Abstract
Current biodetection illumination technologies (laser, LED, tungsten lamp, etc.) are based on spot illumination with additional optics required when spatial excitation is required. Herein we describe a new approach of spatial illumination based on electroluminescence (EL) semiconductor strips available in several wavelengths, greatly simplifying the biosensor design by eliminating the need for additional optics. This work combines EL excitation with charge-coupled device (CCD) based detection (EL-CCD detector) of fluorescence for developing a simple portable detector for botulinum neurotoxin A (BoTN-A) activity analysis. A Förster Resonance Energy Transfer (FRET) activity assay for BoTN-A was used to both characterize and optimize the EL-CCD detector. The system consists of two modules: (1) the detection module which houses the CCD camera and emission filters, and (2) the excitation and sample module, containing the EL strip, the excitation filter and the 9-well sample chip. The FRET activity assay used in this study utilized a FITC/DABCYL-SNAP-25 peptide substrate in which cleavage of the substrate by BoTN-A, or its light chain derivative (LcA), produced an increase in fluorescence emission. EL-CCD detector measured limits of detection (LODs) were similar to those measured using a standard fluorescent plate reader with valves between 0.625 and 1.25 nM (31–62 ng/ml) for LcA and 0.313 nM (45 ng/ml) for the full toxin, BoTN-A. As far as the authors are aware this is the first demonstration of phosphor-based EL strips being used for the spatial illumination/excitation of a surface, coupled with CCD for point of care detection.
Keywords: Electroluminescent excitation, CCD, Fluorescence, Förster resonance energy transfer (FRET), Botulinum neurotoxin A, Biodetection, Point of care
1. Introduction
All fluorescence detectors contain two main elements: (1) the excitation light source and (2) a light detector. There are a number of potential excitation light sources to choose from, all with associated advantages and disadvantages. Tungsten (Taylor et al., 1997), mercury (Krishnan et al., 2004), or xenon lamps, for example, offer the user high intensity and broad wavelength spectrum excitation. However they are mainly used for bench-top devices and are often bulky, expensive and require high voltage supplies, thus limiting the portability of any developed device. Lasers and LEDs, on the other hand, offer narrow wavelength excitation bands and are available in a number of colors that span the UV–vis region of the electromagnetic spectrum. They are generally low cost, highly efficient, have a small footprint, are more durable and consume relatively little power compared to the lamps. These characteristics make lasers (Golden et al., 2005; Ligler et al., 2007; Ligler et al., 2003) and LEDs ideal excitation light sources for portable fluorescent detection systems. However, LEDs and in particular lasers have a very narrow spatial field of illumination and so excitation of a relatively large surface area, requires either scanning (moving the light source or the surface), the use of a panel of excitation sources, or incorporation of a number of optical components such as, waveguides, line generators, mirrors and lenses (Golden et al., 2005; Ligler et al., 2007; Ligler et al., 2003). Such components can rapidly complicate platform design, manufacturing, maintenance and cost.
Phosphor-based electroluminescent (EL) semiconductor strips or panels offer the same advantages discussed for lasers and LEDs, with the added benefit of effortlessly providing spatial illumination over large surface areas. EL materials are widely used for signs and instrument dial illumination (e.g. indigo watches, clock radio, personal organizers, cockpit instrumentation, etc.), however they have not yet been used, to date, for fluorescence excitation and biodetection. EL-based excitation has great potential for a variety of applications based on fluorescence detection (e.g. fluorimeter, microarray, PCR, etc.), as well as illumination-based optical detection (e.g. ELISA reader).
As with the excitation light sources discussed above, light detectors for fluorescent detection take many forms and include photodiodes (Bruno et al., 1997; Capitan-Vallvey et al., 2007; Claycomb and Delwiche, 1998; Mac Sweeney et al., 2004), photomultipliers (Moehrs et al., 2006; Roda et al., 2003; Ruiz-Martinez et al., 1993; Takei et al., 2001; Tibbe et al., 2001; Tsukagoshi et al., 2005) and the charged-coupled device (CCD) (Burkert et al., 2007; Knecht et al., 2004; Ligler et al., 2003; Liu and Danielsson, 2007; Roda et al., 2003; Sohn et al., 2005; Svitel et al., 2001; Tohda and Gratzl, 2006). Photodiodes and photomultipliers are inherently spot detectors, for analysis of limited size areas (unless an array of detectors is used), while CCD-based detectors are spatial detectors, capable of monitoring much larger surfaces. This property makes CCD detectors ideal for multi-channel applications or imaging surfaces with an array of features. To take full advantage of the spatial detection offered by CCD detectors simple spatial illumination of the sample is desired. As mentioned above, the EL semiconductor strip described in this study is inherently a space illuminator, thereby bypassing the need for additional complex optical designs.
There are seven serotypes (A–G) of botulinum neurotoxins (BoNTs) produced and secreted by Clostridium botulinum (Cai et al., 2007). Although a number of in vitro detection tests for BoNTs are commercially available, the gold standard laboratory test for botulinum toxin activity is the mouse bioassay (CDC, 1998; Keller, 2006; Nowakowski et al., 2002). The mouse bioassay is very sensitive and can detect as little as 0.03 ng of botulinum toxin (1 ng = 30 mouse 50% lethal doses) within 6–96 h (Schantz and Johnson, 1990, 1992). Several different immunoassays have been developed for BoTN-A detection including, enzyme-linked immunosorbent assays (ELISA) (Ferreira et al., 2004; Guglielmo-Viret et al., 2005; Han et al., 2007; Keller et al., 1999; Peruski et al., 2002; Sharma et al., 2006), lateral flow (Gessler et al., 2007; Sharma et al., 2005), immunomagnetic beads (Gessler et al., 2006; Rivera et al., 2006), immunoaffinity column (Attree et al., 2007; Gessler et al., 2005) and immunosensors (Golden et al., 2005; Ligler et al., 2007; Ligler et al., 2003). While these immunoassays have similar sensitivities to the mouse bioassay, and are much more rapid, they do not provide information about the activity state of the toxin.
BoTN-A activity can be detected based on the ability of the toxin to cleave the specific SNAP-25 peptide (Blasi et al., 1993; Dong et al., 2004; Keller et al., 1999; Schiavo et al., 1992a,b, 1993; Shone et al., 1993; Sudhof et al., 1993; Trimble, 1993). Such cleavage activity has been detected by mass spectrometry (Barr et al., 2005; Gaunt et al., 2007; Kalb et al., 2006), with a cantilever micromechanical sensor (Liu et al., 2003; Parpura and Chapman, 2005), using fluorescent sensors to detect FRET-based reactions (Dong et al., 2004), and also a microfluidics device (Mangru et al., 2005). Some of the technical issues concerning these protease-based activity assays, including matrix inhibition and non-specific signals, were recently addressed (Kalb et al., 2006). While these activity-based assays are very promising, they are not currently configured in an easy-to-use multi-sample format that is both sensitive and portable. Even the microfluidics application that has been developed requires a microscope for fluorescent detection (Mangru et al., 2005). The EL-CCD detector described in this study was designed with the aim of producing a portable, simple and inexpensive modular-based fluorescence detection system, in a relatively small and compact manner, capable of measuring multiple samples simultaneously. The modular design increases the versatility of the system by allowing the end user simple switching of excitation EL strips, available in various wavelengths, and filter sets when studying different fluorescent systems. The BoTN-A FRET-based activity assay was chosen to demonstrate the successful design and utility of the prototype EL-CCD point of care detector.
2. Material and methods
2.1. Materials and reagents
All chemicals were of reagent grade and used as received from the manufacturer. Bovine Serum Albumin (BSA), zinc chloride (ZnCl), DL-dithiothreitol (DTT) and N-[2-hydroxyethyl] piperazine-N′ [2-ethane sulfonic acid] (HEPES) were obtained from Sigma–Aldrich (St. Louis, MO). The SNAP-25 peptide substrate, internally labeled with the FRET pair fluorescein-thiocarbamoyl (FITC) and 4-(dimethylaminoazo)benzene-4-carboxylic acid (DABCYL), was purchased from List Biological Laboratories (FITC/DABCYL-SNAP-25; Campbell, CA). The positive control FITC-labeled SNAP-25 (FITC-SNAP-25; not labeled with acceptor DABCYL), the recombinant light chain of BoTN-A (LcA), and the full toxin BoTN-A were also obtained from List Biological Laboratories. The 9-well sample chips, used to hold the samples for analysis, were prepared using 1/8 in. black poly(methyl methacrylate) (PMMA) also known as acrylic (Total Plastics, Harrisburg, PA). Glass (LifterSlip, Erie Scientific Company, Portsmouth, NH) and various transparent plastics, including Cyclic Olefin Copolymer (COC) (TekniFlex COC P7P; Teknioplex Europe, Belgium), Polyethylene terephthalate (boPET) polyester film also known as Mylar (Mylar, clear polyester; GE Polymershapes, Devens, MA) and polycarbonate (PC) (Piedimont Plastic Inc., Beltsville, MD), were investigated as bases and tops for the 9-well chip.
2.2. Fluorescence detection
The EL-CCD detector consists of a Royal Blue EL strip cut to 1 in. × 2 in. (Being Seen Technologies, Bridgewater, MA), a blue excitation filter HQ480/20x (Chroma Technology Corp, Rockingham, VT), two green emission filters EmF-A, 535/50/75 (Intor, Soccorro, NM) and EmF-B filter D535/40 m (Chroma Technology Corp Rockingham, VT), and a cooled CCD camera Atik 16 (Adirondack Video Astronomy, Hudson Falls, NY), equipped with a 5 mm Pentax extension and Pentax 12 mm f1.2 lens (Spytown, Utopia, NY). All the system components are enclosed in a black plastic box (Radioshack; www.radioshack.com). The EL requires 110 AC volts supplied by an inverter which converts the 12 V DC input into 110 V AC output (Being Seen Technologies, Bridgewater, MA). The CCD detector employs a Sony ICX-429ALL 752 × 582 pixels CCD and is connected to a Windows XP PC via a USB port and controlled using Artemis ATK Capture software (Adirondack Video Astronomy, Hudson Falls, NY). The CCD image intensities were analyzed using ImageJ software, developed and distributed freely by NIH (http://rsb.info.nih.gov/ij/download.html), and the data generated is then imported into Microsoft Excel (Microsoft, Redmond, WA) for further manipulation. Fluorescence detection using the standard 96-well plates (Greiner, polystyrene) was measured using a PerkinElmer HTS 700 Plus Bio Assay reader (PerkinElmer, Fremont, CA).
2.3. Fabrication of the 9-well sample chips
The 9-well sample chips used in this study were designed in CorelDraw11 (Corel Corp. Ontario, Canada) and micro-machined in 1/8 in. black acrylic using a computer controlled laser cutter Epilog Legend CO2 65 W cutter (Epilog, Golden, CO). Before cutting, both sides of the acrylic sheet were coated with 3 M 9770 adhesive transfer double sided tape (Piedmont Plastics, Beltsville, MD). Initial studies were carried out to determine the optimal materials for sealing the base and top of the 9-well acrylic sample chips. Materials investigated include Mylar, COC, PC and glass. PC was cut using the laser cutter, while Mylar and COC were best cut to size using scissors. Glass was cut using a diamond cutter pen. The appropriate material was first attached to the bottom on the 9-well acrylic sample chip by removing the protective cover on the double sided tape and firmly pressing the material into place. The wells were then cleaned using Versa Clean® (FisherBrand, Pittsburgh, PA) and washed with MilliQ water before being dried with air. The reaction solution was added to the top of the sample wells, if required, attached by again removing the protective cover of the double sided tape and firmly pressing the top material into place.
2.4. Experiments for optimization of the 9-well sample chips materials
To characterize the different combinations of materials used to seal the sample chip base and top, the wells were filled with blank and positive solutions. Blank solutions consisted of 20 mM HEPES + 1 mg/ml BSA (HEPES/BSA buffer). Positive controls were either 62.5 nM FITC-SNAP-25 or a dose response range between 0.25 and 31.25 nM FITC-SNAP-25 in HEPES/BSA buffer. The samples were then loaded onto the 9-well chips (12 μl/well) and images were taken with the EL-CCD detector.
2.5. FRET assay for LC-A and BoTN-A activity detection
The FITC/DABCYL-SNAP-25 FRET-based assay used in this study for the detection of BoTN-A full toxin and its Light Chain (LcA) were optimized following the manufacturers instructions. Note, since Biosafety II conditions are required for use of the full toxin (BoTN-A) initial studies utilized LcA which could be used under the Biosafety I rating of the FDA Office of Science and Engineering Laboratories. Typically, a range of LcA concentrations were first prepared between 0.08 and 20 nM in 20 mM HEPES + 0.3 mM ZnCl + 1 mg/ml BSA and 1.25 mM DTT (HEPES/Zn/BSA/DTT buffer), in MicroAmp tubes (PE Biosystems, Foster City, CA). A negative control (no LcA) was included in each assay. The FITC/DABCYL-SNAP-25 was then added to each MicroAmp tube, to a final concentration of 5 μM per tube. These samples were then incubated, at 37 °C for 2 h, either in the MicroAmp tubes, in cleaned 9-well sample chips, or cleaned 9-well sample chips pre-blocked with 10 mg/mL BSA (overnight at RT). After incubation the samples in the MicroAmp tubes were transferred to both 96-well plates (100 μl), for characterization using a fluorescent plate reader, and the 9-well sample chips (12 μl) for measurement with the EL-CCD detector. Samples incubated in the 9-well sample chips were directly measured using the EL-CCD detector.
Studies using the full toxin BoTN-A were carried out at the Center for Food Safety and Applied Nutrition (CFSAN) under Biosafety II conditions. Unlike LcA, the full toxin required reduction prior to use. This was achieved by preparing 320 nM BoTN A in 20 mM HEPES/0.3 mM ZnCl/10 mM DTT (freshly prepared – note no BSA) and incubating at 37 °C for 30 min. The reduced BoTN-A was then diluted to a range of concentrations between 0.08 and 10 nM using HEPES/Zn/BSA/DTT buffer, in MicroAmp tubes and the activity assays performed exactly as described above for the LcA. This dilution ensures a final DTT concentration no greater than 1.25 mM since concentrations above this level were found to interfere with the assay. All used chips, tips and MicroAmp tubes were properly disposed of in Biohazard reciprocals, sterilized and disposed of in the appropriate manner.
3. Results and discussion
3.1. Electroluminescence-based fluorescence point of care detector design (EL-CCD detector)
The basic configuration of EL-CCD detector is shown schematically in Fig. 1A and B.
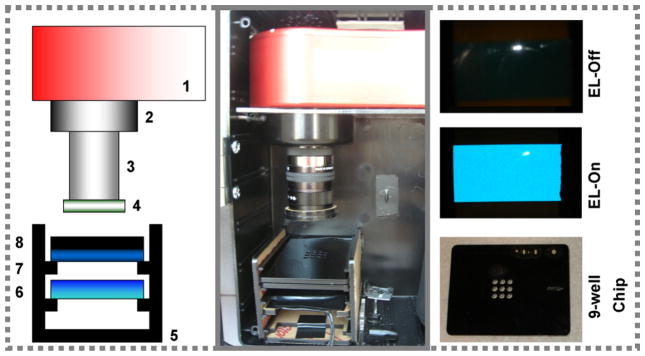
Fig. 1.
The EL-CCD point of care detector. (A) A schematic configuration of the detector and (B) a photograph of the actual detection platform where: Atic-16 CCD camera [1] is equipped with one of two green band pass filters. A 5 mm extension tube [2] is attached to the 12 mm Pentax f1.2 lens [3]. The second green band pass filter B [4] is mounted on the end of the lens. A black acrylic shelf box [5] was designed to hold the EL strip panel [6], the blue band pass filter [7] and the black acrylic 9-well sample chips [8]. (C) Photographs of Royal Blue EL strip off and on and the black acrylic 9-well chips designed to hold aqueous samples for imaging. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
The system consists of two modules, the detection and excitation modules, contained within a black acrylic box (dimensions: H8 in. × L6 in. × W3 in.). The detection module consists of the CCD camera, two green emission filters A and B, a 5 mm extension tube and a Pentax lens. The excitation and sample module was comprised of a black acrylic shelf box designed to hold the interchangeable EL strip panel (a Royal Blue EL used in this work), interchangeable filters (blue band pass excitation filter used in this work) and assay chips. Since the excitation source is in line with the CCD camera the choice of the excitation and emission filters is crucial for blocking excitation light reaching the detector. A cooled astronomy-CCD camera was chosen to reduce the thermal noise generated by the CCD and hence improve the sensitivity by reducing the background signal. The camera was equipped with a 16 bit analog-to-digital converter enabling a dynamic range of 65,536 levels of grayscale. The signal generated from the 9-well sample chips was focused onto the CCD camera using a lens, connected via a C adapter, and a 5 mm extension tube enabling short distance (30 mm) focusing over a small field of view (15 mm × 20 mm). The overall power requirements of the detector were designed to be minimal, with only ~825 mA of consumption and 12 V DC input (power supply or battery) needed to run both the EL and the CCD. The device was designed to be battery operated and 12 V is commonly used for car batteries, cordless drills and other portable devices. Moreover, the CCD camera is also operated on 12 V, so it is a reasonable choice. The use of an external power supply offers several advantages.
First, there are several different AC voltages used internationally (110 V, 220 V, 240 V, etc.), so an external power supply enables the simple change of an outlet adapter as needed. Second, an external power supply may reduce potential electrical noise.
Also, a magnetic levitating fan was used in the camera, to minimize any residual vibration that may occur during longer exposure times.
The BoTN-A FRET-based activity assay used in this study specifically monitors fluorescence from the FITC portion of the FITC/DABCYL-SNAP-25 therefore requiring an excitation wavelength of 494 nm, while monitoring for emission at 523 nm. To achieve 494 nm excitation a Royal Blue EL strip was chosen and Fig. 1C shows an “off” and “on” photo of the EL strip (taken using a Sony Cyber-shot 5.1 MegaPixel color digital camera, minus the blue excitation filter), demonstrating the bright and uniform nature of the EL emitted light. However, unlike the monochromatic light emitted from lasers these EL strips emit over a range of wavelengths, including low levels of green light for our blue EL. Considering the FITC emission is also green such background from the EL could limit the sensitivity, but was readily minimized using a blue band pass excitation filter placed directly above the EL strip. The 9-well sample chip, prepared using the laser cutter in black acrylic, which was used to minimize light scattering and cross talk between wells, is shown in Fig. 1C. The sample chip dimensions are 2.75 in. × 2.125 in. with the nine sample wells confined to a centrally located 0.5 in. × 0.5 in. square. Each well holds 12 μl samples and is loaded using a standard pipette.
3.2. EL-CCD point of care detector performance and 9-Well sample chip optimization
A number of materials were investigated for sealing the 9-well sample chip base and top including glass, COC, Mylar and PC (Table 1). The 9-well sample chips were first prepared with different base materials and the wells filled (12 μl) with either buffer (negative control) or 62.5 nM FITC-SNAP-25 (positive control). Images were captured on the EL-CCD detector using a 60 s exposure and the resulting CCD imaged analyzed in ImageJ. The ImageJ software allows the user to measure the mean intensity from the CCD image of define areas. A study varying the size of the circles used to analyze the images, from ~10 to 60% of the overall well area, revealed a ≤2% variation in the mean intensity determined. A circle encompassing an area 20% of the overall well area was used for the remainder of the data analysis allowing us to avoid the lighter circles that appear around the parameter of the well images and the bubbles that occur when the sample chips are sealed with a top (see later images). Analysis of the mean intensities obtained for the positive and negative sample images determined a relative standard deviation (R.S.D.) between the nine sample wells of 6.2%. Such variation could be further decreased by using improved optics or through a computational approach. Results from the negative control (neg) and positive sample (pos) analysis for the different materials are summarized in Table 1. Mylar was found to have a significant amount of autofluorescence under the excitation conditions resulting in a background signal over double that of the other materials.
Table 1.
Select properties of the materials used in the 9-well sample chip base and top designs
| Material | Total thickness | Fluorescent signal ± R.S.D.a | Optical transparencyb | General comments |
|---|---|---|---|---|
| PC | 125 μm | 309 ± 5.6 (neg) 1020 ± 9.6 (pos) |
≥300 nm | Hard to scratch, cut with laser cutter or scissors. |
| Mylar | 250 μm | 865 ± 4.4 (neg) 1419 ± 4.6 (pos) |
≥400 nm | Easy to scratch, cut with laser cutter or scissors. |
| Glass | 250 μm | 320 ± 7.1 (neg) 962 ± 6.9 (pos) |
≥350 nm | Hard to scratch, however fragile if apply force, cut with diamond cutter. |
| COC | 250 μm | 335 ± 6.3 (neg) 967 ± 5.3 (pos) |
≥250 nm | Easy to scratch, cut with scissors. |
Measured using the EL-CCD detector, from wells filled with buffer only (neg) and positive FITC-SNAP-25 (pos) samples. The 9-well sample chip was modified with the base material only, no top was attached to the chip.
Absorbance ≤0.2 (relative to an air background) as measured using a Amersham Biosciences UltoSpec 2100 pro UV–vis Spectrometer (GE Healthcare Life Sciences, Piscataway, NJ).
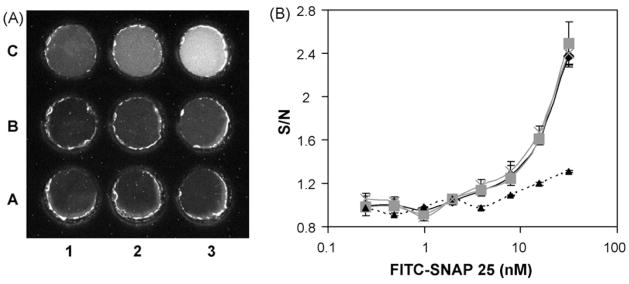
Solutions from the serial dilution of the FITC-SNAP-25 were then loaded into the 9-well sample chip and the resulting EL-CCD image recorded. Fig. 2A shows the CCD image, following a 120 s exposure, and clearly demonstrates a decrease in fluorescence intensity as the concentration of FITC-SNAP-25 decreases in solution. Dose-response curves of the signal-over-noise (S/N) ratio, where the noise is the buffer blank (well A1), are plotted as a function of the FITC-SNAP-25 concentration for the different base materials (Fig. 2B). One-way analysis of variance (ANOVA) was performed (in Excel) at the highest concentration of FITC-SNAP-25 tested to compare the different base materials, using a critical value of 0.05. When compared to the COC there was no significant difference between the COC, PC (P = 0.398) and glass (P = 0.714) bases, however the Mylar base, with its inherent background fluorescence, was much less sensitive, resulting in significantly (P = 5 × 10−5) lower S/N ratios at the higher concentrations of the FITC-SNAP-25. ANOVA analysis is based on the assumption of normally distributed errors and equal variances within each group/condition. If there are outliers in the data, or large differences in error variances between the different groups/conditions, then these may inflate the ANOVA variance estimates and create significant power loss, however, the strong differences shown in Fig. 2 suggest that the significant differences that were detected by the ANOVA are real.
Fig. 2.
The optimization of the 9-well chip base material using FITC-SNAP-25 peptide. (A) CCD image of FITC-SNAP-25 dose response solutions added to the 9-well black chip taken using the EL-CCD detector, 120 s exposure. The COC based wells were loaded with FITC-SNAP-25 at concentrations ranging from 0 nM (A1) to 31.25 nM (C3). (B) The dose response curves plotted versus signal-over-noise (therefore blank = 1) for the different chip base materials; COC (black diamonds/solid line), PC (gray squares/solid line), Mylar (black triangles/dashed line) and glass (gray crosses/dashed line). Standard deviations were determined from n ≥ 3 chips.
In certain applications, for example when measuring toxic substances is desired, the ability to seal the top of the wells may be desirable both to prevent sample evaporation and allow for sample incubation prior to or sample storage after imaging. For this chip top study 9-well chips with COC or PC bases were filled with 62.5 nM FITC-SNAP-25 and then sealed with either glass, COC, PC or Mylar and the mean intensities calculated from the resulting CCD image. As observed in the chip base optimization studies the Mylar top coupled with either the COC or PC base produces a higher signal than the other materials. The COC, PC and glass tops all produce a slight drop (9.5% ± 1.8%) in signal intensity relative to a sample chip with no top.
COC provides the best optical transparency, closely followed by PC, see Table 1. In terms of ease of use and robustness of the base material PC was readily cut to the size of the chips using the laser cutter and was much harder to scratch than the COC material. The optical transparency of the 9-well chip top at wavelengths ≤450 nm is not as crucial as the chip base (since fluorescence emission is at longer wavelengths) and PC is the ideal material for this application given its ease of use and robustness. For the remainder of these studies COC was chosen as the base material and where required PC tops were used to seal the samples in the chip.
3.3. EL-CCD point of care detection of botulinum toxin activity
A schematic of the FRET activity assay, used to demonstrate the utility of the EL-CCD detector, is show in Fig. 3A. A short chain peptide sequence of the BoTN-A binding protein SNAP-25 is labeled with the FRET pair (FITC/DABCYL). The close proximity of the DAB-CYL causes significant quenching of the FITC fluorescence. Binding of BoTN-A or its light chain derivative LcA causes cleavage of the peptide sequence and a concomitant increase in FITC fluorescence at 523 nm.
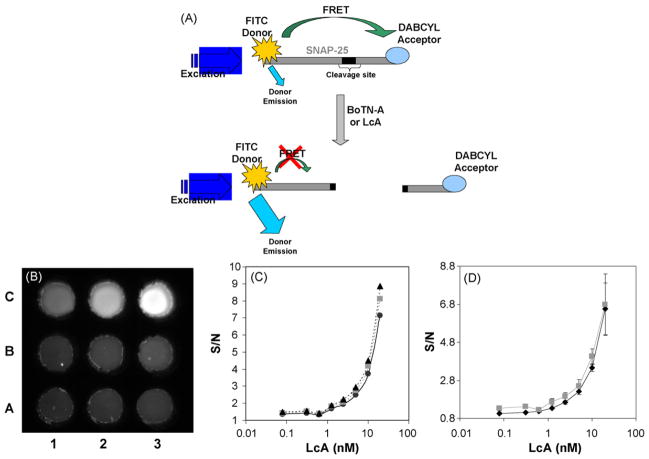
Fig. 3.
The LcA FRET assay. (A) Schematic of the SNAP-25 FRET assay for BoTN-A detection. The SNAP-25 peptide substrate for BoTN-A is labeled with the FITC donor/DABCYL acceptor FRET pair. Interaction of the substrate with either the full toxin BoTN-A or the light chain LcA derivative results in cleavage of the peptide sequence, disrupting the FRET and resulting in increased FITC donor emission. (B) CCD image of quenched 5 μM FITC/DABCYL-SNAP-25 peptide exposed to different concentrations of LcA (ranging from A1: 0 nM to C3: 20 nM), added to the 9-well black chip after a 2 h exposure at 37 °C in MicroAmp tubes, taken using the EL-CCD detector. (C) LcA dose response curves as a function of the CCD image exposure time, 30 s (black circle/solid line), 60 s (gray squares/dashed line), 120 s (black triangles/dashed line), standard deviations were determined from n ≥ 3 readings, on the same chip. (D) LcA dose response curves as determined from the CCD image (gray squares) and measurements taken of the same solutions on a fluorescent plate reader (black diamonds). Standard deviations were determined from n ≥ 4 readings, on different chips.
The FITC/DABCYL-SNAP-25 was prepared at a fixed 5 μM concentration and exposed to LcA at concentrations varying from 0.078 to 20 nM, a negative control with peptide but no LcA was also performed. Following the 2 h incubation at 37 °C in the MicroAmp tubes, the samples were loaded into either 96-well plates for measurement with the fluorescent plate reader or the 9-well COC based chip for detection with the EL-CCD detector. Fig. 3B shows the resulting CCD image taken using the EL-CCD detector. Clearly, as the concentration of the LcA increases from 0 (well A1) through 20 nM (well C3) the fluorescence signal from the FITC label increases, indicating cleavage of the FITC/DABCYL-SNAP-25 by the action of the LcA. The CCD images were analyzed using ImageJ and the resulting dose-response plotted as S/N versus LcA concentration (on a log scale). Fig. 3C shows the dose-response plot as a function of the CCD exposure time. Increasing the exposure time appears to have only limited improvement of the S/N ratio. This observation is contrary to studies using FITC-SNAP-25 and fluorescein solutions, where the S/N was found to significantly improve as a function of the exposure time (data not shown). This apparent discrepancy is not altogether surprising considering the nature of the backgrounds used for the different experiments. In the case of the LcA activity assay the background consists of a fluorescent species, the FITC/DABCYL-SNAP-25 with no LcA, unlike the FITC-SNAP-25 and fluorescein studies where the background is simply buffer (not fluorescent). Therefore the S/N improvement is likely to be more pronounced when the background is non-fluorescent versus fluorescent, since signal from the fluorescent background will also increase as a function of the exposure time. Dose-response curves taken using the EL-CCD detector were compared to those measured with a fluorescent plate reader, both plotted as S/N versus LcA concentration, shown in Fig. 3D. There is no significant (P = 0.843 with ANOVA) difference between the two measurements demonstrating that the EL-CCD detector is equivalent to the plate reader for this particular activity assay using LcA. The limit of detection (LOD) was determined to be 1.25 nM and represents a measured concentration that generates a signal above 3 standard deviations of the background signal. This 1.25 nM LOD is equivalent to 62.5 ng/ml of the LcA and is comparable to similar studies by Schmidt and Stafford (Schmidt and Stafford, 2003) using a SNAP-25 peptide substrate labeled with a different FRET pair (1 nM; 60 ng/ml), measured using a standard fluorimeter. When compared to a number of immunoassay formats including ELISAs (Han et al., 2007), some commercial lateral flow test kits (Gessler et al., 2007) and fluorescent immunoassays (Sapsford et al., 2005) the LOD is in the same nanogram per milliliter range (2–50 ng/ml). Some more sophisticated detection schemes, such as those combining paramagnetic beads and electrochemiluminescent immunoassays, have measured down to 50 pg/ml of BoTN-A (Rivera et al., 2006).
These results suggest that the EL-CCD point of care detector was equivalent to the fluorescent plate reader. Next we performed activity assays using the LcA and full BoTN-A toxin, in these studies the FITC/DABCYL-SNAP-25/toxin incubation was carried out in either: (1) the MicroAmp reaction tubes, (2) COC base/PC top 9-well chips or (3) COC base/PC top 9-well chips pre-blocked with at 10 mg/ml BSA solution. The results of the LcA and full BoTN-A activity assays are summarized in Fig. 4, respectively.
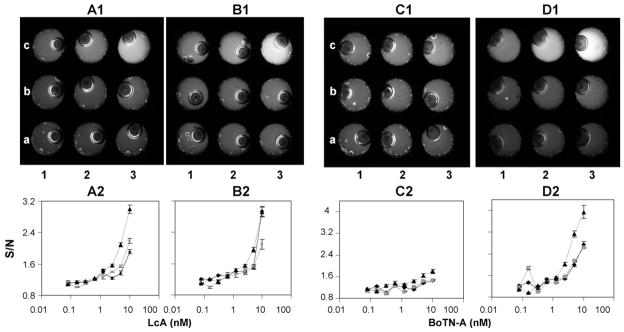
Fig. 4.
FRET assays using COC-base/PC top 9-well chips; study of incubation method. (A1), (B1), (C1) and (D1) are CCD images of the 5 μM FITC/DABCYL-SNAP-25 peptide exposed to different concentrations of LcA (A1 and B1) or BoTN-A (C1 and D1), added to the 9-well black chip after a 2 or 24 h exposure at 37 °C in the MicroAmp tubes. LcA and BoTN-A concentrations ranged from A1: 0 nM to C3: 10 nM. Note, the bubbles observed in the well result when the PC top is attached to the chip. (A2) and (B2) are the LcA dose response curves after 2 and 24 h, respectively, as a function of the FRET reaction incubation method. While (C2) and (D2) are the BoTN-A dose response curves after 2 and 24 h, respectively. Incubation in MicroAmp tubes prior to addition to chips for detection (black triangles/dashed line), incubation in COC base/PC top 9-well black chips, and incubation in COC base/PC top 9-well black chips previously blocked overnight with 10 mg/ml BSA. Standard deviations were determined from n ≥ 3 separate readings of the chips.
In the case of the LcA both the reaction tube and in-chip incubations showed increases in the FITC/DABCYL-SNAP-25 substrate FITC fluorescence as the toxin concentration increased from 0 to 10 nM, following the initial 2 h incubation (Fig. 4A1 and A2). Incubation of the activity assay in the MicroAmp reaction tubes resulted in an LOD of 0.625 nM which decreased to 1.25 nM for the in-chip incubations. The in-chip incubated samples showed a slight improvement in LOD following a 24 h incubation at 37 °C (Fig. 4B1 and B2), increasing to 0.625 nM. The reactions incubated in the MicroAmp tubes, however, showed no improvement following 24 h of incubation. The results reveal a dampening of the S/N ratio for in-chip incubated samples relative to the MicroAmp tubes (Fig. 4A1). One-way ANOVA (critical value 0.05) analysis at the highest LcA concentration revealed a significant difference (P = 0.026) between the two incubation methods. This maybe a result of non-specific binding of either the FITC/DABCYL-SNAP-25 or the toxin inside the chip sample wells leading to inactivation, although the BSA blocked chips did not seem to elevate this issue (P = 0.032 versus tubes). Analogous experiments with the full toxin BoTN-A revealed a limited increase in FITC fluorescence following a 2 h incubation (4C1 and 4C2). Only after the 24 h incubation was a significant response (4D1 and 4D2) for the BoTN-A observed, suggesting the full toxin had a slower interaction with the peptide substrate as compared to the LcA sub unit of the toxin. Incubation of the activity assay in the MicroAmp tubes resulted in a 2 h LOD of 2.5 nM which increased significantly to 0.31 nM after 24 h. As with the LcA, in-chip incubations resulted in a dampening of the S/N ratio and a loss of sensitivity with LODs of 2.5 nM even after a 24 h incubation.
4. Conclusion
An EL-CCD detector platform has been developed to demonstrate the effective combination of EL spatial illumination and CCD detection. In this current study the EL-CCD detector was applied to the detection of BoTN-A using a FRET-based FITC/DABCYL-SNAP-25 activity assay. LODs for LcA were between 0.625 and 1.25 nM (31–62 ng/ml), were achieved after only a 2 h incubation, and were comparable to a standard fluorescent plate reader. The full toxin, BoTN-A, required a 24 h incubation resulting in a LOD of 0.313 nM (45 ng/ml; assuming a MW 150 kDa for the full toxin), thus demonstrating the potential application of EL-based excitation for fluorescent biodetection and providing a simple sensitive method for BoTN-A activity detection.
The prototype EL-CCD detector platform is portable, with all the modular components enclosed in a small plastic container, and controlled using a laptop computer via a USB connection. Black acrylic 9-well sample chips with COC bases and PC tops (available should the wells require sealing) required only 12 μl to fill unlike the 100 μl needed for the plate reader analysis, making the platform suitable for small sample volume analysis. The relative simplicity, ease of use and physical properties of these EL strips makes them ideal candidates for excitation sources for a number of applications, particularly as sensing platforms become smaller and more integrated, such as lab-on-a-chip platforms and point of care detectors. Future plans include using blue, green and red ELs to excite a wider range of commercial fluorophores, including quantum dots to future increase the versatility of the EL-CCD detector.
Acknowledgments
We would like to thank Dr. Nikolay Sergeev and Mr. Sean Wilson for their technical assistance. This work was supported in part by the Office of Public Health emergency Preparedness (OPHEP) IAG 224-05-655 (to A. Rasooly) and by FDA contract HHSF223200610765P (to Dr. Yordan Kostov).
References
- Attree O, Guglielmo-Viret V, Gros V, Thullier P. Journal of Immunological Methods. 2007;325:78–87. doi: 10.1016/j.jim.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Barr JR, Moura H, Boyer AE, Woolfitt AR, Kalb SR, Pavlopoulos A, McWilliams LG, Schmidt JG, Martinez RA, Ashley DL. Emerging Infectious Diseases. 2005;11:1578–1583. doi: 10.3201/eid1110.041279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, De Camilli P, Sudhof TC, Niemann H, Jahn R. Nature. 1993;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- Bruno AE, Barnard S, Rouilly M, Waldner A, Berger J, Ehrat M. Analytical Chemistry. 1997;69:507–513. doi: 10.1021/ac960855n. [DOI] [PubMed] [Google Scholar]
- Burkert K, Neumann T, Wang J, Jonas U, Knoll W, Ottleben H. Langmuir. 2007;23:3478–3484. doi: 10.1021/la063122z. [DOI] [PubMed] [Google Scholar]
- Capitan-Vallvey LF, Asensio LJ, Lopez-Gonzalez J, Fernandez-Ramos MD, Palma AJ. Analytical Chimica Acta. 2007;583:166–173. doi: 10.1016/j.aca.2006.09.052. [DOI] [PubMed] [Google Scholar]
- CDC. Centers for Disease Control and Prevention; Atlanta, GA: 1998. [Google Scholar]
- Claycomb RW, Delwiche MJ. Biosensors & Bioelectronics. 1998;13:1173–1180. doi: 10.1016/s0956-5663(98)00080-3. [DOI] [PubMed] [Google Scholar]
- Dong M, Tepp WH, Johnson EA, Chapman ER. Proceedings of the National Academy Science USA. 2004;101:14701–14706. doi: 10.1073/pnas.0404107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JL, Eliasberg SJ, Edmonds P, Harrison MA. Journal of Food Protection. 2004;67:203–206. doi: 10.4315/0362-028x-67.1.203. [DOI] [PubMed] [Google Scholar]
- Gaunt PS, Kalb SR, Barr JR. Journal of Veterinary Diagnostic Investigation. 2007;19:349–354. doi: 10.1177/104063870701900402. [DOI] [PubMed] [Google Scholar]
- Gessler F, Hampe K, Bohnel H. Applied and Environmental Microbiology. 2005;71:7897–7903. doi: 10.1128/AEM.71.12.7897-7903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler F, Hampe K, Schmidt M, Bohnel H. Diagnostic Microbiology and Infectious Disease. 2006;56:225–232. doi: 10.1016/j.diagmicrobio.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Gessler F, Pagel-Wieder S, Avondet MA, Bohnel H. Diagnostic Microbiology and Infectious Disease. 2007;57:243–249. doi: 10.1016/j.diagmicrobio.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Golden J, Shriver-Lake L, Sapsford K, Ligler F. Methods. 2005;37:65–72. doi: 10.1016/j.ymeth.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Guglielmo-Viret V, Attree O, Blanco-Gros V, Thullier P. Journal of Immunological Methods. 2005;301:164–172. doi: 10.1016/j.jim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Han SM, Cho JH, Cho IH, Paek EH, Oh HB, Kim BS, Ryu C, Lee K, Kim YK, Paek SH. Analytical Chimica Acta. 2007;587:1–8. doi: 10.1016/j.aca.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Kalb SR, Moura H, Boyer AE, McWilliams LG, Pirkle JL, Barr JR. Analytical Biochemistry. 2006;351:84–92. doi: 10.1016/j.ab.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Keller JE, Nowakowski JL, Filbert MG, Adler M. Journal of Applied Toxicology. 1999;19 (Suppl 1):S13–S17. doi: 10.1002/(sici)1099-1263(199912)19:1+<s13::aid-jat607>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Knecht BG, Strasser A, Dietrich R, Martlbauer E, Niessner R, Weller MG. Analytical Chemistry. 2004;76:646–654. doi: 10.1021/ac035028i. [DOI] [PubMed] [Google Scholar]
- Krishnan M, Burke DT, Burns MA. Analytical Chemistry. 2004;76:6588–6593. doi: 10.1021/ac0488356. [DOI] [PubMed] [Google Scholar]
- Ligler FS, Sapsford KE, Golden JP, Shriver-Lake LC, Taitt CR, Dyer MA, Barone S, Myatt CJ. Analytical Sciences. 2007;23:5–10. doi: 10.2116/analsci.23.5. [DOI] [PubMed] [Google Scholar]
- Ligler FS, Taitt CR, Shriver-Lake LC, Sapsford KE, Shubin Y, Golden JP. Analytical and Bioanalytical Chemistry. 2003;377:469–477. doi: 10.1007/s00216-003-1992-0. [DOI] [PubMed] [Google Scholar]
- Liu W, Montana V, Chapman ER, Mohideen U, Parpura V. Proceedings of the National Academy Science USA. 2003;100:13621–13625. doi: 10.1073/pnas.2233819100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Danielsson B. Analytical Chimica Acta. 2007;587:47–51. doi: 10.1016/j.aca.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Mac Sweeney MM, Bertolino C, Berney H, Sheehan M. Conference Proceedings IEEE Engineering in Medicine and Biology Society. 2004;3:1960–1963. doi: 10.1109/IEMBS.2004.1403579. [DOI] [PubMed] [Google Scholar]
- Mangru S, Bentz BL, Davis TJ, Desai N, Stabile PJ, Schmidt JJ, Millard CB, Bavari S, Kodukula K. Journal of Biomolecular Screening. 2005;10:788–794. doi: 10.1177/1087057105278927. [DOI] [PubMed] [Google Scholar]
- Moehrs S, Del Guerra A, Herbert DJ, Mandelkern MA. Physics in Medicine and Biology. 2006;51:1113–1127. doi: 10.1088/0031-9155/51/5/004. [DOI] [PubMed] [Google Scholar]
- Nowakowski A, Wang C, Powers DB, Amersdorfer P, Smith TJ, Montgomery VA, Sheridan R, Blake R, Smith LA, Marks JD. Proceedings of the National Academy Science USA. 2002;99:11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parpura V, Chapman ER. Croatian Medical Journal. 2005;46:491–497. [PubMed] [Google Scholar]
- Peruski AH, Johnson LH, 3rd, Peruski LF., Jr Journal of Immunological Methods. 2002;263:35–41. doi: 10.1016/s0022-1759(02)00030-3. [DOI] [PubMed] [Google Scholar]
- Rivera VR, Gamez FJ, Keener WK, White JA, Poli MA. Analytical Biochemistry. 2006;353:248–256. doi: 10.1016/j.ab.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Roda A, Manetta AC, Portanti O, Mirasoli M, Guardigli M, Pasini P, Lelli R. Luminescence. 2003;18:72–78. doi: 10.1002/bio.708. [DOI] [PubMed] [Google Scholar]
- Ruiz-Martinez MC, Berka J, Belenkii A, Foret F, Miller AW, Karger BL. Analytical Chemistry. 1993;65:2851–2858. doi: 10.1021/ac00068a023. [DOI] [PubMed] [Google Scholar]
- Sapsford KE, Taitt CR, Loo N, Ligler FS. Applied and Environmental Microbiology. 2005;71:5590–5592. doi: 10.1128/AEM.71.9.5590-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schantz EJ, Johnson EA. Lancet. 1990;335:421. doi: 10.1016/0140-6736(90)90263-5. [DOI] [PubMed] [Google Scholar]
- Schantz EJ, Johnson EA. Microbiological Reviews. 1992;56:80–99. doi: 10.1128/mr.56.1.80-99.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Nature. 1992a;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Poulain B, Rossetto O, Benfenati F, Tauc L, Montecucco C. The EMBO Journal. 1992b;11:3577–3583. doi: 10.1002/j.1460-2075.1992.tb05441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Rossetto O, Catsicas S, Polverino de Laureto P, DasGupta BR, Benfenati F, Montecucco C. Journal Biological Chemistry. 1993;268:23784–23787. [PubMed] [Google Scholar]
- Schmidt JJ, Stafford RG. Applied and Environmental Microbiology. 2003;69:297–303. doi: 10.1128/AEM.69.1.297-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Eblen BS, Bull RL, Burr DH, Whiting RC. Applied and Environmental Microbiology. 2005;71:3935–3941. doi: 10.1128/AEM.71.7.3935-3941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Ferreira JL, Eblen BS, Whiting RC. Applied and Environmental Microbiology. 2006;72:1231–1238. doi: 10.1128/AEM.72.2.1231-1238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shone CC, Quinn CP, Wait R, Hallis B, Fooks SG, Hambleton P. European Journal Biochemistry. 1993;217:965–971. doi: 10.1111/j.1432-1033.1993.tb18327.x. [DOI] [PubMed] [Google Scholar]
- Sohn YS, Goodey A, Anslyn EV, McDevitt JT, Shear JB, Neikirk DP. Biosensors & Bioelectronics. 2005;21:303–312. doi: 10.1016/j.bios.2004.08.050. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, De Camilli P, Niemann H, Jahn R. Cell. 1993;75:1–4. [PubMed] [Google Scholar]
- Svitel J, Surugiu I, Dzgoev A, Ramanathan K, Danielsson B. Journal of Materials Science and Material Medicine. 2001;12:1075–1078. doi: 10.1023/a:1012810527291. [DOI] [PubMed] [Google Scholar]
- Takei M, Kida T, Suzuki K. Applied Radiation and Isotopes. 2001;55:229–234. doi: 10.1016/s0969-8043(00)00392-4. [DOI] [PubMed] [Google Scholar]
- Taylor TB, Winn-Deen ES, Picozza E, Woudenberg TM, Albin M. Nucleic Acids Research. 1997;25:3164–3168. doi: 10.1093/nar/25.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbe AG, de Grooth BG, Greve J, Liberti PA, Dolan GJ, Terstappen LW. Cytometry. 2001;43:31–37. [PubMed] [Google Scholar]
- Tohda K, Gratzl M. Analytical Sciences. 2006;22:937–941. doi: 10.2116/analsci.22.937. [DOI] [PubMed] [Google Scholar]
- Trimble WS. Journal Physiology Paris. 1993;87:107–115. doi: 10.1016/0928-4257(93)90004-d. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi K, Jinno N, Nakajima R. Analytical Chemistry. 2005;77:1684–1688. doi: 10.1021/ac040133t. [DOI] [PubMed] [Google Scholar]