Abstract
This essay gives the author’s personal account on the development of concepts underlying single-particle reconstruction, a technique in electron microscopy of macromolecular assemblies with a remarkable record of achievements as of late. The ribosome proved to be an ideal testing ground for the development of specimen preparation methods, cryo-EM techniques, and algorithms, with discoveries along the way as a rich reward. Increasingly, cryo-EM and single-particle reconstruction, in combination with classification techniques, is revealing dynamic information on functional molecular machines uninhibited by molecular contacts.
1. Introduction
Cryo-electron microscope (EM) single-particle reconstruction methods have come a long way, to a level of perfection not imagined only 14 years ago, when 25-Å cryo-EM density maps of the Escherichia coli ribosome obtained with this methods were published (Frank et al. 1995; Stark et al. 1995). The appearance of these articles in my opinion set a milestone (see also Moore, 2009) in the development of single-particle cryo-EM, as the ribosome came to be the testing ground for these techniques, due to its high stability and contrast – both related to the large RNA content – and its relatively large size. Stability is required as it creates conformational uniformity among the many copies of the molecule whose images are combined in a three-dimensional (3D) image. While this requirement has been mitigated to some extent by recent developments of powerful classification methods, to be outlined further on, it was initially a conditio sine qua non. Both contrast and size of the molecule are important determinants of the signal obtained in aligning images by cross-correlation (Saxton & Frank, 1976) and thereby of the accuracy with which the alignment can be achieved. This accuracy, in turn, is a critical factor in determining the resolution of a two-dimensional (2D) average and, by implication, of the density map generated by 3D reconstruction.
Another general remark is in order. As a technique, 3D reconstruction of biological objects is sharply split into two methodologies with different data collection and computational strategies (Frank et al. 2002): following one methodology, an individual object, say a mitochondrion, is reconstructed from multiple projections obtained by tilting the object in the EM (electron tomography), and following the other one (electron crystallography, understood in the most general sense of this term), an object that exists in multiple copies of identical structure is reconstructed from a large number (~103 – 106) of projections originating from any of these multiple copies. The latter approach has the advantage of allowing dose reduction without sacrificing resolution, but beyond a certain size range, biological objects are no longer found in identical ‘copies,’ hence, electron tomography becomes the only resort.
In this essay, I would like to give a personal account retracing the development of single-particle techniques in EM over the span of 30 years, with a focus on the most important challenges encountered and concepts advanced in the field. Due to the explosive development of these techniques in recent years, reflected by the increasing number of relevant methods papers in the Journal of Structural Biology and increasing visibility of ensuing biological results in the general literature, it is impossible to review the field in its entirety – the reader is referred to recent review- or introductory literature (see Frank, 2006; Glaeser et al. 2006). In a way, the present account complements a recent retrospective by the pioneers of cryo-EM, Ken Taylor and Bob Glaeser (Taylor & Glaeser, 2008), which recounts the development of the cryo-EM technique, the problems of radiation damage and the challenges of specimen preparation.
The terms single-particle averaging and single-particle reconstruction refer to quantitative ways of determining the structure of macromolecules from micrographs showing them as a collection of isolated, unattached particles. These terms were coined well before physical methods of probing single molecules (such as force measurements and atomic force imaging) had been developed, and so there is a potential for confusion. As a matter of fact, a typical reconstruction uses not one but tens of thousands of low-dose images of single molecules that occur in many ‘copies,’ all with nearly identical structure.
Three recent studies, one on the ‘unlocking’ action of eEF2 during messenger RNA (mRNA)–transfer RNA (tRNA) translocation triggered by GTP hydrolysis (Taylor et al. 2007), the other two on the GTPase mechanism of EF-Tu (Villa et al. 2009; Schuette et al. 2009), illustrate the current power of cryo-EM combined with single-particle reconstruction: when augmented by classification and flexible fitting, the technique is able to reveal the dynamics of molecular interactions at the functional centers of the ribosome. A look at the recent literature (e.g., Williams et al. 2009) amply demonstrates that challenges posed by the quest to understand other molecular machines with comparable complexity are now in reach, as well.
2. The detection criterion and the limits of cross-correlation alignment
The main tool for aligning two noisy EM images of a biological object is the cross-correlation function (Fig. 1; Frank, 1970; Langer et al. 1970). (Even though some fancier functions have subsequently been introduced, the main arguments to be made below remain valid.) The potential of the single-particle averaging approach in EM can be inferred from the observation that the peak of the cross-correlation function, even though its width is comparable with the resolution of the images, can be located with much higher precision. The reason is that theoretically, this peak is identical with the autocorrelation function of the point spread function associated with the imaging process (Frank, 1975a), which is a smooth centrosymmetric function whose center can be determined with a precision of approximately one-fifth of the resolution distance, if not better. This is, in fact, a corollary of Michael Rossmann’s later observation (Rossmann, 2000) that the precision of fitting an X-ray structure into a cryo-EM density map is much better (an estimated factor of 5) than the resolution.
Fig. 1.
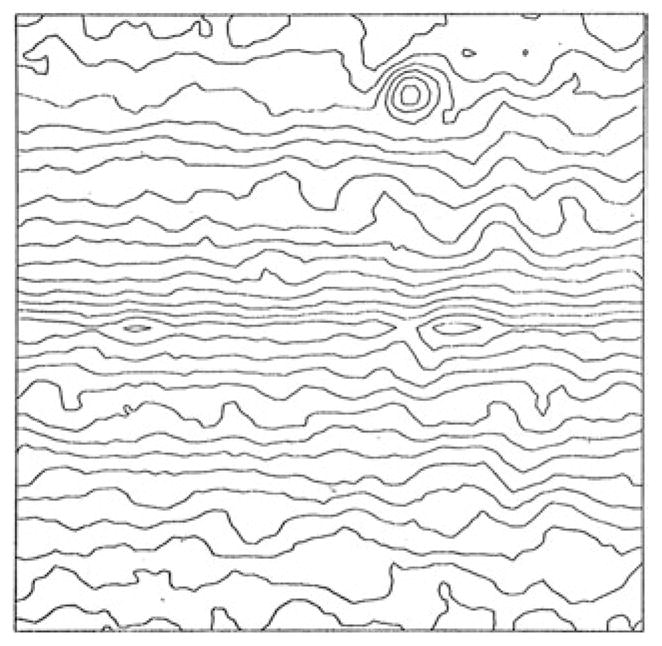
Cross-correlation function, showing a peak indicating the relative position of two successive images taken of the same carbon film. Data reproduced from Frank (1970).
Much of the noise in the image is due to the need to image the molecule at very low doses, to mitigate radiation damage. The extent and deleterious effects of radiation damage were generally recognized (see Beer et al. 1974) following Bob Glaeser’s earlier ‘fade-out’ diffraction studies with α-valine (Glaeser, 1971) and led to Unwin and Henderson’s pioneering study of the purple membrane protein (Unwin & Henderson, 1975; Henderson & Unwin, 1975). As the electron dose is lowered, the signal-to-noise (SNR) ratio of the image decreases and so does the SNR ratio in the cross-correlation function and with it our ability to locate the cross-correlation peak with sufficient certainty. A principal question that had to be resolved in the beginning was the range of parameters affecting the SNR of the cross-correlation function, namely particle size D, contrast c, critical dose pcrit and resolution d (expressed as length), for which particle alignment is feasible. The answer was formulated, as a relationship between these quantities, in a paper I coauthored with Owen Saxton during the time I spent at the Cavendish Laboratory (Saxton & Frank, 1976):
The result clearly indicated that alignment of raw images of two molecules embedded in ice presenting the same orientation was feasible for molecules in a biologically interesting size range, at a resolution that would give significant structural information and with a dose low enough (below pcrit, a value determined from experiments such as Bob Glaeser’s) to avoid radiation damage. What might be called ‘biologically interesting’ is the approximate size range of molecular machines (~150 Å, e.g., the transcription complex, to ~1500 Å, e.g., the nuclear pore complex), that is, complex assemblies formed by multiple binding partners that perform a biological function in a processive manner (Alberts, 1998). It is the large size and function-related conformational variability of such assemblies that pose great hurdles to the application of X-ray crystallography.
This relationship, based on rough estimates, was later revisited and refined by Richard Henderson (1995) as he investigated the feasibility of determining the structure of single molecules lacking symmetry using neutrons, electrons or X-rays, making use of the experimental scattering data in his estimation. However, the much more simplistic relationship I formulated together with Owen Saxton 20 years earlier gave the green light to forge ahead with the development of software that would allow the formulation and execution of complex processing paths for alignment, averaging and reconstruction of biological macromolecules from single-particle images.
3. SPIDER – software for single-particle averaging and reconstruction
In exploring and realizing the single-particle approach, the need for a versatile software system in which procedures could be formulated by stringing together basic commands for operations on images (such as ‘mask,’ ‘shift,’ ‘cross-correlate,’ ‘Fourier transform’) into scripts was obvious. Software systems existing at the time addressing the needs of the EM community, such as ‘EM’ in Martinsried (Hegerl & Altbauer, 1982) and the MRC suite of programs (see later documentation by Crowther et al. 1996), were of the ‘old-school’ kind, in the sense that they were large single-standing programs, where addition of new functionality required changes and recompilation of the entire code. The key new requirement was modularity – each module should address a single operation on the image; it should be small and easily overseeable; it should be independent from all other modules and communicate with them by means of a common database. Such a system existed at the Jet Propulsion Laboratory at the time I worked there as a postdoctoral student, in 1970. VICAR, a modular Fortran-based program (Billingsley, 1970), had been created in the 1960s to process images sent back to earth from space probes (in 1970, Jupiter was the favorite subject). I wrote several subroutines as ‘piggybacks’ of the VICAR system, taking advantage of its highly developed infrastructure in which, among other features, Do-loops and execution-time changes of file names were supported. The result of my effort was, incidentally, a paper on heavy versus light atom discrimination based on a focus series of DNA supplied by Walter Stoeckenius (Frank, 1972). The first incarnation of a truly modular system aimed at the needs of EM was SEMPER, developed by Owen Saxton in the early 1970s but not published until later (Saxton et al. 1979).
The design of SPIDER (System for Processing of Image Data from Electron microscopy and Related fields), whose basic structure was developed by 1978 as a result of a brainstorming with two gifted students, Helen Dowse and Brian Shimkin, addressed the need for modularity and high versatility and included a nested hierarchical calling structure and passing of parameters by keyed document files (Frank et al. 1981a). Extensive contributions to the 3D reconstruction algorithms and the program’s infrastructure were later made by Michael Radermacher (Radermacher et al. 1987; Radermacher, 1988), Pawel Penczek (Penczek et al. 1992, 1994, 1996, 1997), and Ardean Leith in my group. Although written in Fortran, an ancient programming language by today’s standards, SPIDER is still maintained and widely used by the structural biology community.
4. Single molecules versus crystals
Initially, the idea of obtaining structural information from single, i.e., unattached and non-crystallized molecules imaged under conditions of low dose, was greeted with little more than bemused interest. How could structures be solved if not from crystals or highly ordered aggregates of molecules? What could be the possible meaning of results obtained from individual noisy images that had to be forced into a common reference frame?
The prevailing view was that for a molecular structure to be quantitatively characterized, it had to be in crystal, highly ordered form. Indeed, as the technique of cryo-EM was perfected at the EMBL in the early 1980s, it was first exclusively applied to 2D crystals, helices, and spherical viruses. Thus, the community who enthusiastically adopted cryo-EM technology for exploration of native molecular structure made primarily use of concepts and working methods of X-ray crystallography, suitably modified to deal with EM images (e.g., Amos et al. 1982).
In X-ray crystallography, information on amplitudes is concentrated in small spots or layer lines in the Fourier transform, and phases are not supplied directly by the diffractometer but have to be garnered by computation with the aid of additional experiments, employing multiple heavy atom replacements or multiple wavelengths. In contrast, the EM produces actual images, which is tantamount to saying it measures both amplitudes and phases in Fourier space, giving rise to an adage coined by Walter Hoppe’s (see Hoppe, 1983), of the EM as a ‘phase-measuring diffractometer.’
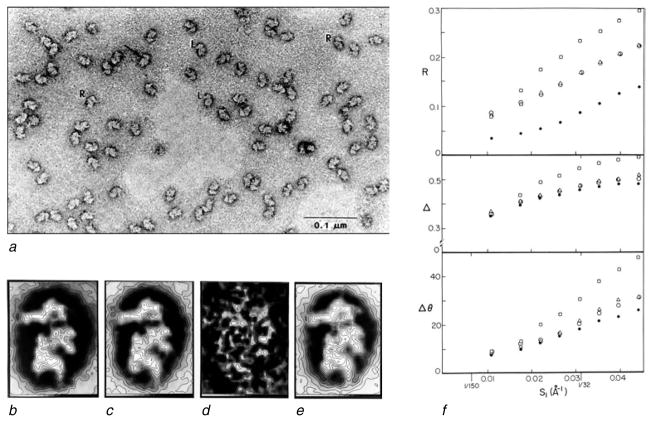
From this Fourier-centric perspective, the idea of applying real-space averaging to images of single molecules (Frank, 1975b) seemed a step back, into territory ruled by subjectivity. It was therefore of crucial importance, in gaining respectability, to demonstrate the reproducibility of the results in a quantitative way, as first done in the study of the HeLa 40S ribosomal subunit (Fig. 2; Frank et al. 1981b). As the outset, two averages from independently drawn images of the negatively stained subunit looked strikingly similar, suggesting a high degree of reproducibility (Fig. 2a, b). The challenge was to measure the reproducibility as a function of spatial frequency. In fact, reproducibility turned out to be a key to the question how spatial resolution could be defined.
Fig. 2.
Averaging of 40S subunits from HeLa cell ribosomes. (a) Electron micrograph of negatively stained subunits, with “L” denoting left-facing views selected for alignment and averaging. (b, c) Averages of halfsets of 40 particles each; (d) variance map; (e) total average; (f) three measures of resolution: R, R factor; Δ, Euclidean distance; δϕ, DPR indicating a resolution of 20 Å at 45°. Data reproduced from Frank et al. (1981b); panels R and Δ in (f) are from unpublished data.
5. The meaning and measurement of resolution
The absence of diffractions spots in a non-periodic structure poses a problem in defining and measuring resolution. Unlike the Fourier transform of periodic structures, where signal-related information is sharply localized, the transform of a non-periodic structure has signal and noise components intermixed in a seemingly indistinguishable way. The answer, still underlying the principle of resolution measurements today, is to split the data set randomly in half and compare the two averages (or 3D reconstructions) over rings (or shells, respectively) with increasing radius in Fourier space using a suitable measure of reproducibility. As to the criterion to be used, I originally proposed three measures (Fig. 2f): the differential phase residual (DPR), which was familiar to X-ray crystallographers and thus had the advantage of being widely appreciated; an ‘R factor’ based on a comparison of Fourier amplitudes; and the (generalized, normalized) Euclidean distance, which is essentially an inverted Fourier ring correlation plot. Of the three measures, two were axed by the editor to enforce the length restrictions of a Science report, keeping only the DPR in the paper, and leaving it to other authors to propose the now-popular Fourier ring/shell correlation (Saxton & Baumeister, 1982; van Heel et al. 1982).
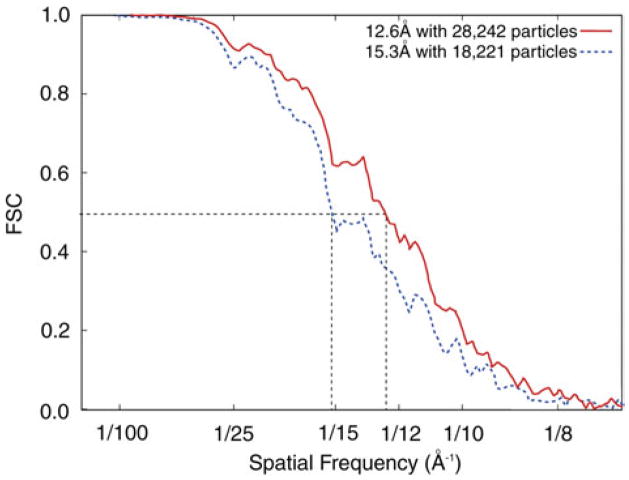
While the typical DPR (Fig. 2f, bottom) starts at 0 and rises like a parabola, indicating increasing phase residuals, the typical Fourier ring or shell correlation curve starts at 1 and falls off in a more or less sigmoidal way, reaching a value of 0 or close to zero asymptotically (Fig. 3). Thus, both curves show that the reproducibility of structural information is generally quite good at low spatial frequencies and poor at high spatial frequencies. To arrive at a single quotable figure, a criterion had to be defined for the acceptable level of reproducibility. Initial controversies (Orlova et al. 1997; Malhotra et al. 1998) have given way to the acceptance of FSC=0·5 as criterion, which was initially introduced by Boettcher et al. (1997) in a study of the hepatitis B virus capsid. (This criterion is, incidentally, quite consistent with the criterion DPR=45° earlier used.)
Fig. 3.
Example of Fourier shell correlation curves, with resolution criterion (FSC=0·5) indicated. Data reproduced from Sengupta et al. (2008).
The half-set-based measurement of resolution has two shortcomings, with opposite effects: one is that it is inherently based on the statistical properties of merely half the data set and thus leads to a pessimistic estimate of resolution, the other is that before being split in half, the entire data set has been processed in the same way, sharing intermediate references in the angular refinement all the way to the end. The halves of the data set are, therefore, not statistically independent, making the resolution estimate overly optimistic (see Grigorieff, 2000). It is unrealistic to expect that the two opposite tendencies, overestimation and underestimation, cancel each other out precisely, but it is comfortable to know that they cooperate to push the result closer to the truth. (To jump ahead in time: there is also now an emerging consensus that evidence of structural features resolved, such as RNA helices (Malhotra et al. 1998), alpha-helices (Boettcher et al. 1997; Conway et al. 1997), beta-sheets (Kong et al. 2004; Baker et al. 2007), or individual phosphorus atoms showing up as ‘bumps’ along RNA strands (LeBarron et al. 2008), are the best criterion for resolution achieved in practice.)
6. Correlation averaging: crystals treated as ordered collections of single particles
Application of correlation functions to EM images of thin crystals proved that most crystals are far from ideal. When a small fragment of the crystal image is cross-correlated with the entire field, the resulting peaks lie on curves, not on straight lines, indicating distortions of the lattice. Occasionally, there are dislocations, as well or changes of local structure. As a result, application of algorithms that assume perfect crystals will yield a blurred average. It is straightforward to see that treatment of the crystal as a collection of single particles, one for each repeating unit, will result in a superior average. First applications of the single-particle approach on images of bacterial surface layers confirmed this expectation (W. Baumeister and J. Frank, 1981, unpublished; Baumeister and Saxton, 1982). One can demonstrate that front and back layers of a collapsed bacterial envelope can be easily separated in the real-space approach (Kessel et al. 1985). However, the most important benefit of the single-particle approach to crystal averaging is that it lends itself to the application of classification (Frank et al. 1988), as further elaborated below.
Another approach to exploit information in images of distorted crystals has become more prevalent, under the name of ‘unbending.’ Instead of using the data directly, boxed out from the image and properly repositioned after alignment, the idea here is to ‘unbend’ the data so that they conform to existing programs that have been designed for ideal lattices (van Heel & Hollenbeck, 1980; Henderson et al. 1986). Unfortunately, computational convenience in this approach leads to the sacrifice of valuable information relating to the clustering of the data due to local changes in conformation (see Frank et al. 1988; Morgan et al. 1992).
In contrast, the single-particle approach has been pursued, with great success, in the characterization and 3D reconstruction of helical polymers such as actins, where changes in local structure appear to be the norm and even seem required for biological function (Egelman, 2007).
7. The need for classification – multivariate data analysis of projections
The need for an objective means of classification was recognized early when 2D averaging of negatively stained biological molecules produced images with somewhat disappointing resolution (Frank et al. 1978; Zingsheim et al. 1980). First of all, classification is a general prerequisite to 2D averaging as it is required to separate molecules in a field into subsets presenting the same view. Since the drum-shaped glutamine synthetase molecules, the subject of the first application of single-particle methods, had a tendency to stand on their circular bases, there was no need for this step in this particular case. However, inspection of these images showed clear evidence of conformational variability, some of it related to radiation damage (Kessel et al. 1980).
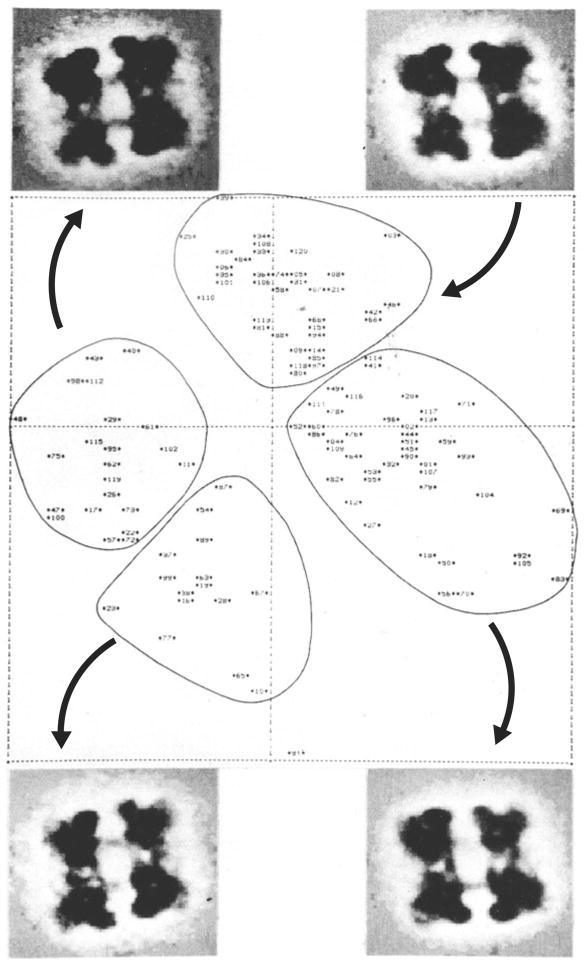
A fortuitous osmosis of know-how, recounted in a recent profile article (Mossman, 2007), led to the introduction of multivariate statistical analysis in the form of correpondence analysis (see Lebart et al. 1984) – a standard technique in the field of laboratory medicine at the time – into single-particle analysis. The reason is that once aligned, images of a molecule can be regarded as comparable multicomponent vectors whose closeness is a reflection of their similarity. Hence, clustering in factorial space reflected the existence of cohesive subsets of molecules with identical or closely similar structure and presenting similar orientations. Hemocyanin molecules, which are oligomeric assemblies of one or more building block proteins, present a fascinating variety of views and thus were ideally suited for development and testing of algorithms of classification. Half-molecules of Limulus polyphemus hemocyanin are composed of four hexamers arranged in rhombus. In one of the first demonstrations of the power of the classification technique, images of this molecule were separated into four groups, according to their ‘flip’ versus ‘flop’ views on the grid and the finer division into two rocking positions in both of these views (Fig. 4; van Heel & Frank, 1981; see Frank, 1984). The unique features of the molecule’s 3D architecture, the twisted arrangement of the four hexamers, which were revealed in a surprising way from patterns of variation in a 2D factor map, were later confirmed and elaborated in much higher-resolution detail (for most recent works, see Martin et al. 2007; Cong et al. 2009).
Fig. 4.
Factor map (factors 1 versus 2) resulting from correspondence analysis of L. polyphemus hemocyanin half molecules. Averages of images falling into demarcated clusters are shown in the four corners (a–d). (a) and (c) are interpreted as two rocking positions of molecules lying on one (“flip”) side, while (b) and (d) relate to rocking positions on the other (“flop”) side. Adapted from Frank (1984).
Following this proof of concept, this and related methods of multivariate data analysis and automated computer techniques of classification based upon them have become indispensible tools in single-particle reconstruction (Frank, 1990; van Heel, 1987; van Heel, 1989; see Frank, 2006).
8. The jump into the third dimension
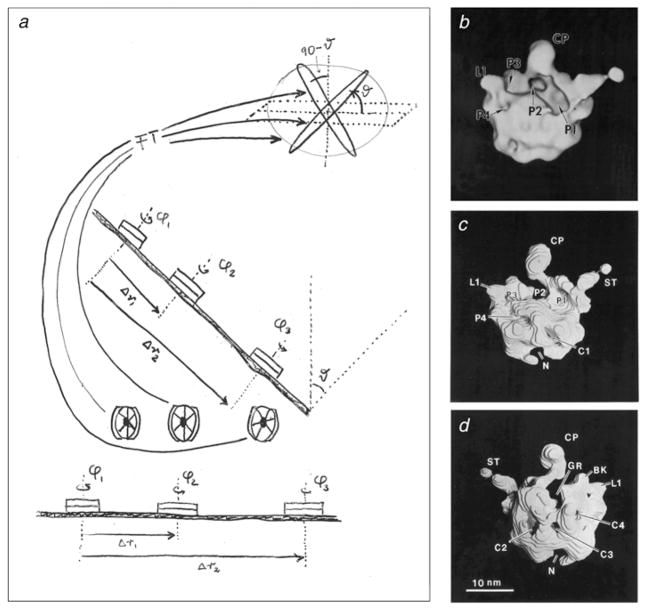
The main difficulty in realizing 3D reconstruction of single particles is to obtain an initial reliable assignment of angles to the particle images. The random–conical reconstruction method (Fig. 5a) (Frank et al. 1978; Radermacher et al. 1987; Radermacher, 1988) achieved this initial assignment by taking advantage of one or more preferential orientations of the particles on the grid and by making use of an additional tilt of the whole field of particles. At once, the extensive book-keeping required in this approach proved worth the investment, made in the years before, in the formal language aspects of the SPIDER software; in particular, its nested calling capability, its versatility in execution-time file naming, and its capability to pass keyed sets of parameters from one operation to the other. First reconstructions of the ribosome following this protocol were obtained for negatively stained specimens in 1987 (Fig. 5b–d; Radermacher et al. 1987) and for cryo-EM specimens in 1991 (Frank et al. 1991; Penczek et al. 1992).
Fig. 5.
Random–conical reconstruction. (a) Principle of the random–conical data collection method. Two images are taken of the same field of molecules. Only molecules are considered that present the same view on the grid. Azimuthal angles are obtained by aligning the images of the untilted micrograph. Thus, with both azimuth and tilt angles known, the Fourier transform of each projection can be properly placed into the 3D Fourier reference frame of the molecule. From J. Frank (unpublished hand-drawing on overhead transparency, 1979). (b–d) Density map of the 50S ribosomal subunit from E. coli, the first 3D reconstruction using the random–conical data collection method. (a) Surface representation of intersubunit face; (b, c) higher-threshold solid model obtained by stacking of contoured slices, viewed from front and back. The subunit was negatively stained with uranyl acetate and air-dried, which accounts for the partial flattening. The ridge of the deep groove running horizontally, termed interface canyon, is created by the helix 69 of 23S rRNA, as later recognized when the X-ray structure of the large subunit was solved. Annotations refer to morphological details; for example, pocket ‘P2’ was suggested to be the peptidyl transferase center and CP the central protuberance. Data reproduced from Radermacher et al. (1987).
An alternative approach to obtaining a de novo assignment of angles is by forming class averages where each class represents particles showing the same view. Their relative angles can then be determined by finding their common lines in Fourier space, following a method first proposed for virus particles (Crowther et al. 1970), or, equivalently, by finding their common one-dimensional projections in real space (‘angular reconstitution’ – van Heel, 1987). Another variant of the common-lines approach was later developed by Penczek et al. (1996). A common thread underlying these closely related approaches is found in the Radon transform whose value in EM-related image processing and 3D reconstruction has been explored by Radermacher (1997).
Evidently, once angles are determined, reconstruction can proceed along paths well treaded before, except for one complication: weighted back-projection, the non-iterative, hence rapid reconstruction technique, required even-spaced angles while single-particle projections are randomly distributed in angular space. (The availability of a fast algorithm was a particularly important consideration at that time.) This problem was solved by the introduction of general weights designed ad hoc for a given geometry (Radermacher et al. 1987; Radermacher, 1988).
Both methods for de novo assignment of angles to projections of an unknown structure will yield an approximate, low-resolution estimate. For an exhaustive exploitation of the data, iterative angular refinement (Penczek et al. 1994) is subsequently used, in the course of which angle assignments are refined on progressively finer angular grid, until stabilized. One of the major problems that had to be solved is the merging of data with different contrast transfer functions (CTFs), toward a final, CTF-corrected reconstruction (Grigorieff, 1998; Penczek et al. 1997; van Heel et al. 2000; Zhu et al. 1997).
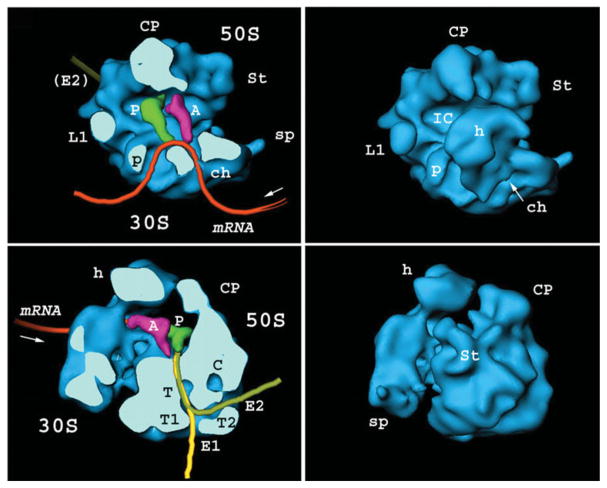
As to the progress in the study of the ribosome structure, further milestones were reached with 25 Å (Fig. 6) (Frank et al. 1995; Stark et al. 1995), 11·5 Å (Gabashvili et al. 2000) and eventually the range of 6–7 Å (LeBarron et al. 2008; Villa et al. 2009; Schuette et al. 2009). The first discoveries reflecting on the binding of tRNA (Agrawal et al. 1996; Stark et al. 1997a) and elongation factors (Agrawal et al. 1998; Stark et al. 1997b) and the conformational changes accompanying mRNA–tRNA translocation (Frank & Agrawal, 2000) – essentially snapshots of a molecular machine in motion – were made just before the X-ray structures of ribosomal subunits appeared (Ban et al. 2000; Wimberly et al. 2000).
Fig. 6.
Cryo-EM reconstruction of the (empty) E. coli ribosome from 4300 particle images, at 25-Å resolution. Cut-open density maps depict the way mRNA, tRNAs, and the polypeptide chain were thought to be positioned during protein synthesis, a model that has stood up to the test of time. Landmarks on the large subunit (50S): CP, central protuberance; St, stalk base; L1, L1 stalk; IC, interface canyon; T, polypeptide exit tunnel; T1, T2, two putative branches of the tunnel exit; E1, E2, corresponding exit sites. Landmarks on the small subunit (30S): h, head; p, platform; ch, mRNA channel. A, P, putative positions of A- and P-site tRNA. Data reproduced from Frank et al. (1995).
Comprehensive protocols are now available for all steps of single-particle reconstruction: specimen preparation (Grassucci et al., 2007), cryo-EM imaging (Grassucci et al., 2008), and image processing using SPIDER (Shaikh et al., 2008).
9. Classification of data with entangled viewing angles and class memberships
Classification in cryo-EM applications was initially concerned with two different, very specific situations: grouping into classes was to be achieved either for different copies of a presumably invariable molecule presenting different orientations – and here the objective of classification was the separation of molecule images according to view – or different conformers of a molecule coexisting in the sample and occurring in essentially the same orientation. The latter situation is typical for negatively stained samples, which often show strong preferences of orientation.
As we have seen, a powerful approach to solve either of these two separate problems was found early on by the introduction of multivariate data analysis into EM. Yet cryo-EM of molecules from a heterogeneous sample presents a more formidable problem as the molecules lack preferences for distinct orientations, so that the variation in the appearance of the projections is the result of two intermingled effects, one being the variation in view angle over the entire angular sphere, the other the coexistence of different states of the molecule (which includes binding states of functional ligands). In this case, application of multivariate data analysis is not helpful, and another solution is needed to be found.
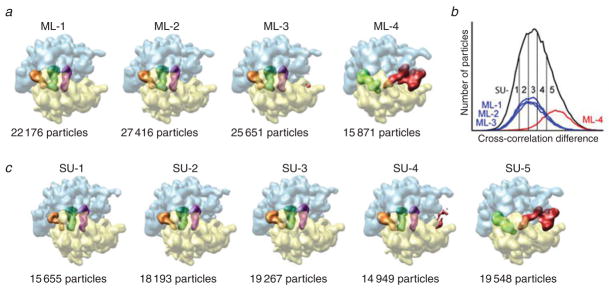
The approaches to the solution fall in two categories, referred to as supervised and unsupervised classification. In the first case, each experimental image is compared with two or more 3D reference maps, and assignment to a class is based on the similarity to those references, as measured by the cross-correlation coefficient (see Valle et al. 2002). In the second case, data are clustered according to their self-similarity and consistency, under the assumption that they originate from one of a known number of 3D structures. The second kind of approaches is obviously much preferred, as the outcome is independent of model assumptions. Among several ones recently developed, I should mention Scheres et al.’s (2007) maximum likelihood approach, which produced remarkable agreement with the result of supervised classification in the case of a well-characterized problem: of dividing a data set of a ribosome complex according to its conformational state (macrostate I versus II; see Frank et al. 2007) (Fig. 7). However, to develop new algorithms, one would ideally require a data set generated from a phantom, such that the class membership of each projection is known. Only in that case can the performance of the algorithm be characterized by the number of true and false positives and so forth. Generation of such a data set requires the simulation of the EM imaging and noise processes. A step in this direction has recently been done by Baxter et al. (2009). Ultimately, unsupervised classification of data created by cryo-EM of molecular machines engaged in all steps of processing may allow us to create ‘movies’ in which ligands and other moving parts come to life.
Fig. 7.
Classification of ribosome complexes with or without EF-G by two methods: supervised and unsupervised classification. The experimental data set contains two states of the ribosome, distinguished as follows: (one) contains no EF-G; the A and P sites are occupied by tRNAs in the classic state; and the ribosome is in macrostate I (i.e., unratcheted), the other has EF-G bound; there is a single tRNA in P/E position; and the ribosome is in macrostate II (i.e., ratcheted). (a) Results of classification with the maximum likelihood (ML) method, with four classes specified. (b) Cross-reference between classes from ML and those derived from dividing the correlation histogram obtained by supervised classification. (c) Results of supervised classification using two reference maps, in ribosome macrostates I and II but without EF-G. Data reproduced from Scheres et al. (2007).
10. Interpretation of EM density maps by flexible fitting
Most density maps obtained in EM are of insufficient resolution to allow ab initio chain tracing or even a determination of the atomic structure as practiced in X-ray crystallography. In a few cases, this has been achieved for highly symmetric molecules where the number of repeats in a single molecule boosts the statistics (e.g., Ludtke et al. 2008; see review by Lindert et al. 2009). The situation more frequently encountered is that the resolution is in a range (~6–12 Å) where the map can be used as one of several constraints for flexible fitting, and the resulting structural model, far from being fixed with all of its side chains, must be understood as a plausible representative of an entire ensemble of structures all consistent with the cryo-EM data (see a discussion of this point by Trabuco et al. 2008). The terms quasi-atomic or pseudo-atomic structure have been used to refer to coordinates that have this residual uncertainty, but these terms are somewhat negatively loaded, as they contrast structural models obtained through interpretation of cryo-EM maps with a purportedly ‘real’ structure obtained by interpretation of X-ray density maps. A few words are in order on the rapidly shrinking difference between models of one kind and the other.
There is frequently a situation where the building of atomic models with tentative positioning of side chains is desired to characterize the binding of a ligand to a target molecule. Even though the density map cannot pin down the side chains, the choices are drastically narrowed down since steric and chemical constraints come into play (Volkman et al. 2001). In a certain sense, the building of a structural model from a 3D EM density map by flexible fitting can be compared with the building of atomic models from an electron density map obtained by X-ray crystallography, since in both cases known structural primitives (i.e., entire component structures in the former case and the known structures of an enumerable set of residues in the latter) are assembled to explain the observed density, following rules of stereochemistry (see, for instance, Topf et al. 2008).
Again, the ribosome, with its large size and complexity and existence of large-scale conformational changes such as the ratchet motion (Frank & Agrawal, 2000), proves to be the most challenging testing ground for the development of algorithms for flexible fitting. As a molecular machine with large-scale conformational changes and multiple binding interactions, the ribosome seen by cryo-EM is ever-changing, proving that the underlying atomic structure possesses intrinsic instabilities and that it visits several thermally accessible states in its working cycle (Frank et al. 2007). Three approaches to flexible fitting and their application to the ribosome should be mentioned: real-space refinement (Chapman, 1995; Gao et al. 2003), normal-mode analysis flexible fitting (Tama et al. 2004) and molecular dynamics flexible fitting (Trabuco et al. 2008). Their relative merits have been discussed by Trabuco et al. (2008) and Lindert et al. (2009). Arguably, fitting tools employing molecular dynamics, such as molecular dynamics flexible fitting, appear to come closest to a structural interpretation bestowing the atomic model with realistic stereochemical properties.
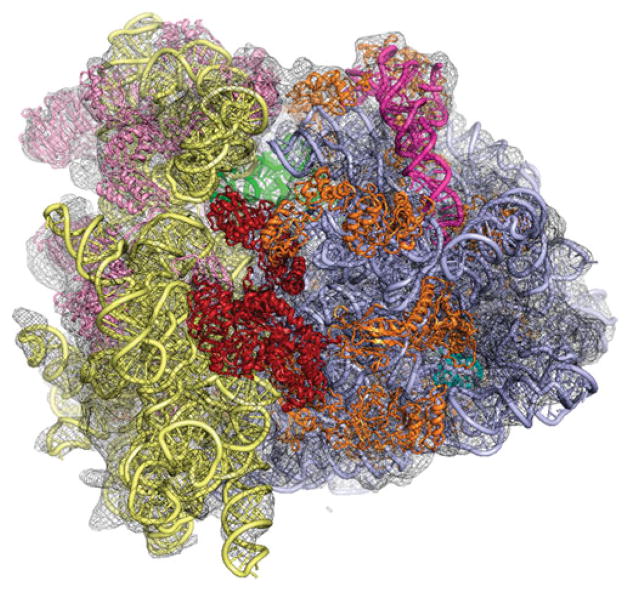
One of the most challenging problems in modeling is presented by the 80S eukaryotic ribosome, for which no structural data exist except for density maps by cryo-EM. While the functional core, composed of rRNAs, is highly conserved, peripherally located expansion segments acquired in the course of evolution have largely unknown structure and function. Here the challenge is to build a model de novo conforming with RNA sequence, local engagement rules, and observed density. Second, there are proteins with eubacterial counterparts that require homology modeling. Lastly, there are a number of proteins entirely without any eubacterial counterparts, which require de novo modeling. Two models exist at the present time, one for yeast (Spahn et al. 2001; Fig. 8), the other for dog (Chandramouli et al. 2008). Another, more comprehensive atomic model for yeast based on a cryo-EM map of the ribosome from the closely related species T. lanuginosus is forthcoming (Taylor et al. 2009). Thus, already today, the eukaryotic ribosome, recently highlighted as one of the ‘structures of desire’ by X-ray crystallographers (Bhattacharya, 2009), presents a rich trove of structural information thanks to the improvements both of cryo-EM as a technique and of the tools for mining the resulting density maps.
Fig. 8.
Homology modeling of the proteins and RNA of the 80S eukaryotic ribosome from the fungus T. lanuginosus, based on the sequence of a close relative, Saccharomyces cerevisiae, and on the X-ray structure of E. coli (Taylor et al. 2009). Structure in red seen in front is eEF2.
11. The potential and limits of single-particle reconstruction
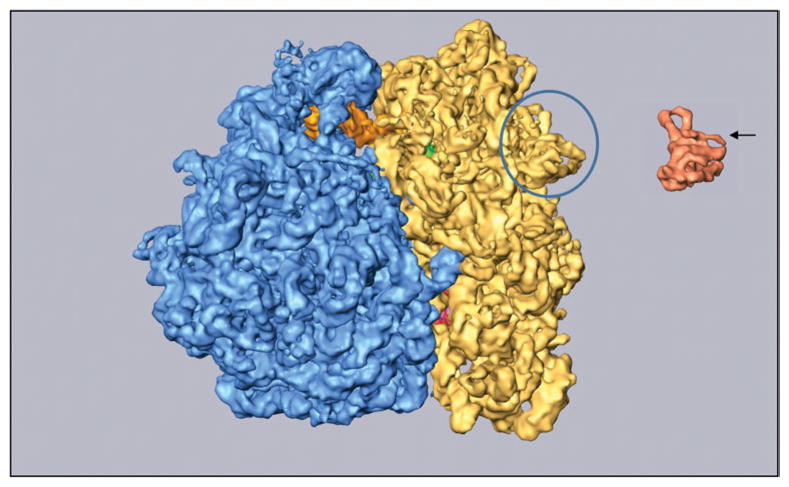
Glancing over the peripatic history of the single-particle reconstruction method, from its obscure beginning in 1978 to its present wide acceptance makes me realize that the technique has succeeded in a way nobody imagined. For one, I would not have predicted that a molecule, after the harsh treatment of freeze-plunging and without support of companions in a crystal matrix, would allow us to see molecular detail at its very periphery (Fig. 9). Apparently, only the outer-most alpha-helix of protein S2 has been affected in this reconstruction of the ribosome.
Fig. 9.
Cryo-EM reconstruction of the 70S ribosome of E. coli, with Phe-tRNAPhe·EF-Tu·GDP bound in the presence of kirromycin (LeBarron et al. 2008). Yellow: 30S subunit; blue: 50S subunit. Highlighted peripheral ribosomal protein S2 (encircled) is compared with its X-ray structure (orange) presented in the same orientation. Arrow points to an outermost helix that is missing in the density map, presumably because of disorder, while the rest of the structure is intact.
Foremost, single-particle reconstruction provides a way to image macromolecular complexes too large and too flexible to allow application of X-ray crystallography. The technique’s strength lies in its ability to yield 3D images of ‘molecular machines’ in a native environment, in a variety of biochemically well-characterized states, unconstrained by intermolecular contacts characteristic for a crystal. It is difficult, however, to find recipes for improving the resolution beyond what has been achieved, except for brute-force increasing data collection in ways that severely challenge the current means of storage and processing. The fact that close to atomic resolution has been actually achieved for some molecules bearing high symmetry offers hope that the same can be done for molecules lacking it, but it gives daunting estimates on the number of particles to be collected and processed. Ultimately, what needs to happen is the establishment of a data collection and processing pipeline in which smart scrutiny based on existing knowledge is applied at the very start.
Acknowledgments
This work was supported by HHMI and NIH R37 GM29169. I thank Lila Iino-Rubenstein for assistance with the artwork, and both Derek Taylor and Michael Radermacher for supplying me with illustrations.
References
- Agrawal RK, Penczek P, Grassucci RA, Li Y, Leith A, Nierhaus KH, Frank J. Direct visualization of A-, P-, and E-site transfer RNAs in the Escherichia coli ribosome. Science. 1996;271:1000–1002. doi: 10.1126/science.271.5251.1000. [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proceedings of the National Academy of Sciences USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of molecular machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Amos LA, Henderson R, Unwin PNT. Three-dimensional structure determination by electron microscopy of 2-dimensional crystals. Prog Biophys Mol Biol. 1982;39:183–231. doi: 10.1016/0079-6107(83)90017-2. [DOI] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2·4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Baxter WT, Grassucci RA, Gao H, Frank J. Determination of signal-to-noise ratios and spectral SNRs in cryo-EM low-dose imaging of molecules. Journal of Structural Biology. 2009;166:126–132. doi: 10.1016/j.jsb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer M, Frank J, Hanszen KJ, Kellenberger E, Williams RC. The possibilities and prospects of obtaining high-resolution (below 30 Å) on biological material using the electron microscope. Some comments and reports inspired by an EMBO workshop held at Gais, Switzerland, October 1973. Quarterly Reviews of Biophysics. 1974;7:211–238. doi: 10.1017/s0033583500001414. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A. Structures of desire. Nature. 2009;459:24–27. doi: 10.1038/459024a. [DOI] [PubMed] [Google Scholar]
- Billingsley FC. Applications of digital image processing. Applied Optics. 1970;9:289–299. doi: 10.1364/AO.9.000289. [DOI] [PubMed] [Google Scholar]
- Boettcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;36:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8·7 Å resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MS. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron density function. Acta Crystallographica. 1995;A51:69–80. [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- Cong Y, Zhang Q, Woolford D, Schweikardt T, Khant H, Daugherty M, Ludtke SJ, Ck W, Decker H. Structural mechanism of SDS-induced enzyme activity of scorpion hemocyanin revealed by electron cryomicroscopy. Structure. 2009;17:749–758. doi: 10.1016/j.str.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, DeRosier DJ, Klug A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proceedings of the Royal Society of London. 1970;317:319–340. doi: 10.1098/rspb.1972.0068. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith J. MRC image processing programs. Journal of Structural Biology. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- DeRosier D. Who needs crystals anyway? Nature. 1997;386:26–27. doi: 10.1038/386026a0. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. Journal of Structural Biology. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Frank J. PhD thesis. Technical University Munich; 1970. Untersuchungen von elektronenmicroskopischen Aufnahmen hoher Auflösung mit Bilddifferenz- und Rekonstruktionsverfahren [Studies of Electron Micrographs Using Image Difference and Reconstruction Methods] [Google Scholar]
- Frank J. A study on heavy/light atom discrimination in bright-field electron microscopy using the computer. Biophysical Journal. 1972;12:484–511. doi: 10.1016/s0006-3495(72)86099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Controlled focusing and stigmating in the conventional and scanning transmission electron microscope. Journal of Physics E: Scientific Instruments. 1975a;8:582–587. [Google Scholar]
- Frank J. Averaging of low exposure electron micrographs of nonperiodic objects. Ultramicroscopy. 1975b;1:159–162. doi: 10.1016/s0304-3991(75)80020-9. [DOI] [PubMed] [Google Scholar]
- Frank J. The role of multivariate statistical analysis in solving the architecture of the Limulus polyphemus hemocyanin molecule. Ultramicroscopy. 1984;13:153–164. doi: 10.1016/0304-3991(84)90065-2. [DOI] [PubMed] [Google Scholar]
- Frank J. Classification of macromolecular assemblies studied as single particles. Quarterly Reviews of Biophysics. 1990;23:281–329. doi: 10.1017/s0033583500005564. [DOI] [PubMed] [Google Scholar]
- Frank J, Penczek P, Grassucci R, Srivastava S. Three-dimensional reconstruction of the 70S E. coli ribosome in ice: the distribution of ribosomal RNA. Journal of Cell Biology. 1991;115:597–605. doi: 10.1083/jcb.115.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in their Native State. New York: Oxford University Press; 2006. [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Goldfarb W, Eisenberg D, Baker TS. Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy. 1978;3:283–290. doi: 10.1016/s0304-3991(78)80038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Shimkin B, Dowse H. SPIDER – a modular software system for electron image processing. Ultramicroscopy. 1981a;6:343–358. [Google Scholar]
- Frank J, Verschoor A, Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981b;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Frank J, Chiu W, Degn L. The characterization of structural variations within a crystal field. Ultramicroscopy. 1988;26:345–360. doi: 10.1016/0304-3991(88)90234-3. [DOI] [PubMed] [Google Scholar]
- Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata RK, Agraewal RK. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like intersubunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Wagenknecht T, McEwen BF, Marko M, Hsieh CE, Mannella CA. Three-dimensional imaging of biological complexity. Journal of Structural Biology. 2002;138:85–91. doi: 10.1016/s1047-8477(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA–tRNA translocation. Proceedings of the National Academy of Sciences USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CMT, Grassucci RA, Frank J, Penczek P. Solution structure of the E coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J. Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- Glaeser RM. Limitations to significant information in biological electron microscopy as a result of radiation damage. Journal of Ultrastructure Research. 1971;36:466–482. doi: 10.1016/s0022-5320(71)80118-1. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Downing KH, DeRosier D, Chiu W, Frank J. Electron Crystallography of Biological Macromolecules. New York: Oxford University Press; 2006. [Google Scholar]
- Grassucci RA, Taylor D, Frank J. Visualization of macromolecular complexes using cryo-electron microscopy with FEI Technai transmission electron microscopes. Nature Protocols. 2008;3:330–339. doi: 10.1038/nprot.2007.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassucci RA, Taylor D, Frank J. Preparation of macromolecular complexes for cryoelectron microscopy. Nature Protocols. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. Three-dimensional structure of bovine NAHD: ubiquinone oxireductase (complex I) at 22 Å in ice. Journal of Molecular Biology. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Resolution measurement in structures derived from single particles. Acta Crystallographica. 2000;D56:1270–1277. doi: 10.1107/s0907444900009549. [DOI] [PubMed] [Google Scholar]
- Hegerl, Altbauer The “EM” program system. Ultramicroscopy. 1982;9:109–116. doi: 10.1016/0304-3991(82)90233-9. [DOI] [PubMed] [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons, and X-rays for atomic resolution microscopy of unstained biological molecules. Quarterly Reviews of Biophysics. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- Henderson R, Unwin PNT. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F. Structure of purple membrane from Halobacterium halobium: recording, measurement and evaluation of electron micrographs at 3·5 Å resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- Hoppe W. Electron diffraction with the transmission microscope as a phase-determining diffractometer – from spatial frequency filtering to the three-dimensional structure analysis of ribosomes. Angewandte Chemie International Edition in English. 1983;22:456–485. [Google Scholar]
- Kessel M, Frank J, Goldfarb W. Low dose electron microscopy of individual biological macromolecules. In: Baumeister W, editor. Electron Microscopy at Molecular Dimensions State of the Art and Strategies for the Future. Berlin: Springer; 1980. pp. 154–160. [Google Scholar]
- Kessel M, Radermacher M, Frank J. The structure of the stalk surface layer of a brine pond microorganism: correlation averaging applied to a double layered lattice structure. Journal of Microscopy. 1985;139:63–74. doi: 10.1111/j.1365-2818.1985.tb04662.x. [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhang X, Baker TS, Ma J. A structural–informatics approach for tracing beta-sheets: building pseudo-C(alpha) traces for beta-strands in intermediate-resolution density maps. Journal of Molecular Biology. 2004;339:117–130. doi: 10.1016/j.jmb.2004.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer R, Frank J, Feltynowski A, Hoppe W. Kohlefolien bei Elektronenbestrahlung [Application of the image subtraction method to study structural changes in thin carbon films] Berichte der Bunsen-Gesellschaft für Physikalische Chemie. 1970;74:1120–1126. [Google Scholar]
- LeBarron J, Grassucci RA, Shaikh TR, Baxter WT, Sengupta J, Frank J. Exploration of parameters in cryo-EM leading to an improved density map of the E. coli ribosome. Journal of Structural Biology. 2008;164:24–32. doi: 10.1016/j.jsb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebart L, Morineau A, Warwick KM. Multivariate Descriptive Statistical Analysis. New York: John Wiley; 1984. [Google Scholar]
- Lindert S, Stewart PL, Meiler J. Hybrid approaches: applying computational methods in cryo-electron microscopy. Current Opinion in Structural Biology. 2009;19:1–8. doi: 10.1016/j.sbi.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Malhotra A, Penczek P, Agrawal RK, Gabashvili IS, Grassucci RA, Jünemann R, Burkhardt N, Nierhaus KH, Frank J. Escherichia coli 70S ribosome at 15 Å resolution by cryo-elecytron microscopy: localization of fMet-tRNAfmet and fitting of L1 protein. Journal of Molecular Biology. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- Martin AG, Depoix F, Stohr M, Meissner U, Hagner-Holler S, Hammouti K, Burmester T, Heysd J, Wriggers W, Markl J. Limulus polypemus hemocyanin: 10A cryo-EM structure, sequence analysis, molecular modeling and rigid-body fitting reveal the interfaces between the eight hexamers. Journal of Molecular Biology. 2007;366:1332–1350. doi: 10.1016/j.jmb.2006.11.075. [DOI] [PubMed] [Google Scholar]
- Morgan DG, Grant RA, Chiu W, Frank J. Patch averaging of electron images of GP3*I crystals with variable thickness. Journal of Structural Biology. 1992;108:245–256. doi: 10.1016/1047-8477(92)90025-6. [DOI] [PubMed] [Google Scholar]
- Moore P. The ribosome returned. Journal of Biology. 2009;8:1–10. doi: 10.1186/jbiol103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman K. Profile of Joachim Frank. Proceedings of the National Academy of Sciences USA. 2007;104:19671–19678. doi: 10.1073/pnas.0710323105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova EV, Dube P, Harris JR, Beckman E, Zemlin F, Markl J, van Heel M. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 resolution by electron cryomicroscopy and angular reconstitution. Journal of Molecular Biology. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- Penczek P, Radermacher M, Frank J. Threedimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Penczek P, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Zhu J, Frank J. A common-lines based method for determining orientations for N>3 particle projections simultaneously. Ultramicroscopy. 1996;63:205–218. doi: 10.1016/0304-3991(96)00037-x. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Zhu J, Schröder R, Frank J. Three-dimensional reconstruction with contrast transfer function compensation from defocus series. Scanning Microscopy Supplement. 1997;11:1–10. [Google Scholar]
- Radermacher M. Radon transform techniques for alignment and three-dimensional reconstruction from random projections. Scanning Microscopy Supplement. 1997;11:171–177. [Google Scholar]
- Radermacher M. The three-dimensional reconstruction of single particles from random and non-random tilt series. Journal of Electron Microscopy Technique. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random–conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. Journal Microscopy. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallographica. 2000;D56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. Journal of Microscopy. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Frank J. Motif detection in quantum noise-limited electron micrographs by cross-correelation. Ultramicroscopy. 1976;2:219–227. doi: 10.1016/s0304-3991(76)91385-1. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Pitt TJ, Horner M. Digital image processing: the SEMPER system. Ultramicroscopy. 1979;4:343–354. [Google Scholar]
- Scheres SHW, Goa H, Valle M, Herman GT, Eggermont PPB, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nature Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- Schuette JC, Murphy FV, IV, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO Journal. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Kjeldgaard M, Nissen P, Frank J. Visualization of the eEF2–80S ribosome transition-state complex by cryo-electron microscopy. Journal of Molecular Biology. 2008;382:179–187. doi: 10.1016/j.jmb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nature Protocols. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S Ribosome from Saccharomyces cerevisiae – tRNA-ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Stark H, Müller F, Orlova EV, Schatz M, Dube P, Erdemir T, Zemlin F, Brimacombe R, van Heel M. The 70S Escherichia coli at 23 Å resolution: fitting the ribosomal RNA. Structure. 1995;3:815–821. doi: 10.1016/s0969-2126(01)00216-7. [DOI] [PubMed] [Google Scholar]
- Stark H, Orlova EV, Rinke-Appel J, Jünke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. Arrangement of tRNAs in pre- and posttranslocational ribosomes revealed by electron cryomicroscopy. Cell. 1997a;88:19–28. doi: 10.1016/s0092-8674(00)81854-1. [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997b;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- Tama F, Miashita O, Brooks CL., III Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. Journal of Structural Biology. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO Journal. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Glaeser RM. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on the opportunities for the future. Journal of Structural Biology. 2008;163:214–223. doi: 10.1016/j.jsb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Devkota B, Huang A, Topf M, Narayanan E, Sali A, Harvey S, Frank J. Comprehensive atomic model of the eukaryotic ribosome. 2009. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Ciu W, Sali A. Protein structure fitting and refinement guided by cryoEM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin PNT, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. Journal of Molecular Biology. 1975;94:425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO Journal. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–124. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- van Heel M. Classification of very large electron microscopical data sets. Optik. 1989;82:114–126. [Google Scholar]
- van Heel M, Frank J. Use of multivariate statistical analysis in analyzing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- van Heel M, Hollenbeck J. On the stretching of distorted images of two-dimensional crystals. In: Baumeister W, Vogell W, editors. Electron Microscopy at Molecular Dimensions. Berlin: Springer-Verlag; 1980. pp. 256–260. [Google Scholar]
- van Heel M, Keegstra W, Schutter WG, van Bruggen EFJ. Arthropod hemocyanin studied by image analysis. Life Chemistry Report Supplement. 1982;1:69–73. [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy: towards atomic resolution. Quarterly Review of Biophysics. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysids. Proceedings of the National Academy of Sciences USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, Amann KH, Stollova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Structure of Arp 2/3 comoplex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals a helices and insight into DNA binding. Structure. 2009;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Zhu J, Penczek PA, Schröder R, Frank J. Three-dimensional reconstruction with contrast transfer function correction from energy-filtered cryoelectron micrographs: procedure and application to the 70S Escherichia coli ribosome. Journal of Structural Biology. 1997;118:197–219. doi: 10.1006/jsbi.1997.3845. [DOI] [PubMed] [Google Scholar]
- Zingsheim HP, Neugebauer DC, Barrantes FJ, Frank J. Structural details of membrane-bound acetylcholine receptor from Torpedo marmorata. Proceedings of the National Academy of Sciences USA. 1980;77:952–956. doi: 10.1073/pnas.77.2.952. [DOI] [PMC free article] [PubMed] [Google Scholar]