Abstract
During follow-up in antimalarial drug trials, treated subjects can be newly infected. PCR correction is used to distinguish this re-infection from drug failure (recrudescence) and to adjust final drug efficacy estimates. The epidemiological, biological and technical limitations of PCR correction and how this may lead to misclassification in drug trial outcomes are underappreciated. This article considers these limitations and proposes a framework for reporting, interpreting, and improving PCR correction of antimalarial trials.
Clinical trials of antimalarial drugs: the role of PCR correction
Because of the high prevalence of chloroquine (CQ)- and sulfadoxine-pyrimethamine (SP)-resistant Plasmodium falciparum infections, artemisinin-containing combination therapies (ACT) are now being increasingly deployed. However, resistance to artemisinins and ACTs is emerging in Southeast Asia [1, 2]. In order to develop new ACTs, Phase III studies comparing new regimens with existing regimens are needed. However, particular difficulties arise with study design and the end points used for these trials (see Box 1) [3-5]. A conventional ‘superiority’ trial design does not apply when the novel regimen of interest is being tested against an existing regimen of high efficacy, like an ACT; thus non-inferiority trials are more appropriate. However, non-inferiority trials tend to be large, so the choice of endpoint is critical (See Box 1). Typically, antimalarial trials use ‘PCR-corrected’ parasitological failure as an endpoint [5, 6], but the estimate of drug efficacy thus obtained is vulnerable to misclassification errors [4].
How well does ‘PCR correction’ correctly correct? In this paper, we will consider the role of PCR correction of P. falciparum antimalarial trials in the ACT era, and present its epidemiological, biological and technical limitations. We will also discuss the consequences of misclassification by PCR correction and propose a framework for reporting, interpreting, and improving PCR correction of antimalarial trials.
Multiplicity of Infection (MOI)
Individuals infected with P. falciparum typically harbour a mixture of genetically distinct variants (also called genotypes, clones or strains). These variants represent genetically distinct P. falciparum parasite populations resulting from multiple infected bites or bites from mosquitoes bearing multiple variants. Using nested PCR amplification with gel electrophoresis of the polymorphic parasite genes encoding merozoite surface protein 1 (msp-1), merozoite surface protein 2 (msp-2) and glutamate-rich protein (glurp), the first recognition of the genetic complexity of falciparum infections was made by Snounou and colleagues in the 1990s [7, 8]. Although these have become the standard methods for studying parasite diversity and multiplicity, other genotyping techniques have also been described [9-15]. These include tri-nucleotide repeats (microsatellite) genotyping, capillary electrophoresis, Southern blotting, single-strand conformational polymorphisms (SSCP), PCR-RFLP-SSCP and heteroduplex tracking assays (HTAs). Each of these methods has advantages and disadvantages, but it is clear that none of them is capable of detecting all parasite variants within a sample [10]. The range of MOI has been reported from a single variant to >10 variants in an isolate and can vary over time in an individual [16-19].
PCR correction – an oxymoron?
Antimalarial efficacy trials assess drug cure rates by following patients after treatment for sufficient time to ‘capture’ all or most treatment failures [5]. This period of follow-up can range from 28 days (recommended as a minimum duration by WHO), up to 63 days, depending primarily on the elimination kinetics of the drug being studied [5, 20]. Recurrent parasitemia observed during this follow-up period is of three broad types: (i) parasites that were present in the blood before or during treatment;(ii) parasites inoculated prior to initiation of treatment which emerge from the liver after completion of treatment; (iii) parasites derived from a post-treatment inoculation. Although it would be useful for an antimalarial treatment to prevent any type of recurrent infection (a post prophylactic effect) (see Box 1), prevention of recrudescent infections is generally considered the hallmark of antimalarial efficacy. But how do you distinguish recrudescence from recent liver emergence in the face of highly complex infections and frequent reinfections? This is the role of PCR correction (Box 2).
In order to standardize methodology for PCR correction, the WHO convened a meeting of experts in May 2007 in Amsterdam, The Netherlands, where a number of definitions were agreed and Guidelines were produced for genotyping to define reinfections and recrudescences [21] (http://apps.who.int/malaria/docs/drugresistance/MalariaGenotyping.pdf). In theory, the procedure is straightforward. Comparisons of the pre- and post-treatment msp1, msp2, and glurp electrophoresis patterns are used to distinguish recrudescences from reinfections (Box 2). However, in practice, the actual implementation of PCR correction is affected by five potential sources of error: (i) asynchronous sequestration of distinct genotypes, removing them from the peripheral blood; (ii) lack of sufficient sensitivity of methods to detect minority variants (<10–20% of the parasite population); (iii) over-amplification of abundant DNA sequences, particularly with nested PCR, drowning out competing sequences; (iv) direct effects of antimalarial treatment on the abundance and composition of the within-host parasite population; and (v) the possibility of re-infection by a new parasite with the same genotype (at the locus or loci examined) as pre-treatment parasites.
The first three of these sources of error synergise, making it is almost impossible to sample exhaustively the genotypes present in any P. falciparum infection, particularly when parasite isolates are typically collected in clinical trials from a single finger-prick blood sample of less than 100 μl. The major effect of this will be to miss genotypes in the pre-treatment sample and then incorrectly classify a recurrence as a re-infection when this band appears post-treatment. This will lead to an overestimation of ‘new infections’. Although no current method captures the complete MOI within an individual, heteroduplex tracking assays (HTAs) have been shown to have sufficient sensitivity to identify minority variants in mixed malaria infections [10, 12, 13, 22-25]. This technique has been validated and used extensively to study other polyclonal infections, such as HIV [26-28]. Recently, an evaluation of six recurrent parasitaemias from trials in Southeast Asia provided the first direct evidence of this type of misclassification. These samples were classified by conventional genotyping as ‘new infections’. However, using a HTAs for msp-1 and msp-2, the investigators classified 5 of 6 samples as recrudesences [10]. The shared alleles were confirmed in one patient by cloning and sequencing. These results are consistent with the clinical history: the patients were either kept in hospital during follow-up (and thus protected from additional mosquito bites), or they stayed in a region where there is extremely low transmission and a small chance of re-infection. This type of misclassification may also explain the results of Mugittu et al. in a trial in Uganda comparing SP to Artesunate-SP. Using standard genotyping methods, these investigators found a high rate of early ‘new infections’ (5 of 26 [19.2%] failures at Day 7) [29] despite the fact that the typical pre-patent period for malaria infections is 6-12 days. The new genotypes, detected at Day 7, could have derived from unseen minority variants present before treatment.
Minority variant drug-tolerant parasites, which are present (but undetected) before treatment, can grow up to higher densities after the removal of dominant drug-sensitive genotypes by drugs. In animal models with mixed wild-type and resistant parasites, the drug-resistant parasites are competitively released by drug, growing to a higher density than the mixture does in the absence of drug [30]. There is indirect evidence that this occurs in human malaria infections [16, 31-34]. For example, in six imported falciparum malaria cases in the UK treated with either atovaquone-proguanil or quinine-SP, recrudescences consisting of drug-resistant parasites occurred 21 and 42 days after treatment, respectively [35, 36]. These infections could not have been caused by intervening inoculations. In addition, a recent report described the emergence of drug resistant parasites in pregnant women consistent with competitive release of drug resistance variants after intermittent preventive therapy [32]. Thus, a ‘new’ variant that appears after treatment could simply be an unseen minority variant with increased fitness (from the therapy) that was present before treatment. This is best illustrated by the demonstration that artemether-lumefantrine ‘selects’ particular pfmdr1 genotypes among ‘new infections’ in in vivo studies [37, 38].
The errors described above lead to overestimation of drug efficacy. The final source of error, coincidental re-infection by the same genotype present before treatment, can lead to underestimation of efficacy. The basis for this lies in the fact that, for any given genotype present in a patient before treatment, there is a finite possibility that a patient might be re-infected with that genotype after treatment, even if the patient is initially cured. Both Greenhouse et al. and Kwiek et al. measured the prevalence of individual genotypes among all patients in clinical trials in Malawi and Uganda, as an estimate of the genotype prevalence within the communities in which the studies were performed [12, 39]. Both studies were conducted in areas of high transmission and high genetic diversity, and estimated that ~20–40% of PCR-corrected recrudescences were actually re-infections. This type of error can occur both in areas with high transmission and in areas of low transmission. In the first case, a high inoculation rate increases the likelihood of re-infection with the same variant, even if it is uncommon. In the second case, although MOI will tend to be low, reinfection with a highly prevalent genotype is likely. In all areas, each subject's home or micro-environment is likely to contain mosquitoes with the same variants present before he or she was treated.
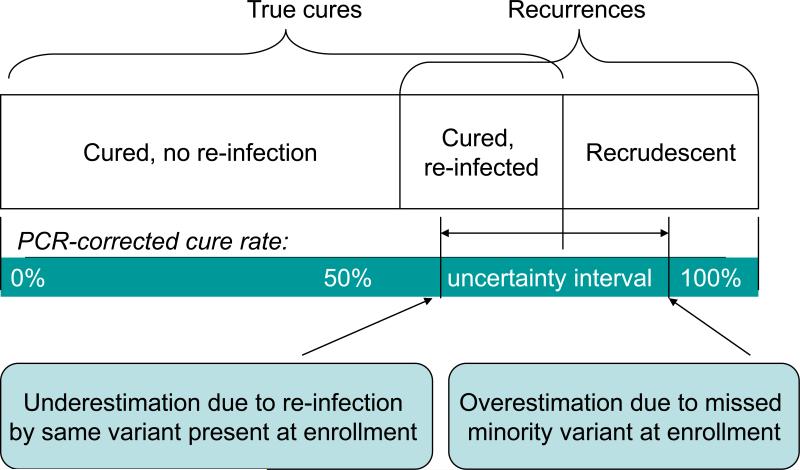
These opposite sources of error, and a theoretical model to describe their influence, are illustrated in Figure 1. The question remains: how important is this misclassification? There is still little direct evidence of the extent of overestimation of efficacy rates. Correctly reclassifying the four Cambodian re-infections as recrudescences in Juliano et al. would change the PCR corrected failure rate for Artesunate–Mefloquine from 18.8% to 21%, similar to the 21.4% unadjusted failure rate [10, 40]. In terms of underestimating efficacy rates, correcting for coincidental reinfection by the same variant present before infection requires more complex statistical methods [12, 41]. Kwiek et al. used a binomial probability algorithm to adjust SP treatment failures in an IPTp trial. They found an adjusted failure rate of 21.5%, instead of the unadjusted rate of 24.8%. Thus, either type of misclassification probably only accounts for a few percentage points of error.
Figure 1. Theoretical model for influence of misclassification on PCR correction.
Misclassification in antimalarial trials will result in erroneous estimates of the clinical efficacy of a drug. Underestimation of efficacy occurs due to the re-infection of the patient with a common shared variant which appears to be a recrudescence. This will shift the efficacy rate to the left on the model. The inability to detect a minor variant at enrolment that later appears as a recrudescence leads to overestimation of the efficacy rate. These patients will be considered re-infections rather than ‘failures’ and the efficacy rate will shift to the right on the model.
Does a 2–3% error mean anything for the results of a trial? The likely answer is ‘yes’. In the case of a Phase III non-inferiority trial, where the delta margin (Box 1) might be set at 5%, this amount of error could significantly impact the interpretation of results. In the case of monitoring drug efficacy by control programs, as the WHO currently recommends changing first-line antimalarials if clinical efficacy drops below 90%, errors in this range could have a profound impact on policy decisions [20]. Furthermore, the relative importance of opposite sources of error will differ between trial sites as transmission intensity, levels of acquired immunity, prevalence of parasite resistance, the choice of antimalarial used and the genetic complexity of the parasite population vary. This jeopardises the validity of multisite comparisons.
Due to these potential sources of error, some investigators have suggested that PCR correction in clinical trials should be abandoned, especially in areas of high transmission [5, 42]. The primary argument behind this is the belief that re-infections and recrudesences are of equal importance in these settings. Is the correct identification of recrudescent parasitemia always important? Yes and no. In a phase III trial, where approval of a drug might depend on small differences in efficacy, the accurate assessment of re-infections and recrudescences is going to be important. For national malaria control programs, the situation is more complex. The presence of any parasitemia after treatment, whatever the cause, will have important public health impacts and affect the ability for continued transmission. However, ignoring PCR correction would discriminate against the development of drugs with poor post-prophylactic effect (Box 1) [5], and certainly have implications for the WHO recommendation that at least 90% efficacy is required to retain a front-line antimalarial drug. For now, however, until more evidence is produced about the relative importance of re-infection and recrudescences for malaria control, PCR correction will continue to be used in clinical trials and drug efficacy monitoring. In the following section, we suggest possible solutions to the problems we have highlighted.
Potential solutions
Should PCR-correction be abandoned? Will it ever be possible to measure true antimalarial efficacy? The answer to both questions is a qualified ‘no’. There are five actions that can be taken to facilitate the appropriate measurement of PCR-dependent clinical endpoints: (i) educate policy makers about the strengths and limitations of PCR correction; (ii) improve study design and statistical methods to compensate for misclassification and add credibility intervals; (iii) develop improved genotyping methods; (iv) develop new efficacy endpoints, such as parasite clearance times [35], that are not affected by super-infection; and (v) conduct trials in non-endemic regions or isolate patients to prevent the chance of re-infection.
The results of antimalarial efficacy studies can change treatment policies and impact hundreds of thousands of people. Currently, policy makers use PCR-corrected data and ignore the raw data on recurrent infections. Better-informed policy decisions could be made using both sources of data, and scientific journals, editors and reviewers should insist that, when PCR-corrected efficacy is estimated, uncorrected estimates of efficacy are always reported with a similar emphasis from the trial, and comparisons of corrected rates between different trials are avoided.
Rather than giving a single ‘PCR-corrected’ cure rate, statistical analyses can be developed to help minimize these errors. Several investigators have published potential methods for statistically correcting for the chance of re-infection due to the same variant, but these do not account for all the types of error described above [12, 41]. One possibility would be to develop tools for investigators capable of providing certainty or credibility intervals for PCR correction. Along the same lines, Bayesian statistics and uncertainty analyses are employed by forensic scientists doing paternity tests and DNA matches [43]. In order to do this, ‘priors’ (a set of data that reflects certain assumptions) would need to be established for different epidemiological settings. Although these explicit assumptions about the degree of misclassification required by uncertainty analyses may make some researchers uncomfortable, this approach is preferable to assuming misclassification is entirely absent.
New genotyping technologies can improve the ability to discriminate variants and measure minority variants. For example, the use of capillary electrophoresis instead of agarose gels improves the ability to distinguish variants and identify newly infecting genotypes [44]; HTAs are also an improvement in that they can distinguish variants based on sequence as well as size diversity and because they can recognize minority variants [10, 12, 13]. New methods such as Massively Parallel Pyrosequencing, which have revolutionized studies on in-host diversity of HIV and other viruses, can be used in malaria [45]. All of these methods have limitations, especially in cost. Given the rapid progress being made in studying the genetics of malaria and in genetic technologies, new field-friendly genotyping methods could be achievable in the near future. If they are, they will have major public health impact.
Equally important are new approaches to the definition of primary efficacy in terms of parasite clearance rates, which can be measured either microscopically [46] or by quantitative PCR [35]. These approaches focus on changes in parasite density on Days 1–3, and, as they only evaluate parasites present at the time of treatment, have the potential to revolutionise the way we think about antimalarial drug resistance.
Concluding remarks
Policy makers and developers of new antimalarial compounds should continue to use PCR correction, as long as its limitations are recognized. We also call for continued development of improved molecular and statistical methods for the analyses of malaria clinical trial results. It is our belief that uncorrected failure estimates should always be presented, and comparisons of PCR-corrected failure estimates between different studies should be strongly discouraged.
Box 1. Clinical Trial Design for Antimalarial Drugs.
Superiority Trial
A superiority trial is designed to detect a difference between two treatment regimens. This approach was previously used to compare new antimalarial treatments against failing drugs such as CQ and SP. In these trials, competing interventions are evaluated for a significant difference in efficacy for treatment that must be deemed clinically relevant.
Equivalence Trial
An equivalence trial is designed to confirm the absence of a meaningful difference between treatments. This approach is less commonly used in Phase III development than non-inferiority trials.
Non-inferiority Trial
A non-inferiority trial is designed to show that a new treatment is no less effective than an existing treatment - it may be more effective or have a similar effect. This trial approach is particularly useful when comparing two highly efficacious drug regimens, such as comparing ACT therapies. In this design a ‘delta margin’ (which determines the efficacy benchmark below which the new drug fails to be ‘non-inferior’), must be set a priori. The measured efficacy of the comparator drug minus the delta margin must be above the benchmark for the drug to be considered ‘non-inferior’ [4].
Primary Endpoints
The main result, set a priori in the study protocol, which is measured at the end of a study to see if a given treatment worked. For antimalarial trials, these could include [3]:
Chemotherapeutic efficacy- How well does it clear parasitemia?
Combination chemotherapeutic and post treatment prophylactic- How well does it cure primary parasitemia and prevent secondary infection?
Clinical risk reduction - How well does treatment reduce post-treatment incidence of malaria and its complications?
PCR Correction
The use of genetic fingerprinting to determine whether P. falciparum parasites recurring in a patient's blood after antimalarial therapy are identical or different than those which occurred prior to therapy [4]. This is used in malaria drug efficacy trials as they typically occur in endemic regions where during the follow-up period (usually 28-63 days) patients are at significant risk of reinfection. Fingerprinting occurs by detection of size differences of polymorphic antigens by PCR and gel electrophoresis or capillary electrophoresis [21]. If a single parasite variant in the recurrent sample is identical to the pre-treatment sample at any one of three genetic loci, the recurrent parasitemia is considered to be a recrudescence (drug failure). If the recurrent parasites differ at all loci from the initial parasitemia, the recurrent parasitemia is considered a reinfection (not a drug failure).
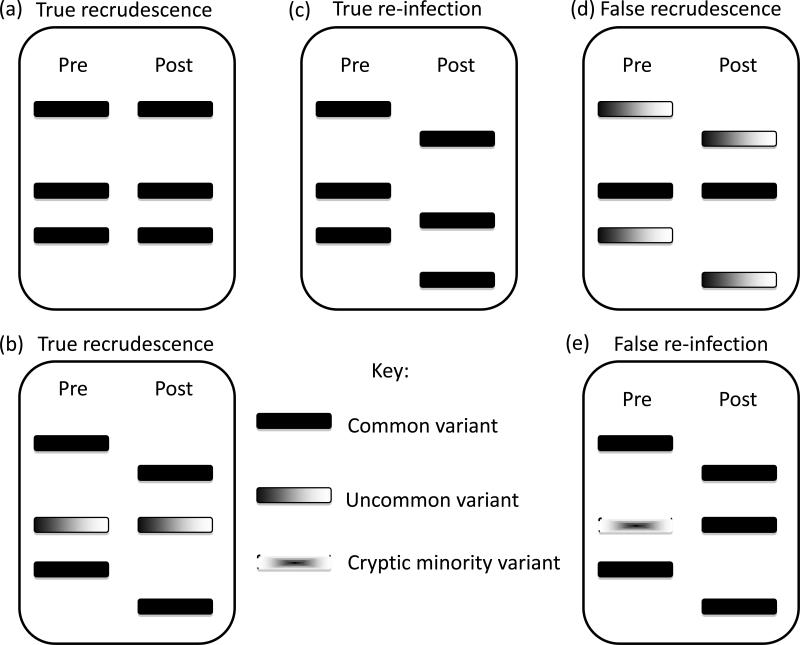
Box 2. Electrophoretic Patterns and Misclassification of Recurrent Parasitemiaa.
Panels (a) and (b) (of Figure I) represent true recrudescent parasitemias. In Panel (a), all alleles are shared between pre- and post-treatment samples. In Panel (b), the recurrent parasitemia shares a single variant with the initial parasitemia. Panel (c) shows a true reinfection, where no alleles are shared between pre- and post-treatment samples. Panel (d) shows a false recrudescence. In this case the post-treatment sample technically shares an allele with the pre-treatment. However, this represents a common variant in the population causing a reinfection. Panel (e) represents a false reinfection, where a cryptic minority variant is missed in the initial parasitemia and emerges to a detectable level in the recurrent parasitemia.
A recrudescence is defined as a parasitaemia occurring subsequent to treatment in which at least one allele at each locus examined is common to both paired samples (Figure I, Panels (a) and (b)).
A reinfection is defined as a parasitaemia occurring subsequent to treatment in which all alleles in the post-treatment sample of a patient are completely different from those in the admission sample, for one or more loci tested (Figure I, Panel (c)).
The indeterminate category of previous schemes has been dropped, with the general principle being that, if a recurrent infection does not differ from the pre-treatment infection at all alleles of a particular marker, it should be considered a recrudescence.
Figure I for Box 2.
Acknowledgements
Financial Support: J.J.J. is currently supported by Award Number KL2RR025746 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the NIH. CJS is supported by the UK Health Protection Agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Definitions from Ref. [21]
References
- 1.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia–Thailand border. Emerg. Infect. Dis. 2008;14:716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrmann S, et al. Revisiting the design of phase III clinical trials of antimalarial drugs for uncomplicated Plasmodium falciparum malaria. PLoS Med. 2008;5:e227. doi: 10.1371/journal.pmed.0050227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutherland CJ. Comparing highly efficacious antimalarial drugs. PLoS Med. 2008;5:e228. doi: 10.1371/journal.pmed.0050228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stepniewska K, White NJ. Some considerations in the design and interpretation of antimalarial drug trials in uncomplicated falciparum malaria. Malar J. 2006;5:127. doi: 10.1186/1475-2875-5-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins WJ, et al. The use of genotyping in antimalarial clinical trials: a systematic review of published studies from 1995-2005. Malar J. 2006;5:122. doi: 10.1186/1475-2875-5-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snounou G, Beck HP. The use of PCR genotyping in the assessment of recrudescence or reinfection after antimalarial drug treatment. Parasitol. Today. 1998;14:462–467. doi: 10.1016/s0169-4758(98)01340-4. [DOI] [PubMed] [Google Scholar]
- 8.Viriyakosol S, et al. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull. WHO. 1995;73:85–95. [PMC free article] [PubMed] [Google Scholar]
- 9.Greenhouse B, et al. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am. J. Trop. Med. Hyg. 2006;75:836–842. [PMC free article] [PubMed] [Google Scholar]
- 10.Juliano JJ, et al. Misclassification of Drug Failures in Plasmodium falciparum Clinical Trials in Southeast Asia. J. Infect. Dis. 2009;200:624–628. doi: 10.1086/600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kain KC, et al. Single-strand conformational polymorphism analysis differentiates Plasmodium falciparum treatment failures from re-infections. Mol. Biochem. Parasitol. 1996;79:167–175. doi: 10.1016/0166-6851(96)02656-4. [DOI] [PubMed] [Google Scholar]
- 12.Kwiek JJ, et al. Estimating true antimalarial efficacy by heteroduplex tracking assay in patients with complex Plasmodium falciparum infections. Antimicrob. Agents Chemother. 2007;51:521–527. doi: 10.1128/AAC.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngrenngarmlert W, et al. Measuring allelic heterogeneity in Plasmodium falciparum by a heteroduplex tracking assay. Am. J. Trop. Med. Hyg. 2005;72:694–701. [PubMed] [Google Scholar]
- 14.Nyachieo A, et al. Plasmodium falciparum genotyping by microsatellites as a method to distinguish between recrudescent and new infections. Am. J. Trop. Med. Hyg. 2005;73:210–213. [PubMed] [Google Scholar]
- 15.Ohrt C, et al. Determination of failure of treatment of Plasmodium falciparum infection by using polymerase chain reaction single-strand conformational polymorphism fingerprinting. Clin. Infect. Dis. 1999;28:847–852. doi: 10.1086/515204. [DOI] [PubMed] [Google Scholar]
- 16.Daubersies P, et al. Rapid turnover of Plasmodium falciparum populations in asymptomatic individuals living in a high transmission area. Am. J.Trop. Med. Hyg. 1996;54:18–26. doi: 10.4269/ajtmh.1996.54.18. [DOI] [PubMed] [Google Scholar]
- 17.Farnert A, et al. Extensive dynamics of Plasmodium falciparum densities, stages and genotyping profiles. Malar J. 2008;7:241. doi: 10.1186/1475-2875-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jafari S, et al. Plasmodium falciparum clonal population dynamics during malaria treatment. J. Infect. Dis. 2004;189:195–203. doi: 10.1086/380910. [DOI] [PubMed] [Google Scholar]
- 19.Jafari-Guemouri S, et al. Plasmodium falciparum genotype population dynamics in asymptomatic children from Senegal. Microbes Infect. 2006;8:1663–1670. doi: 10.1016/j.micinf.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 20.World Health Organization . Guidelines for the treatment of malaria. World Health Organization; Geneva: 2006. 2006. [Google Scholar]
- 21.World Health Organization . Methods and techniques for clinical trials on antimalarial drug efficacy: Genotyping to identify parasite populations. World Health Organization; Geneva: 2007. 2007. [Google Scholar]
- 22.Juliano JJ, et al. Novel dhps and pfcrt polymorphisms in Plasmodium falciparum detected by heteroduplex tracking assay. Am. J. Trop. Med. Hyg. 2009;80:734–736. [PMC free article] [PubMed] [Google Scholar]
- 23.Juliano JJ, et al. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg. Infect. Dis. 2007;13:872–827. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 24.Juliano JJ, et al. Nonradioactive heteroduplex tracking assay for the detection of minority-variant chloroquine-resistant Plasmodium falciparum in Madagascar. Malar. J. 2009;8:47. doi: 10.1186/1475-2875-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juliano JJ, et al. Detection of the dihydrofolate reductase-164L mutation in Plasmodium falciparum infections from Malawi by heteroduplex tracking assay. Am. J. Trop. Med. Hyg. 2008;78:892–894. [PMC free article] [PubMed] [Google Scholar]
- 26.Delwart E, et al. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. Aids. 2002;16:189–195. doi: 10.1097/00002030-200201250-00007. [DOI] [PubMed] [Google Scholar]
- 27.Harrington PR, et al. Cross-sectional characterization of HIV-1 env compartmentalization in cerebrospinal fluid over the full disease course. Aids. 2009;23:907–915. doi: 10.1097/QAD.0b013e3283299129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell G, et al. Identification and recovery of minor HIV-1 variants using the heteroduplex tracking assay and biotinylated probes. Nucleic Acids Res. 2008;36:e146. doi: 10.1093/nar/gkn713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mugittu K, et al. Molecular genotyping in a malaria treatment trial in Uganda - unexpected high rate of new infections within 2 weeks after treatment. Trop. Med. Int. Health. 2007;12:219–223. doi: 10.1111/j.1365-3156.2007.01813.x. [DOI] [PubMed] [Google Scholar]
- 30.Wargo AR, et al. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19914–19919. doi: 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruce MC, et al. Cross-species interactions between malaria parasites in humans. Science. 2000;287:845–848. doi: 10.1126/science.287.5454.845. [DOI] [PubMed] [Google Scholar]
- 32.Harrington WE, et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9027–9032. doi: 10.1073/pnas.0901415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastings IM, et al. Malaria control and the evolution of drug resistance: an intriguing link. Trends Parasitol. 2003;19:70–73. doi: 10.1016/s1471-4922(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 34.Mercereau-Puijalon O, et al. Revisiting host/parasite interactions: molecular analysis of parasites collected during longitudinal and cross-sectional surveys in humans. Parasite Immunol. 1996;18:173–180. doi: 10.1046/j.1365-3024.1996.d01-79.x. [DOI] [PubMed] [Google Scholar]
- 35.Sutherland CJ, et al. Novel pfdhps haplotypes among imported cases of Plasmodium falciparum malaria in the UK. Antimicrob. Agents Chemother. 2009;53:3405–3410. doi: 10.1128/AAC.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland CJ, et al. Mutations in the Plasmodium falciparum cytochrome b gene are associated with delayed parasite recrudescence in malaria patients treated with atovaquone-proguanil. Malar. J. 2008;7:240. doi: 10.1186/1475-2875-7-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphreys GS, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sisowath C, et al. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop. Med. Int. Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 39.Greenhouse B, et al. Impact of transmission intensity on the accuracy of genotyping to distinguish recrudescence from new infection in antimalarial clinical trials. Antimicrob. Agents Chemother. 2007;51:3096–3103. doi: 10.1128/AAC.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers WO, et al. Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar. J. 2009;8:10. doi: 10.1186/1475-2875-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatton ML, Cheng Q. Can estimates of antimalarial efficacy from field studies be improved? Trends Parasitol. 2008;24:68–73. doi: 10.1016/j.pt.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slater M, et al. Distinguishing recrudescences from new infections in antimalarial clinical trials: major impact of interpretation of genotyping results on estimates of drug efficacy. Am. J. Trop. Med. Hyg. 2005;73:256–262. [PubMed] [Google Scholar]
- 43.Curran JM. An Introduction To Bayesian Credible Intervals for Sampling Error in DNA Profiles. Law, Probability and Risk. 2005;4:115–126. [Google Scholar]
- 44.Liljander A, et al. Optimization and validation of multi-coloured capillary electrophoresis for genotyping of Plasmodium falciparum merozoite surface proteins (msp1 and 2). Malar. J. 2009;8:78. doi: 10.1186/1475-2875-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushman FD, et al. Massively parallel pyrosequencing in HIV research. Aids. 2008;22:1411–1415. doi: 10.1097/QAD.0b013e3282fc972e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wootton DG, et al. Open-label comparative clinical study of chlorproguanil-dapsone fixed dose combination (Lapdap) alone or with three different doses of artesunate for uncomplicated Plasmodium falciparum malaria. PLoS ONE. 2008;3:e1779. doi: 10.1371/journal.pone.0001779. [DOI] [PMC free article] [PubMed] [Google Scholar]