Abstract
Background
The relation of γ-aminobutyric acid (GABA) to alcohol dependence (AD) has been widely studied. Several previous studies suggest that GABA may be involved in alcohol withdrawal, tolerance, and the symptoms that form an AD diagnosis. The genes coding for glutamate decarboxylase (GAD), the rate-limiting enzyme in GABA synthesis, are of potential interest for their association to ethanol consumption and AD. There are two isoforms of GAD, GAD1 and GAD2, which were reported to be associated with AD in males of Han Taiwanese (GAD1) and Russian (GAD2) ancestry. The present study examined the association of the two GAD isoforms with AD and relevant alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence [Prescott, C.A., Sullivan, P.F., Myers, J.M., Patterson, D.G., Devitt, M., Halberstadt, L.J., Walsh, D., Kendler, K.S., 2005. The Irish Affected Sib Pair Study of Alcohol Dependence: study methodology and validation of diagnosis by interview and family history. Alcohol.-Clin. Exp. Res. 29 (3) 417–429].
Methods
Participants were recruited in Ireland, including 575 independent cases who met DSM-IV AD criteria and 530 controls, screened for heavy drinking. We first conducted case-control analyses of the GAD genes with AD and, within the cases, examined associations with age at onset of AD, withdrawal symptoms, and two quantitative measures: initial sensitivity and tolerance (based on scales from the Self-Rating of the Effects of Ethanol) [Schuckit, M.A., Smith, T.L., Tipp, J.E., 1997. The self-rating of the effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction 92, 979–988]. A total of 29 SNPs were genotyped for GAD1 and GAD2 using the Illumina GoldenGate protocols. Statistical procedures were implemented to control for false discovery rates (FDR).
Results
Nine of 29 markers with minor allele frequencies less than 0.01 were removed from standard analysis; the remaining 20 markers were all in Hardy-Weinberg equilibrium. Three markers in the intronic regions of GAD1 were associated with initial sensitivity to alcohol (P = 0.002); the associations remained significant after a FDR based correction for multiple testing. In addition, one marker located 3 kb upstream of GAD1 exhibited association with age at onset of AD (P = 0.0001). Gender specific effects were observed in results of both single marker and haplotype analyses.
Conclusion
We found no evidence for the association of GAD genes with AD but significant association of GAD1 with initial sensitivity and age at onset of AD. Our findings suggest that the underlying pathophysiology regulated by genes like GAD1 may be more directly related to the component processes that form AD than to the clinical disorder.
Keywords: GABA, Initial sensitivity, Response to ethanol, Withdrawal, Gender difference
1. Introduction
In the central nervous system, γ-aminobutyric acid (GABA) is the major inhibitory neurotransmitter. Results from both human and animal studies suggest that neuroadaptation in the GABAergic system and pharmacological sensitivity of GABA receptors play important roles in the behavioral and functional neuronal changes associated with ethanol dependence (Crabbe et al., 2006; Follesa et al., 2006). Acute ethanol consumption enhances GABA neurotransmission and inhibits the glutamatergic system (Nevo and Hamon, 1995). The reduction in excitatory effect and the enhancement of inhibitory GABA neurotransmission account for the sedating and dose-dependent depressant effects of ethanol intoxication. However, prolonged ethanol exposure produces neruoadaptation in these neurotransmitter systems, resulting in reduced inhibitory activity of GABA receptors and a sensitization of N-methyl-d-aspartic acid receptors (see reviews by Crews et al., 1996; Davis and Wu, 2001). Sudden cessation of ethanol intake produces a hyperexcitable neuronal state that results in mild withdrawal symptoms just as the “shakes” which, in more severe cases, leads to seizures, delirium, and excitotoxic neuronal death (Tsai and Coyle, 1998).
The biosynthesis of GABA largely depends on the enzyme glutamate decarboxylase (GAD) (Petroff, 2002). Two isoforms of GAD have been identified, GAD1 and GAD2, which previously were called GAD67 and GAD65, respectively. These enzymes couple with pyridoxal phosphate as a cofactor and are responsible for catalyzing glutamate to produce GABA in the brain. In humans, GAD1 maps to chromosome 2q31 and spans about 45 kb, and GAD2 maps to chromosome 10p11.23 with a size about 88 kb. GAD1 and GAD2 are highly similar in their catalytic domains, regional expression, and subcellular localization (Erlander et al., 1991; Feldblum et al., 1995), but are associated with different modes of GABA release. GAD1 is involved in cytosolic GABA synthesis and is responsible for maintaining basal GABA levels, whereas GAD2 is predominately involved in synaptosomal GABA release, and can be rapidly activated when there is high demand for GABA (Soghomonian and Martin, 1998). Rodent studies suggest that GAD1 is important in maintaining GABA levels in the brain, and that the GAD1 knockout is usually lethal. In contrast, GAD2 knockout mice maintain normal brain GABA levels, but are susceptible to seizure, show increased anxiety behavior, and have decreased response to sedative drugs such as benzodiazepines and pentobarbital (Asada et al., 1996; Kash et al., 1999).
Most prior studies of the relation between GAD genes and substance addictive behaviors have used animal models. Mice and rat studies have shown that GAD genes expression was altered during diazepam withdrawal (Izzo et al., 2001), alcohol withdrawal or dependence (Eravci et al., 2000), or chronic exposure to high-dose methamphetamine (Zhang et al., 2006). However, the evidence of expression alteration of GAD genes is not consistent (Fehr et al., 2003), and gene expression levels appear to vary by brain region in response to drug treatment. We are aware of only two published reports examining the association of GAD genes with alcohol-related phenotypes in humans. Loh et al. (2006) examined the association of 9 SNPs for GAD1 and 3 SNPs for GAD2 in 140 alcoholic cases and 146 controls in Han Taiwanese men. They found evidence of association with AD in GAD1 but not GAD2. Lappalainen et al. (2007) genotyped 13 SNPs for GAD2 in a sample of 113 Russian males with AD and 100 controls and found modest association for a functional marker, rs2236418 (−243 A > G). However, in the same report, these authors reported they were unable to replicate this association using two other samples: European–American with AD and U.S. college students with drinking problems. Therefore, the role of GAD genes in AD and alcohol-related problems requires further study.
The majority of previous reports, in both animals and humans, included only male subjects. Gender is an important element when studying substance use. For instance, in humans males begin drinking regularly and heavily much earlier than females; men also consume larger amount of alcohol daily than women (Prescott et al., 2005). Although women have a lower incidence of AD than men, on average, they develop alcohol dependence after a shorter duration of chronic drinking and show withdrawal following consumption of smaller volumes of ethanol. Some studies suggest that male and female alcoholics show differences in brain changes and negative consequences due to alcohol abuse (Pfefferbaum et al., 2001; Wuethrich, 2001). However, lack of genetic association data in female alcoholism is a weakness in the literature. The first aim of the present study was to test the association of GAD genes with AD in a different and larger independent sample. Given the differences in the effects of ethanol between males and females noted above, we tested whether there are gender specific effects in the association of GAD genes with AD.
In addition, results from quantitative trait locus (QTL) studies in animals suggest that specific loci may influence different alcohol-related behaviors, such as acute/chronic withdrawal, preference drinking, and blood level after acute ethanol dose (see Crabbe et al., 1999). One such alcohol-related trait is the level of response to ethanol, measured by the dose of ethanol needed to produce a specified effect. Level of response at the time of initial use is a risk factor for the development of alcoholism (Schuckit and Smith, 2001). Other alcohol-related traits, including tolerance and withdrawal, reflect physiological adaptation to chronic alcohol use (Schuckit et al., 1998). Withdrawal is an especially pivotal physical symptom for AD diagnosis (Langenbucher et al., 2000) and is associated with worse prognosis (Hasin et al., 2000). In addition to these physiological traits, variation in age at onset of AD may also reflect differences in the magnitude of the underlying AD liability (Johnson et al., 2000). These quantitative alcohol-related traits represent different aspects of the dependence process, and may provide greater insight into the genetic contributions to the risk to develop AD. A second aim of our study, therefore, was to examine evidence that GAD genes are involved with quantitative alcohol-related traits including level of response to ethanol, tolerance, withdrawal and age at onset.
We conducted a genetic association study of both GAD genes in a sample of Western European origin that is more culturally and genetically homogeneous than samples used in previous studies. We examine the association of the GAD1 and GAD2 genes with AD and relevant alcohol-related traits as well as possible gender specific effects in these genetic associations.
2. Methods
2.1. Subjects and phenotype assessment
Participants in this study were recruited in Ireland and Northern Ireland between 1998 and 2002. Details of the study design, sample ascertainment, and clinical characteristics of this sample are described elsewhere (Prescott et al., 2005). In brief, ascertainment of probands was mainly conducted in community alcoholism treatment facilities and public and private hospitals. Probands were eligible for study inclusion if they met the current DSM-IV criteria for AD and if all four grandparents had been born in Ireland, Northern Ireland, Scotland, Wales or England. After a prospective family was identified through probands, parents and potentially affected siblings whom the probands provided permission to contact were recruited.
Interviews were conducted by clinically trained research interviewers, most of whom had extensive clinical experience with alcoholism. The interview included demographic characteristics, lifetime history of AD and alcohol-related traits, and comorbid conditions. DSM-IV AD was assessed using the SSAGA (Semi-Structured Assessment of the Genetics of Alcoholism) interview (version II, Bucholz et al., 1994) modified to reduce assessment time by omitting items that address onset age of each symptom.
All participants provided informed consent. There were 1238 individuals meeting DSM-IV AD diagnosis from 591 families. Controls were recruited in the Northern Ireland from volunteers of blood donating (88.5%) and in Ireland from national police force and army reserve. Controls were screened and their samples excluded if they reported a history of heavy drinking or problematic alcohol use. In the present case-control study design, we included 530 controls and 575 independent AD cases from the IASPSAD families (for detailed case and control sample description see Kuo et al., 2008). Samples were selected based on high yield of high quality DNA for genotyping and only one case per family was included.
In the present study, we are interested in several alcohol-related phenotypes, which are hypothesized to be associated with GAD genes, including age-at-onset of AD, subjective response to ethanol, and withdrawal symptoms. Age-at-onset (ONSET) was defined as the age at which the first criteria for DSM-IV AD was met. Subjective response to ethanol was measured using the Self-Rating of the Effects of Alcohol, SRE (Schuckit et al., 1997) to form two scores, initial sensitivity (ISENS) and tolerance/maximum drinking (TOLMX). The SRE asks how many drinks were required for an individual to experience effects (e.g. feel dizzy, begin stumbling, or pass out) from alcohol consumption at different stages of alcohol use. ISENS is based on “the first 5 times you ever drank” and items contributing to TOLMX concern the “period when you drank the most”. The score of each measure was computed by summing the number of drinks required to produce an effect and dividing by the numbers of effects endorsed. The SRE has been shown to have good internal consistency and test–retest reliability, provide good validity for identifying people who had low response to alcohol in a laboratory challenge test, and has been associated with AD diagnosis in several populations. A withdrawal severity factor score (WDSFS) was based on ten symptoms in the SSAGA interview (such as hands trembling, feeling anxious following cessation or reduction of drinking). To account for the possible non-equal contribution of each symptom to withdrawal severity, a factor analysis was conducted (for details see Kuo et al., 2006). A factor score of withdrawal severity for each individual was derived based on the item loadings on one major factor, which accounted for 70% of the variance in these symptoms based on the entire IASPSAD sample.
2.2. Genotyping
Genotypes for a total of 29 SNPs in the GAD1 (N = 12) and GAD2 (N = 17) genes were obtained as part of a large candidate gene study using an Illumina custom genotyping array designed in Dr. David Goldman's Laboratory of Neurogenetics, National Institute on Alcohol Abuse and Alcoholism. Detailed information of array design, genotyping and SNP selection are described elsewhere (Hodgkinson et al., 2008). In brief, for each gene selected, a genomic region including 5 kb upstream and 1 kb downstream was retrieved from NCBI Human Genome Build 35.1 and minimum index SNPs that represented maximum haplotype information for each gene were selected. The performance of the initially selected SNP set was validated by the manufacturer and replacements made where necessary. None of the selected 12 SNPs in GAD1 are coding SNPs, and two of them are at 5′ of the gene (around 1 kb and 3 kb upstream, respectively). One selected SNP in GAD2 is a coding SNP (rs2839673, locates in exon 6; A/G polymorphism codes for Glu/Gly) with a low minor allele frequency (0.01) in the present sample.
All genotyping was conducted in Dr. David Goldman's Laboratory. Genotyping was performed using the Illumina GoldenGate genotyping protocols on 96-well format Sentrix® arrays and 500 ng of sample DNA were used per assay. All pre-PCR processing was performed using a TECAN liquid handling robot running Illumina protocols. Arrays were imaged using an Illumina Beadstation GX500 and the data analyzed using GenCall v6.2.0.4 and GTS Reports software v5.1.2.0 (Illumina).
2.3. Statistical methods
Case-control association analyses for AD were conducted using PLINK (Purcell, http://pngu.mgh.harvard.edu/∼purcell/plink/) at both single marker and haplotype levels for GAD1 and GAD2 genes. For quantitative alcohol-related traits measured within the cases, we used regression based models implemented in PLINK to calculate effect size and significance level for each marker. To examine the gender specific effects in the association of GAD genes with AD and alcohol-related traits, the aforementioned case-control binary and case-only quantitative association analyses were conducted in the overall sample and in the female and male samples separately. Among 1105 total samples, there were 668 males (370 cases), 433 females (204 cases), and 4 samples with unspecified sex1 which were excluded from gender specific analysis.
To address multiple testing issues in these analyses, a FDR (false discovery rate) based procedure (Benjamini and Hochberg, 1995) was implemented using the SAS package to obtain adjusted P-values. The FDR is estimated as the proportion of expected false rejections over the total number of rejections and is determined from the observed P-value distribution.
Haploview 4.0 was used to calculate the strength of linkage disequilibrium (LD, i.e. the non-random association of alleles at two or more loci) using the D′ index. Relative location of SNPs within each GAD gene and the LD plot are shown in Fig. 1 (detailed information of both D′ and r2 indexes of pair-wise SNPs for each GAD gene is listed in supplement Table 1). Haplotype blocks were defined using the default method in Haploview by Gabriel et al. (2002). In general, there was high LD within each GAD gene.
Fig. 1.
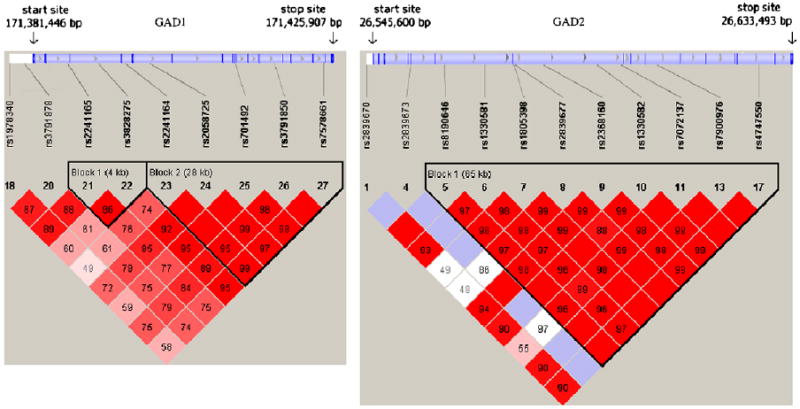
The LD plot (D′) and SNPs location within each GAD gene. Haplotype blocks were defined using the default method in Haploview by Gabriel et al. (2002).
3. Results
Summary information of alcohol-related traits and comorbid major depressive disorder by gender is shown in Table 1. In general, men exhibited more severe alcohol-related problems compared to women. Men had younger age-at-onset of AD, drank more to feel the effects of ethanol during both early exposures and the period of heaviest drinking and had more withdrawal symptoms than women (detailed description of alcohol-related behaviors for the whole IASPSAD family sample is in Prescott et al., 2005).
Table 1.
Descriptive characteristics for alcohol-related traits by gender among cases with alcohol dependence (AD).
| Trait | Male | Female | ||||
|---|---|---|---|---|---|---|
| N | Mean ± SD | Range | N | Mean ± SD | Range | |
| ONSETa | 801 | 24.8 ± 7.9 | 13–64 | 437 | 28.9 ± 10.3 | 12–67 |
| ISENSa | 745 | 6.5 ± 3.2 | 1.0–27.8 | 415 | 5.5 ± 2.5 | 1.3–23.3 |
| TOLMXa | 748 | 13.6 ± 6.7 | 2.7–44.0 | 414 | 10.3 ± 5.1 | 2.0–38.3 |
| WDSFSa | 809 | −0.01 ± 0.80 | −1.85 to 1.02 | 437 | −0.20 ± 0.84 | −1.84 to 1.02 |
| MDsx | 791 | 5.7 ± 3.6 | 0–9 | 431 | 6.0 ± 3.4 | 0–9 |
| N | n | Proportion (%) | N | n | Proportion (%) | |
| MDD | 791 | 554 | 70.0 | 431 | 317 | 73.6 |
Note: ONSET, age-at-onset of AD; ISENS, initial sensitivity; TOLMX, tolerance/maximum drinking; WDSFS, withdrawal severity factor score; MDsx, symptom counts for major depressive diagnosis; MDD, major depressive disorder.
Test for gender differences, P < 0.0001.
3.1. Genotyping completion
Genotyping was completed for 12 SNPs in GAD1 and 17 SNPs in GAD2. Among the 29 genotyped SNPs we excluded 1 with a low genotyping rate (<80%) and 8 with MAF < 0.01, leaving 9 in GAD1 and 11 in GAD2. The average genotyping call rate for these 20 SNPs analyzed was ranged from 94.3 to 95.5%. All SNPs retained in the analysis were in Hardy-Weinberg equilibrium (using a cutoff P-value of 0.001) in the overall sample and in controls alone. Among the 1105 genotyped individuals, 16 (9 cases, 7 controls) were removed from analysis because of genotyping call rates less than 50%. The sample included in the final analysis thus consisted of 566 AD cases and 523 controls. The average genotyping rate among these individuals is 95.84%.
3.2. Single marker association
Marker information, allele frequency and single marker allelic association results are displayed in Table 2 for 20 SNPs in the two GAD genes. In the total sample, no SNP in either GAD gene was associated with AD (other than allelic association test, genotypic tests including general genotype test, dominant & recessive models, and Cochran-Armitage trend test were also performed. No genotypic associations were found and results were not presented in Table 2). Among females, several SNPs in GAD2 showed modest association (P = 0.04) with AD, although the effects become non-significant after adjusting for false discoveries (Table 2).
Table 2.
Markers information of GAD1 and GAD2 genes in cases and controls; both genes show no association with alcohol dependence (AD).
| GeneSymbol | SNP | bp | Location | Call rate | Allele frequency in total sample | P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF | Case | Control | Total sample | Females | Males | |||||
| GAD1 (Chromosome 2) | rs1978340 | 171495628 | 5′ near gene | 0.955 | 0.325 | 0.327 | 0.322 | 0.82 | 0.29 | 0.48 |
| rs3791878 | 171497698 | 5′ near gene | 0.954 | 0.337 | 0.328 | 0.347 | 0.36 | 0.89 | 0.27 | |
| rs2241165 | 171503886 | intron 2 | 0.955 | 0.238 | 0.234 | 0.243 | 0.63 | 0.27 | 0.55 | |
| rs3828275 | 171508247 | intron 3 | 0.955 | 0.431 | 0.443 | 0.418 | 0.24 | 0.62 | 0.33 | |
| rs2241164 | 171512066 | intron 4 | 0.955 | 0.330 | 0.325 | 0.336 | 0.59 | 0.89 | 0.47 | |
| rs2058725 | 171515628 | intron 5 | 0.955 | 0.237 | 0.235 | 0.239 | 0.82 | 0.48 | 0.54 | |
| rs701492 | 171527987 | intron 9 | 0.954 | 0.289 | 0.280 | 0.298 | 0.35 | 0.94 | 0.18 | |
| rs3791850 | 171533607 | intron 12 | 0.955 | 0.242 | 0.239 | 0.245 | 0.74 | 0.54 | 0.65 | |
| rs7578661 | 171540640 | intron 15 | 0.955 | 0.288 | 0.279 | 0.298 | 0.35 | 0.92 | 0.15 | |
| GAD2 (Chromosome 10) | rs2839670 | 26544346 | 5′ near gene | 0.955 | 0.180 | 0.165 | 0.196 | 0.06 | 0.04* | 0.38 |
| rs2839673 | 26553557 | exon 6 (A/G) | 0.951 | 0.011 | 0.009 | 0.014 | 0.28 | 0.85 | 0.31 | |
| rs8190646 | 26560513 | intron 7 | 0.953 | 0.092 | 0.102 | 0.081 | 0.10 | 0.66 | 0.13 | |
| rs1330581 | 26568841 | intron 7 | 0.943 | 0.285 | 0.278 | 0.292 | 0.45 | 0.05 | 0.78 | |
| rs1805398 | 26574815 | intron 7 | 0.955 | 0.100 | 0.108 | 0.092 | 0.22 | 0.80 | 0.10 | |
| rs2839677 | 26574943 | intron 8 | 0.954 | 0.099 | 0.107 | 0.091 | 0.22 | 0.99 | 0.14 | |
| rs2368160 | 26580777 | intron 8 | 0.950 | 0.289 | 0.285 | 0.294 | 0.68 | 0.09 | 0.55 | |
| rs1330582 | 26592120 | intron 8 | 0.955 | 0.186 | 0.172 | 0.200 | 0.10 | 0.04* | 0.58 | |
| rs7072137 | 26596973 | intron 8 | 0.955 | 0.107 | 0.113 | 0.101 | 0.36 | 0.87 | 0.20 | |
| rs7900976 | 26604439 | intron 11 | 0.955 | 0.187 | 0.174 | 0.200 | 0.13 | 0.04* | 0.65 | |
| rs4747550 | 26625608 | intron 15 | 0.955 | 0.185 | 0.173 | 0.198 | 0.13 | 0.04* | 0.65 | |
Note: MAF, minor allele frequency.
P < 0.05 (FDR adjusted P becomes non-significant).
Quantitative association results of single markers for each alcohol-related trait are listed in Table 3. A positive regression coefficient in the regression models indicates that individuals with the minor allele have higher scores for the examined trait, and vice versa. For instance, the negative value (−1.24) for ONSET in the total sample at the first marker listed (rs1978340) indicates that individuals with the minor allele of rs1978340 have earlier AD onset than those with the common allele. This association of marker rs1978340 with ONSET exhibits a strong effect (regression coefficient = −2.28 with a FDR adjusted P < 0.001) in male samples. Three other markers in GAD1 showed association with ISENS in the total sample (FDR adjusted P = 0.015). These three markers showed modest association in male subgroup, but were not significant after adjusting for false discoveries. The negative regression coefficients of the three markers with ISENS indicate higher response to ethanol (lower number of drinks needed to feel subjective effects) with the minor allele, which suggests that a common allele of these markers may increase the risk for developing AD. Although a few markers in the GAD2 gene have P-values < 0.05 for their associations with TOLMX and WDSFS, they became non-significant after adjusting for false discoveries, which may result from small effect size of those markers or insufficient power to detect their effects with the current sample size of cases.
Table 3.
Effect size and direction of single marker association between GAD1 and GAD2 genes and alcohol-related traits among cases.
| GeneSymbol | SNP | ONSET | ISENS | TOLMX | WDSFS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Female | Male | Total | Female | Male | Total | Female | Male | Total | Female | Male | ||
| GAD1 (Chromosome 2) | rs1978340 | −1.24* | 1.01 | −2.28£ | 0.31 | 0.04 | 0.39 | 0.47 | 0.31 | 0.33 | 0.01 | 0.09 | −0.03 |
| rs3791878 | −0.63 | 1.13 | −1.48* | 0.38 | 0.26 | 0.36 | 0.27 | 0.15 | 0.09 | −0.02 | 0.00 | −0.04 | |
| rs2241165 | 0.74 | −0.60 | 1.32* | −0.66† | −0.51 | −0.70* | −0.27 | 0.22 | −0.39 | −0.02 | 0.04 | −0.05 | |
| rs3828275 | 0.19 | 0.56 | 0.05 | 0.23 | 0.13 | 0.29 | 0.32 | −0.38 | 0.73 | 0.02 | −0.09 | 0.07 | |
| rs2241164 | −0.61 | 0.57 | −0.91 | 0.29 | 0.22 | 0.25 | −0.17 | −0.09 | −0.52 | 0.03 | −0.01 | 0.04 | |
| rs2058725 | 0.21 | −1.38 | 0.87 | −0.66† | −0.44 | −0.71* | −0.65 | −0.21 | −0.60 | −0.03 | 0.10 | −0.10 | |
| rs701492 | −0.72 | 0.20 | −0.93 | 0.39 | 0.33 | 0.35 | −0.13 | 0.18 | −0.48 | 0.01 | 0.04 | −0.01 | |
| rs3791850 | 0.27 | −1.21 | 0.86 | −0.68† | −0.49 | −0.72* | −0.76 | −0.29 | −0.74 | −0.04 | 0.08 | −0.10 | |
| rs7578661 | −0.82 | 0.01 | −1.07 | 0.40 | 0.35 | 0.37 | −0.10 | 0.13 | −0.39 | 0.01 | 0.04 | −0.01 | |
| GAD2 (Chromosome 10) | rs2839670 | −0.63 | 0.20 | −0.67 | 0.46 | 0.17 | 0.51 | 0.11 | −0.89 | 0.30 | 0.11 | 0.22 | 0.06 |
| rs2839673 | −1.99 | −8.09 | 0.66 | 0.66 | 0.49 | 1.18 | −1.54 | 0.98 | −2.85 | −0.34 | 0.24 | −0.59* | |
| rs8190646 | 0.64 | 0.71 | 1.04 | 0.11 | −0.10 | 0.10 | 0.69 | 0.63 | 0.33 | −0.04 | 0.06 | −0.10 | |
| rs1330581 | −0.09 | 0.62 | 0.17 | 0.34 | −0.12 | 0.41 | 0.32 | −0.33 | 0.16 | 0.07 | 0.15 | 0.02 | |
| rs1805398 | 0.16 | −0.70 | 1.01 | 0.27 | −0.15 | 0.34 | 0.89 | 0.81 | 0.50 | −0.02 | 0.11 | −0.09 | |
| rs2839677 | 0.13 | −0.83 | 1.04 | 0.33 | 0.05 | 0.35 | 0.81 | 0.68 | 0.47 | −0.03 | 0.09 | −0.10 | |
| rs2368160 | 0.06 | 0.83 | 0.26 | 0.32 | −0.11 | 0.39 | 0.43 | 0.12 | 0.19 | 0.07 | 0.13 | 0.04 | |
| rs1330582 | −0.25 | 1.29 | −0.45 | 0.35 | 0.10 | 0.36 | −0.21 | −1.11 | −0.14 | 0.11 | 0.11 | 0.09 | |
| rs7072137 | 0.60 | 0.09 | 1.21 | 0.29 | −0.17 | 0.41 | 1.10 | 1.61* | 0.55 | −0.02 | 0.07 | −0.08 | |
| rs7900976 | −0.30 | 1.32 | −0.49 | 0.33 | 0.03 | 0.35 | −0.11 | −1.12 | −0.02 | 0.12* | 0.12 | 0.11 | |
| rs4747550 | −0.31 | 1.24 | −0.47 | 0.33 | 0.03 | 0.36 | −0.10 | −1.12 | −0.01 | 0.12 | 0.13 | 0.10 | |
Note: ONSET, age-at-onset of AD; ISENS, initial sensitivity; TOLMX, tolerance/maximum drinking; WDSFS, withdrawal severity factor score.
FDR adjusted P < 0.05 are in bold type.
P < 0.05 (FDR adjusted P becomes non-significant).
P = 0.002 (FDR adjusted P = 0.015).
P = 0.0001 (FDR adjusted P < 0.001).
3.3. Linkage disequilibrium structure
For GAD1, two blocks were defined consisting of 2 and 5 markers, respectively. However, LD between these two blocks is strong and yields a highly correlated haplotype distribution for the common haplotypes. The most common haplotype in block 1 (frequency 42.8%) occurs with the most common haplotype in block 2 (43.1%); the second and third most common haplotypes in block 1 are similarly related to the corresponding haplotypes in block 2. The one-to-one correspondence of the three common haplotypes results in a high correlation of 0.84 between the two blocks. Therefore, we collapsed the two blocks together, forming a long haplotype block with seven markers. For GAD2, LD was extremely high among 9 out of 11 genotyped markers, with a mean pair-wise D′ of 0.983 (D′ estimates for adjacent SNPs were greater than 0.95 for 97% of the comparisons). We used Tagger with r2 of 0.8 and MAF of 0.2 (de Bakker et al., 2005) to select the minimum numbers of markers to capture maximum haplotype variation within blocks for the GAD1 and GAD2 genes. Consequently, four (rs2241165, rs3828275, rs2058725, rs701492) and two (rs2839677, rs7900976) tagging SNPs were used for haplotype analyses in GAD1 and GAD2, respectively.
3.4. Haplotypic association
We examined association of haplotypes using only the Tagger-defined tagging SNPs (Table 4). For GAD1, the four SNPs form seven haplotypes with frequency greater than 0.01. Three common haplotypes together represent 90% of haplotypes observed. A significant association was found for ISENS with haplotype 2111 (frequency = 0.01) in females, which consisted of a minor allele of marker rs2241165 and common alleles of rs3828275, rs2058725, and rs701492. Again, the negative regression coefficient of this haplotype with ISENS and TOLMX indicated fewer drinks needed to feel subjective effects, in both drinking initiation and heaviest drinking periods, among individuals with this haplotype. In males, the 1121 haplotype (frequency = 0.02) showed association with WDSFS (FDR adjusted P < 0.05) with a negative regression coefficient, indicating a less severe withdrawal syndrome among individuals with this haplotype. However, both associated haplotypes had small frequency (representing 13–25 chromosomes each among our samples), thus the significant haplotypic findings and their true contributions should be taken in caution and larger study is required to confirm these findings. For GAD2, none of the three common haplotypes were significantly associated with alcohol-related traits using criteria of FDR adjusted P < 0.05.
Table 4.
Effect size and direction of haplotype-specific associations between GAD1 or GAD2 genes and alcohol-related traits among cases.
| Haplotype-block | Frequency | ONSET | ISENS | TOLMX | WDSFS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Female | Male | Total | Female | Male | Total | Female | Male | Total | Female | Male | ||
| GAD1 | |||||||||||||
| 1211 | 0.421 | 0.695 | 0.500 | 0.802 | 0.245 | 2.669 | −1.102 | 0.280 | 2.201 | −0.787 | 0.001 | −0.101 | 0.057 |
| 1112 | 0.269 | −1.238 | 0.366 | −1.775 | −1.560 | −3.258 | −0.831 | −2.424 | −4.273 | −1.770 | 0.009 | 0.047 | −0.017 |
| 2121 | 0.211 | 0.592 | −0.929 | 1.267 | 2.042 | 3.916 | 1.005 | 2.766 | 5.369 | 1.437 | −0.012 | 0.072 | −0.055 |
| 1111 | 0.043 | 0.423 | 1.220 | 0.156 | −1.466 | −14.390* | 4.938 | −0.084 | −12.920 | 6.199 | 0.089 | 0.018 | 0.120 |
| 1121 | 0.019 | −3.373 | −4.796 | −3.431 | 3.125 | 2.515 | 4.498 | 0.807 | 1.397 | 1.777 | −0.257 | 0.170 | −0.708† |
| 2112 | 0.011 | 4.855 | 0.974 | 6.982 | 2.829 | −0.055 | 4.444 | 3.112 | −0.058 | 4.904 | −0.218 | −0.321 | −0.160 |
| 2111 | 0.011 | 1.014 | 4.671 | −2.623 | −14.970* | −42.660£ | 6.065 | −13.040 | −40.850† | 8.625 | −0.058 | −0.232 | 0.101 |
| GAD2 | |||||||||||||
| 11 | 0.715 | 0.551 | −0.612 | 0.351 | −0.673 | −3.358 | 0.483 | −0.499 | −2.294 | 0.723 | −0.077 | −0.153 | −0.030 |
| 12 | 0.185 | −0.469 | 1.420 | −0.696 | −0.258 | 3.651 | −1.964 | −0.442 | 3.415 | −2.467 | 0.125* | 0.123 | 0.111 |
| 21 | 0.101 | −0.473 | −0.788 | 0.257 | 1.747 | 1.427 | 1.781 | 1.648 | −0.304 | 1.994 | −0.017 | 0.124 | −0.094 |
Note: ONSET, age-at-onset of AD; ISENS, initial sensitivity; TOLMX, tolerance/maximum drinking; WDSFS, withdrawal severity factor score.
GAD1 Haplotype-block consists of SNPs: rs2241165-rs3828275-rs2058725-rs701492.
GAD2 Haplotype-block consists of SNPs: rs2839677-rs7900976.
FDR adjusted P < 0.05 are in bold type.
P < 0.05 (FDR adjusted P becomes non-significant).
P < 0.01 (FDR adjusted P < 0.05).
P < 0.001 (FDR adjusted P < 0.01).
4. Discussion
Several lines of animal and human research have linked the GABAergic neurotransmitter system to various aspects of addictive behaviors and pharmacotherapies (Ikemoto, 2005; Kalivas, 2007; Koob, 2004; Roberts, 2005). The GAD genes are of special interests because GAD is the rate-limiting enzyme for synthesis of GABA in the brain. Animal QTL studies of alcohol-related phenotypes have consistently identified murine genomic regions which mapped to GABA receptor and the GAD genes (Buck and Finn, 2001; Crabbe et al., 1999, 2006). Past rodent studies have also produced valuable information for understanding genetic effects on specific components of alcohol-related phenotypes relative to the heterogeneous performance of AD clinical diagnosis in humans. The present study examined the association between GAD genes with AD as well as relevant alcohol-related phenotypes in humans.
Our main association findings were for the quantitative alcohol-related traits, initial sensitivity to alcohol and age at onset of AD. Several markers in GAD1 showed significant associations, and the effects were stronger in males than in females. For the three markers in GAD1 (Table 3) that exhibited significant associations with initial sensitivity to alcohol for which the common allele is risk allele, we calculated genotype means of alcohol intake to reach subjective effect at the time of initial use. Individuals with homozygote common vs. rare alleles required mean drinks of 6.9 vs. 5.2 in males, and 5.6 vs. 4.6 in females to feel ethanol effects. Initial sensitivity to alcohol may be more related to acute response to ethanol. Animal studies have shown that acute response to ethanol results in increase of GABA release in the brain and requires more GAD to transfer glutamic acid to produce GABA. For age at onset of AD, marker rs1978340 in GAD1 gene exhibited significant association. Males with homozygote common vs. rare alleles had mean onset age of 26.3 vs. 22.6 years, while onset age was not differed by genotypes in females.
Contrary to our expectations, neither the GAD1 nor the GAD2 genes exhibited significant associations with AD after the significance level was adjusted to control for false discoveries. Other than two previous reports for the associations of GAD1 and GAD2 in Han Taiwanese men (Loh et al., 2006) and GAD2 in Russian men (Lappalainen et al., 2007), the current study is the first to include both male and female subjects with relatively larger sample size to examine the associations of AD and several relevant alcohol-related traits with the two GAD genes in a sample from Ireland that is culturally and genetically homogeneous. In different populations, allele or genotype frequencies may exhibit variation across ethnic groups that will influence genetic associations. For GAD1, one marker (rs701492) in Loh et al. (2006) report was also genotyped in our sample with very similar MAF in both studies (In Loh's Taiwanese and our Irish samples: case = 0.29, control = 0.32). However, this marker exhibited genotypic difference (P = 0.01) between their male AD cases and controls, but did not show such difference (P = 0.42) in our study. For GAD2, marker rs2839670 which is located near 5′ of the gene was genotyped in all three studies with varying MAF distribution across populations (Among AD cases, MAF were 0.12, 0.18, 0.28 for Russian, Irish, Taiwanese samples; among controls, MAF were 0.19, 0.20, 0.23 for the three populations). This marker showed significant genotypic association in the Russian sample (P = 0.01) but not in the Irish and Taiwanese samples. With the sample size in the current study, in most of the situations we shall have power of 0.8 to detect an effect size (such as odds ratio) greater than 1.3. Therefore, the negative association findings with AD diagnosis are unlikely due to inadequate power if loci have notable effects on AD. Overall, although we did not observe direct effects of single markers in the GAD genes on the risk to develop AD, they may have impact via their influences on initial sensitivity to ethanol and the onset age of developing AD.
Several markers exhibited gender specific association effects. For instance, markers rs1978340 & rs3791878 (at 5′ near GAD1 gene) showed effects in males only with minor allele as the risk allele (has early AD age-at-onset), while the same markers showed no such effects in females. Females with minor allele had later age-at-onset of AD (Table 3, males and females have coefficients in opposite directions) though not statistically significant. Similarly, marker rs2839673, which is a coding SNP (in exon 6 of GAD2) was modestly associated with withdrawal severity showing opposite direction of coefficients in females and males. Haplotypic analyses results (Table 4) showed similar patterns of bi-direction effects by genders and the associations mainly restricted to one gender. For instance, haplotype 2111 in GAD1 exhibited strong association with initial sensitivity to ethanol and tolerance/maximum drinking only in females, and haplotype 1121 exhibited strong association with withdrawal severity only in males. Because both haplotypes are not common haplotypes (frequency around 1–2%, which represents 13–25 chromosomes in our sample), we did not test for gender interactions directly in the present study. The real contribution of rare haplotypes in genetic association studies and possible gender differences need to be further tested and confirmed in larger studies.
Many reasons can contribute to gender differences, such as hormone levels (e.g. estrogen, progesterone), innate differences in neuronal system adaptations, and pharmacokinetics of abused drugs in body (Becker, 1999; Carroll et al., 2004; Russo et al., 2003). Significant gender differences were reported in self-administration of drugs during pharmacological treatment using GABA receptor agonist in rats and kappa opioid receptor agonist in monkeys, which suggest sex-related differences in neurotransmission system. Rodent studies have shown that gender impacts on adaptations in GABA receptors. Female ethanol withdrawn rats showed greater sensitization to endogenous neuroactive steroids, which have selective effects at GABA and N-methyl-d-aspartate receptors (Devaud et al., 1998). In addition, GABAAα4 subunit expression in hippocampus and cerebral cortex were significantly increased in males, but not in females after chronic ethanol administration (Devaud and Alele, 2004). Effects of ethanol withdrawal on gene expression were also different by gender, thus female rats tend to have higher GAD1 level than males in the hippocampus and cerebral cortex, while male rats exhibited higher level of GAD2 than females in both brain areas (Alele and Devaud, 2005). Overall, gender differences may occur at many levels of alcohol addiction, from observed behavioral changes, neuroadaptive functions, to underlying genetic variations. Evaluation of gender specific effects in physiological and genetic studies may assist in understanding of underlying mechanisms of compulsive addicted behaviors and have clinical implications on the influences of drug treatment response among addictive individuals. Potential gender differences in GAD genetic associations we reported may concert with behavioral and gene expression findings reported in the literature, although the underlying physiological mechanism requires further study.
This study has some limitations. First, alcohol-related traits were measured only among cases but not controls. This is not a problem for age-at-onset of AD and withdrawal, which are only meaningful in AD cases. For initial sensitivity, without information from a general population sample, our association findings have limited generalizability. It would be ideal to test for gene effects for initial sensitivity on alcohol use in the general population. Second, our association results do not directly address whether there is functional variation in the GAD genes that is related to expression level change of GAD. Addressing this question requires a different study design. Third, although the LD patterns of both GAD genes are compatible with Hapmap CEPH population data, it remains possible that we do not have a full coverage set of SNPs to detect potential association in these genes, especially rare functional polymorphisms. Finally, whereas GAD genes are our main focus in the present study, we did not include genes encoding for the downstream product GABA, and several GABA receptor genes. In future studies we plan to examine the roles of GABA receptor genes and to conduct pathway analysis for genes involved in the inhibitory neurotransmission system.
Supplementary Material
Acknowledgments
We thank Halberstadt (VCU), Margaret Devitt (HRB) and Victor Robinson (U of Ulster) for supervising data collection. Lisa Halberstadt (VCU), Margaret Devitt (HRB) and Victor Robinson (U of Ulster) supervised data collection. Ruth Barrington, Ros Moran and Carol Cronin of the HRB provided administrative support. F.A. O'Neill, the Northern Ireland Blood Transfusion Service of the British National Health Service and the Irish Gardai provided assistance in obtaining control blood samples. At VCU, Helen Wang provided computer assistance, Indrani Ray, John Myers and Cheryl Smith assisted with data management, and Stacey Garnett, Jill Opalesky, and Melissa Hayes provided administrative support.
Role of funding source: This work was supported by National Institutes of Health grant R01-AA-11408 and the Irish Health Research Board for original data collections and data analysis. Writing of this manuscript was also supported by a Young Investigator award from the National Alliance for Research on Schizophrenia and Depression to Po-Hsiu Kuo.
Footnotes
Most of the controls were obtained from blood donation centers and the sex of the donors was not available to us directly. Therefore, the sex of subjects was determined experimentally by genotyping 3 X-specific and 3 Y-specific SNPs. We cannot determine sex of four subjects either due to failure of genotyping or the genotyping results were not conclusive.
Contributors: P.H. Kuo conducted analyses and wrote this manuscript. G. Kalsi, C.A. Hodgkinson, D. Goldman, J. Alexander and B.P. Riley assisted for experimental design and genotyping. C.A. Prescott, Patrick F. Sullivan, Diana G. Patterson, Dermot Walsh, and K.S. Kendler helped for data collection and management. C.A. Prescott, B.P. Riley, E.J. van den Oord and K.S. Kendler also assisted for study design, statistical methods as well as provided useful discussion. All authors contributed to and have approved the final manuscript.
Conflict of interest: None declared.
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.drugalcdep.2008.11.009.
References
- Alele PE, Devaud LL. Differential adaptations in GABAergic and glutamatergic systems during ethanol withdrawal in male and female rats. Alcohol Clin Exp Res. 2005;29:1027–1034. doi: 10.1097/01.alc.0000167743.96121.40. [DOI] [PubMed] [Google Scholar]
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding R, Ji FY, Kanbara N, Kuzume H, Sanbo M, Yagi T, Obata K. Mice lacking the 65 kDa isoform of glutamic acid decarboxylase (GAD65) maintain normal levels of GAD67 and GABA in their brains but are susceptible to seizures. Biochem Biophys Res Commun. 1996;229:891–895. doi: 10.1006/bbrc.1996.1898. [DOI] [PubMed] [Google Scholar]
- Becker JB. Gender differences in dopaminergic function in striatum and nucleus accumbens. Pharmacol Biochem Behav. 1999;64:803–812. doi: 10.1016/s0091-3057(99)00168-9. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;85:289–300. [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JIJ, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Finn DA. Genetic factors in addiction: QTL mapping and candidate gene studies implicate GABAergic genes in alcohol and barbiturate withdrawal in mice. Addiction. 2001;96:139–149. doi: 10.1046/j.1360-0443.2001.96113910.x. [DOI] [PubMed] [Google Scholar]
- Carroll ME, Lynch WJ, Roth ME, Morgan AD, Cosgrove KP. Sex and estrogen influence drug abuse. Trends Pharmacol Sci. 2004;25:273–279. doi: 10.1016/j.tips.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Buck KJ, Cunningham CL, Belknap JK. Identifying genes for alcohol and drug sensitivity: recent progress and future directions. Trends Neurosci. 1999;22:173–179. doi: 10.1016/s0166-2236(99)01393-4. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crews FT, Morrow AL, Criswell H, Breese G. Effects of ethanol on ion channels. Int Rev Neurobiol. 1996;39:283–367. doi: 10.1016/s0074-7742(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Davis KM, Wu JY. Role of glutamatergic and GABAergic systems in alcoholism. J Biomed Sci. 2001;8:7–19. doi: 10.1007/BF02255966. [DOI] [PubMed] [Google Scholar]
- de Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Devaud LL, Alele P. Differential effects of chronic ethanol administration and withdrawal on gamma-aminobutyric acid type A and NMDA receptor subunit proteins in male and female rat brain. Alcohol Clin Exp Res. 2004;28:957–965. doi: 10.1097/01.alc.0000128225.83916.40. [DOI] [PubMed] [Google Scholar]
- Devaud LL, Fritschy JM, Morrow AL. Influence of gender on chronic ethanol-induced alterations in GABA(A) receptors in rats. Brain Res. 1998;796:222–230. doi: 10.1016/s0006-8993(98)00357-6. [DOI] [PubMed] [Google Scholar]
- Eravci M, Schulz O, Grospietsch T, Pinna G, Brodel O, Meinhold H, Baumgartner A. Gene expression of receptors and enzymes involved in GABAergic and glutamatergic neurotransmission in the CNS of rats behaviourally dependent on ethanol. Br J Pharmacol. 2000;131:423–432. doi: 10.1038/sj.bjp.0703596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- Fehr C, Rademacher BLS, Buck KJ. Evaluation of the glutamate decarboxylase genes Gad1 and Gad2 as candidate genes for acute ethanol withdrawal severity in mice. Progr Neuro-Psychopharmacol Biol Psychiatry. 2003;27:467–472. doi: 10.1016/S0278-5846(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Feldblum S, Dumoulin A, Anoal M, Sandillon F, Privat A. Comparative distribution of GAD65 and GAD67 mRNAs and proteins in the rat spinal cord supports a differential regulation of these two glutamate decarboxylases in vivo. J Neurosci Res. 1995;42:742–757. doi: 10.1002/jnr.490420603. [DOI] [PubMed] [Google Scholar]
- Follesa P, Biggio F, Talani G, Murru L, Serra M, Sanna E, Biggio G. Neurosteroids, GABAA receptors, and ethanol dependence. Psychopharmacology (Berl) 2006;186:267–280. doi: 10.1007/s00213-005-0126-0. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Hasin D, Paykin A, Meydan J, Grant B. Withdrawal and tolerance: prognostic significance in DSM-IV alcohol dependence. J Stud Alcohol. 2000;61:431–438. doi: 10.15288/jsa.2000.61.431. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CE, Xu K, Yuan Q, Shen PH, Heinz E, Ehlers C, Roy A, Gelerntner J, Mann J, Riley B, Tabakoff B, Todd R, Zhou Z, Goldman D. Addictions biology: haplotype based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto S. The supramammillary nucleus mediates primary reinforcement via GABA(A) receptors. Neuropsychopharmacology. 2005;30:1088–1095. doi: 10.1038/sj.npp.1300660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo E, Auta J, Impagnatiello F, Pesold C, Guidotti A, Costa E. Glutamic acid decarboxylase and glutamate receptor changes during tolerance and dependence to benzodiazepines. Proc Natl Acad Sci USA. 2001;98:3483–3488. doi: 10.1073/pnas.051628698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Cloninger CR, Roache JD, Bordnick PS, Ruiz P. Age of onset as a discriminator between alcoholic subtypes in a treatment-seeking outpatient population. Am J Addict. 2000;9:17–27. doi: 10.1080/10550490050172191. [DOI] [PubMed] [Google Scholar]
- Kalivas PW. Neurobiology of cocaine addiction: implications for new pharmacotherapy. Am J Addict. 2007;16:71–78. doi: 10.1080/10550490601184142. [DOI] [PubMed] [Google Scholar]
- Kash SF, Tecott LH, Hodge C, Baekkeskov S. Increased anxiety and altered responses to anxiolytics in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA. 1999;96:1698–1703. doi: 10.1073/pnas.96.4.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. A role for GABA mechanisms in the motivational effects of alcohol. Biochem Pharmacol. 2004;68:1515–1525. doi: 10.1016/j.bcp.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcohol Clin Exp Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, Kendler KS, Prescott CA. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol Clin Exp Res. 2006;30:1807–1816. doi: 10.1111/j.1530-0277.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- Langenbucher J, Martin CS, Labouvie E, Sanjuan PM, Bavly L, Pollock NK. Toward the DSM-V: the withdrawal-gate model versus the DSM-IV in the diagnosis of alcohol abuse and dependence. J Consult Clin Psychol. 2000;68:799–809. [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Kranzler HR, Luo X, Remizov M, Pchelina S, Taraskina A, Zvartau E, Rasanen P, Makikyro T, Somberg LK, Krystal JH, Stein MB, Gelernter J. Mutation screen of the GAD2 gene and association study of alcoholism in three populations. Am J Med Genet B Neuropsychiatr Genet. 2007;144:183–192. doi: 10.1002/ajmg.b.30377. [DOI] [PubMed] [Google Scholar]
- Loh EW, Lane HY, Chen CH, Chang PS, Ku LW, Wang KHT, Cheng ATA. Glutamate decarboxylase genes and alcoholism in Han Taiwanese men. Alcohol Clin Exp Res. 2006;30:1817–1823. doi: 10.1111/j.1530-0277.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- Nevo I, Hamon M. Neurotransmitter and neuromodulatory mechanisms involved in alcohol abuse and alcoholism. Neurochem Int. 1995;26:305–336. doi: 10.1016/0197-0186(94)00139-l. discussion 337-342. [DOI] [PubMed] [Google Scholar]
- Petroff OA. GABA and glutamate in the human brain. Neuroscientist. 2002;8:562–573. doi: 10.1177/1073858402238515. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Rosenbloom M, Deshmukh A, Sullivan E. Sex differences in the effects of alcohol on brain structure. Am J Psychiatry. 2001;158:188–197. doi: 10.1176/appi.ajp.158.2.188. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Myers JM, Patterson DG, Devitt M, Halberstadt LJ, Walsh D, Kendler KS. The Irish Affected Sib Pair Study of Alcohol Dependence: study methodology and validation of diagnosis by interview and family history. Alcohol Clin Exp Res. 2005;29:417–429. doi: 10.1097/01.alc.0000156085.50418.07. [DOI] [PubMed] [Google Scholar]
- Roberts DC. Preclinical evidence for GABAB agonists as a pharmacotherapy for cocaine addiction. Physiol Behav. 2005;86:18–20. doi: 10.1016/j.physbeh.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Festa ED, Fabian SJ, Gazi FM, Kraish M, Jenab S, Quinones-Jenab V. Gonadal hormones differentially modulate cocaine-induced conditioned place preference in male and female rats. Neuroscience. 2003;120:523–533. doi: 10.1016/s0306-4522(03)00317-8. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. The clinical course of alcohol dependence associated with a low level of response to alcohol. Addiction. 2001;96:903–910. doi: 10.1046/j.1360-0443.2001.96690311.x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Daeppen JB, Eng M, Li TK, Hesselbrock VM, Nurnberger JI, Bucholz KK. Clinical relevance of the distinction between alcohol dependence with and without a physiological component. Am J Psychiatry. 1998;155:733–740. doi: 10.1176/ajp.155.6.733. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Tipp JE. The self-rating of the effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction. 1997;92:979–988. [PubMed] [Google Scholar]
- Soghomonian JJ, Martin DL. Two isoforms of glutamate decarboxylase: why? Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- Tsai G, Coyle JT. The role of glutamatergic neurotransmission in the pathophysiology of alcoholism. Annu Rev Med. 1998;49:173–184. doi: 10.1146/annurev.med.49.1.173. [DOI] [PubMed] [Google Scholar]
- Wuethrich B. Neurobiology—does alcohol damage female brains more? Science. 2001;291:2077–2079. doi: 10.1126/science.291.5511.2077. [DOI] [PubMed] [Google Scholar]
- Zhang XW, Lee TH, Xiong XY, Chen Q, Davidson C, Wetsel WC, Ellinwood EH. Methamphetamine induces long-term changes in GABA(A) receptor alpha 2 subunit and GAD(67) expression. Biochem Biophys Res Commun. 2006;351:300–305. doi: 10.1016/j.bbrc.2006.10.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


