located within 40 nt of each other. The region between the 3′ ss AG and the next upstream AG dinucleotide is defined as the AG exclusion zone (AGEZ). In introns with distant BPs, there is no apparent constraint on the distance between the PPT and 3′ ss as long as it is free of AGs. (B) Four exons of mammalian serotonin receptor (HTR4) genes have extended AGEZ. The position of the first AG upstream of the 3′ ss is shown for each exon in five different vertebrates. For dog and chicken, the genomic sequence coverage is not sufficient to determine their AGEZ of exon g (n/a) and in rat only the alternative 3′ ss variant e is present (*). (C) HTR4 isoforms: Exons are depicted as filled black boxes, 3′UTRs as white boxes, introns as lines, large AGEZ (>100 nt) in thick gray lines. Known alternative splicing events are indicated by dashed lines. (D) Experimental strategy: Each exon with a large AGEZ was cloned along with upstream and downstream intronic sequences into the GFP-based exon trapping construct driven by chicken β-actin promoter (cBA) for tissue culture transfection. The AGEZ (in gray) with some additional upstream intronic region was cloned into the PY7 in vitro splicing construct to replace the BP, PPT, and 3′ ss elements of TPM1 exon 3 (TM3).
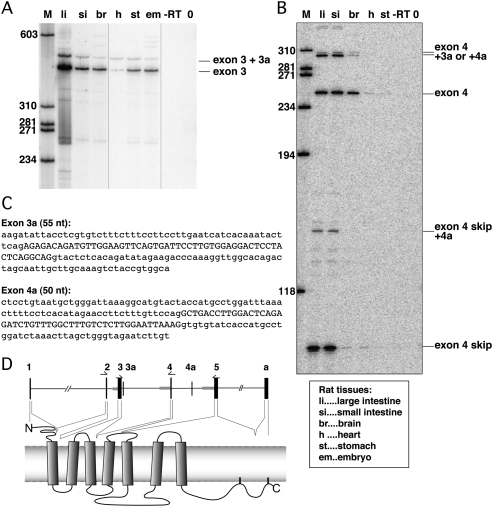
FIGURE 6.
HTR4 alternative splicing in rat tissues. (A) Alternative splicing between HTR4 exons 2 and 4 detected by RT-PCRs using primers in exons 2 and 4. No exon 3 skipping is detected but inclusion of a novel exon 3a was detected and verified by cloning and sequencing. Labels indicating rat tissues are explained beneath panel B. (B) Alternative splicing between HTR4 exons 3 and 5 by RT-PCRs using primers in exons 3 and 5. Exon 4 skipping as well as inclusion of exon 3a and another novel exon 4a were detected. All RT-PCR products were cloned and validated by sequencing. (C) Sequences of novel exons detected in A and B. Exons in upper case, surrounding intronic regions in lower cases. Position of exon 3a according to UCSC rat assembly Nov. 2004: chr18:58,418,797–58,418,851 and position of 4a chr18:58,442,685–58,442,734. (D) Schematic drawing of the HTR4 gene and protein structure. Position of primers used for A and B are depicted above according exons, novel exons 3a and 4a are inserted into the gene structure. The parts of HTR4 protein encoded by each exon are indicated by dashed lines.
Here we report a systematic analysis of the BPs used by the four HTR4 exons with large AGEZs. We mapped the dBPs by in vitro splicing followed by primer extensions, and found that all but exon g used dBPs. Exons 4 and 5 were able to utilize several BPs, some also in canonical positions. All of these BPs were analyzed by the effect of mutagenesis upon splicing in tissue culture reporter constructs. We were able to validate dBPs for exon 3. Exons 4 and 5 might use additional cryptic BPs but showed a significant decrease in exon inclusion upon disruption of its BPs. Exon g actually used a dBP under these conditions. We also further show here for the first time that exon 4 is alternatively spliced, and we discovered two novel exons. Our results highlight the necessity of combining several methods for accurate BP mapping, and emphasizes the importance of taking into account large AGEZs when carrying out resequencing for SNP discovery.
RESULTS
Analyzing predicted branch points in vitro and in vivo
Of the three common methods used to map BPs, two permit precise identification of the nucleotide involved in lariat formation. The “gold standard” is to in vitro splice RNA and to exploit the block of primer extension by reverse transcriptase at the BP. This, however, is only possible if constructs splice efficiently in vitro (Ruskin et al. 1984). Method number two takes advantage of the fact that certain reverse transcriptases are able to extend—albeit with low and variable efficiency—across the 2′– 5′ bond of the lariat at the BP. The resulting cDNAs are then used for PCR amplification and sequencing, allowing mapping of the junction of the BP with the 5′ ss. The unusual RNA structure at the lariat also makes the reverse transcriptases more prone to misincorporate nucleotides at the position of the BP, which gives another useful indication of its exact position (Vogel et al. 1997; Gao et al. 2008). In the third method, minigene reporter constructs are used to monitor changes in splicing upon mutation of potential BPs. This approach only gives indirect evidence for BPs, as it generally identifies nucleotides important for exon inclusion in the context of cell culture based transfections. For the four HTR4 exons we have employed a combination of two of these methods: BP mapping by primer extension of in vitro splicing reactions and validation of these BPs by mutagenesis in exon-trapping constructs containing the HTR4 exons and their surrounding introns (Fig. 1D). These constructs were transfected into HeLa cells and the resulting splice products were analyzed by RT-PCR.
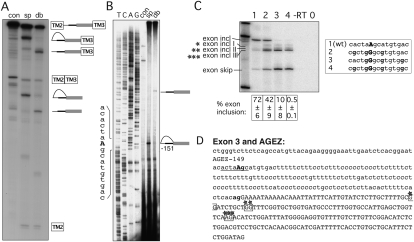
Exon 3 has a dBP at –151
Exon 3 of HTR4 has an AGEZ of 149 nt (Fig. 1B) within which there were several positions that fit the consensus for BPs with PPTs (see Materials and Methods). The size of the AGEZ is maintained between 149 and 154 nt in several mammalian species (Fig. 1B). To analyze this intron by in vitro splicing we cloned the AGEZ and additional 150 upstream nucleotides replacing the BP and PPT of exon 3 of a minigene reporter containing the rat α-tropomyosin (TPM1) exons 2 and 3 (Fig. 1D). This was necessary, as a minigene construct containing the HTR4 exon 3 with surrounding introns did not splice efficiently, precluding further analysis by primer extension (data not shown). In HeLa nuclear extracts this reporter went efficiently through the first and second step of splicing as both the lariat intermediate and the lariat accumulate after 1 h (Fig. 2A, lane sp). Due to its looped structure lariat RNA migrates at a higher apparent size than the linearized, debranched RNA (Fig. 2A, lane db), which migrates according to its size. To determine the site of branch formation we performed primer extensions using a primer hybridizing to the most 3′ end of the intron. The major stop in the extension in Figure 2B, lane sp, was found at nucleotide position –150. Comparing it with the sequencing lanes at the left-hand side of the panel, it aligned with a G, but as the reverse transcriptase stops 1 nt before the BP it maps to an adenosine within a good consensus (ctaAgc) at position −151 (underlined in Fig. 2D, with BP-adenosine in bold). Here the BP adenosine coincides with the first AG upstream of the AGEZ. Moreover, it lies just upstream of a 44-nt PPT uninterrupted by purines.
FIGURE 2.
Mapping BP of exon 3. (A) In vitro splicing of construct containing the HTR4 exon 3 AGEZ. Splicing precursor, intermediates, and products are depicted to the right of the gel, with the α-Tropomyosin exons TM2 and TM3 in boxes, the AGEZ in gray, and introns in black. Looped RNA-forms correspond to the lariat and lariat intermediates. Lane con, unspliced control; sp, spliced (2-h time point); db, spliced and then debranched. (B) Primer extension map BP to position –151. Left panel shows sequencing lanes, right panel primer extension with control RNA in the splicing buffer (con), spliced transcripts (sp), and RNA that was debranched after splicing (db). Sequence surrounding the BP is depicted on the left side of the gel. (C) Mutagenesis of BP in exon trapping construct: lane 1 shows WT splicing pattern with mainly exon inclusion. Upon mutation (lanes 2–4) of more As into Gs (sequence changes in bold in box beside the gel) splicing pattern changes to exon skipping and usage of alternative 3′ ss (exon incl I– III). Percentage of exon inclusion (n = 3) ± standard deviation is shown underneath the gel. Control lanes are reverse transription reactions without an enzyme (−RT) and PCRs without a template (0). (D) The sequence of exon 3 and its AGEZ: intron sequence in lower case and the exon sequence in upper case; first upstream AG dinucleotide and 3′ ss AG in bold. Positions of BP are underlined and BP-A is in upper case. Alternative 3′ ss highlighted in boxes with stars correspond to the three different alternative 3′ ss products in C).
We next analyzed the effect of exon 3 BP mutagenesis in an exon-trapping vector. We cloned the exon with the AGEZ plus an additional 150 upstream nucleotides and 220 nt of the downstream intron into a GFP-based two-exon minigene reporter construct (Fig. 1D). When transfected into HeLa cells, exon 3 was included over 70% in the wild-type (WT) construct (Fig. 2C, lane 1). Because nearby adenosines can compensate for mutated BPs we also eliminated potential cryptic sites. Upon mutation of A's to G's at positions −155, −152, −151, and −148, exon inclusion levels dropped from 70% to 40% (Fig. 2C, lane 2). Mutation of positions –152, −151, −148, and −143 led to inclusion levels below 10%, suggesting that position −143 is a major cryptic BP (Fig. 2C, lane 3). Only mutating all five A's into G's abolished exon inclusion completely (Fig. 2C, lane 4). Interestingly, sequence alignments between six mammalian species (human, cow, dog, macaque, gorilla, mouse, and rat data not shown) show that adenosine −151 and uracil –153 at the BP are conserved in five out of seven species and that the cryptic BP adenosine at position –143, together with a U at –145, also shows 100% conservation. Strikingly, any manipulation of the BP regions led not only to exon skipping but also to increased use of cryptic 3′ ss within exon 3 (Fig. 2C, exon incl I, exon incl II, exon incl III; Fig. 2D, highlighted in boxes with stars). These splice sites were validated by sequencing. Due to their location, these 3′ ss are expected to be used in conjuction with the associated cryptic BPs and PPTs.
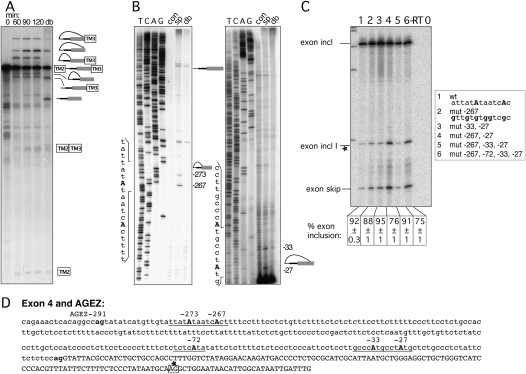
Exon 4 uses distant and canonical BPs
Exon 4 of HTR4 has a very extended AGEZ of 291 nt, and this arrangement is conserved in other mammals (Fig. 1B, AGEZs 288–313 nt). Using a construct making TPM1 exon 3 splicing dependent on the intronic region of HTR4 exon 4 containing the AGEZ and another 120 upstream nucleotides, we analyzed the progression of its splicing in the nuclear extract. Over a time period from 0 to 120 min, an increasing amount of splice intermediates and products appeared (Fig. 3A). Intriguingly, four different lariat species were observed. These correspond to two different populations of lariat products and intermediates, which then collapse into two linear forms in the debranched lane (Fig. 3A, lane db), suggesting the use of two different BPs. Lariats of BPs in canonical positions have larger loops than lariats in more distant positions, and therefore migrate more slowly. The primer extensions verified the use of two pairs of neighboring BPs. Two dBPs were mapped at positions –267 and –273 (Fig. 3B, left panel) and two in canonical positions –33 and –27 (Fig. 3B, right panel). Due to the large AGEZ it was necessary to use a second primer further upstream within the intron (positions −195 to −216) to map the dBPs (Fig. 3B, left panel). In the cell transfection assay with the GFP-based reporter, exon 4 of HTR4 was included to 92% (Fig. 3C, lane 1). Mutation of six adenosines surrounding the dBP at −267 and −273 (Fig. 3C, lane 2), or disruption of the two canonical BPs (Fig. 3C, lane 3) had a negligible effect on exon inclusion. Combining mutations at the dBP and BP at position –27 reduced the exon inclusion levels to 76% (Fig. 3C, lane 4), although surprisingly, additional disruption of the −33 BP led to 91% inclusion (Fig. 3C, lane 5). Introducing mutations of three adenosines at positions –72 to –68 in combination with all the mutations in Figure 3C, lane 5, resulted in a reduction of exon inclusion down to 75% (Fig. 3C, lane 6). These sites were chosen as we had some evidence from primer extensions that the adenosine at –72 was used as a BP, but not reproducibly (data not shown). Interestingly, these manipulations of BPs again led to an increased usage of a suboptimal 3′ ss within the sequence of exon 4 (Fig. 3C, exon incl I), as well as in the expected exon skipping event. The position of this splice site was estimated by calculating the PCR-product size according to its migration in correlation with a molecular weight marker, and it is highlighted by a box with stars in Figure 3D. The modest response to the combined mutations (Fig. 3C, lane 6) suggests that further cryptic BPs are able to compensate in vivo.
FIGURE 3.
Mapping BPs of exon 4. (A) In vitro splicing of construct containing the HTR4 exon 4 AGEZ. Splicing reactions were incubated from 0 to 120 min. A sample from the 120-min time point was subsequently debranched (lane db). Increasing amounts of splicing intermediates and products accumulate over the 120-min time course. Two different populations of lariats and lariat intermediates correspond to differently positioned BPs. (B) Primer extension maps BPs to positions −27, −33, −267, and −273. Left panel shows primer extension and sequencing with an upstream primer (position −195 to −216) due to the large size of the AGEZ. Right panel primer extension and sequencing with primer hybridizing to the most 3′ end of the intron. Sequences surrounding the BPs are depicted on the left side of each gel. (C) Mutagenesis of BPs in exon trapping construct: lane 1 shows WT splicing pattern with mainly exon inclusion. Upon mutation (lanes 2–6) of As into Gs (sequence changes in bold in box beside the gel) splicing pattern changes to exon skipping and usage of a cryptic 3′ ss (exon incl I). Percentage of exon inclusion (n = 3) ± standard deviation is shown underneath the gel. Control lanes are reverse transription reactions without enzyme (−RT) and PCRs without a template (0). (D) Sequence of exon 4 and its AGEZ: intron sequence in lower case and exon sequence in upper case; first upstream AG dinucleotide and 3′ ss AG in bold. Positions of BPs underlined and BP-As in bold upper case. Cryptic 3′ ss observed in C highlighted in box with star.
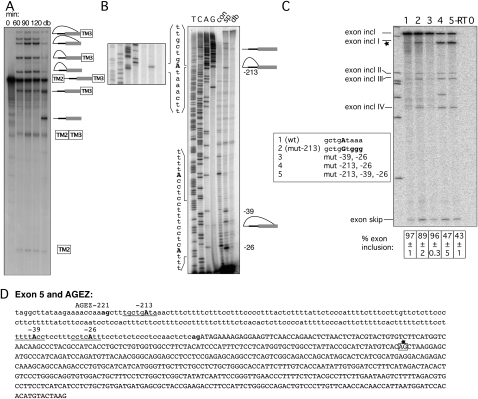
Exon 5 has distant and canonical BPs
The 569-nt exon 5 is the largest of the four HTR4 exons, and the AGEZs range from 221 nt in humans to 255 nt in the rat (Fig. 1B). In the in vitro splicing reporter we observed accumulation of splice products over a period of 2 h (Fig. 4A). Once more, multiple lariat species collapsed to two linear forms upon debranching, suggesting the use of multiple widely separated branch points. This was confirmed by direct primer extension mapping of BPs to positions –213, −39, and – 26 (Fig. 4B). BPs –213 and –26 lie within a good BP consensus, ctgAta and ctcAtt, respectively (underlined in Fig. 4D), and have extended PPTs on their downstream side. Mutagenesis of the four neighboring A's at the −213 dBP in the exon trapping construct led to no reduction of exon inclusion levels in comparison with the WT (Fig. 4C, lanes 1,2). Likewise, mutation of the BPs at positions –39 and –26 had no effect (Fig. 4C, lane 3). However, combinations of the −213 dBP mutations and mutation of either –26 alone (Fig. 4C, lane 4), or additionally, of –39 (Fig. 4C, lane 5) reduced inclusion levels to 47% and 43%, respectively. The residual exon inclusion presumably depends on additional cryptic BPs. Interestingly, rather than exon skipping, the predominant effect of these BP mutations was the use of one major, and several minor, cryptic 3′ ss within exon 5 (Fig. 4C, exon incl I, exon incl II, exon incl III, exon incl IV). The identity of the major cryptic 3′ ss—highlighted in Figure 4D in a box with a star—was verified by sequencing.
FIGURE 4.
Mapping BPs of exon 5. (A) In vitro splicing of construct containing the HTR4 exon 5 AGEZ. Splicing reactions were incubated from 0 to 120 min. A sample from the 120-min timepoint was subsequently debranched (lane db). Increasing amounts of splicing intermediates and products accumulate over the 120-min time course. Two different populations of lariats and lariat intermediates correspond to differently positioned BPs. (B) Primer extensions map BPs to positions −26, −39, and −213. Left panel shows detail of primer extension and sequencing surrounding the BP; right panel shows primer extension and sequencing with primer hybridizing to the most 3′ end of the intron. Sequences surrounding the BPs are depicted on the left side of the gel. (C) Mutagenesis of BPs in exon trapping construct: lane 1 shows WT splicing pattern with mainly exon inclusion. Upon mutation (lanes 2–5) of As into Gs (sequence changes in bold in box beside the gel) splicing pattern changes to exon skipping and usage of cryptic 3′ ss (exon incl I–IV). Percentage of exon inclusion (n = 3) ± standard deviation is shown underneath the gel. Control lanes are reverse transription reactions without enzyme (−RT) and PCRs without a template (0). (D) Sequence of exon 5 and its AGEZ: intron sequence in lower case and exon sequence in upper case; first upstream AG dinucleotide and 3′ ss AG in bold. Positions of BPs underlined and BP-As in bold upper case. Alternative 3′ ss highlighted in box with star that corresponds to the major alternative 3′ ss product in C that also was validated by sequencing.
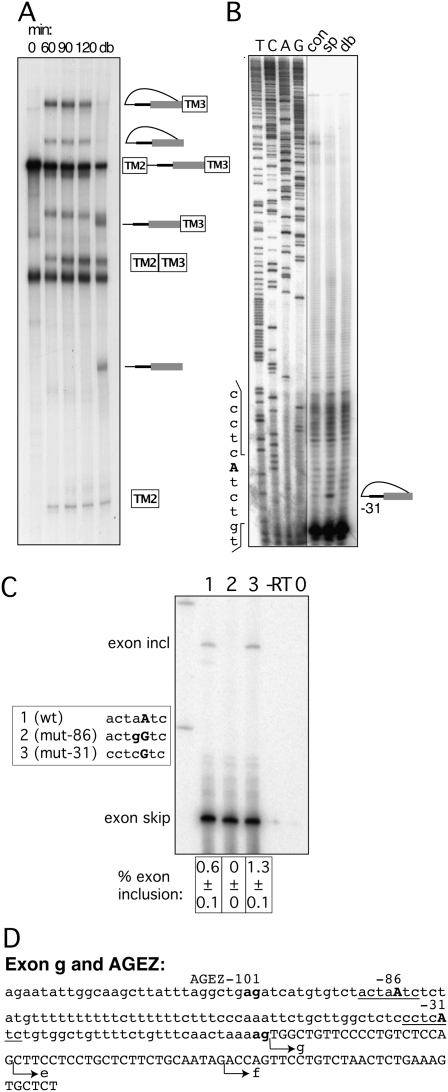
Exon g uses a dBP in vivo but a canonical BP in vitro
Exon g is known alternatively as a spliced exon. It is mainly included in brain tissues, but it is skipped in intestinal samples (Medhurst et al. 2001). The AGEZ of 101 nt including its own suboptimal AAG 3′ ss was cloned upstream of exon 3 in the TPM1-based reporter. We monitored its splicing over a time period of 2 hs and found that, despite the weak 3′ ss, the construct spliced efficiently. It went through steps 1 and 2 of splicing, and we observed a single lariat population indicating the use of a single BP (Fig. 5A). A BP at position –31 was determined by primer extension (Fig. 5B). In stark contrast, mutagenesis of this site in the GFP-based reporter did not interfere with exon inclusion (Fig. 5C, lanes 1,3). However, mutation of a more distant BP at position –86 led to complete exon skipping (Fig. 5C, lane 2). The discrepancy between the in vitro and in vivo results could be explained by the fact that exon 3 of TPM1 has several exonic splicing enhancers that might facilitate use of the weaker –31 canonical BP in the in vitro assay. In contrast, in the exon trapping vector the weak −31 BP might not be viable in the context of its own exon, leaving its splicing dependent only on the more distant stronger BP at position –86 with a strong, very U-rich PPT.
FIGURE 5.
Mapping BPs of exon g. (A) In vitro splicing of construct containing the HTR4 exon g AGEZ. Splicing reactions were incubated from 0 to 120 min. A sample from the 120-min time point was subsequently debranched (lane db). Over the 120-min time course splice products and a single population of lariat and lariat intermediate accumulate. (B) Primer extensions map BP to position −31. Sequences surrounding the BP are depicted on the left side of the gel. Sequencing panel obtained from the same gel but with a different exposure. (C) Mutagenesis of BP in exon trapping construct: lane 1 shows WT splicing pattern with mainly exon skipping and a small amount of exon inclusion. Lane 2—upon disruption of dBP −86 exon inclusion is completely abolished. Lane 3—upon mutation of BP −31 no reduction in exon inclusion is detected. Sequence changes in bold in box beside the gel. Percentage of exon inclusion (n = 3) ± standard deviation is shown underneath the gel. Control lanes are reverse transription reactions without enzyme (−RT) and PCRs without a template (0). (D) Sequence of exon g and its AGEZ: intron sequence in lower case and exon sequence in upper case; first upstream AG dinucleotide and 3′ ss AG in bold. Positions of BPs underlined and BP-As in bold upper case. Alternative 3′ ss of isoforms e and f are indicated by arrows.
Novel HTR4 alternative splicing events
The initial bioinformatics study revealed that exons with larger AGEZs had a higher frequency of alternative splicing, and the extended region between the BP and 3′ ss can serve as a binding platform for factors involved in splicing regulation (Gooding et al. 2006). However, from the literature and expressed sequence tag (EST) data, HTR4 exons 3, 4, and 5 are annotated as constitutive. This suggests either that the large AGEZ arrangement can be advantageous for some constitutive exons, or that exons 3–5 may be alternatively spliced under some circumstances. To address whether exon 3 or 4 can be skipped, we carried out RT-PCR analysis of RNA from various rat tissues, including the large and small intestines, brain, heart, stomach, and embryo using primers in the corresponding flanking exons (Fig. 6D).
Exon 3 showed the expected exon inclusion band of 411 base pairs (bp), but no exon-skipping product was detectable. Surprisingly, an additional band of ∼460 bp was visible (Fig. 6A). A novel exon was verified by sequencing and mapped to a region 100 nt downstream from exon 3 (chromosome position according to UCSC rat assembly Nov. 2004: chr18:58,418,797–58,418,851). The sequence of the 55-nt-long exon 3a (our nomenclature)—in capital letters in Figure 6C—is not represented in any ESTs. Its consensus 3′ and 5′ ss sequences and the overall exon splicing enhancer (ESE) profile make it look like a bona fide exon rather than a pseudoexon (Zhang and Chasin 2004). All these elements are also present in six other vertebrate species, although the overall exon sequence is less conserved (data not shown).
Exon 4 showed a more complex splicing pattern. In addition to the 249 bp band that represents exon 4 inclusion, we also detected an exon skipping 95-bp band (Fig. 6B). Additional bands were observed that were intermediate in size between the exon 4 included and skipped products, and that were also larger than the exon 4 included (Fig. 6B, lanes li, si). By sequencing RT-PCR products we not only identified exon 4 inclusion and skipping, but also exon 4 skipping in combination with a novel 50-nt-long exon 4a (our nomenclature) 2500 bp upstream of exon 5 (chromosome position according to UCSC rat assembly Nov. 2004: chr18:58,442,685–58,442,734). A dublet (∼300-bp) was visible, and these bands corresponded to inclusion of exon 4 together with either exon 3a or 4a. We also have evidence from our sequencing data that both of these exons can be included together with exon 4. Again, exon 4a shows all the key characteristics of a bona fide exon, although we cannot make any statements about interspecies conservation since we failed to find corresponding regions in other vertebrates. The approximate position of exons 3a and 4a within the rat HTR4 transcript is indicated in Figure 6D.
We also looked at exon 5 alternative splicing using primers in exon 4 and reverse primers in exon a or b—the two alternative terminal exons annotated in rat—and could not detect any exon 5 skipping in any tissues. But, again, we detected inclusion of exon 4a and we validated this event by sequencing (data not shown).
All of the newly characterized alternative splicing events share the property of leading to frame shifts in the resulting transcript, which would introduce premature termination codons (PTC) and could lead either to translation of truncated proteins or to nonsense-mediated decay (NMD). The reduced level of various PTC-containing isoforms in total brain RNA seems particularly interesting, suggesting a tissue-specific regulation of these events (Fig. 6B, lane br). We cannot prove that this PTC-causing nonproductive splicing would be targeted by NMD as we currently lack a cell line expressing HTR4 at sufficient levels.
DISCUSSION
Conserved HTR4 AGEZs harbor distant and canonical BPs
HTR4 caught our attention as a gene with known disease associations within which four exons have conserved, extended AGEZs. Moreover, some of the disease-associated SNPs are in the region of the large AGEZ exons. Of the four exons, only exon g was annotated to be alternatively spliced, suggesting either that exons 3, 4, and 5 are, in fact, alternatively spliced under some circumstances, or that the extended AGEZ arrangement can be of advantage to some constitutively spliced exons. Using a combination of mapping BPs in vitro by primer extension, followed by mutagenesis of mapped BPs in exon-trapping transfection constructs, we confirmed that all four exons had dBPs toward the 5′ end of their AGEZs. This validated our use of the AGEZ as a minimal upstream boundary for cloning of regions containing probable dBPs (Gooding et al. 2006). However, our results also yielded a number of surprises, including the observation that exons 4 and 5 use multiple BPs in both distant and canonical locations.
What could be the function, implied by conservation, of the extended AGEZs upstream of the HTR4 exons? The original examples of extended AGEZs were upstream of mutually exclusive exons where the distant BPs were sufficiently close to the upstream 5′ ss to prevent the two exons from splicing together (Smith and Nadal-Ginard 1989; Southby et al. 1999). A more general role of dBPs in alternative splicing appears to be to accommodate negative regulatory elements between the BP and exon (Helfman et al. 1990; Wollerton et al. 2004). Alternative splicing of HTR4 exons 4 and g might be facilitated by such an arrangement. By contrast, there is currently no evidence for alternative splicing of either exon 3 or 5.
Exons 4 and 5 show the most extraordinary arrangement of BPs. Both exons not only used dBPs, but in addition, they used two canonically positioned BPs with very weak consensus contexts and relatively short PPTs. From the intensity of the different lariat bands, and their apparent redundancy upon mutation (Figs. 3, 4) the distant and canonical BPs are used with comparable efficiencies. Use of multiple BPs in association with a single 3′ ss was originally observed in SV40 T-antigen splicing, where 5 BPs between −18 and −32 were used (Noble et al. 1987). More recently, Gao et al. (2008) found that in 36 out of 52 introns analyzed, the introns had more than one BP with a maximum separation of 28 nt between BPs. These examples of closely spaced BPs represent alternative sites for recognition by SF1 and U2 snSNP, adjacent to a common PPT recognized by U2AF65. In contrast, the distant and canonical BPs of HTR4 exons 4 and 5 are separated by much larger distances of 237 and 174 nt, respectively. In these cases the widely spaced BPs are each associated with their own adjacent U2AF65 binding PPTs. While the dBPs will be recognized as AG-independent splice site units (Reed 1989), recognition of the canonically positioned BPs are also likely to involve U2AF35 interaction with the nearby 3′ ss AG (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). In the case of exon 5, both the dBP at −213 and the major canonical BP at −26 (but not at −39) are clearly conserved across several species, suggesting that not only the large AGEZ/dBP, but also the dual distant/canonical BP, arrangement are both functionally important. One possible explanation of this apparent redundancy is that without the dual canonical and dBPs, HTR4 exon 5 would be skipped, under some circumstances. The dual BP arrangement may, in this case, help to enforce constitutive splicing using the same 3′ ss, even under conditions where either the distant or canonical BPs are not efficiently recognized. The observation that for exons 3, 4, and 5 a common consequence of BP mutation was activation of cryptic 3′ ss within the exon may provide a further clue to the need for the dBP arrangements. The presence of 3′ ss-like sequences within the exon may be unavoidable due to protein coding or other constraints. This might make an AG-independent recognition of strong dBP and PPTs necessary to ensure correct splicing.
BP mapping and prediction
With the exception of exon g, our two methods for identifying branch points were largely in agreement or they complemented each other. The key difference between the approaches is that in vitro reverse transcription directly identifies the BPs that are being used, whereas mutagenesis of the exon-trapping vector relies on prior experimental or computational identification of potential BPs. The drawbacks of the mutagenesis approach are twofold. Formally, it can only suggest that an element has a positive effect upon splicing, but does not prove that it is a BP. Second, mutagenesis can be without effect, either due to activation of otherwise silent cryptic BPs or because multiple BPs are normally used. HTR4 exons 4 and 5 (Figs. 3, 4) were particularly resilient to multiple mutations of potential cryptic BPs. The general pyrimidine richness of the extended AGEZs might facilitate such activation of cryptic BPs. Unfortunately, direct BP mapping by reverse transcriptase can also be problematic. In preliminary experiments we found that the HTR4 exons did not splice efficiently in vitro. Therefore, we substituted the BP-PPT and 3′ ss in an efficiently spliced construct with the test AGEZs from each of the HTR4 exons. The contradictory results for the exon g BPs in our two assays could be related to this design. Strong ESEs can favor the use of suboptimal PPTs or BPs (Buvoli et al. 1997). It is plausible that the primer extension mapped –31 BP of exon g was activated by the proximity of ESEs in TPM1 exon 3 that were stronger than those in exon g itself. By contrast, in the normal context of HTR4 exon g, only the –86 branch point—with its stronger consensus and adjacent U-rich PPT—is active.
Another method that has been used with some success (Vogel et al. 1997; Gao et al. 2008) has the advantage of mapping BPs precisely from cellular RNA by reverse transcription across the BP 2′–5′ bond, followed by PCR and sequencing. This method relies on some assumptions about the general location of branch points but it does not require identification of precise candidates. Gao et al. (2008) successfully mapped 101 BPs in 52 introns, but their approach would not have allowed discovery of very distant BPs, since the forward PCR-primers were 100 nt upstream of the 3′ ss and the most distant branch point was mapped 65 nt upstream of its 3′ss. It would, however, be simple to use the AGEZ as a criterion in the primer design of this method. However, a key unanswered question for this approach is the degree to which the efficiency of reverse transcriptase extension across 2′–5′ bonds biases the ability to detect different BP sequences.
Although the optimal metazoan BP sequences resemble the invariant yeast UACUAAC consensus (Zhuang et al. 1989), BP sequences can vary widely from this optimum and prediction of metazoan BPs remains a challenging task. Moreover, while splice site consensus can be compiled by aligning huge numbers of EST/mRNA and genomic sequences, the number of mapped BPs is relatively modest. BP scores, based only on a position weight matrix for the BP consensus (Gooding et al. 2006), showed variable success in predicting the mapped HTR4 BPs. For exons 5 and g the mapped −213 and −86 BPs were the highest scoring predicted BPs, while the mapped dBP −151 nt upstream of exon 3 was ranked second in its AGEZ. However, none of the exon 4 mapped BPs at −273, −267, −33, and −27 even scored above threshold. Surprisingly, a better consensus BP (−270: ataAtc) lies between the −273 and −267 doublet (tatAta and atcAct); however, it is not used. One feature common to all of the mapped dBPs is an extended PPT just downstream. This reflects the known interdependence of BP, PPT, and ESE sequences (Buvoli et al. 1997). Ideally, improved BP prediction programs will include terms for all of these elements combined with their relative locations (Kol et al. 2005), as well as considering AGEZs.
HTR4 alternative splicing and consequences
To date, 10 isoforms of the human HTR4 have been described, most of which are caused by alternative terminal exons. In addition, two internal cassette exons are annotated (Bender et al. 2000; Hiroi et al. 2001). Most HTR4 variants affect the most C-terminal part of the protein, but no functional differences have been determined, although some phosphorylation sites are affected (Blondel et al. 1998). Here, we detected not only skipping of exon 4 but we also discovered two novel exons flanking exon 4. There has been possible evidence for exon 4 skipping in cultured pituitary gland cells (Papageorgiou and Denef 2007). Upon estradiol treatment, HTR4 expression was upregulated, and RT-PCR analysis indicated two bands, one of which appeared to correspond in size to exon 4 skipping. The boundaries of exons 3–5 all fall into the loop regions between trans-membrane regions (Fig. 6D). Skipping of any of these exons would lead to loss of one or more transmembrane domains, and would therefore be highly deleterious to the resulting protein. In light of this, it is perhaps unsurprising that skipping of any of these exons, or inclusion of the novel exons 3a and 4a, would introduce a PTC that would be expected to lead to NMD. Although it has been argued that the majority of splicing events that lead to PTC introduction are unregulated (Pan et al. 2006), there are clearly many examples where such splicing events are regulatory (McGlincy and Smith 2008). It is therefore possible that HTR4 levels might be regulated via these NMD-linked splicing events in a rapid manner to modulate signal transduction. Notably, we observed lower levels of these nonproductive splicing events in brain in comparison with intestine, potentially indicating tissue-specific regulation (Fig. 6B).
SNPs and AGEZs
Several studies have analyzed HTR4 for SNPs associated with mental pathologies and even looked into intronic changes affecting potential splice regulatory elements (Ohtsuki et al. 2002; Suzuki et al. 2003; Li et al. 2006). Suzuki et al. (2003) speculated that a T/C SNP 36-nt upstream of exon 5 might affect a potential BP, thereby impairing splicing of the exon, and consequently causing a predisposition for schizophrenia in these patients. This SNP is located 3-nt downstream from the mapped BP at –39 (tttAcct or tttAccc). On the other hand, our data show that even elimination of this BP adenosine has no effect on the splicing of exon 5. Of more general relevance, the previous exercises to identify new HTR4 SNPs were limited to resequencing the extreme 3′ end of the intron, with a maximum of a 61-nt upstream sequence in the case of exon 5. In the case of exon 3, where both assays agreed on the nonredundant use of a BP at −151 (Fig. 2), SNP sequencing was only carried out from position −35.
Our results therefore emphasize the need for exon resequencing SNP projects to take into account the more extended intronic regions, as indicated by extended AGEZs, which are crucial for correct splicing of some exons. We can readily envisage at least three consequences of SNPs within extended AGEZs. First, sequence variants of a distant BP may affect the degree of exon inclusion, as they do for canonically positioned BPs (Kralovicova et al. 2004, 2006). Second, splicing regulatory elements may be affected, with possible tissue-specific consequences on alternative splicing. Third, and most catastrophically, introduction of an AG dinucleotide at almost any location in this region would create an efficiently used cryptic 3′ ss. In extreme cases, the additional sequences introduced into the mRNA can lead to efficient degradation such that the alteration in structure is not readily apparent in cellular mRNA (Gooding et al. 2006). Our data on the novel exons 3a and 4a also have implications for SNP analysis, as any further strengthening of these exons could be deleterious for the overall HTR4 expression. Increase of inclusion of either of these exons would either lead to truncated proteins or, in the worst case, to degradation of the RNA by NMD. Indeed, it seems plausible that a number of pseudoexons that have been associated with clinical conditions might, like exons HTR4 3a and 4a, be genuine alternative exons that are usually used at a low level.
MATERIALS AND METHODS
Data output files and explanations
Based on the analysis by Gooding et al. (2006) we obtained the data output files for the four exons of the HTR4 gene from http://bioinformatics.org.au/dBP/. We manually annotated three additional BP predictions for exon 4 (positions –300, –270, and –193) from analysis on http://www.ebi.ac.uk/asd-srv/wb.cgi?method=2, which uses a similar BP scoring system. The first part of this paragraph gives explanations for abbreviations, and the following paragraphs show output files for each exon.
>IDB #: altExtron identifier for the gene
GB_MAP: mapping to the GenBank entry
PROD: Gene name
AGEZ: AG “exclusion zone” (number of nucleotides upstream of the end of the intron to the first AG other than the acceptor splice site itself)
ROI: “Region of interest” (from three AGs upstream of the splice site to two AGs downstream)
AG: Relative positions of the AG nucleotides in the ROI, including the splice site itself at −2
PPT: Positions of pyrimidine-rich tracts that we take as putative PPTs
U2BP: Putative U2 branch point sequences. The number in the [ ] brackets is the bit score generated for this site based on a weight matrix analysis
SEQ1: Intronic sequence part of the ROI with the putative PPTs in upper case
SEQ2: Exonic sequence part of the ROI
Full details may be found under http://bioinformatics.org.au/dBP/agez_format.txt.
HTR4 exon 3
>IDB1090103.1894
GB_MAP: IDB1090103 = AJ243213.1 (9122 . . 94502)
PROD: 5-HT4 receptor
AGEZ: 149
ROI: 1711 . . 1991 −> −184 . . 96
AG: −184, −181, −151, −2, 87, 97,
PPT: −142 . .−72, −69 . .−5, 16 . . 40,
U2BP: −151 [5.23], −70 [6.32], −42 [3.87], −18 [4.64], −8 [4.09], −4 [5.07], 45 [3.13], 68 [3.78], 87 [4.85],
SEQ1: agaaggggaaattgaatctcacggaatacactaagcatgtgaCTTTTTCTTTCCTCTTTCCCCCTCCCTTCTTTTCTTCTTTCTTTGTTTCCCTTTTCCCTGTTTTTTCTCTTgaTCTCTTCTCCCTCCCCTTTTTCCCTTCATCCCTCCCCCTGCCTCCTCTCTTACACTTTTTCACTCacag
SEQ2: gaaaataaaaacaaaTTATTTCATTGTATCTCTTGCTTTTgcggatctgctggtttcggtgctggtgatgccctttggtgccattgagctggttcaag
HTR4 exon 4
>IDB1090103.27415
GB_MAP: IDB1090103 = AJ243213.1 (9122 . . 94502)
PROD: 5-HT4 receptor
AGEZ: 291
ROI: 27108 . . 27450 −> −308 . . 34
AG: −308, −298, −293, −2, 21, 35,
PPT: −291 . .−272, −269 . .−143, −140 . .−130, −127 . .−3, 2 . .20,
U2BP: −300 [4.67], −270 [3.66], −193 [3.61], −174 [3.24], −166 [4.18], −160 [3.11], −141 [3.98], −72 [4.62],
SEQ1: agaaactcacaggccagTATATCATGTTGTATTATATaaTCACTTTTCCTTTCCTCTGTTCTTTTCTCTCTCTTCCTTCTTTTTCCCTTCCTCTGCCACTTGCTCTCCTCTTTTTACCCTGTATTCTTTCTTTTATTTCCTTATTTTTATTCTCTGCTTCCCCTCCgaCTCTTCTCCTCaaTGTTTGCTGTTCTCTATCCCTTGCTCCATCCCCTCTTCCTCCCCTTTTCTCTCTCATATATTCTCTTTTCTCTCCCTCCCATTCCTCCTTGCCCATGCCTATGCTCTGCCCTCTATTCTCTCTCCag
SEQ2: gTATTACGCCATCTGCTGCCagcctttggtctatag
NB BPs mapped at −267, −273, −33, and −27 (Fig. 3) are not scored above threshold.
HTR4 exon 5
>IDB1090103.40737
GB_MAP: IDB1090103 = AJ243213 . 1 (9122. .94502)
PROD: 5-HT4 receptor
AGEZ: 221
ROI: 40496. .40745 −> −242. .7
AG: −242, −223, −223, −2, 3, 8,
PPT: −208. .−128, −125. .−10,
U2BP: −213[6.84], −183 [3.89], −175 [3.24], −134 [3.11], −89 [4.99], −57 [4.09], −39 [3.61], −26 [4.65], −8 [3.46], −2 [3.23], 1 [4.01],
SEQ1: aggcttataagaaaaccaaagctttgcctgataaaCTTTCTTTTCTTTCCTTTCCCCTTTACTCTTTTATTCTCCCATTTTCTTTCCTTGTTCTCTTCCCCTTCTTTTTATCTTCCaaTCCTCCACTTTCTTCCTTTTCCCTTTTTCTTTTCTCACACTCTTCCCCATTTTTCCCACTTCTTTTTCACTTTTTCTTTCCTTTTTACCTCCTTTCCTCATTTCCTCTCTCCCTCCaactctcag
SEQ2: atagaaaag
HTR4 exon g
>IDB1090103.84829
GB_MAP: IDB1090103 = AJ243213 . 1 (9122. .94502)
PROD: 5-HT4 receptor
AGEZ: 101
ROI: 84703. .84864 −> −118. .43
AG: −118, −109, −103, −2, 20, 44,
PPT: −116. .−110, −100. .−88, −85. .−54, −50. .−43, −40. .−26, −23. .−10, 4. .19, 22. .38,
U2BP: −103[3.21], −86[6.5], −51 [3.64], −31 [4.78],
SEQ1: agCTTATTTaggctgagaTCATGTGTCTACTaaTCTCTATGTTTTTTTTTTCTTTTTTCTTTCCCaaaTTCTGCTTggCTCTCCCTCATCTGTggCTGTTTTCTGTTTCaactaaaag
SEQ2: tggCTGTTCCCCTGTCTCagCTTCCTCCTGCTCTTCTgcaatag
Constructs and mutagenesis
HTR4 exons and surrounding introns were PCR-amplified from genomic DNA isolated from HeLa cells using Pfu polymerase, A-overhangs were introduced with Taq polymerase and construct ligated into pGEM-Teasy (Promega), which served as a shuttle vector.
PCR primers for these four exons were as follows:
Exon 3 forward: 5′-GCAGGGAAACTCAGATTAGGG-3′;
Exon 3 reverse: 5′-GTTATAGGTCTTACTAATCTGTGC-3′;
Exon 4 forward: 5′-GGAGCTCCTCCAGATATTGGG-3′;
Exon 4 reverse: 5′-CAAGACCTAGATGGGACAAGG-3′;
Exon 5 forward: 5′-CCCTGGGATATGATCATGTTCC-3′;
Exon 5 reverse: 5′-CAAACTCTATGCAGGTTTCAGG-3′;
Exon g forward: 5′-GTTGTGCAATAATTATGCTCCCC-3′; and
Exon g reverse: 5′-GTGCCAACAATTGTGCACCC-3′.
Branch point mutations were carried out using QuikChange Site-Directed Mutagenesis (Stratagene) in the Teasy clones. Wild-type and mutant clones were subcloned using standard cloning techniques (Sambrook and Russell 2001) from the shuttle vector as a NotI fragment into pCAGGsEGFP under a chicken β-actin promoter (cBA) (Okabe et al. 1997; Wollerton et al. 2004).
AGEZs were cloned with the following primers into PY7 vector (Scadden and Smith 1995) via XhoI and PvuII (restriction sites are underlined):
Exon 3 forward: 5′-GTGCTCGAGGAATTGAATCTCACGGAATAC-3′;
Exon 3 reverse: 5′-GTGACAGCTGTGAGTGAAAAAGTGTAAGAGAGG-3′;
Exon 4 forward: 5′-GTGCTCGAGCTGCAGGCAATGATTCTCAG-3′;
Exon 4 reverse: 5′-GTGACAGCTGGAGAGAGAATAGAGGGCAG-3′;
Exon 5 forward: 5′-GTGCTCGAGCATCAAGGTTCTTTGTATTGTAGGC-3′;
Exon 5 reverse: 5′-GTGACAGCTGAGAGTTGGAGGGAGAGAGG-3′;
Exon g forward: 5′-GTGCTCGAGGCTTATTTAGGCTGAGATCATGTG-3′; and
Exon g reverse: 5′-CTTTTAGTTGAAACAGAAAACAGCC-3′ (used for blunt end cloning to preserve suboptimal 3′ ss).
Cell culture, transfections, and analysis of cellular RNA
HeLa cells were grown in Dulbecco's modified Eagles medium containing 10% fetal calf serum. Transient transfection was carried out using Lipofectamine 2000 (Invitrogen), total RNA was isolated using TRI reagent (Sigma) and RT-PCR was carried out as previously described (Wollerton et al. 2001). Primers for pCAGGsEGFP-based constructs were:
RT primer 3′CGRT: 5′-TAGTTGTACTCCAGCTT-3′;
Forward 5′CGTM: 5′-GGCAAAGAATTCGCCACCA-3′;
Reverse 3′CGTM: 5′-GGGTGTCGCCCTCGAACTT-3′; or
Reverse EGFP-N1-R: 5′-CGTCGCCGTCCAGCTCGACCAG-3′.
Conditions for the PCR were 30 cycles of 94°C for 30 sec, an annealing temperature of 58°C for 30 sec followed by an extension at 72°C for 1 min using MgCl2 concentrations of 1.5 mM. Reactions were analyzed by a PhosphorImager after denaturing polyacrylamide gel electrophoresis and quantified with ImagequantTL software (GE).
In vitro splicing and primer extensions
Py7 constructs were linearized with XbaI prior to in vitro transcription with SP6 polymerase. In vitro transcription, splicing, and debranching were carried out as previously described (Ruskin and Green 1985; Gooding et al. 1998; Southby et al. 1999; Wollerton et al. 2001). A total of 100 fmol of 32P-5′-labeled primer were hybridized to a 100 fmol of spliced, debranched, or control RNA template at the most 3′end of the intron, and annealing was allowed for 30 min at 42°C. Lariat branch points were mapped by extending with 10 units of AMV reverse transcriptase (Promega) for 45 min at 42°C and by comparing the resulting terminations in the RT to the ones of the debranched and control RNAs. Primer extension reactions were loaded on 8% denaturing polyacrylamide gels side by side with sequencing reactions with the same primers and appropriate plasmid templates using T7 DNA polymerase.
Cloning a novel exon from rat tissue
RNAs from rat heart, large and small intestine, brain, and stomach were isolated according to Ellis et al. (2004). RNA from rat embryo was purchased from Ambion. RT was performed using oligo-dT and Superscript II (Invitrogen). PCR was carried out using [32P]-end label primers and was analyzed by a PhosphorImager after gel electrophoresis on denaturing polyacrylamide gels, as well as subcloning PCR products, and subsequent sequencing and aligning to the rat genome.
Exon 3 forward: 5′-CACGTTCTTCGCAATGGTTA-3′;
Exon 3 reverse: 5′-CCTTGCATTATGGGGAGAAA-3′;
Exon 4 forward: 5′-TAGGGCTTGTTGACCATGAA-3′; and
Exon 4 reverse: 5′-TCACCTCTGCTGCATTTCC-3′.
ACKNOWLEDGMENTS
We thank Miriam Llorian for critical comments on the manuscript. This work was supported by the Wellcome Trust (program grant 077877) and by EC grant EURASNET-LSHG-CT-2005-518238.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2013110.
REFERENCES
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Bender E, Pindon A, van Oers I, Zhang YB, Gommeren W, Verhasselt P, Jurzak M, Leysen J, Luyten W. Structure of the human serotonin 5-HT4 receptor gene and cloning of a novel 5-HT4 splice variant. J Neurochem. 2000;74:478–489. doi: 10.1046/j.1471-4159.2000.740478.x. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Blondel O, Gastineau M, Dahmoune Y, Langlois M, Fischmeister R. Cloning, expression, and pharmacology of four human 5-hydroxytryptamine 4 receptor isoforms produced by alternative splicing in the carboxyl terminus. J Neurochem. 1998;70:2252–2261. doi: 10.1046/j.1471-4159.1998.70062252.x. [DOI] [PubMed] [Google Scholar]
- Buvoli M, Mayer SA, Patton JG. Functional crosstalk between exon enhancers, polypyrimidine tracts, and branch point sequences. EMBO J. 1997;16:7174–7183. doi: 10.1093/emboj/16.23.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Anderson K, Moore MJ. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc Natl Acad Sci. 2000;97:593–598. doi: 10.1073/pnas.97.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compan V, Zhou M, Grailhe R, Gazzara RA, Martin R, Gingrich J, Dumuis A, Brunner D, Bockaert J, Hen R. Attenuated response to stress and novelty and hypersensitivity to seizures in 5-HT4 receptor knock-out mice. J Neurosci. 2004;24:412–419. doi: 10.1523/JNEUROSCI.2806-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis PD, Smith CW, Kemp P. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by α-tropomyosin regulatory elements in transgenic mice. J Biol Chem. 2004;279:36660–36669. doi: 10.1074/jbc.M405380200. [DOI] [PubMed] [Google Scholar]
- Fontana DJ, Daniels SE, Wong EH, Clark RD, Eglen RM. The effects of novel, selective 5-hydroxytryptamine (5-HT)4 receptor ligands in rat spatial navigation. Neuropharmacology. 1997;36:689–696. doi: 10.1016/s0028-3908(97)00055-5. [DOI] [PubMed] [Google Scholar]
- Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding C, Roberts GC, Moreau G, Nadal-Ginard B, Smith CW. Smooth muscle-specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding C, Roberts GC, Smith CW. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Gooding C, Clark F, Wollerton MC, Grellscheid SN, Groom H, Smith CW. A class of human exons with predicted distant branch points revealed by analysis of AG dinucleotide exclusion zones. Genome Biol. 2006;7:R1. doi: 10.1186/gb-2006-7-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman DM, Roscigno RF, Mulligan GJ, Finn LA, Weber KS. Identification of two distinct intron elements involved in alternative splicing of β-tropomyosin pre-mRNA. Genes and Dev. 1990;4:98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- Hiroi T, Hayashi-Kobayashi N, Nagumo S, Ino M, Okawa Y, Aoba A, Matsui H. Identification and characterization of the human serotonin-4 receptor gene promoter. Biochem Biophys Res Commun. 2001;289:337–344. doi: 10.1006/bbrc.2001.5979. [DOI] [PubMed] [Google Scholar]
- Kol G, Lev-Maor G, Ast G. Human–mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- Kralovicova J, Houngninou-Molango S, Kramer A, Vorechovsky I. Branch site haplotypes that control alternative splicing. Hum Mol Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- Kralovicova J, Lei H, Vorechovsky I. Phenotypic consequences of branch point substitutions. Hum Mutat. 2006;27:803–813. doi: 10.1002/humu.20362. [DOI] [PubMed] [Google Scholar]
- Letty S, Child R, Dumuis A, Pantaloni A, Bockaert J, Rondouin G. 5-HT4 receptors improve social olfactory memory in the rat. Neuropharmacology. 1997;36:681–687. doi: 10.1016/s0028-3908(96)00169-4. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Y, Zhou R, Wang B, Zhang H, Yang L, Faraone SV. Association of attention-deficit/hyperactivity disorder with serotonin 4 receptor gene polymorphisms in Han Chinese subjects. Neurosci Lett. 2006;401:6–9. doi: 10.1016/j.neulet.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Liu ZR, Laggerbauer B, Luhrmann R, Smith CW. Crosslinking of the U5 snRNP-specific 116-kDa protein to RNA hairpins that block step 2 of splicing. RNA. 1997;3:1207–1219. [PMC free article] [PubMed] [Google Scholar]
- Lucas G, Rymar VV, Du J, Mnie-Filali O, Bisgaard C, Manta S, Lambas-Senas L, Wiborg O, Haddjeri N, Pineyro G, et al. Serotonin4 (5-HT4) receptor agonists are putative antidepressants with a rapid onset of action. Neuron. 2007;55:712–725. doi: 10.1016/j.neuron.2007.07.041. [DOI] [PubMed] [Google Scholar]
- Marchetti E, Chaillan FA, Dumuis A, Bockaert J, Soumireu-Mourat B, Roman FS. Modulation of memory processes and cellular excitability in the dentate gyrus of freely moving rats by a 5-HT4 receptors partial agonist, and an antagonist. Neuropharmacology. 2004;47:1021–1035. doi: 10.1016/j.neuropharm.2004.06.033. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Medhurst AD, Lezoualc'h F, Fischmeister R, Middlemiss DN, Sanger GJ. Quantitative mRNA analysis of five C-terminal splice variants of the human 5-HT4 receptor in the central nervous system by TaqMan real-time RT-PCR. Brain Res Mol Brain Res. 2001;90:125–134. doi: 10.1016/s0169-328x(01)00095-x. [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Noble JC, Pan ZQ, Prives C, Manley JL. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987;50:227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Ishiguro H, Detera-Wadleigh SD, Toyota T, Shimizu H, Yamada K, Yoshitsugu K, Hattori E, Yoshikawa T, Arinami T. Association between serotonin 4 receptor gene polymorphisms and bipolar disorder in Japanese case–control samples and the NIMH Genetics Initiative Bipolar Pedigrees. Mol Psychiatry. 2002;7:954–961. doi: 10.1038/sj.mp.4001133. [DOI] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes and Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou A, Denef C. Estradiol induces expression of 5-hydroxytryptamine (5-HT) 4, 5-HT5, and 5-HT6 receptor messenger ribonucleic acid in rat anterior pituitary cell aggregates and allows prolactin release via the 5-HT4 receptor. Endocrinology. 2007;148:1384–1395. doi: 10.1210/en.2006-1198. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes and Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Green MR. An RNA processing activity that debranches RNA lariats. Science. 1985;229:135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning. A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Scadden ADJ, Smith CW. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Smith CW, Nadal-Ginard B. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: Implications for constitutive splicing. Cell. 1989;56:749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith CW, Porro EB, Patton JG, Nadal-Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southby J, Gooding C, Smith CW. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Iwata N, Kitamura Y, Kitajima T, Yamanouchi Y, Ikeda M, Nishiyama T, Kamatani N, Ozaki N. Association of a haplotype in the serotonin 5-HT4 receptor gene (HTR4) with Japanese schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003;121:7–13. doi: 10.1002/ajmg.b.20060. [DOI] [PubMed] [Google Scholar]
- Vogel J, Hess WR, Borner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes and Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang YA, Goldstein AM, Weiner AM. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]