Abstract
We combined single molecule fluorescence resonance energy transfer (smFRET) with single particle tracking in live cells to detect the in vivo conformation of individual proteins. We site-specifically labeled recombinant SNARE proteins with a FRET donor and acceptor before microinjecting them into cultured cells. We observed that individual proteins rapidly incorporated into folded complexes at the cell membrane, demonstrating the potential of this method to reveal dynamic interactions within cells.
Keywords: protein folding, SNAP-25, SNAP-29, SNARE complex, intrinsically unstructured protein, disordered, FRET
Dynamic conformational changes are important for function in many proteins. X-ray crystallography and NMR provide atomic-level details of the averaged behavior of an ensemble of molecules, but protein motions are not commonly revealed. Single molecule spectroscopic methods allow access to the conformational dynamics and transient associations of biomolecules that can be fundamental to biological function1. Unfortunately, biomolecular function in purified assays can be different than in native environments. Molecular crowding, regulatory interactions and cellular feedback networks all obscure the physiological relevance of many quantitative, in vitro studies. Applying structural and biochemical methodologies in an in vivo setting would address these issues, but such live cell applications are challenging2. Ensemble fluorescence resonance energy transfer (FRET) studies are widely used to measure interactions and conformations averaged over many molecules inside cells. Extending this approach to the single molecule level will enable observations hidden by ensemble averaging. Interrogating individual molecules can reveal heterogeneous subpopulations within the ensemble and can detect multiple pathways or intermediate states during dynamic transitions that are often unsynchronizable across populations inside cells. Here we combined single molecule FRET (smFRET) measurements, which are capable of reporting molecular conformations at nanometer resolution, with single particle tracking to observe individual SNARE proteins entering into membrane-tethered complexes in live cells.
Single molecule fluorescence observations are used extensively in live cells for tracking3. Extensions to smFRET detection in unperturbed live cell contexts have been limited to detection of small molecule ligands binding protein targets on the exterior surface4 or inside cells5. Neither of these applications used smFRET to address protein conformational changes related to function.
Ensemble FRET studies of molecular function are common in live cells using transfected protein chimeras with fluorescent proteins as a donor-acceptor pair. Variable expression levels make single molecule application of this approach challenging. Additionally, the large fluorescent moieties limit the choice of attachment location and may inhibit close approach of linked domains. We circumvented these limitations by using cysteine mutation in bacterially-expressed, recombinant proteins to allow site-specific attachment of small, organic fluorophores. Unnatural amino acid approaches for site-specific labeling can be used for proteins not amenable to cysteine mutation6. High-resolution structures guided our designs for labeling sites such that smFRET could confidently report conformation. We microinjected dilute solutions of labeled proteins into cells and viewed them using objective-type total internal reflection fluorescence microscopy (TIRFM) with spectrally-resolved detection for smFRET analysis (Methods).
We demonstrate our approach with SNARE proteins involved in cellular membrane fusion. SNAREs on vesicles form a complex with other SNAREs on membranes targeted for fusion. Many SNARE proteins, including those we studied, are unstructured when monomeric and undergo a dramatic folding transition upon entering into SNARE complex, a 10 nm-long coiled-coil of four α-helicies7. We designed dual-labeled SNAREs that emit low FRET when not in a complex, but could either yield low or high FRET depending on dye location when folded into a SNARE complex (Fig. 1a-b).
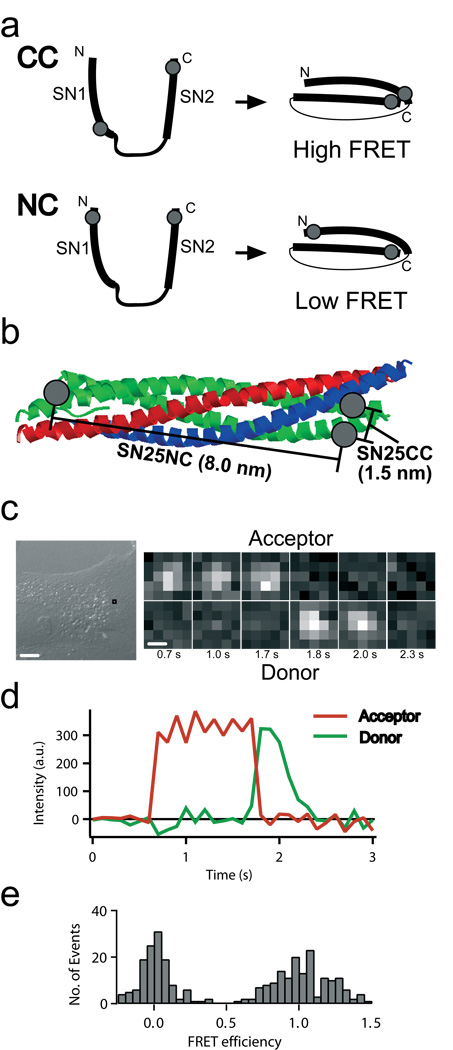
Figure 1. smFRET detection in live cells.
(a) Schematic of the SN25CC and NC label site locations. Isolated SNAP-25 (left) is unstructured, resulting in low FRET due to the large separation of the label attachment sites. When SNAP-25 enters SNARE complex (right, synaptobrevin and syntaxin not shown) the CC and NC constructs yield high and low FRET states, respectively. Gray dots represent the approximate dye-labeling sites (Methods). (b) Location of CC and NC label sites in assembled SNARE complex (PDB ID: 1sfc; syntaxin – red, synaptobrevin – blue, SNAP-25–green). CC (or NN) constructs were designed such that dye distances are less than 2 nm apart in the final SNARE complex and give high FRET. NC was designed so dyes are 8 nm apart in parallel complex and give low FRET. (c) Detail of a movie of fluorescence emission in the donor (Cy3, lower) and acceptor (Cy5, upper) channel taken from the boxed location indicated in the differential interference contrast image for SN25CC in a BS-C-1 cell. Time after injection in seconds is below the frames. (d) Intensity timecourse for smFRET example from c. Green illumination is active for entire interval plotted. (e) FRET efficiency histogram for single molecule events occurring in Supplementary Video 1 using Cy3-Cy5 labeled SN25CC in a BS-C-1 cell accumulated for 0.2 seconds (FRET<0.5) and 1.0 seconds (FRET>0.5) to sufficiently populate both peaks. Scalebars: 10 µm (c-left), .5 µm (c-right).
Our first experiments used the neuronal SNARE SNAP-25. Upon forming SNARE complex with its partner SNAREs synaptobrevin (on vesicles) and syntaxin (on the plasma membrane), the SNARE domains of SNAP-25 (SN1 and SN2) assemble into parallel alpha-helicies such that initially well-separated parts of the protein are brought close (Fig. 1a). We labeled SNAP-25 with a donor (Cy3 or Alexa555) and an acceptor (Cy5 or Alexa647), one in SN1 and the other in SN2 (similar to another ensemble, genetically-encoded FRET study8). High FRET occurs if both label sites are at the C-terminal (CC) or N-terminal ends (NN) of SN1 and SN2, whereas low FRET occurs if one dye is in the N-terminal end of SN1 and the other dye is in the C-terminal end of SN2 (NC) (Fig. 1a-b). Isolated SNAP-25 is unstructured, yielding low FRET. We verified these behaviors in vitro (Methods and Supplementary Fig. 1).
We injected these double-labeled soluble SNAP-25s (SN25CC and SN25NC) into BS-C-1 adherent cells while imaging with objective-type TIRFM. A diffuse fluorescence emission rapidly spread throughout the cell (Supplementary Videos 1-3). Immediately after injection, discrete points of emission in the donor and acceptor channels for SN25CC (Fig. 1c-e) indicated that some molecules became membrane-localized. Diffusing soluble molecules appear blurred with our 100 msec exposures (stopped motion imaging can image soluble molecules9), but the membrane-tethered SNAREs appear as diffraction-limited spots. Single molecule locations and spatial trajectories were compared to images of the cell obtained before and after fluorescence imaging (Fig. 2a-b). High FRET events spreading from the injection site indicated diffusion of the soluble protein in the cell (Fig. 2a inset).
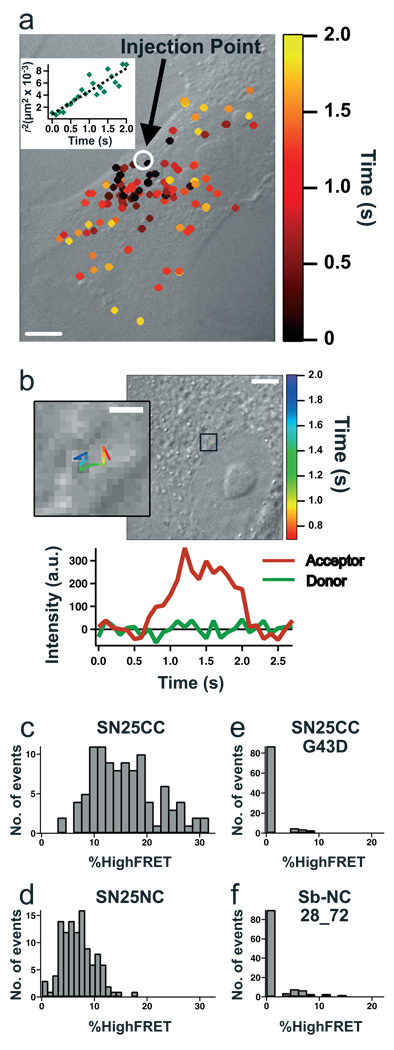
Figure 2. smFRET studies of SNARE proteins in BS-C-1 cells.
(a) high FRET events for Cy3-Cy5 labeled SN25CC overlaid onto a differential interference contrast cell image and color-coded by time after injection. The inset plots squared distance from the injection site of smFRET events vs. time. (b) Single-particle tracking of a high FRET, mobile Cy3-Cy5 labeled SN25CC with intensity timecourse. The inset is a zoom of the boxed region in the larger image. (c-f) histograms of %HighFRET measurements of multiple cells (N=10, 11, 10, 12) injected with proteins as indicated. SNAP-25 has a preference for parallel alignment (c vs. d). Also, the controls using the G43D SNAP-25 mutant and the Sb-NC do not yield high FRET. Scalebars: 10 µm (a), 5 µm (b), 1 µm (b-inset).
Examination of fluorescence intensities for membrane-bound molecules revealed sudden steps with anti-correlated donor and acceptor emission that are characteristic of smFRET (Fig. 1c-d and Supplementary Fig. 2). The emission of high FRET indicates the folding of SN25CC into SNARE complexes (Fig. 1e). Control experiments injecting mixtures of donor-only and acceptor-only SNAP-25 (SN25C) showed no high FRET (Supplementary Table 1 and Supplementary Fig. 3).
Precise injection efficiencies varied, so we normalized high acceptor emission events to the total single molecules imaged per cell. Comparing histograms of this normalized %HighFRET for experiments across many cells revealed that SN25CC (Fig. 2c) results in about twice as much %HighFRET as SN25NC (Fig. 2d). High FRET from SN25NC indicates in vivo anti-parallel SNARE complex assembly can occur10.
Several experiments confirmed that the high FRET signal results from SNARE complex formation. SN25CC with a mutation (G43D) that prevents SNARE complex formation in vitro and in vivo11 yielded no high FRET events above background levels (Fig. 2e, Supplementary Video 3 and Supplementary Table 1). Furthermore, injections of synaptobrevin lacking a transmembrane domain labeled with donor and acceptor dyes at opposite ends of the helix-forming region, keeping them 7 nm apart in complex (Sb-NC), also produced no high FRET events above background levels (Fig. 2f, Supplementary Table 1 and Supplementary Fig. 3).
We compared SNAREs from distinct trafficking pathways (SNAP-25 and SNAP-29) within two different cell types. SNAP-29 is widely expressed in most tissues, whereas SNAP-25 is specific for neurons. Therefore, we compared neuron-like PC-12 (neuroendocrine) cells to BS-C-1 green monkey kidney epithelial cells, which do not express SNAP-25. Interestingly, we observed both proteins participating efficiently in SNARE complex formation in both cell lines (Supplementary Table 1 and Supplementary Fig. 3). Despite some variability, we found SNAREs from distinct cellular pathways can substitute for each other in assembled complexes in living cells. This lack of specificity is not completely unanticipated because SNAREs are well-known to assemble promiscuously in vitro with many non-cognate partners7. Our in vivo observation of SNARE promiscuity supports other findings that implicate non-SNARE factors in maintaining proper vesicular trafficking in cells7,12.
We observed smFRET due to SNARE complex assembly immediately following injection (Fig. 2a), indicating that binding sites are available to quickly accept SNAREs in the natural resting state of these cells. Immunoassay12 and binding studies in permeabilized cells13,14 indicate that ~90% of the native syntaxin at the plasma membrane is available for forming SNARE complex while exogenous SNAP-25 and synaptobrevin both rapidly assemble into SNARE complex on plasma membranes. Our studies extend these conclusions to real-time analysis of fully intact, living cells.
Applying smFRET in live cells allows observations not possible with ensemble approaches, particularly revealing behaviors of small subpopulations. For example, our experiments determined from the duration of individual, high FRET binding events that SNAP-25 separates into two populations (10% and 90%) with exponentially distributed lifetimes differing by a factor of 4 (Supplementary Note and Supplementary Fig. 4). The two lifetimes could reflect two distinct bound states in the high FRET conformation – possibly the binary and ternary SNARE complexes – that would not be differentiated in ensemble measurements. Single particle tracking also allowed us to individuate molecules based upon mobility. Most high FRET (>0.7) emitting SNAP-25 molecules did not move, but 3.5% ± 1.5% of molecules (N=10 cells) diffused with mobility near .25 µm2·sec−1 (Fig. 2b and Supplementary Note), possibly due to attachment on intracellular vesicles. Co-injection of doubly-labeled SN25CC with excess (25 µM) unlabeled soluble synaptobrevin (1-96) increased the fraction of high FRET emitting SNAP-25 molecules that were mobile to 10.5% ± 3.4% (N=10 cells), demonstrating that increased synaptobrevin abundance alters the distribution of SNAP-25 acceptor sites within the cell’s trafficking networks in ways that our method can resolve.
smFRET in live cells presents many challenges. Dye-labeled protein concentrations must be sufficiently low to allow single molecules to be optically resolved. We used microinjection to introduce low concentrations of labeled protein into cells, but other approaches such as protein transfection reagents or attachment of membrane translocation domains may be useful in other applications. Detecting fluorescence emission from single molecules is difficult because cellular autofluorescence competes with emission from exogenous dyes. Our studies used the dramatic folding transition of SNAP-25 during SNARE complex formation to generate a large change in FRET efficiency. Cellular autofluorescence will be a more substantial obstacle in other applications where FRET changes are more subtle. Photobleaching the autofluorescence before injection of the labeled protein can avoid this problem. While we found no difference in %HighFRET following extensive photobleaching (Supplementary Fig. 5), we sought to avoid this approach due to phototoxicity concerns. Yet, assessing cells before injection sometimes involved 10–60 sec. of illumination. Other methods to minimize the impact of autofluorescence included optimizing cell growth conditions, microinjection of selected low-autofluorescent cells and TIRFM to observe membrane-localized events. Development of brighter fluorophores, two-photon excitation and infrared dye use could minimize the impact of autofluorescence and improve our method. Fluorophore lifetime currently prevents long-term observations, although some media additives can delay photobleaching in vivo (Methods and Supplementary Fig. 6).
In summary, we present an approach to follow conformational changes of individual proteins in living cells using smFRET and single particle tracking. We have found that SNARE proteins form promiscuous pairings with the large population of acceptor SNAREs natively present within two cell types. Extensions of this method using alternating two-color excitation to directly assess acceptor photoactivity and imaging with nanometer localization15 will further expand the utility of this approach.
Supplementary Material
Acknowledgements
We thank Brandon Choi and Trevor Anderson for assistance with protein preparation, Guilhem Ribeill and Eric Cady for software development, Guoping Feng and Zhonghua Lu for assistance with PC-12 cells, and Mark Bowen, James Ernst and George Augustine for useful discussions. Keith Weninger is supported in part by a CASI award from the Burroughs Wellcome Fund. This work was funded by the NIH (GM076039).
Footnotes
Author Contributions:
J.S. performed all measurements and data analysis. J.S. and K.W. contributed substantially to all other aspects.
LITERATURE CITATIONS
- 1.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Annu Rev Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 2.Inomata K, et al. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 3.Wieser S, Schutz GJ. Methods. 2008;46:131–140. doi: 10.1016/j.ymeth.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Sako Y, Minoghchi S, Yanagida T. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 5.Murakoshi H, et al. Proc Natl Acad Sci U S A. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brustad EM, Lemke EA, Schultz PG, Deniz AA. J Am Chem Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahn R, Scheller RH. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 8.An SJ, Almers W. Science. 2004;306:1042–1046. doi: 10.1126/science.1102559. [DOI] [PubMed] [Google Scholar]
- 9.Xie XS, Yu J, Yang WY. Science. 2006;312:228–230. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 10.Weninger K, Bowen ME, Chu S, Brunger AT. Proc Natl Acad Sci U S A. 2003;100:14800–14805. doi: 10.1073/pnas.2036428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loranger SS, Linder ME. J Biol Chem. 2002;277:34303–34309. doi: 10.1074/jbc.M202125200. [DOI] [PubMed] [Google Scholar]
- 12.Bajohrs M, Darios F, Peak-Chew SY, Davletov B. Biochem J. 2005;392:283–289. doi: 10.1042/BJ20050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-On D, et al. FEBS Lett. 2008;582:3563–3568. doi: 10.1016/j.febslet.2008.08.040. [DOI] [PubMed] [Google Scholar]
- 14.Lang T, Margittai M, Holzler H, Jahn R. J Cell Biol. 2002;158:751–760. doi: 10.1083/jcb.200203088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz A, et al. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 16.Bowen ME, Weninger K, Brunger AT, Chu S. Biophys J. 2004;87:3569–3584. doi: 10.1529/biophysj.104.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen ME, Weninger K, Ernst J, Chu S, Brunger AT. Biophys J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steegmaier M, et al. J Biol Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- 20.Su Q, Mochida S, Tian JH, Mehta R, Sheng ZH. Proc Natl Acad Sci U S A. 2001;98:14038–14043. doi: 10.1073/pnas.251532398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Augustine GJ, Weninger K. Biophys J. 2007;93:2178–2187. doi: 10.1529/biophysj.106.101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boukobza E, Sonnenfeld A, Haran G. Journal of Physical Chemistry B. 2001;105:12165–12170. [Google Scholar]
- 23.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuurman N, Vale R. In: Live Cell Imaging: A laboratory manual. Goldman RD, Spector DL, editors. Woodbury: Cold Springs Harbor Laboratory Press; 2005. pp. 585–602. [Google Scholar]
- 25.Cordes T, Vogelsang J, Tinnefeld P. Journal of the American Chemical Society. 2009;131:5018–5019. doi: 10.1021/ja809117z. [DOI] [PubMed] [Google Scholar]
- 26.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 28.Rasnik I, McKinney SA, Ha T. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.