Abstract
Although memory allocation is a subject of active research in computer science, little is known about how the brain allocates information within neural circuits. There is an extensive literature on how specific types of memory engage different parts of the brain, and how neurons in these regions process and store information. Until recently, however, the mechanisms that determine how specific cells and synapses within a neural circuit (and not their neighbors) are recruited during learning have received little attention. Recent findings suggest that memory allocation is not random, but rather specific mechanisms regulate where information is stored within a neural circuit. Novel methods that allow tagging, imaging, activation and inactivation of neurons in behaving animals, promise to revolutionize studies of brain circuits, including memory allocation. Results from these studies are likely to have a considerable impact on both computer science as well as on the understanding of memory and its disorders.
Introduction
How and where specific items are stored has a lot to do with how easily they can be retrieved and used. Organized storage saves space (for example superfluous or duplicate items can be eliminated), it minimizes search times and reduces errors during retrieval. Memory allocation refers to a set of processes that determine where information is specifically stored in a neural circuit. Are there neurobiological processes that determine which cells and synapses within a given circuit are engaged during learning, or is this random? Does memory allocation involve competition between different cells (or synapses) activated during learning? Do memory allocation processes take place at different time scales? The allocation of information is an especially important problem for the brain because of the enormous number of related memories stored through out a lifetime. Without a mechanism that appropriately groups and separates memories, how would the brain store so many complex memories of both similar and discrete events? One possibility is that the brain stores related memories in overlapping populations of neurons and in synapses within the same dendritic branch, so that activation of one component of the memory increases the likelihood of retrieval of other related components. This strategy would require mechanisms that regulate where memory is stored in neural networks.
The principles and mechanisms of memory allocation are not only fascinating because of the insights they provide into how the brain stores information, but they also have far reaching implications for the design of artificial intelligence systems and for cognitive disorders. The central question that we will address in this review is whether memory allocation is random or whether specific mechanisms determine where information is stored within a neural network. Recent findings suggest that there are mechanisms that regulate allocation of memory to specific neurons in a network and modulate the synaptic selection processes that determine where these memories are stored (Fig. 1).
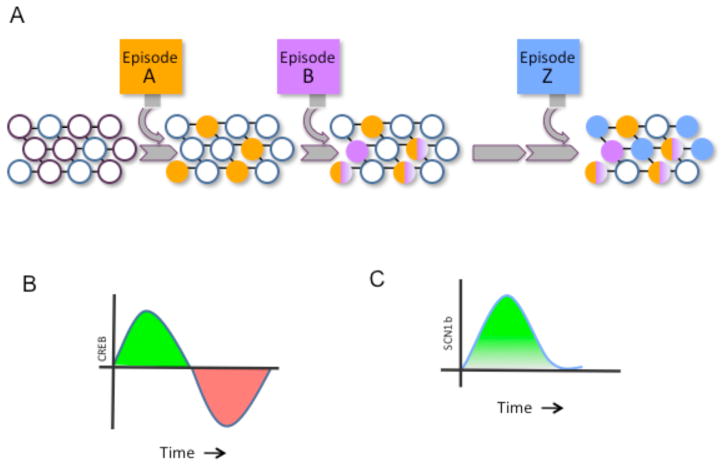
Fig 1. Model of memory allocation in neuronal populations.
(A) Neurons in a naïve neural circuit (open circles) are recruited into encoding episode A (orange). This increases their excitability so that shortly thereafter they are very likely to also be involved in encoding episode B (purple). With time, the increase in excitability wanes and consequently episode Z (blue) no longer is stored in the same neurons. A consequence of this pattern of storage is that recall of episode A will also result in the recall of episode B (and vice versa), while recall of episode Z becomes unrelated to the other two episodes.
(B) Training causes an temporary increase in the activity and levels of CREB. The CREB gene family of transcription factors includes both activators (they increase transcription) and repressors. Following the initial increase in activators (green section of the curve), CREB repressors are expressed that decrease the overall levels of CREB activity, thus eventually bringing them below basal levels (red section of the curve). (C) Higher levels of CREB in a specific cell population result in increases in the levels of specific proteins (e.g. Scn1b), which in turn increase the excitability of neurons. Thus, two memories acquired while CREB levels are high would be stored in overlapping populations of neurons. Since the change in excitability would involve transcription and require the stability of transcribed molecules, such as channels, the time scale of this memory allocation mechanism would be in the order of hours (or perhaps even days), so that one memory could affect the allocation of proceeding memories for many hours.
CREB and memory allocation
The cAMP Responsive Element Binding protein (CREB) regulates the transcription of other genes and has a well-known role in the stability of synaptic potentiation and memory (1, 2). A recent series of papers (3–5) provided compelling evidence that there are molecular and cellular processes that determine which cells are recruited to store information within a neural circuit. In particular, this work suggested the hypothesis that CREB activated during learning triggers changes in the cell (i.e. increase in excitability) that then affect whether that cell participates in subsequent memories. This idea was tested by artificially increasing the levels of CREB in amygdala neurons using a replication defective Herpes viral vector. The initial results showed that higher CREB levels increase the probability (~3 fold) that amygdala neurons participate in memory for tone fear conditioning (3). In this form of Pavlovian conditioning, animals learn to associate a tone with an aversive event, such as a mild foot shock. The amygdala is a subcortical brain structure with a well-known role in emotional memories such as fear memory (6, 7). ARC (Activity-Regulated Cytoskeletal-associated protein) RNA, a gene required for synaptic function and memory, was used to identify the neurons encoding a tone conditioning memory. ARC has been extensively used to determine which neuronal populations are activated by specific behavioral stimuli (8),(9). Immediately after a memory test, amygdala cells transfected with the viral CREB (identified through its fluorescent tag) were three times more likely to express ARC RNA (i.e., be involved in memory) than neighboring cells (3). Thus, CREB levels seem to bias which neurons encoded tone conditioning in the amygdala. It is conceivable that an interaction between the effects of the viral vectors used and the genes transfected could have affected the results of these early studies. To address this and other possible confounding interpretations of the results, cell lesion and inactivation strategies were also used to probe the role of CREB in memory allocation (4, 5).
A targeted cell lesion strategy was used to explore further the possibility that CREB biases the allocation of memory for tone conditioning in the amygdala (4). The authors took advantage of a transgenic mouse with a silenced diphtheria toxin receptor to specifically kill the cells with the virally-delivered CREB in mice. A replication defective Herpes viral vector carried CREB and a recombinase that could activate the silenced diphtheria receptor gene by deleting a RNA translation STOP sequence. CREB biased memory allocation since killing the cells transfected with the viral CREB disrupted memory for tone conditioning, while ablating the cells transfected with the control virus had no effect. A series of elegant control experiments showed that ablating the CREB cells did not prevent the animals from making new amygdala-dependent memories, and nor did it affect memories acquired before viral infection. Instead, killing the cells with the viral CREB only affected the memory for tone conditioning acquired after viral transfection.
Targeted killing of nearly 20% of cells in a neural circuit may have had unintended effects that could confound the interpretation of behavioral experiments. However, this does not seem to have been the case since similar results (5) to those just described were obtained with another approach (10) that allows for reversible neuronal inactivation. This strategy takes advantage of a Drosophila receptor (the allatostatin receptor) that can be functionally linked to potassium channels in mouse neurons. When activated by the allatostatin receptor, these channels silence neurons (keep them from firing action potentials). Since mice lack the ligand for the allatostatin receptor, only neurons that have the exogenously expressed receptor can be inactivated by treatments with the allatostatin peptide. The results (5) show that inactivating amygdala cells expressing a virally provided CREB, results in a pronounced amnesia for tone conditioning. This amnesia could be readily reversed simply by retesting the mice a day later in the absence of allatostatin, demonstrating both the reversibility of the allatostatin effects, and the link between activity in the CREB cells and recall. As before, if CREB was replaced with a control gene in the viral vectors, then inactivation of the transfected cells had no impact on memory. Further, the authors showed that CREB manipulations could also affect the allocation of memory for conditioned taste aversion (CTA) (5), another form of memory that engages the amygdala. Animals that experience intestinal malaise (i.e. nausea) after eating or drinking a novel food learn to avoid that food since they associate it with the subsequent malaise. Inactivating the neurons transfected with the CREB virus had a very specific impact on memory, since inactivation affected a CTA memory acquired after viral transfection, but did not disrupt in the same animals a tone conditioning memory acquired before viral transfection.
Each of the strategies used to study the role of CREB in memory allocation have their own limitations. For example, the expression of the allatostatin receptor could interfere with other receptors in amygdala neurons and thus alter the behavioral results described above. Nevertheless, the convergence of findings with three different strategies (ARC cell tagging, diphtheria and allatostatin inactivation) confirms that CREB has a role in the allocation of memory in the amygdala.
One of the byproducts of the inactivation experiments described above(4, 5) is the finding that deleting or inactivating 15–20% of cells in an amygdala memory trace does not seem to affect that memory, while deleting 50–60% of those cells does cause a substantial amnesia. This resiliency could be a mechanism for preserving memories in the face of neuronal death or other events that would otherwise erase memory traces.
The studies described above also suggest that there is a competition process that keeps the number of neurons encoding a given memory constant (3, 4) (but see (11)). Despite 60–70% of CREB transfected cells being ARC positive and hence engaged in memory (versus 15–20% normally), the overall number of ARC positive cells in the amygdala did not increase. Therefore, it is possible that there are memory allocation mechanisms that control the size of memory traces so as to economize storage space. For example, changes in synaptic inhibition in response to changes in overall levels of circuit excitability could provide a dynamic way to control the overall number of neurons involved in a specific memory, thus modulating the storage space allocated to any one memory.
Memory allocation: physiological mechanisms
Electrophysiological studies (5) uncovered a possible mechanism for how CREB manipulations affect memory allocation. Consistent with previous studies(12), the findings showed that the cells transfected with the viral CREB were more excitable than neighboring cells or cells transfected with a virus carrying a control gene: the CREB cells fired more action potentials and were activated more easily(5). Perhaps because CREB cells are more excitable, they are more likely to respond to sensory inputs and therefore be activated during conditioning. Previous studies had suggested that learning triggers a temporary increase in neuronal excitability (13, 14). This excitability increase could define a window of time in which related memories are co-stored in overlapping populations of neurons.
The synaptic inputs carrying information about tone and shock are known to change the strength of synapses of neurons in the lateral amygdala(15). Electrophysiology studies showed that following tone conditioning, the potentiation of lateral amygdala inputs is considerably larger in the cells transfected with viral CREB than in their neighbors (5). A previous study (16) suggested that following conditioning as many as 4% of all synapses can become potentiated in neurons recruited into amygdala memory traces! All together these findings indicate that amygdala cells transfected with viral CREB are more excitable than their neighbors and that they express higher levels of synaptic potentiation following training, a result consistent with the hypothesis that they are preferentially chosen during tone conditioning. However, it is unlikely that CREB alone regulates which cells are involved in a given memory. Instead, it is plausible that the recent activation history of the cell (Fig. 1), as well as a myriad of molecular processes, including CREB targets, modulate memory allocation. As outlined in fig. 1, CREB-dependent changes in excitability would involve a time-scale of hours, so that one memory could affect the allocation of proceeding memories for hours afterwards. It is possible that other cellular allocation mechanisms would have different time-scales. Next, we will review a series of findings that suggest that neurogenesis in the dentate gyrus may affect memory allocation with a time-scale of days.
New neurons and the temporal integration of hippocampal memories
The adult brain continues to make new neurons (neurogenesis) and this seems to be important for a number of processes including learning and memory(17). Similar to the CREB expressing amygdala neurons (5), newly born neurons in the dentate gyrus of rodents show high levels of excitability(18, 19) and synaptic potentiation (20). Neurobehavioral (21) and computational (22) studies proposed that upon joining dentate gyrus circuits, these newly born neurons are more likely to be recruited into emerging memory traces, thus suggesting that neurogenesis has a role in memory allocation in the dentate gyrus!
A recent study (21) used BrdU, a permanent stain that intercalates dividing DNA, to label new neurons, and genes known to be activated by training (c-fos and ARC) to determine whether these newly born neurons were engaged in spatial learning. The results (21) demonstrated that 4–8 week-old neurons (BrdU +) are 2–3 times more likely than old neurons to be recruited into spatial memory (c-fos+ or ARC+). In contrast, 2-week old neurons integrate into spatial memory circuits with lower efficiencies, and one-week old neurons did not integrate at all(21). These and other results showed that the timing of neuronal development relative to training is crucial, and they are consistent with previous studies showing that 4-, but not 1-, week old neurons have the required synaptic structures and physiology to support mature connections with established hippocampal circuits(23). Once mature, the newly born neurons are thought to receive connections from the entorhinal cortex and send connections to the CA3 region. These results suggest that neurogenesis may affect memory allocation: Young dentate gyrus neurons are preferentially recruited following spatial learning!
Recent computational modeling studies also suggested that neurogenesis has a role in memory allocation (22, 24). The authors designed a model network that incorporated key specific neuronantomical features of the dentate gyrus as well as immature neurons that matured progressively over time, became connected and fully functional. As in the dentate gyrus, new neurons in the model were temporarily endowed with the highest levels of excitability (18, 19) and synaptic plasticity (20, 25). When the excitability and plasticity of a specific set of new neurons is high, they are very likely to participate in the encoding of incoming memories. Thus, a defined population of newly born neurons would encode and therefore integrate a set of memories occurring within a limited window of time. Later, activation of those new neurons could activate that specific set of memories.
Once the excitability and plasticity of a specific group of newly born neurons wane, the window for memory integration closes and consequently a newer cluster of memories would be integrated into another group of maturing neurons. This process continues, with each subsequent set of newly born dentate neurons incorporating a newer set of memories, each set with a distinct time stamp. Thus, new neurons with high excitability and plasticity could endow the dentate gyrus with pattern integration abilities. This idea for the temporal integration of memories does not necessarily contradict a role for the dentate gyrus as a pattern separation devise: Memories could be both made distinct from other similar memories in the dentate gyrus by the million-plus mature neurons in this structure (whether they were born during development or in adult animals (26)), at the same time that distinct memories are given a specific time stamp (i.e. integrated) by newly connected young neurons (21). Altogether the neuroanatomical and modeling studies described suggest that neurogenesis has a role in memory allocation. Nevertheless, the findings reviewed could have alternative explanations. For example, the connectivity of new neurons, rather than dynamic memory cell allocation mechanisms, could be responsible for their preferential incorporation into new memories. This connectivity, however, could also have a role of its own on memory allocation, with only a subpopulation of some of the synapses initially involved eventually being recruited into memory storage. Below, we will review recent results that suggest a synaptic allocation mechanism with a time-scale of minutes.
Synaptic selection and memory allocation
Although our descriptions of memory allocation so far focused on individual neurons, memories are thought to be stored in the synapses of those neurons (27, 28). Thus, single neurons are likely to be involved in multiple memories. Previous studies in the amygdala suggested that individual memories can recruit as much as 20% of all synapses of neurons engaged in that memory (16)! Is the allotment of this synaptic space random or are there mechanisms that determine synaptic selection during memory allocation? Previous synaptic tagging studies showed that there are molecular physiological rules that determine which synapses are stably potentiated, suggesting that the commitment of specific synapses to memory is not random. Molecular tags may temporarily mark synapses activated during learning so that transcriptional products coming from the cell body stabilize synaptic changes specifically in those synapses. (29–31).
As predicted by clustered plasticity models(31–33), long-term potentiation at a specific synapse in a CA1 pyramidal neuron increases the probability for potentiation at neighboring synapses. This affects synapses within 10 micrometers of the potentiated synapse, and the effect may lasts at least ten minutes (Fig. 2) (34). Potentiation of one synapse did not change the responses or structure of nearby synapses. Instead, it simply lowered the threshold for the induction of these changes in those synapses. In addition, potentiation of single synapses broadened the time window effective at inducing potentiation in nearby synapses (34). These findings may have implications for memory allocation since they suggest that events closely associated in time (within 10 minutes of each other) are likely to be allocated (i.e., trigger synaptic modifications) to neighboring synapses in recruited CA1 neurons (Fig. 2). How could the potentiation of one synapse affect other synapses nearby?
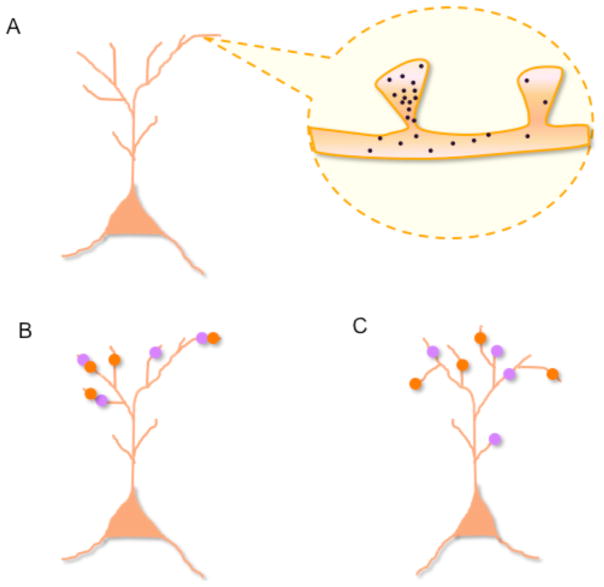
Fig 2. Model of memory allocation within dendritic trees.
(A) Following learning and subsequent potentiation of synapses in encoding neurons, molecular components diffuse to nearby (10 microns) unpotentiated spines for a limited time (10 minutes), thus resulting in a temporary increase in the probability that these spines will participate in subsequent learning (become potentiated themselves). (B) Two memories acquired within minutes of each other may be stored in similar populations of cells (Fig. 1) and in nearby synapses (orange and purple circles), thus resulting in strong co-recall. (B) In contrast, two memories acquired within hours of each other may be stored in overlapping cellular populations, but perhaps not in nearby synapses, which would result in weaker co-recall. Lack of neuronal or dendritic co-localization would prevent automatic co-recall. Thus, unlike the cellular allocation mechanism discussed in fig. 1, the time-scale of synaptic allocation mechanisms would be in the order of minutes.
Studies that fluorescently labeled and imaged specific biochemical changes in potentiated synapses, demonstrated that some activated molecular components from one potentiated synapse can reach other nearby synapses (35), while others are spatially restricted to the activated synapses ((36), but see (37)). Because only some of the activated molecular components of potentiated synapses are shared, this could account for why potentiation is synapse-specific while affecting the probability of potentiation at nearby synapses. Branch specific changes in excitability(38) could also affect synaptic selection mechanisms, and therefore modulate memory allocation.
The results reviewed above are exciting, but it is important to note that the ground-breaking methods used to potentiate and image synapses could have affected the very biochemistry that they were intended to study. Furthermore, it remains to be shown whether similar mechanisms are at play during learning and memory. Nevertheless, these and other related results suggest that there are specific rules that regulate synaptic selection. Thus, besides mechanisms that determine which cells are engaged in a given memory, working with time scales of hours and days (fig. 1), there may be also mechanisms, functioning with time-scales of minutes, that determine which synapses are recruited. What would be the implications of having allocation mechanisms with different time-scales? One possibility is that cell and synaptic allocation mechanisms could set up gradients of memory relatedness: Memories with both overlapping cellular and synaptic domains would be expected to be more interconnected (more often co-retrieved) than memories that only share similar cell populations (Fig. 2B). The very different time-scales of these processes would limit memory integration at the synaptic and cellular levels. Accordingly, synaptic integration would require closer juxtaposition of events (seconds and minutes), than cellular allocation (hours, days).
The allocation of episodic memories
Episodic memories usually involve extended temporal interconnections between complex stimuli. Memory allocation mechanisms could potentially be used to set up partially overlapping populations of neurons that could account for these extended temporal interconnections. However, previous studies showed that very specific features of complex stimuli activate only briefly neurons in the human hippocampus and associated structures (the medial temporal lobe or MTL). Activation normally lasts 300 to 600 ms, and rarely continues beyond 1 to 2 s following stimulus onset. However, episodic memories can be much longer, and it is unknown how they are encoded in MTL. Are different components of an episodic memory allocated to overlapping populations of neurons, or are they encoded in separate MTL neural circuits? Consistent with the former possibility, prolonged activations of MTL neurons in response to single episodes have now been reported (39). The subjects in these experiments were epileptic patients with implanted electrodes in the MTL to help locate the source of seizures. To trigger more extended episodic memories, subjects were initially exposed to 5–10 sec clips of movies, while implanted electrodes recorded responses from the MTL.
Nearly 10% of the 847 units recorded showed prolonged responses throughout the entire replay of the movie clip (i.e. during the whole “episode”). Each episode included numerous related but changing features that were nevertheless continuously recognized by single MTL neurons! Although there are many potential explanations for this prolonged firing, one possibility is that different episodic components present in different parts of the clip were allocated to the same neurons! Another possibility is that firing of the units recorded was not specific, which could account for continued firing during the entire clip. However, this was not the case; For example, one of the units in the entorhinal cortex fired specifically in response to a clip (amongst 48 clips) of an episode of the television series “The Simpsons”. Could the rules of cellular and synaptic memory allocation emerging from studies in rodents also apply to human memory? Could CREB-dependent mechanisms, neurogenesis and synaptic selection have a role in human episodic memory formation? Studies like the one mentioned above could start to address these important questions.
Conclusions
Newly developed strategies for imaging, activating and inactivating specific neurons in a given brain region are changing the way neuroscientists tackle traditional problems, and more importantly, they are opening up new areas of investigation, such as memory allocation. The results reviewed here represent the first steps towards uncovering the molecular and cellular strategies used by circuits to allocate information during encoding. They suggest that mechanisms working at different time-scales, including CREB signaling, neurogenesis and synaptic selection, modulate the allocation of memory to specific cells and synapses within a neural network. Memory allocation will likely involve a plethora of synaptic, cellular and systems mechanisms regulated by a myriad of molecular processes, including CREB. Beyond CREB, it is possible that other molecular components involved in the acquisition and consolidation of memory, may also regulate not only which cells are allocated to a given memory, but also which synapses in those cells are eventually recruited into encoding that information.
The results reviewed here suggest that there are competitive mechanisms that affect memory allocation. For example, new dentate gyrus neurons, amygdala cells with higher excitability, and synapses near previously potentiated synapses seem to have the competitive edge over other cells and synapses, and thus affect memory allocation with time scales of weeks, hours and minutes. Are all memory allocation mechanisms competitive, or are there mechanisms of memory allocation that do not involve competition? Even though at this point it is difficult to resolve this question, it is important to note that most mechanisms of memory allocation in computers do not involve competition.
Studies of memory allocation may also have an impact on the study of memory deficits. For example, a number of studies have shown that aging decreases both the excitability of neurons in the hippocampal formation and the rates of neurogenesis. Consequently, aging very likely disrupts hippocampal memory allocation, which could possibly contribute to age-related cognitive decline. Disruptions of inhibition and modulatory neurotransmission in psychiatric conditions are also likely to affect memory allocation and therefore be partially responsible for cognitive impairments associated with these conditions. Inappropriate associations between unrelated events could conceivably be a reflection of abnormal memory allocation in psychiatric conditions such as schizophrenia.
Although memory allocation has only been recently studied in neuroscience, there is a long history of studies of memory allocation in computer science. Insights into the strategies that allocate information in neuronal networks may provide not only useful insights for the treatment of memory disorders, but also suggest novel design principles for effective allocation of memory in artificial networks.
Acknowledgments
We would like to thank Silva lab members, especially S. Josselyn, S. Kushner, J. Won, M. Zhou, M. Karlsson, as well as our colleagues F. Gage, D. Buonomano, P. Golshani and P Poirazi, for discussions that helped shape the ideas summarized here. This work was supported by grants from the NIH (AG013622 and P50-MH0779720) to AJS.
References and Notes
- 1.Silva AJ, Kogan JH, Frankland PW, Kida S. Annu Rev Neurosci. 1998;21:127. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 2.Carlezon J, William A, Duman RS, Nestler EJ. Trends in Neurosciences. 2005;28:436. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Han JH, et al. Science. 2007;316:457. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 4.Han JH, et al. Science. 2009;323:1492. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, et al. Nature Neuroscience. 2009 in press. [Google Scholar]
- 6.Maren S, Quirk GJ. Nat Rev Neurosci. 2004;5:844. doi: 10.1038/nrn1535. [DOI] [PubMed] [Google Scholar]
- 7.Phelps EA, LeDoux JE. Neuron. 2005;48:175. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Tzingounis AV, Nicoll RA. Neuron. 2006;52:403. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Miyashita T, Kubik S, Lewandowski G, Guzowski JF. Neurobiol Learn Mem. 2008;89:269. doi: 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan EM, et al. Neuron. 2006;51:157. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 11.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Science. 2007;317:1230. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 12.Viosca J, Lopez de Armentia M, Jancic D, Barco A. Learn Mem. 2009;16:193. doi: 10.1101/lm.1254209. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Kang J. Rev Neurosci. 2005;16:311. doi: 10.1515/revneuro.2005.16.4.311. [DOI] [PubMed] [Google Scholar]
- 14.Disterhoft JF, Oh MM. Trends Neurosci. 2006;29:587. doi: 10.1016/j.tins.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Sah P, Westbrook RF, Luthi A. Ann N Y Acad Sci. 2008;1129:88. doi: 10.1196/annals.1417.020. [DOI] [PubMed] [Google Scholar]
- 16.Rumpel S, LeDoux J, Zador A, Malinow R. Science. 2005;308:83. doi: 10.1126/science.1103944. [DOI] [PubMed] [Google Scholar]
- 17.Zhao C, Deng W, Gage FH. Cell. 2008;132:645. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 18.Esposito MS, et al. J Neurosci. 2005;25:10074. doi: 10.1523/JNEUROSCI.3114-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mongiat LA, Esposito MS, Lombardi G, Schinder AF. PLoS ONE. 2009;4:e5320. doi: 10.1371/journal.pone.0005320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt-Hieber C, Jonas P, Bischofberger J. Nature. 2004;429:184. doi: 10.1038/nature02553. [DOI] [PubMed] [Google Scholar]
- 21.Kee N, Teixeira CM, Wang AH, Frankland PW. Nat Neurosci. 2007;10:355. doi: 10.1038/nn1847. [DOI] [PubMed] [Google Scholar]
- 22.Aimone JB, Wiles J, Gage FH. Neuron. 2009;61:187. doi: 10.1016/j.neuron.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge S, Sailor KA, Ming GL, Song H. J Physiol. 2008;586:3759. doi: 10.1113/jphysiol.2008.155655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aimone JB, Wiles J, Gage FH. Nat Neurosci. 2006;9:723. doi: 10.1038/nn1707. [DOI] [PubMed] [Google Scholar]
- 25.Ge S, Yang C-h, Hsu K-s, Ming G-l, Song H. Neuron. 2007;54:559. doi: 10.1016/j.neuron.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clelland CD, et al. Science. 2009;325:210. doi: 10.1126/science.1173215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YS, Silva AJ. Nat Rev Neurosci. 2009;10:126. doi: 10.1038/nrn2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidiropoulou K, Kyriaki E, Pissadaki, Poirazi P. EMBO. 2006;7:886. doi: 10.1038/sj.embor.7400789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin KC, Kosik KS. Nat Rev Neurosci. 2002;3:813. doi: 10.1038/nrn942. [DOI] [PubMed] [Google Scholar]
- 30.Morris RG. Eur J Neurosci. 2006;23:2829. doi: 10.1111/j.1460-9568.2006.04888.x. [DOI] [PubMed] [Google Scholar]
- 31.Govindarajan A, Kelleher RJ, Tonegawa S. Nat Rev Neurosci. 2006;7:575. doi: 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- 32.Poirazi P, Mel BW. Neuron. 2001;29:779. doi: 10.1016/s0896-6273(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 33.Mehta MR. Trends Neurosci. 2004;27:69. doi: 10.1016/j.tins.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Harvey CD, Svoboda K. Nature. 2007;450:1195. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey CD, Yasuda R, Zhong H, Svoboda K. Science. 2008;321:136. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Nature. 2009;458:299. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose J, Jin SX, Craig AM. Neuron. 2009;61:351. doi: 10.1016/j.neuron.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Losonczy A, Makara JK, Magee JC. Nature. 2008;452:436. doi: 10.1038/nature06725. [DOI] [PubMed] [Google Scholar]
- 39.Gelbard-Sagiv H, Mukamel R, Harel M, Malach R, Fried I. Science. 2008;322:96. doi: 10.1126/science.1164685. [DOI] [PMC free article] [PubMed] [Google Scholar]