Summary
Quorum sensing (QS) bacteria assess population density through secretion and detection of molecules called autoinducers (AIs). We identify and characterize two Vibrio harveyi negative feedback loops that facilitate precise transitions between low-cell-density (LCD) and high-cell-density (HCD) states. The QS central regulator LuxO autorepresses its own transcription and the Qrr small regulatory RNAs (sRNAs) posttranscriptionally repress luxO. Disrupting feedback increases the concentration of AIs required for cells to transit from LCD to HCD QS modes. Thus, the two cooperative negative feedback loops determine the point at which V. harveyi has reached a quorum and control the range of AIs over which the transition occurs. Negative feedback regulation also constrains the range of QS output – by preventing sRNA levels from becoming too high and preventing luxO mRNA levels from reaching zero. We suggest that sRNA-mediated feedback regulation is a network design feature that permits fine-tuning of gene regulation and maintenance of homeostasis.
Keywords: Quorum sensing, Vibrio harveyi, sRNA, negative feedback loop
Introduction
Bacteria can communicate to coordinate gene expression on a population-wide scale, and in so doing, they synchronize group behaviors. This cell-to-cell communication process, referred to as quorum sensing (QS), is a mechanism by which bacteria assess their population density through the production, secretion, and detection of diffusible signal molecules called autoinducers (AIs) (Waters and Bassler, 2005). QS underpins behaviors that are productive only when carried out by large numbers of bacteria acting in unison. QS-controlled behaviors include bioluminescence, biofilm formation, and virulence factor production (Davies et al., 1998; McFall-Ngai and Ruby, 2000; Novick, 2003). How QS bacteria accurately link external AI levels to internal regulation of downstream target gene expression remains an open question and is the main focus of our study.
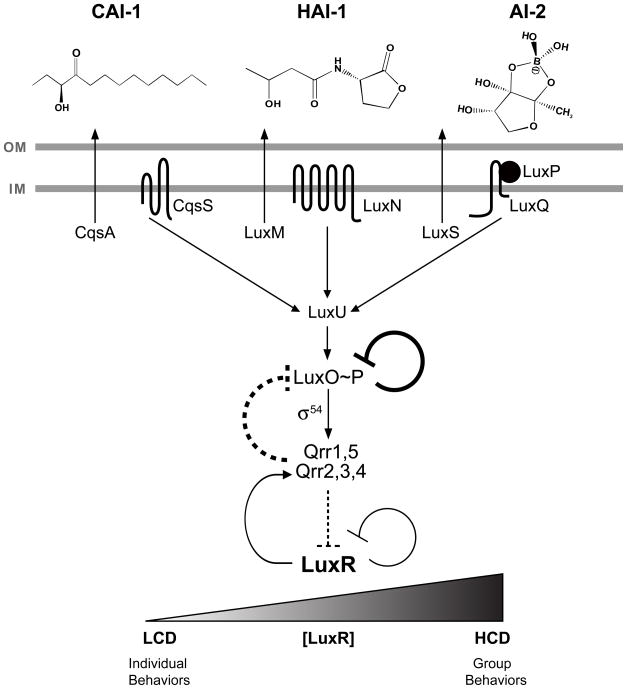
The Gram-negative bioluminescent marine bacterium Vibrio harveyi has emerged as an ideal model organism to study the molecular features underlying QS signal integration and the regulation of downstream QS-dependent genes. V. harveyi produces three AIs (CAI-1, HAI-1, and AI-2), and each is detected by a cognate membrane-bound two-component histidine-kinase receptor (CqsS, LuxN, LuxPQ) (Figure 1). At negligible AI concentrations, i.e., at low cell density (LCD), the three V. harveyi sensors act as kinases and transfer phosphate to the shared histidine-phosphotransfer protein LuxU, which subsequently transfers the phosphate to LuxO (LuxO~P) (Freeman and Bassler, 1999a, b). LuxO is a DNA-binding response regulator that, in conjunction with the alternative sigma factor σ54, activates the expression of genes encoding five highly conserved small regulatory RNAs (sRNAs) called Qrr1–5 (for Quorum Regulatory RNA) (Lenz et al., 2004; Tu and Bassler, 2007). The Qrr sRNAs bind to and destabilize the luxR mRNA transcript. LuxR is the master regulator of QS target genes, and it acts as both a transcriptional activator and a repressor (Miyamoto et al., 1994; Pompeani et al., 2008; Swartzman et al., 1992). Thus, at LCD, expression of the Qrr sRNAs represses the production of LuxR and V. harveyi does not display QS behaviors. When AIs are present, i.e., at high cell density (HCD), they bind to their cognate receptors. These events switch the receptors from kinases to phosphatases, leading to dephosphorylation of LuxO and termination of transcription of the qrr genes. As a result, luxR mRNA is stabilized, LuxR protein accumulates, and LuxR activates and represses numerous genes (Waters and Bassler, 2006). Importantly, one LuxR-activated target is the luxCDABE operon, which is required for bioluminescence (Meighen, 1991). In order for QS to be productive, communication between cells must occur with high fidelity despite the unpredictability of the environments in which bacteria reside as well as variations among genetically identical cells due to gene expression noise (Rosenfeld et al., 2005). We propose that QS circuits are designed to promote accurate signaling and, thus, facilitate precise population-wide transitions between LCD and HCD modes in response to changing AI concentrations.
Figure 1. Model of the V. harveyi quorum-sensing circuit.
V. harveyi produces and detects three autoinducers, CAI-1, HAI-1, and AI-2, to control individual and group behaviors through modulation of the level of the master transcriptional regulator, LuxR. The five Qrr sRNAs negatively regulate LuxR by destabilizing the luxR mRNA, as indicated by the dotted cross bar. There are four negative feedback loops in the QS circuit. 1. LuxR autorepresses its own transcription. 2. LuxR activates expression of qrr2, qrr3, and qrr4. 3. LuxO autorepresses its own transcription. 4. Qrr sRNAs repress luxO translation. The latter two loops are highlighted in bold and are the focus of the present work. Together, the feedback loops control the proper timing and levels of QS-regulated target gene expression and they set the AI level at which V. harveyi initiates quorum sensing. (OM), outer membrane; (IM), inner membrane; (LCD), low cell density; (HCD), high cell density.
Here we identify and characterize two negative feedback loops that act in the V. harveyi QS circuit. Both feedback loops involve the central QS response regulator LuxO. We find that LuxO negatively autoregulates its own transcription irrespective of its phosphorylation state and the Qrr sRNAs posttranscriptionally control LuxO production by pairing with and repressing the luxO mRNA transcript. Thus, both feedback loops reduce LuxO production which, in turn, reduces qrr expression. Disruption of these negative feedback loops results in increased LuxO~P and, as a consequence, an alteration in the timing and levels of the expression of QS-regulated genes. We propose that the two feedback loops exist to precisely control LuxO protein levels in order to tightly regulate Qrr production. This arrangement primes the Qrr sRNAs to respond accurately to changes in AI levels and, therefore, permits a faithful QS response and properly aligned QS-regulated gene expression with AI concentration. We suggest that sRNA-mediated feedback regulation could be a common signaling network design feature that permits fine-tuning of gene regulation and maintenance of homeostasis.
Results
LuxO represses its own transcription independent of its phosphorylation state
LuxO functions at the core of the V. harveyi QS cascade to integrate inputs from the three AI signals (Figure 1). Given its central role in QS, we wondered if LuxO was subject to additional regulation to restrict its activity. Our suspicion is that unchecked LuxO activity could render the QS circuit susceptible to large variations in target gene expression. To test this idea, we examined the region upstream of the luxO gene where a regulator might act. The V. harveyi qrr1 gene is transcribed divergently from the luxO gene (Figure 2A). We showed previously that LuxO recognizes a TTGCAwwwTGCAA enhancer sequence upstream of each qrr gene (Lenz et al., 2004; Tu and Bassler, 2007). At this site, LuxO~P interacts with the σ54-RNAP complex to regulate transcription. In the case of the qrr1-luxO region, the LuxO binding site is located ~100 bp upstream of the qrr1 transcriptional start site. Because of the proximity of the qrr1 gene to the luxO gene, the promoter regions are situated such that the LuxO-binding site overlaps the −35 site in the luxO promoter (Figure 2A), suggesting that, if LuxO is bound to this site, it could repress its own transcription by competing with RNAP for access to the DNA.
Figure 2. LuxO directly represses its own transcription.
(A) The V. harveyi qrr1-luxO promoter region. qrr1 is transcribed divergently from luxO. The luxO transcriptional start site is indicated by +1, and was determined by 5′ RACE analysis. The LuxO-binding site required for activation of qrr1 expression is shown in the black box. This site overlaps the −35 region of the luxO promoter (underlined). Black dots denote predicted base-pairing between the luxO 5′UTR and the Qrr sRNAs. (RBS), ribosome-binding site; (Start), translational start site. (B) LuxO autorepression does not depend on the phosphorylation state of LuxO. luxO-gfp expression was measured in the following V. harveyi strains: WT, wildtype; luxO, luxO null mutant; LuxO D47E, mimic of LuxO~P; LuxO D47A, mimic of dephosphorylated LuxO; luxU, luxU null mutant. (C) DNA binding gel-mobility-shift analyses with purified LuxO, LuxO D47E, and LuxO D47A isoforms and the qrr1 promoter region, luxOqrr1. A probe containing a randomized LuxO-binding site, luxOscrambled, was used as the negative control. (D) Disruption of the LuxO autorepression loop. Fluorescence production was measured for a LuxO-YFP protein fusion under the control of the endogenous V. harveyi luxO promoter (WT) or under the control of the promoter with the LuxO-binding site mutated (LuxO Loop−) in E. coli. Sequences of the two luxO promoters are shown: In the WT sequence, the black box indicates the LuxO-binding site; In the LuxO Loop− sequence, the line represents the mutations (highlighted in bold) in the LuxO-binding site that eliminate LuxO autorepression. For (B) and (D), cultures were grown in triplicate and error bars denote the standard deviation of the mean.
To determine whether LuxO regulates its own transcription, we constructed a reporter plasmid containing a gfp transcriptional fusion to the luxO promoter. Expression of the luxO-gfp fusion is 30-fold higher in a ΔluxO mutant than in wildtype V. harveyi (Figure 2B), suggesting that LuxO does indeed repress its own transcription. Because LuxO only activates transcription of the qrr genes when it is phosphorylated, we next tested whether phosphorylation is required for LuxO autorepression. To do this, we introduced the luxO-gfp transcriptional fusion into V. harveyi strains containing luxO D47E or luxO D47A on the chromosome. The luxO D47E allele mimics constitutively active LuxO~P, and the luxO D47A allele mimics constitutively unphosphorylated LuxO (Freeman and Bassler, 1999a). Expression of luxO-gfp in the luxO D47E and luxO D47A strains is similar to that in the wildtype, and again, is significantly lower than in the ΔluxO strain (Figure 2B). This result suggests that both the LuxO D47E and LuxO D47A proteins are fully capable of repressing luxO-gfp expression. Consistent with this, the luxO-gfp fusion is repressed in a ΔluxU mutant strain (Figure 2B). LuxU is required to transfer phosphate to LuxO, thus no LuxO~P is present in a ΔluxU mutant strain (Figure 1). Together, the results in Figure 2B indicate that LuxO represses its own transcription in vivo, and this repression does not require that LuxO be phosphorylated.
To confirm that unphosphorylated LuxO can indeed bind DNA, we used gel mobility shift assays. We purified C-terminal HIS-tagged LuxO, LuxO D47E, and LuxO D47A, and assayed each protein’s ability to bind to the qrr1-luxO promoter region (Figure 2C). Wildtype LuxO protein, which presumably purifies in the unphosphorylated state, binds the luxOqrr1 DNA probe (left panel). As a control, we randomized the order of the bases in the LuxO-binding site and showed that wildtype LuxO protein no longer binds to the probe (Figure 2C, second panel from left). Both the purified LuxO D47E and the LuxO D47A proteins bind equally well to the wildtype qrr1-luxO region (Figure 2C, right panels), supporting our in vivo genetic evidence that both phosphorylated and unphosphorylated forms of LuxO readily bind DNA. Gel shift assays show that LuxO binds readily to all the qrr promoters in the unphosphorylated state (data not shown).
To examine the role of LuxO autorepression on LuxO production, we needed to extract this negative feedback loop from the remainder of the V. harveyi QS circuit. To accomplish this, we engineered the LuxO autorepression loop into E. coli and we measured LuxO protein levels using a LuxO-YFP protein fusion. Additionally, we disabled the loop and measured LuxO-YFP levels. To disable the LuxO autoregulatory loop, we randomized a portion of the LuxO-binding site. Our strategy maintained the −35 site intact, which is essential for expression of luxO, but disrupted LuxO’s ability to recognize the site. From here forward, we call this construct LuxO Loop− (Figure 2D). The LuxO Loop− mutation is identical to the alteration in the luxOrandomized construct shown in the gel shift assays in Figure 2C (second panel), and thus, in vitro, LuxO cannot bind to this altered LuxO-binding site. We verified that the mutated LuxO Loop− construct has the same promoter strength as does the WT (Figure S1). We compared production of LuxO-YFP driven from the native luxO promoter and the promoter with the non-functional LuxO-binding site. Approximately 7-fold more LuxO-YFP is produced in the strain carrying the LuxO Loop− construct than in the strain harboring the wildtype luxO promoter (Figure 2D), indicating that elimination of LuxO autorepression significantly increases production of LuxO-YFP.
The V. harveyi Qrr sRNAs repress luxO translation
The V. harveyi Qrr sRNAs repress luxR by base-pairing with the 5′UTR of the luxR mRNA over an approximately 26 bp region upstream of the start codon. Close inspection of the 5′UTR of the luxO mRNA shows that there exists significant complementarity to the conserved region of the V. harveyi Qrr sRNAs (Figure 2A, black dots), suggesting that the Qrr sRNAs could also repress luxO. To test this possibility, we again used E. coli. We transformed the above LuxO-YFP protein fusion into E. coli and subsequently measured LuxO-YFP protein levels in the presence and absence of expression of a Qrr sRNA (Figure 3). LuxO-YFP levels are significantly reduced in the presence of a vector overexpressing Qrr4 (Figure 3, first two bars). Introduction of a vector overexpressing the Qrr4 antisense sRNA had no effect on LuxO protein levels (Figure 3, third bar), indicating that Qrr4 specifically base-pairs with luxO mRNA to repress translation. We also constructed overexpression vectors of the V. harveyi qrr1, qrr2, qrr3, and qrr5 genes and showed that they too repress luxO (data not shown).
Figure 3. The V. harveyi Qrr sRNAs repress luxO translation.
Predicted Qrr4-luxO duplex formation and complementary base-pairing regions are indicated by black dots. (RBS), ribosome-binding site, (AUG), LuxO start codon. Fluorescence production from a LuxO-YFP protein fusion (WT), was measured in E. coli in the presence of an empty vector, a plasmid overexpressing Qrr4, and a plasmid overexpressing the Qrr4 antisense RNA. Disruption of the Qrr4-luxO duplex, also known as sRNA Loop−, was achieved by engineering a U-to-A point mutation in the luxO promoter, as indicated. Fluorescence production from the LuxO-YFP fusion (luxO U->A) was measured in the presence of an empty vector, a plasmid overexpressing Qrr4, and a plasmid overexpressing Qrr4 with an A->U compensatory mutation, as indicated. Cultures were grown in triplicate and error bars denote the standard deviation of the mean.
Analogous to our strategy for the LuxO autoregulatory loop, we disabled the sRNA-LuxO loop in vivo to analyze its effects on LuxO production. To do this, we systematically engineered single base-pair mutations in the 5′UTR of luxO mRNA that we predicted pair with the Qrr sRNAs. Our strategy was to identify a key nucleotide residue that when altered, abolished binding with the Qrr sRNAs. Indeed, a U-to-A mutation five nucleotides downstream of the luxO RBS did not affect basal expression of LuxO-YFP (Figure 3, fourth bar from the left), however, when we overexpressed Qrr4 in E. coli carrying the luxO U-to-A mutation, Qrr4 no longer repressed luxO (Figure 3, second bar from the right). We call this construct sRNA Loop−. To demonstrate that this loss in Qrr repression of LuxO is due to impairment of sRNA-luxO base-pairing, we engineered a compensatory mutation in the Qrr4 sRNA (Qrr4 A to U). This mutation restored repression of luxO by Qrr4 (Figure 3, rightmost bar). These results reveal a new target of the V. harveyi Qrr sRNAs: luxO, and moreover, this interaction forms a posttranscriptional negative feedback loop that regulates LuxO levels.
Disruption of the feedback loops alters LuxO~P levels
The above results show that we have identified two negative feedback loops that function to repress LuxO production. First, LuxO negatively regulates its own transcription independent of its phosphorylation state (Figures 1 and 2) and, second, the Qrr sRNAs posttranscriptionally repress LuxO translation by binding to the luxO 5′ UTR (Figures 1 and 3). More importantly, we were able to make mutations in the promoter region of luxO to disrupt each feedback loop individually and also in combination: the mutation in the LuxO-binding site, LuxO Loop− (Figure 2D), abolishes LuxO autorepression; the single point mutation near the luxO translational start site, sRNA Loop− (Figure 3), abolishes base-pairing between the Qrr sRNAs and luxO; and finally, the combination of the LuxO Loop− and sRNA Loop− mutations abolishes both LuxO regulatory feedback loops. We call this final construct Double Loop−. Our next goal was to introduce these mutations into the V. harveyi chromosome and study the consequences of misregulation of LuxO production on QS-dependent gene expression.
Because LuxO protein levels increase in the absence of each LuxO feedback loop, we first wondered what effect this has on LuxO activity. The qrr genes are activated by LuxO~P at LCD, so we can use qrr expression as a direct readout of LuxO~P activity in V. harveyi. We used a V. harveyi strain that is locked in the LCD state, specifically, the AI synthases for HAI-1 and AI-2 and the sensor kinase for CAI-1 are deleted (ΔluxM, ΔluxS, ΔcqsS) (Long et al., 2009). This locked LCD strain makes high levels of LuxO~P because the two intact AI sensors are locked in kinase mode and because the CqsS sensor is deleted, the CAI-1 system provides no input into the QS circuit. We engineered the feedback loop mutations individually and in combination into this locked LCD strain, transformed a qrr4-gfp transcriptional fusion reporter into the mutants, and quantified fluorescence. Figure 4A shows that qrr4-gfp expression is high in the wildtype LCD strain (red bar). In the LuxO Loop− strain, qrr4-gfp expression is increased compared to wildtype (green bar), and a similar increase in qrr4-gfp expression occurs in the sRNA Loop− strain (blue bar). These results suggest that, in the absence of either the LuxO or sRNA negative feedback loop, total LuxO~P increases at LCD. Moreover, in the Double Loop− strain, there is a much greater increase in qrr4-gfp expression (purple bar), indicating that the two feedback loops cooperate to control LuxO~P levels.
Figure 4. The negative feedback loops make V. harveyi more sensitive to AIs.
(A) Disruption of the negative feedback loops increases LuxO~P. A qrr4-gfp transcriptional fusion was transformed into a V. harveyi ΔluxM, ΔluxS, ΔcqsS strain, denoted WT (red bar). Into this strain, the following mutations were engineered: LuxO Loop− (green bar), sRNA Loop− (blue bar), and Double Loop− (purple bar). Fluorescence production was measured in overnight cultures grown in triplicate and error bars denote the standard deviation of the mean. (B) Dose-response curves of the QS-activated luciferase operon. WT (red squares), LuxO Loop− (green triangles), sRNA Loop− (blue circles), and Double Loop−(purple diamonds). (C) Dose-response curves of the QS-repressed gene qrgA. Strains in (B) were transformed with a qrgA-gfp transcriptional fusion to generate: WT (red squares), LuxO Loop− (green triangles), sRNA Loop− (blue circles), and Double Loop− (purple diamonds). Fluorescence production was measured using a flow cytometer. Data in (B) and (C) were fit with variable-slope sigmoidal dose-response curves to determine the concentration of half-maximal response ( EC ) and the slope of dose-response curve (Hill Coefficient) for each strain. (RLU), relative light units in counts min−1 ml−1/OD600.
The feedback loops affect the timing of QS target gene expression
Our next question was whether alteration in LuxO~P levels affects the ability of V. harveyi to properly control downstream QS gene expression in response to AIs. To examine this, we measured the V. harveyi bioluminescence output response to HAI-1 and AI-2 in our mutants (Figure 4B). Because the strains lack the HAI-1 and AI-2 synthases, this strategy allowed us to precisely control the AI inputs and measure the corresponding QS outputs (Long et al., 2009). The EC50 values for the wildtype, LuxO Loop−, sRNA Loop−, and Double Loop− strains are 25 nM, 61 nM, 64 nM, and 192 nM respectively (Figure 4B). Thus, in the absence of either the LuxO or sRNA feedback loop, V. harveyi becomes two to three times less sensitive to AIs, whereas in the absence of both feedback loops V. harveyi becomes eight times less sensitive to AIs (Note, a two-fold change in the EC50 values is at the margin of significance). We conclude that the two feedback loops cooperate to significantly increase V. harveyi’s sensitivity to AIs. Importantly, Figure 4B shows that disruption of the feedback loops does not substantially alter the slopes of the dose-response curves (Hill coefficient values for the wildtype, LuxO Loop−, sRNA Loop−, and Double Loop− strains are 2.4, 2.8, 2.7, and 2.2, respectively), suggesting that these two feedback loops do not impinge on the downstream amplitude of the QS response, but rather, adjust the AI concentration at which V. harveyi detects that it has reached a quorum and, therefore, the two feedback loops control the timing of QS gene expression.
Many genes are activated and repressed by QS in V. harveyi. To prove that the feedback loops act universally in the QS cascade, we performed an analogous experiment to that in Figure 4B with a known QS-repressed target gene, qrgA (Waters and Bassler, 2006). Dose-response curves for the same V. harveyi strains carrying a qrgA-gfp fusion are shown in Figure 4C. The data are essentially the reciprocal of those for the QS-activated lux operon. The EC50 values for the wildtype, LuxO Loop−, sRNA Loop−, and Double Loop− strains for the repressed qrgA target gene are 82 nM, 198 nM, 192 nM, and 315 nM with Hill coefficients 1.0, 1.2, 1.1, and 1.3, respectively (Figure 4C). Therefore, in the absence of the two feedback loops, four times more AI is required to repress expression of this target gene compared to the wildtype strain. Together, the results in Figure 4 show that disruption of the individual feedback loops increases LuxO~P levels, and as a consequence, affects the threshold for response to AIs of both QS-activated (lux) and QS-repressed (qrgA) target genes. The alteration in QS gene expression and the reduction in AI sensitivity are magnified when both feedback loops are simultaneously eliminated. Thus, the two feedback loops are essential for the proper timing and precise regulation of downstream QS target gene expression.
Function of the feedback loops in V. harveyi containing only one Qrr sRNA
To examine the role of an individual Qrr sRNA in feedback regulation, we engineered a V. harveyi strain deleted for four of the five qrr genes, leaving only the qrr4 gene remaining in the chromosome (we denote this strain qrr4+). We chose to leave Qrr4 intact because qrr4 is the most highly expressed qrr gene at LCD, and thus we suspected it might have the most important role in feedback regulation. qrr4-gfp expression is significantly higher in the qrr4+ strain than in wildtype V. harveyi (compare red bars in Figures 4A and 5A). This result is expected because elimination of four of the five Qrrs reduces sRNA-mediated repression of both luxO and luxR mRNAs, and the resulting higher levels of LuxO~P and LuxR lead to increased activity of the qrr4 promoter (Figure 1) (Svenningsen et al., 2009). The green bar in Figure 5A shows that qrr4-gfp expression is further increased in the LuxO Loop− strain. However, there is no increase in qrr4-gfp expression in the sRNA Loop− strain (Figure 5A, compare red bar to blue bar). Disruption of the sRNA loop does, however, cause a sizable increase in qrr4-gfp level in the qrr4+ strain already lacking the LuxO feedback loop (Figure 5A, Double Loop−, purple bar). We interpret these results to mean that qrr4 levels are not sufficient for repression of LuxO in the qrr4+ strain possessing an intact LuxO feedback loop (Figure 5A, compare red bar and purple bar). However, in the context of the LuxO Loop− background, Qrr4 levels are sufficiently high to repress LuxO translation via the sRNA negative feedback loop, and hence elimination of this loop leads to an increase in LuxO and a concomitant increase in qrr4 expression. One possible explanation for the apparent threshold of qrr4 expression required to repress luxO is that Qrr4 may have a higher binding affinity for luxR than luxO mRNA, and thus, luxR is the primary target and luxO is the secondary target of regulation by Qrr4. In this scenario, Qrr4 only represses luxO, and, in turn, reduces its own expression after luxR has been repressed and only if there remain excess Qrr4 sRNAs to do so. The results in Figure 5 support this model. Specifically, the LuxO-feedback keeps LuxO levels low, which in turn, reduce qrr expression. When this feedback loop is disrupted, qrr4 expression increases, and so increased Qrr4 is produced and available to repress luxO mRNA. If this is indeed the case, the different affinities of the sRNAs for their target mRNAs could allow limited sRNAs to prioritize the regulation of different target mRNAs, which, in our case, ultimately dictates the order and level of QS gene regulation. The bioluminescence and qrgA-gfp dose-response curves in the qrr4+ strain also follow the pattern shown in Figure 5A for qrr4-gfp fluorescence (Figures 5B and 5C and supplemental information).
Figure 5. The role of the feedback loops when only a single Qrr sRNA is present.
(A) The qrr4-gfp fusion from Figure 4A was transformed into the following V. harveyi strains: qrr4+ (red bar), LuxO Loop− (green bar), sRNA Loop− (blue bar), and Double Loop− (purple bar). Fluorescence production was measured in overnight cultures grown in triplicate and error bars denote the standard deviation of the mean. (B) Dose-response curves measuring bioluminescence were obtained as in Figure 4B in the following V. harveyi strains: qrr4+ (open red squares), LuxO Loop− (open green triangles), sRNA Loop− (open blue circles), and Double Loop− (open purple diamonds). (C) Dose-response curves measuring qrgA-gfp were obtained as in Figure 4C in the following V. harveyi strains: qrr4+ (open red squares), LuxO Loop− (open green triangles), sRNA Loop− (open blue circles), and Double Loop− (open purple diamonds). Data in (B) and (C) were fit and coefficients obtained as in Figure 4.
Determination of LuxR protein levels at single-cell resolution
The behavior of the QS population is ultimately dictated by the behavior of individual cells. Up to this point, we have focused on the regulation of QS target genes at the population level. However, our goal is to relate the individual cell behavior to that of the population. Toward this end, we explored how individual cells respond to AI inputs and determined their cell-to-cell variation with and without the LuxO regulatory feedback loops. We focused on quantifying LuxR levels because LuxR is the master regulator of QS target genes, and LuxR ultimately dictates the pattern of QS gene expression. We constructed a full-length LuxR-mCherry protein fusion and introduced it onto the V. harveyi chromosome at the native luxR locus in the wild type and the feedback-loop mutants. Subsequently, we measured LuxR-mCherry in individual cells under different AI input conditions. The individual cell dose-response profiles of LuxR in the various feedback-loop mutants mirror the patterns we observed at the population level with bioluminescence (Figure 6A). The calculated EC50 values for the wildtype, LuxO Loop−, sRNA Loop−, and Double Loop− strains are 36 nM, 57 nM, 62 nM, and 271 nM with Hill coefficients 1.3, 1.1, 1.2, and 1.0, respectively (Figure 6A). Thus, with regard to LuxR levels in individual cells, in the absence of either the LuxO or sRNA feedback loop, V. harveyi becomes roughly two times less sensitive to AIs, whereas in the absence of both feedback loops, V. harveyi becomes eight times less sensitive to AIs. The Hill coefficients of the LuxR dose-response curves are approximately 1, supporting our previous results that LuxR protein levels increase in a graded manner in response to increasing AI concentrations (Waters and Bassler, 2006). The present results show that there is roughly a five-fold difference between LuxR protein levels under minimal and saturating AI levels (Figure 6A), which is the range of LuxR that determines the temporal order of expression of QS-repressed and QS-activated target genes.
Figure 6. LuxR single-cell dose-response curves and relative noise.
(A) Individual-cell dose-response curves were obtained using the same V. harveyi strains as in Figure 4B but containing a LuxR-mCherry protein fusion integrated in the native luxR locus: WT (red squares), LuxO Loop− (green triangles), sRNA Loop− (blue circles), and Double Loop− (purple diamonds). (A.U.), arbitrary units. (B) LuxR dose-response curves in qrr4+ background. The V. harveyi strains from Figure 5B, but containing a LuxR-mCherry protein fusion integrated in the native luxR locus, were analyzed: qrr4+ (open red squares), LuxO Loop− (open green triangles), sRNA Loop− (open blue circles), and Double Loop− (open purple diamonds). Data were fit and coefficients obtained as in Figure 4. Each symbol in (A) and (B) represent the average mCherry value obtained from microscopy images of 100 cells. (C) The relative noise, i.e. the standard deviation (SD) of the population divided by the mean, versus the mean LuxR-mCherry fluorescence intensity for wildtype V. harveyi with various feedback loop mutations (closed symbols) and for the qrr4+ strain with various feedback loop mutations (open symbols).
We also measured LuxR protein levels in response to AIs in single cells containing only qrr4. The calculated EC50 values for the qrr4+ strain, the LuxO Loop− strain, the sRNA Loop− strain, and the Double Loop− strain are 20 nM, 43 nM, 16 nM, and 46 nM with Hill coefficients 1.1, 1.5, 1.7, and 1.5, respectively (Figure 6B). As in the case of bioluminescence, these results show that in a strain containing qrr4 only, disruption of the sRNA feedback loop alone has no significant effect on the LuxR dose-response curve. We note that the basal level of LuxR protein in the various qrr4+ strains at low AI is approximately two-fold higher than that in the corresponding strains containing all five qrr genes. This finding confirms our expectation based on Figure 5, that qrr4 alone cannot fully repress luxR translation to wildtype levels (see Supplemental Information for details). Due to this increased LuxR level in the qrr4+ strain at LCD, there is only a three-fold difference between LuxR protein concentrations under minimal and saturating AI levels, giving V. harveyi a reduced window of QS target gene expression. Thus, the changes in lux and qrgA expression in Figures 4 and 5 can be directly attributed to corresponding changes in LuxR protein levels, suggesting that AI-directed alterations in LuxR concentrations are faithfully transduced into alterations in downstream QS target gene expression.
Our measurements of LuxR protein at single-cell resolution enabled us to quantify the cell-to-cell variation in LuxR in the presence of increasing AI concentrations. Figure 6C shows the relative noise in LuxR protein in the eight different V. harveyi mutants. At low AI concentrations, the standard deviation over the mean is approximately 0.4. Thus, when minimal LuxR is present (LCD), the system is, not surprisingly, somewhat noisy. However, at high AI concentrations, the standard deviation over the mean is approximately 0.2, indicating that there is little LuxR variation between single cells at HCD. Surprisingly, there is no significant difference in the relative noise observed in cells containing or lacking the various feedback loops or in cells that contain multiple versus one Qrr sRNA (Figure 6C).
The feedback loops do not affect the kinetics of the HCD to LCD transition
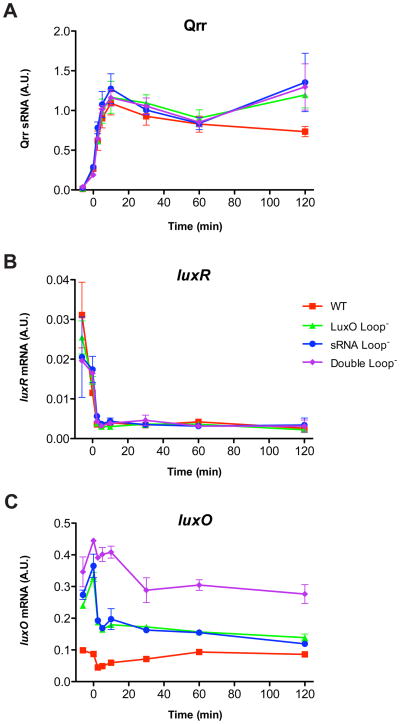
Figures 4 and 5 indicate that the presence of the feedback loops makes wildtype V. harveyi more sensitive to AIs. We previously identified a negative feedback loop in the V. harveyi QS circuit in which LuxR activates the expression of the qrr2, qrr3, and qrr4 genes, leading to a rapid transition out of HCD mode into LCD mode (Tu et al., 2008). To determine whether the feedback loops involving LuxO and the Qrr sRNAs likewise play a role in controlling the dynamics of the HCD to LCD transition, we grew the same V. harveyi strains as in Figure 4B in saturating AI concentrations to simulate HCD mode, washed away the AIs with fresh medium to simulate an immediate transition to LCD mode, and subsequently measured cellular Qrr sRNA levels (Figure 7A). Qrr levels increase to maximal values within 10 minutes after the removal of AIs in all four V. harveyi strains. There do not appear to be significant differences in the rates at which Qrr sRNAs increase or the maximum values reached in the feedback loop mutants, suggesting that the feedback loops do not play a significant role in the kinetics of this transition.
Figure 7. Cellular sRNA and mRNA levels during the HCD to LCD transition.
(A) The feedback loops do not affect dynamics of Qrr sRNAs during the transition from HCD to LCD. The identical V. harveyi strains shown in Figure 4B were grown in duplicate in the presence of saturating HAI-1 and AI-2: WT (red squares), LuxO Loop− (green triangles), sRNA Loop− (blue circles), and Double Loop− (purple diamonds). AIs were washed away with fresh medium and sRNA levels were quantified using a QuantiGene Plex Reagent System (Panomics). Error bars are standard deviations of samples in duplicate. (A.U.), arbitrary units. (B) luxR mRNA levels in the cells from panel A decline rapidly during the HCD to LCD transition for all four V. harveyi strains. (C) luxO mRNA levels show different patterns than luxR mRNA levels during the HCD to LCD transition.
Qrr sRNA induction eliminates luxR, but not luxO, mRNAs
We have previously demonstrated that the Qrrs base-pair with luxR mRNA and destabilize it to prevent further translation (Lenz et al., 2004). To determine whether repression is identical for luxO mRNA, we focused on the cellular mRNA levels of luxR and luxO during the HCD to LCD transition shown in Figure 7A. Consistent with previous observations, luxR mRNA levels decline from the maximal amount present at HCD to a low basal amount within 3 minutes of the transition into LCD mode (Figure 7B). The rapid decrease in luxR mRNA levels matches exactly the fast increase of Qrr sRNA levels (Figure 7A), confirming that luxR mRNA is indeed completely degraded as a consequence of Qrr sRNA repression. Surprisingly, the pattern for luxO mRNA is different (Figure 7C): there is a small decrease in cellular luxO mRNA levels in all mutant strains during the first 3 minutes after the HCD to LCD transition. However, 30 minutes after the transition, luxO mRNAs remain at levels either identical to or only slightly lower than the original HCD levels. Figure 7C also shows that the relative amounts of luxO mRNA in the wild-type and mutant strains are consistent with the relative LuxO protein levels inferred from qrr4-gfp expression (Figure 4A). The initial modest decrease in luxO mRNA levels following the HCD to LCD transition cannot be attributed to sRNA repression, since the sRNA Loop− and the Double Loop− strains display the same pattern. Moreover, the subsequent maintenance of steady luxO mRNA levels cannot be attributed to limited availability of Qrr sRNAs, since the Qrr sRNAs remain abundant even after all luxR mRNAs are degraded (Figure 7AB). Taken together, these results suggest that the Qrr sRNAs do not have the dramatic on/off effect on luxO mRNA like they have on luxR mRNA. Nevertheless, the Qrr sRNAs clearly do repress LuxO protein production via direct basepairing with luxO mRNA (Figure 3). One possibility is that Qrr sRNAs repress luxO mRNA by sequestration of the mRNA without causing mRNA degradation. Consistent with this notion, Northern blot analysis shows that deletion of all qrr genes does not alter the luxO mRNA half-life, and overexpression of Qrr4 extends luxO mRNA half-life slightly (Figure S2 and Supplemental Materials). By contrast, the Qrr sRNAs decrease luxR mRNA half-life by 15-fold. Furthermore, deletion or overexpression of the Qrrs minimally alters total cellular luxO mRNA levels, while causing dramatic changes in total cellular luxR mRNA levels (Figure S2). These results support the idea that the Qrrs sequester luxO mRNA and block its translation. Finally, we note that sequestration alone cannot fully account for the differences in luxO mRNA levels observed at HCD in the strains containing and lacking the sRNA loop (Figure 7C).
Discussion
Coordination of QS processes often involves multi-step signal transduction pathways that control the expression of master transcription factors, which in turn, regulate genes responsible for group behaviors. QS networks must be designed to precisely translate external AI concentrations into proper patterns of internal QS target gene expression. The molecular components making up the V. harveyi QS circuit have been defined and characterized, allowing us to undertake an analysis of the regulatory features of the network that optimize the QS output response. Here, we identified two cooperative negative regulatory feedback loops involving LuxO and the Qrr sRNAs. Specifically, LuxO negatively regulates its own transcription by virtue of a LuxO-binding site in the luxO promoter (Figure 2A). Negative regulation does not depend on the phosphorylation state of LuxO (Figure 2B,C), suggesting that LuxO competition with RNAP for DNA-binding at this LuxO/−35 site sets the level of luxO transcription. LuxO is also subject to posttranscriptional feedback regulation by the Qrr sRNAs (Figure 3); thus luxO mRNA constitutes a new target of the Qrr sRNAs in V. harveyi. The two feedback loops function to make V. harveyi more sensitive to changes in external AI concentrations and thereby set the threshold population density at which cells reach a quorum (Figures 4–6).
The negative feedback loop is a common network-design motif in signal transduction pathways and gene regulatory networks. Negative feedbacks are generally believed to play three major roles: reducing “noise”, i.e. temporal fluctuation or cell-to-cell variation (Becskei and Serrano, 2000; Thattai and van Oudenaarden, 2001); decreasing response rise times (Alon, 2007; Rosenfeld et al., 2002; Savageau, 1974); and producing graded responses (Nevozhay et al., 2009). All three roles have been demonstrated for negatively auto-regulated transcription factors (TF). First, high concentration of a TF will result in repression, while low concentration will result in increased production, leading to a narrower distribution of protein levels than for proteins that are not auto-regulated. This feature reduces expression noise of the TF. Second, a TF subject to negative feedback with a stronger promoter than a non-self-regulated one, can nonetheless reach the identical steady-state level. The negatively regulated TF will increase rapidly until it reaches the threshold level required for repressing its own expression. This feature results in a shorter gene-expression rise time (Rosenfeld et al., 2002). Third, a particular amount of inducer will result in a lower level of a negatively auto-regulated TF than a non-regulated one due to auto-repression of expression. This feature decreases the slope of the dose-response curve and generates a graded response.
We determined whether the two negative feedback loops under study play any of these three canonical roles. In order to measure noise in the QS response, we used single-cell fluorescence microscopy to quantify LuxR levels of V. harveyi in individual cells at various AI concentrations. The relative noise, namely the standard deviation over the mean, is only 20% at high AI concentrations (Figure 6C). Thus, V. harveyi cells act in unison at HCD. However, surprisingly, there is no discernable difference in relative noise in the various feedback loop mutants, whether in the presence of a single or multiple sRNAs. A possible explanation for this observation is that the two central transcriptional regulators in the QS circuit – LuxO and LuxR – are both subject to transcriptional and posttranscriptional negative feedback regulation. The feedback loops on LuxO may primarily contribute to noise reduction in LuxO expression levels, while the feedback loops on LuxR filter out noise originating from various sources, including noise in LuxO levels, to stabilize the LuxR expression level. In any event, we find that the two feedback loops on LuxO do not noticeably contribute to the low relative noise in LuxR expression.
To test whether the translational repression of LuxO by Qrr sRNAs decreases rise time relative to steady-state levels of the Qrrs during the HCD to LCD transition, we measured Qrr sRNA levels in both wildtype cells and in the engineered strains with the feedback loops disabled (Figure 7A). Our results show that the transition happens rapidly – Qrr levels reach maxima within 10 minutes – in all strains. There is no discernible difference either in the rise times or the plateau levels of Qrrs in the presence or absence of the feedback loops. Therefore, the LuxO-Qrr feedback loop does not function to allow a fast transition out of HCD mode.
We also considered whether the feedback loops involving LuxO and Qrr sRNAs might function to generate a graded response. However, as shown in Figure 6A, the slopes of the LuxR dose-response curves are not substantially different for wildtype and the various loop mutants. All the Hill coefficients are ~1 from the fitting curves, suggesting a non-cooperative graded QS response irrespective of whether or not the negative feedbacks were present. With only a single Qrr present, the Hill coefficients for the loop mutants are modestly larger than when both loops are present (Figure 6B, 1.5–1.7 vs. 1.1), but the difference is within our experimental error. The dose-response curves of LuxR target genes (bioluminescence and qrgA) also showed constancy in the Hill coefficients for wildtype and various loop mutants. Thus, we conclude that the negative feedback loops do not make the QS response to autoinducers more graded.
In summary, the two negative feedback loops reported here do not appear to play any of the usual roles of negative feedback loops in gene-regulatory networks. However, the feedback loops clearly do function to tune the sensitivity of the QS response to AI concentrations, and in so doing, establish the point at which the population of V. harveyi reaches a quorum, and thus set the timing of downstream QS target gene expression. It is puzzling why this fine-tuning of the QS threshold is achieved via negative feedbacks rather than, e.g., by adjusting parameters of reactions in the signaling and gene regulatory circuit.
If these two negative feedback loops were not wired into the QS circuit, we speculate that there would be a loss of homeostatic control due to unchecked LuxO and LuxR levels. Specifically, we suggest that the two negative feedback loops have evolved to couple the natural dynamic range of AI concentrations with the desired dynamic range of downstream QS-regulated gene expression. Likewise, negative feedback regulation increases signaling fidelity in the mating pheromone pathway in yeast (Yu et al., 2008). In particular, the specific architecture of the LuxO-Qrr negative feedback loop prevents over-accumulation of Qrrs, and thus prevents the targets of Qrrs from “bottoming out”, i.e. declining to extremely low levels. Intuitively, the obligate activation of Qrr sRNAs by LuxO~P/LuxR places a cap on Qrr expression: if Qrr levels start to become too high, luxO/luxR mRNA is decreased due to the excess of Qrrs, and as a consequence, qrr expression declines due to decreased levels of LuxO~P/LuxR. Therefore, all targets of the Qrrs are protected from “bottoming out” (approaching zero) by the feedback loops in which the Qrrs negatively regulate their own activators, LuxO~P and LuxR. Consider the following simplified kinetic equation for a target mRNA (e.g. luxR or luxO):
| (1) |
Where γ is a constant transcription rate of the mRNA, μ is the coefficient for the rate of mutual degradation (or sequestration) of sRNA and mRNA, and δ is the rate of ordinary degradation/dilution of mRNA. At steady state, the time derivative of the mRNA concentration must be zero, and so the cellular mRNA level is:
| (2) |
If the sRNA level becomes very high, the denominator in Equation (2) will be extremely large and result in a “bottoming out” of the mRNA. It is obvious that by setting an upper limit on Qrr sRNA levels, mRNA levels are thereby prevented from approaching zero. Physiologically, it is likely to be important to maintain a basal level of translation of the mRNAs of key QS players – including LuxO and LuxR – otherwise these proteins will be diluted out during cell growth and division. Thus, the multiple negative feedback loops function to keep LuxO, the Qrrs, and LuxR levels in check, and therefore aid in maintaining the expression of both Qrrs and QS-regulated target genes within restricted windows. We suggest that sRNA-based regulation of mRNAs of the essential QS components is indispensible for proper functioning of QS signaling.
Our results suggest that there are roles negative feedback loops can play in signaling and gene regulatory networks beyond those already recognized. While the LuxR-Qrr feedback loop accelerates the HCD to LCD transition (Tu et al., 2008), the topologically identical LuxO-Qrr feedback loop appears to play a different role – namely fine-tuning the threshold where the bacterial population reaches a quorum while controlling the dynamic range of expression of QS target gene expression. We suggest that bacterial sRNA posttranscriptional negative regulatory feedback loops, and potentially, eukaryotic miRNA negative feedback loops may prove to be a common network design motif. They provide features such as noise reduction, rise time shortening, grading of responses, as well as new ones including the dynamic range control proposed here in the context of the V. harveyi quorum-sensing circuit.
Experimental Procedures
DNA manipulations
All DNA manipulations were performed using standard procedures (Sambrook et al., 1989). Herculase Enhanced DNA polymerase (Stratagene) was used for PCR cloning reactions, and Taq polymerase (Roche) was used for all other PCR reactions. dNTPs, restriction endonucleases, and T4 DNA ligase were obtained from New England BioLabs. DNA purification kits were provided by QIAGEN. QuikChange II XL Site-Directed Mutagenesis Kit and XL-10 Gold Competent Cells (Stratagene) were used for all site-directed mutageneses. Primer sequences are available upon request. Strains and plasmids used in this study are listed in Supplemental Table S1.
Fluorescence analysis and bioluminescence assays
All gfp and yfp expression analyses were performed on a Becton Dickinson FACS Aria cell sorter, and data were analyzed using FACS Diva software as described previously (Tu et al., 2008). For dose-response curve experiments, cultures were grown for 14 h in LM medium and subsequently diluted 1:1000 in fresh medium. In a 96-well microtiter plate, 90 μl of culture was added to 5 μl of 100 μM HAI-1 and 5 μl of 100 μM AI-2, and serial dilutions were made to final concentrations of 10 pM total of both HAI-1 and AI-2. The plates were incubated for 6 h and then each sample was transferred to a 5 mL round-bottom tube (BD Biosciences) and fluorescence was measured. For dose response curves measuring bioluminescence, an identical procedure was used except that cultures were grown in AB medium in quadruplicate. Bioluminescence and OD600 were measured using a Perkin Elmer EnVision plate reader.
Single-cell fluorescence microscopy
For measurements of LuxR-mCherry in individual cells, wildtype, qrr4+, and the corresponding feedback loop mutant V. harveyi strains (KT841 KT843 KT845 KT847 KT849 KT851 KT853 KT855) were grown in AB medium for 16 hrs. Growth was monitored by measuring optical density at 600 nm. Cultures were diluted to OD600 = 0.0005 and exogenous autoinducers were added at the specified concentrations. After growing for 6 hrs, cells were concentrated by centrifugation and maintained on ice until measurements were made. 1 μl of cell culture was spread on a glass slide and covered with a 1% AB agarose pad as well as a coverslip. Phase-contrast and fluorescent images were taken at room temperature using a Nikon TE-2000U inverted microscope. Custom Basic code was written to control the microscope. Images were acquired using a 100x oil objective and a cooled CCD camera (−65°C, Andor iXon). Segmentation of individual cells was performed on phase-contrast images. Background and cellular auto-fluorescence values were subtracted from the red channel. Total fluorescence intensity of each cell was obtained by summing all pixels and fractions of pixels in the segmented cell region. Cell volume was estimated by considering each cell as a cylinder plus two semi-spheres. LuxR-mCherry concentration in each cell was calculated as the total mCherry fluorescence intensity over the volume of that cell.
Quantification of cellular RNA levels
V. harveyi cell cultures grown in LM medium for 14 hrs were diluted to OD600 = 0.005 in 5ml AB medium in duplicate with 0.5 μM HAI-1 and 0.5 μM AI-2 added to the cultures. After 6 hrs growth at 30°C, cells from 1 ml of culture were pelleted, transferred into 2 ml fresh AB medium, and allowed to shake at 30°C. At specified time points, 20 μl cell culture was removed and combined with 180 ul RNAlater® (Ambion) solution. Quantification of RNA levels was performed using QantiGene Plex 2.0 Reagent System (Panomics) that quantifies RNA directly from crude cell lysates using complimentary oligonucleotides attached to beads without any RNA purification, reverse transcription, or target amplification. The assay was performed according to the manufacturer’s recommendations.
Supplementary Material
Acknowledgments
We thank Pankaj Mehta for helpful discussions. This work was supported by HHMI, NIH grant 5R01GM065859, NIH grant 5R01AI054442, NSF grant MCB-0639855 to B.L.B., and the Burroughs Wellcome Fund Graduate Training Program (T.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. An introduction to systems biology: design principles of biological circuits. Boca Raton, FL: Chapman & Hall/CRC; 2007. [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999a;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. Mol Microbiol. 1999b;31:665–677. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Developmental biology in marine invertebrate symbioses. Curr Opin Microbiol. 2000;3:603–607. doi: 10.1016/s1369-5274(00)00147-8. [DOI] [PubMed] [Google Scholar]
- Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto CM, Smith EE, Swartzman E, Cao JG, Graham AF, Meighen EA. Proximal and distal sites bind LuxR independently and activate expression of the Vibrio harveyi lux operon. Mol Microbiol. 1994;14:255–262. doi: 10.1111/j.1365-2958.1994.tb01286.x. [DOI] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci U S A. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savageau MA. Comparison of classical and autogenous systems of regulation in inducible operons. Nature. 1974;252:546–549. doi: 10.1038/252546a0. [DOI] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. Embo J. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman E, Silverman M, Meighen EA. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol. 1992;174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci U S A. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Waters CM, Svenningsen SL, Bassler BL. A small-RNA-mediated negative feedback loop controls quorum-sensing dynamics in Vibrio harveyi. Mol Microbiol. 2008;70:896–907. doi: 10.1111/j.1365-2958.2008.06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.