Summary
Adherence to host tissues mediated by pili is pivotal in the establishment of infection by many bacterial pathogens. Corynebacterium diphtheriae assembles on its surface three distinct pilus structures. The function and the mechanism of how various pili mediate adherence, however, have remained poorly understood. Here we show that the SpaA-type pilus is sufficient for the specific adherence of corynebacteria to human pharyngeal epithelial cells. The deletion of the spaA gene, which encodes the major pilin forming the pilus shaft, abolishes pilus assembly but not adherence to pharyngeal cells. In contrast, adherence is greatly diminished when either minor pilin SpaB or SpaC is absent. Antibodies directed against either SpaB or SpaC block bacterial adherence. Consistent with a direct role of the minor pilins, latex beads coated with SpaB or SpaC protein bind specifically to pharyngeal cells. Therefore, tissue tropism of corynebacteria for pharyngeal cells is governed by specific minor pilins. Importantly, immunoelectron microscopy and immunofluorescence studies reveal clusters of minor pilins that are anchored to cell surface in the absence of a pilus shaft. Thus, the minor pilins may also be cell wall anchored in addition to their incorporation into pilus structures that could facilitate tight binding to host cells during bacterial infection.
Introduction
Corynebacterium diptheriae, the causative pathogen of diphtheria in humans, infects the nasopharynx or skin (Holmes, 2000). Diphtheria is a rapidly developing, acute and feverish infection; it is manifested by bacterial colonization and production of diphtheria toxin (Love and Murphy, 2006). Although diphtheria has been extensively studied, little is known about the adherence mechanism that allows colonization of C. diphtheriae. We have recently shown that corynebacteria assemble three distinct pilus structures, designated as the SpaA-, SpaD- and SpaH-type pili (Spa for sortase-mediated pilus assembly) (Ton-That and Schneewind, 2003; Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). It is likely that these pili are involved in corynebacterial attachment to host tissues, as pilus-mediated adherence is a critical step in the establishment of infection by many Gram-negative pathogens (Pizarro-Cerda and Cossart, 2006).
Corynebacterium diphtheriae shares with many Gram-positive bacteria, including Actinomyces, group A and B streptococci and Streptococcus pneumoniae, a mechanism of pilus assembly that requires sortase (Ton-That and Schneewind, 2004). Sortase is a cysteine transpeptidase conserved in all Gram-positive bacterial genomes (Mazmanian et al., 1999; Ilangovan et al., 2001; Comfort and Clubb, 2004; Dramsi et al., 2005; Marraffini et al., 2006). Sortase cleaves pilin precursors at the conserved LPXTG motif between threonine and glycine and tethers the threonine carboxyl group of the precursor protein to the amino group of cell wall cross-bridges within the lipid II precursor, or to the amino group of the conserved pilin motif within the surface protein precursor (Mazmanian et al., 2001; Ton-That and Schneewind, 2004). The latter reaction leads to the formation of pilus fibres on the bacterial surface (Ton-That and Schneewind, 2004).
In C. diphtheriae, the genes encoding pilin subunits with sorting signals and five sortases (srtA–E) are organized in three separate clusters [(Ton-That and Schneewind, 2003; Fig. 1]. The sixth sortase (srtF) is encoded elsewhere on the chromosome. All three pilus types have similar components, a major pilin subunit and two minor pilus proteins. For example, the SpaA-type pilus is composed of a major pilin subunit SpaA forming the pilus shaft, a possible tip protein SpaC and a minor pilin SpaB decorating along the pilus shaft (Ton-That and Schneewind, 2003). SpaA is essential for the formation of the pilus structure, whereas SpaB and SpaC are dispensable (Ton-That et al., 2004). Two elements within the SpaA pilin precursor, the pilin motif and the sorting signal, are together necessary and sufficient to promote pilus polymerization by a process requiring the function of a pilus-specific sortase (SrtA) (Ton-That and Schneewind, 2003; Ton-That et al., 2004). The absence of other sortase genes (srtB–F) does not abrogate the synthesis or assembly of the SpaA-type pili on the bacterial surface (Ton-That and Schneewind, 2003; Ton-That et al., 2004). SrtA cleaves SpaA precursor proteins at the sorting signal and utilizes the side-chain amino group of the conserved lysine within the SpaA pilin motif sequence to generate covalent cross-links between pilin subunits. Substitution of this lysine with arginine or alanine abolishes the polymerization of SpaA pilins (Ton-That and Schneewind, 2004). SpaC is displayed at the pilus tip presumably by a similar mechanism via the SpaC sorting signal and the SpaA pilin motif (Ton-That and Schneewind, 2003). The incorporation of SpaB into SpaA pili requires a glutamic acid residue within the E box motif of SpaA, a feature that is found conserved in other Gram-positive pathogens that encode sortase and pilin subunit genes with sorting signals and pilin motifs (Ton-That et al., 2004). The precise mechanism of pilus assembly from these pilins, however, still remains to be investigated.
Fig. 1.
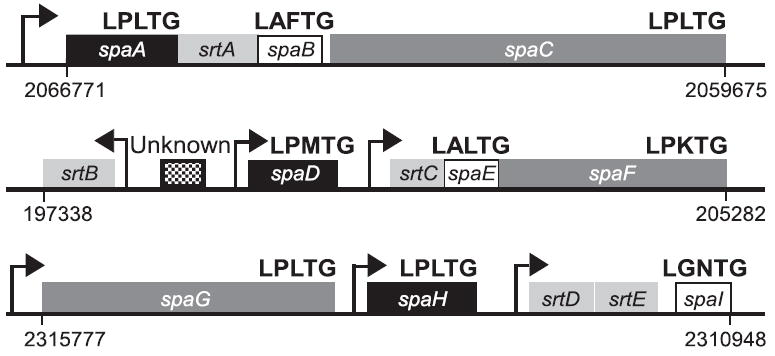
Three gene clusters in the chromosome of C. diphtheriae NCTC13129 that encode sortase genes (srtA–E) and the sortase-mediated pilus assembly genes (spaA–I). A sixth sortase (srtF) is located between positions 2363388 and 2364209 in the chromosome. Arrows indicate the position of predicted promoters as well as direction of transcription. Similarity and homology between spa gene products are indicated as shared patterns. Numbers show the location of pilus gene clusters in the chromosome.
Pilus gene clusters containing sortases have been found in many important Gram-positive pathogens including Clostridium perfringens, enterococci and many streptococcal species (Ton-That and Schneewind, 2003; Nallapareddy et al., 2006; Scott and Zahner, 2006; Telford et al., 2006). There is evidence that some of these filamentous organelles facilitate interactions of the pathogens with different host cells. For example, Actinomyces type 1 fimbriae mediate bacterial adherence to the tooth enamel (Yeung and Ragsdale, 1997), while type 2 fimbriae interact with oral streptococci as well as various host cells, thus culminating the formation of plaques in human mouth (Yeung et al., 1998). Actinomyces mutants lacking fimbriae are defective in binding to host cells or infectious partners (Cisar et al., 1988; Yeung, 1999). Likewise, piliated pneumococci are more virulent than a non-piliated deletion mutant (Barocchi et al., 2006). In Enterococcus faecalis, pili are required for biofilm formation, and a non-piliated mutant was significantly attenuated in an endocarditis model (Nallapareddy et al., 2006). Using in vitro cell cultures, it has been shown that streptococcal pili are required for efficient binding to epithelial cells (Dramsi et al., 2006). Moreover, the presence of pili also stimulates the host inflammatory response (Barocchi et al., 2006). In fact, antigens generated by a combination of pilus proteins efficiently protect mice from re-infection by streptococci in a rodent model, suggesting that pili of Gram-positive bacteria can be valuable vaccine candidates (Maione et al., 2005; Mora et al., 2005).
Previously, C. diphtheriae has been shown to adhere to a few mammalian cells (Colombo et al., 2001; Hirata et al., 2004), but little is known about factors that are required for this process. This study reports the first evidence that the SpaA-type pilus of corynebacteria is responsible for their differential binding to pharyngeal epithelial cells, and only the minor pilins SpaB and SpaC are essential for this process. Importantly, in a non-piliated mutant, the minor pilins are anchored to the cell wall that allows the pathogen to adhere tightly to host cells. We propose a molecular model of how specific pilins might orchestrate intimate bacterial attachment during infection.
Results
Immunoelectron microscopic detection of three different pilus structures on the surface of the wild-type C. diphtheriae
As said above, the pilus genes and sortases of C. diphtheriae are organized in three clusters (also see Fig. 1), each of which expresses a distinct pilus structure, designated as SpaA-, SpaD- and SpaH-type pili. Using specific antibody raised against purified SpaA, SpaD and SpaH as well as gold particles conjugated to IgG, we viewed corynebacteria by electron microscopy and observed the deposition of immunoreactive signals along short fibres (Fig. 2A–C). Throughout this article, we present representative electron microscopic images that were typically observed in majority of the cells in various samples (95% or greater). The deletion of sortase srtA abolished the assembly of SpaA-type, but not SpaD- and SpaH-type pili (Ton-That and Schneewind, 2003). Likewise, deletion of sortases srtB and srtC or srtD and srtE abolished only the assembly of SpaD-type pili or SpaH-type pili respectively (Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). In a strain that lacks all but srtA (designated as SpaABC+) (Ton-That et al., 2004), we observed only the SpaA-type pili on the bacterial surface, whereas SpaD-type and SpaH-type pili were not observed (Fig. 2D–F). Similarly, in a strain that expresses only sortases B and C or D and E, we observed the SpaD-type (SpaDEF+) or SpaH-type (SpaHIG+) pili respectively (Fig. 2G–L). Together, these results confirm our previous claims that corynebacteria assemble three different pilus structures, and that the assembly of each pilus type requires specific sortases encoded within the respective pilus gene cluster. The mutants missing or displaying specific types of pili enabled us to examine their respective contribution to host cell adherence reported below.
Fig. 2.
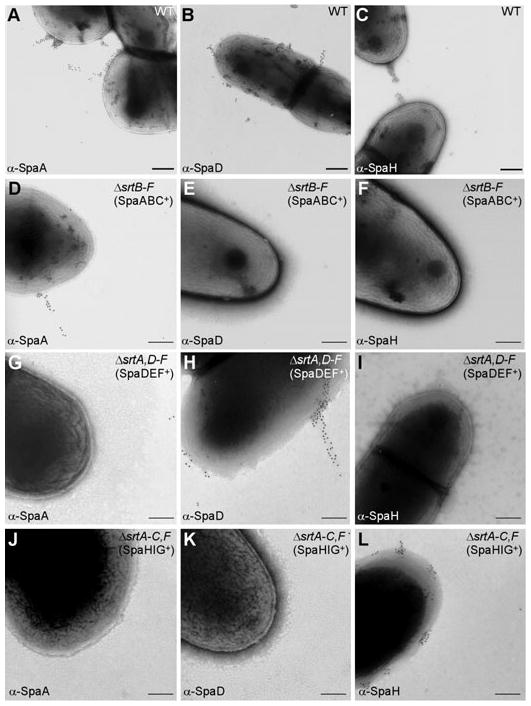
A pilin-specific sortase required for pilus assembly. Wild-type C. diphtheriae (A–C) and its isogenic derivatives ΔsrtB–F (designated as SpaABC+) (D–F), ΔsrtA,D–F (designated as SpaDEF+) (G–I) or ΔsrtA-C,F (designated as SpaHIG+) (J–L) were immobilized on carbon grids, stained with a specific antiserum against SpaA, SpaD or SpaH and goat anti-rabbit IgG conjugated to 12 nm gold particles. Samples were viewed by transmission electron microscopy. Scale bars indicate the length of 0.2 μm.
Differential binding to human epithelial cells mediated by distinct corynebacterial pili
Nasopharynx is the major site of C. diphtheriae infection (Love and Murphy, 2006). To determine whether pili are involved in corynebacterial adherence to this tissue, we examined the relative association of corynebacterial variants with pharyngeal cells (D562) as well as laryngeal (HEp2) and lung (A549) epithelial cells in culture. These cells were grown in multiwell culture plates in an appropriate culture medium to confluence and infected with corynebacterial variants. The infected cells were washed to remove the unbound bacteria, detached from wells, and then plated on agar plates to enumerate bacteria absorbed by the epithelial cells (see Experimental procedures). Wild-type corynebacteria adhered to lung, laryngeal and pharyngeal epithelial cells with varying efficiency, 20%, 25% and 9%, respectively. This result is consistent with previous studies that showed that C. diphtheriae binds to laryngeal cells with an efficiency equal to or greater than 22% (Hirata et al., 2002; Moreira Lde et al., 2003).
The binding of corynebacterial variants to a particular cell line was then compared with that of the wild-type strain, which was set to 100% (Fig. 3A, first column). With a strain, ΔsrtA–F, that lacks all six sortases and thus fails to assemble all pili as well as many cell wall-anchored surface proteins (Ton-That and Schneewind, 2003), we observed about threefold reduction in adherence to lung or laryngeal cells (Fig. 3A, second columns). By comparison, adherence of this strain to pharyngeal cells was drastically decreased (Fig. 3A, second columns). This attenuated phenotype might result from the absence of SpaA-type pili as the deletion of sortase srtA alone strikingly reduced bacterial adherence (Fig. 3A, third columns). In the absence of SpaA-type pili, corynebacteria had only a slight defect in their binding to lung and laryngeal cells (Fig. 3A, third columns). Similarly, with a strain that lacks sortases specific for SpaD-type pili, ΔsrtBC, 2.5-fold reduction in adherence to laryngeal cells was observed, whereas only a marginal reduction was observed for the binding to lung and pharyngeal cells. Optimal adhesion of corynebacteria to the laryngeal cells required the presence of either the SpaD-type pili (Fig. 3A, fourth columns) or the SpaH-type pili (Fig. 3A, fifth columns). These results suggested that specific pili are involved in the binding of corynebacteria to different host cells.
Fig. 3.

Pilus-mediated adherence of C. diphtheriae to human epithelial cells.
A. Adherence of corynebacterial variants to human lung (A549), laryngeal (HEp2) and pharyngeal (D562) epithelial cells. Confluent cell monolayers were infected with the wild-type C. diphtheriae and its isogenic deletion mutants, and adherent bacteria were then enumerated. Data are presented as percentage of adhesion relative to that of wild type. Mean adhesion percentage of wild-type to A549, HEp2 and D562 cells was ~20%, ~25% and ~9% respectively. The results are presented as averages (with standard deviations ± SD) from at least three independent experiments performed in triplicates.
B. Differential binding of corynebacterial pili to epithelial cells. Similar experiments were carried out as described in (A) with strains ΔsrtB–F, ΔsrtA,D–F and ΔsrtA–C,F designated as SpaABC+, SpaDEF+ and SpaHIG+ respectively.
We next carried out a reciprocal set of experiments to examine whether each pilus is sufficient to confer adherence to a particular cell line. We generated strains that assembled only SpaA-, SpaD- or SpaH-type pilus (designated as SpaABC+, SpaDEF+ or SpaHIG+ respectively) (Fig. 2). The ΔsrtB–F strain that assembles only the SpaA-type pilus (Fig. 2D–F) adhered efficiently to lung and pharyngeal epithelial cells and slightly less to laryngeal cells (Fig. 3B, second column). Similarly, the presence of either SpaD- or SpaH-type pilus was sufficient for the binding to the lung and laryngeal cells, but allowed little binding to pharyngeal ones (Fig. 3B, third and fourth column). Altogether, our results indicate that specific corynebacterial pili mediate efficient binding to epithelial cells and that the SpaA-type pili preferentially adhere to pharyngeal cells, which constitute the major site for corynebacterial infection.
Minor pilins of the SpaA-type pili are required for corynebacterial adherence to human pharyngeal cells
The question now arises as to which component of the multiprotein SpaA pilus is mediating host cell adhesion. As stated before, the SpaA-type pili are composed of SpaA, SpaB and SpaC, with SpaA forming the pilus shaft, where SpaB decorates and SpaC is largely located at the tip (Ton-That and Schneewind, 2003). To determine which pilin is important for specific adherence to epithelial cells, we engineered non-polar, in-frame deletion mutants of spaA, spaB or spaC gene in the ΔsrtB–F parental strain, which does not assemble the SpaD- and SpaH-type pili. Pilus polymerization in these strains was first examined by a biochemical assay, whereby corynebacteria are first treated with muramidase to hydrolyse the cell wall (Ton-That and Schneewind, 2003). Muramidase-solubilized proteins are separated via electrophoresis in polyacrylamide gels after boiling cell extracts in sodium dodecyl sulphate (SDS) sample buffer with reducing agent and then identified by immunoblotting with α-SpaA, α-SpaB or α-SpaC antibodies. In the ΔsrtB–F bacteria, the SpaA monomer migrated at 55 kDa (calculated mass of 56 kDa); in addition, a ladder of immunoreactive high-molecular-weight bands of SpaA (labelled SpaAHMW) were detected with an apparent mass greater than 200 kDa (Fig. 4A, lane 2). As expected, deletion of six sortase genes (ΔsrtA–F) abolished the polymerization of SpaA precursor into high molecular weight species; only the monomeric forms of SpaA, faint SpaB and SpaC were observed (Fig. 4A–C, lane 1). The deletion of spaA abrogated the synthesis and assembly of SpaA pilins (Fig. 4A, lane 3), which was overcome by a SpaA-encoding plasmid (Fig. 4A, lane 4). In contrast, neither ΔspaB nor ΔspaC affected the polymerization of SpaA precursor into high-molecular-weight species, although in the spaB deletion mutant, SpaAHMW bands appeared to be more intense (Fig. 4, lane 5). Thus, spaA is essential for the formation of pilus structure, and spaB and spaC are dispensable, in agreement with our previous studies (Ton-That et al., 2004). Importantly, specific high-molecular-weight species of SpaB and SpaC were observed in the spaA deletion mutant (Fig. 4B and C, lane 3), hinting that SpaB and SpaC might be anchored to the cell wall. This was further investigated by immunoelectron microscopy (IEM) and fluorescence microscopy (see below).
Fig. 4.

Pilus polymerization of corynebacteria. The wild-type strain and its isogenic derivatives carrying deletions of srt or spa genes were treated with muramidase prior to extraction with hot SDS sample buffer. Proteins were separated on 4–12% Tris-Glycine gradient gels and detected by immunoblotting with the specific antisera α-SpaA (A), α-SpaB (B) and α-SpaC (C). Strain ΔspaA, ΔspaB or ΔspaC strain was generated from the parental strain ΔsrtB–F (designated as SpaABC+). A recombinant plasmid harbouring the wild-type SpaA, SpaB or SpaC was used to complement a respective deletion mutant. The positions of molecular mass markers (M) are indicated. Note the slower mobility of the SpaC band in ΔspaA strain (C, lane 3).
Next, to determine the role of SpaA, SpaB and SpaC pilins in adherence, the corynebacterial variants were subjected to adhesion assays with pharyngeal and lung epithelial cells as described above. Surprisingly, only a slight reduction in adherence was observed with the spaA deletion mutant as compared with that of the parental strain (Fig. 5A, second column). Thus, the SpaA pilus is not essential for corynebacterial adherence to host cells. Interestingly, the deletion of spaB or spaC strikingly reduced bacterial adherence specifically to the pharyngeal cells. In fact, a more significant reduction (~90%) in this adherence was observed with the double spaB/spaC deletion mutant (Fig. 5A). Complementation of the ΔspaB or ΔspaC mutant with wild-type spaB or spaC on a multi-copy plasmid restored not only SpaB or SpaC polymerization (Fig. 4B and C, lanes 6 and 8), but also bacterial adherence (Fig. 5A). It is noteworthy that none of these deletion mutants affected bacterial adherence to lung epithelial cells as compared with that of the parental strain (Fig. 5B). Thus, the defect of the ΔspaB/ΔspaC mutants is specific for adherence to pharyngeal cells.
Fig. 5.

Minor pilins SpaB and SpaC required for bacterial adherence.
A and B. The parental strain SpaABC+ (ΔsrtB–F) and its isogenic deletion mutants were subjected to adhesion assays using pharyngeal epithelial cells (A) or lung epithelial cells (B). Data are presented as percentage of adhesion relative to that of the parental strain. The results are presented as averages (with standard deviations ± SD) from at least three independent experiments performed in triplicates. Statistical analysis was performed using Student’s t-test.
C. In a similar experiment as described in (A), monolayer of pharyngeal epithelial cells were infected with the parental strain SpaABC+ pre-incubated with a specific antiserum at a dilution of 1:100 for 1 h at 37°C. Pre-immune sera and an unrelated antibody against a pilus protein of Streptococcus agalactiae, α-GBS59, were used as controls.
The above results indicate that the SpaB and SpaC pilins act independently to promote the effective binding of the pathogen to pharyngeal cells. If so, antibodies against these pilins may prevent bacterial adherence. To test this possibility, we pre-incubated the SpaABC+ strain with a specific antibody against each pilus protein, i.e. SpaA, SpaB or SpaC, prior to infecting the epithelial cells (see Experimental procedures). SpaA antibody (α-SpaA) did not block bacterial adherence as expected (Fig. 5C, second column). However, α-SpaB or α-SpaC effectively prevented bacterial binding as compared with the respective pre-immune antisera (Fig. 5C). As a further control, antibody against a major subunit of group B streptococci pili (GBS59; Lauer et al., 2005) gave the same result as the corynebacterial α-SpaA (Fig. 5C, last lane).
SpaB- or SpaC-mediated adherence of latex beads to pharyngeal cells
To further address whether SpaB and SpaC pilins independently mediate adherence to host tissues, we expressed and purified recombinant pilus proteins from Escherichia coli. The recombinant proteins SpaA and SpaB included the mature N-terminus and lacked the cell wall anchor region; the truncated SpaC protein contained the Glu19–Asn344 region that includes the Von Willebrand factor (VWF) domain. Purified SpaA, SpaB or SpaC protein was bound to fluorescent latex beads. Beads coated with protein were blocked in bovine serum albumin (BSA), washed and added to cell monolayers grown on glass coverslips. The monolayers were incubated for 1 h at 37°C, washed, fixed and examined using fluorescence microscopy (see Experimental procedures). Adherence was quantified and presented as the average number of beads bound per 200 epithelial cells from three independent experiments (Fig. 6).
Fig. 6.
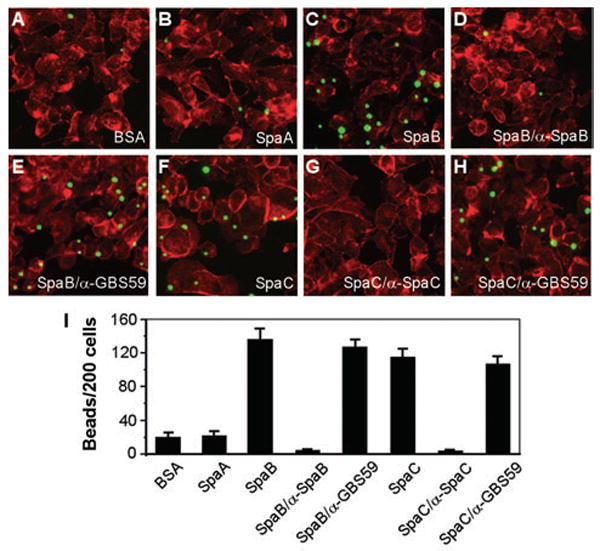
SpaB- or SpaC-mediated adherence of latex beads to pharyngeal cells.
Semi-confluent cells grown on coverslips were infected with protein-coated beads for 1 h.
The washed cells were fixed and stained with Texas-Red-X phalloidin.
A–H. Shown here are the fluorescence images of cells incubated with fluorescent beads bound to BSA (A), SpaA (B), SpaB (C), SpaB blocked with α-SpaB (D), SpaC (F) and SpaC blocked with α-SpaC (G). Beads with bound protein incubated with an unrelated antibody against a pilin of Streptococcus agalactiae, α-GBS59, were used as controls (E and H).
I. The results are shown as an average of the numbers of beads bound per 200 cells of three independent experiments (with standard deviation ± SD).
Remarkably, beads with SpaB- or SpaC-bound protein readily adhered to the pharyngeal epithelial cells, a process that was inhibited by polyclonal antibodies against SpaB or SpaC, respectively, but not by an unrelated antibody against a pilus subunit of group B streptococci (α-GBS59) (Fig. 6). As expected, SpaA-coated beads bound poorly to the epithelial cells at a level similar to that of BSA-coated beads (Fig. 6A, B and I), consistent with the observation above that the SpaA pilus is dispensable for bacterial adherence. Together, these data demonstrate that minor pilus proteins SpaB and SpaC act as specific adhesins, and that each pilin is necessary and sufficient for corynebacterial adherence to pharyngeal epithelial cells.
Minor pilins are anchored on the cell wall in the absence of pilus structures
To explain how the minor pilins cause bacterial adherence in the absence of surface pili, we hypothesized that SpaB and SpaC pilins might be displayed on the bacterial surface independent of a pilus structure. To test this idea, we employed IEM to examine whether SpaB and SpaC are present on bacterial surface. Indeed, when spaA was deleted in the ΔsrtB–F background, both SpaB and SpaC labels were detected abundantly as clusters on the cell surface (Fig. 7A–D). As controls, no labelled SpaB or labelled SpaC was observed in the strain ΔsrtA–F that lacks all six sortases or the strains that lack these proteins (data not shown). The display of SpaB/SpaC minor pilins in the absence of SpaA was observed in a majority of the cell population (> 95%). In otherwise wild-type background, we also observed SpaB and SpaC labels on the bacterial surface when only the spaA gene was deleted (Ton-That and Schneewind, 2003, and data not shown). In fact, SpaB and SpaC were detected on the cell surface as well as on pilus shafts in wild-type corynebacteria (data not shown). Importantly, the surface display of SpaB and SpaC pilins is independent of each other as the clusters of labelled SpaB or SpaC were observed in a strain that lacks spaAC or spaAB respectively (Fig. 7E–H).
Fig. 7.

Assembly of minor pilins SpaB and SpaC on bacterial surface. Corynebacteria were immobilized on carbon grids, stained with specific antiserum against SpaB (α-SpaB) (A, F and G) or SpaC (α-SpaC) (B, E and H) and goat anti-rabbit IgG conjugated to 12 nm gold particles. For double labelling (C), cells were first reacted with α-SpaC, followed by IgG-conjugated 18 nm gold particles (open arrow in D), and then reacted with α-SpaB, followed by IgG-conjugated 12 nm gold particles (filled arrow in D). Samples were viewed by transmission electron microscopy. An enlarged area of (C) is shown in (D). Scale bars indicate the length of 0.2 μm.
To further assess that SpaB and SpaC are distributed on bacterial surface in the absence of the pilus shaft, we employed immunofluorescence microscopy. This assay allows the detection of molecules on the cell surface because, as a highly cross-linked network, the cell wall of corynebacteria is impermeable to proteins such as antibodies (Schleifer and Kandler, 1972; Navarre and Schneewind, 1999). Corynebacteria were treated with α-SpaA, α-SpaB or α-SpaC antibodies, followed by the AlexaFluor 488-stained chicken anti-rabbit IgG, and cells were observed by confocal microscopy. The strain SpaABC+ displayed abundant fluorescence for both SpaB and SpaC, in addition to SpaA fluorescence (Fig. 8A). Similar fluorescence for SpaB and SpaC was still observed when pilus assembly was obliterated by the absence of SpaA (Fig. 8B).
Fig. 8.
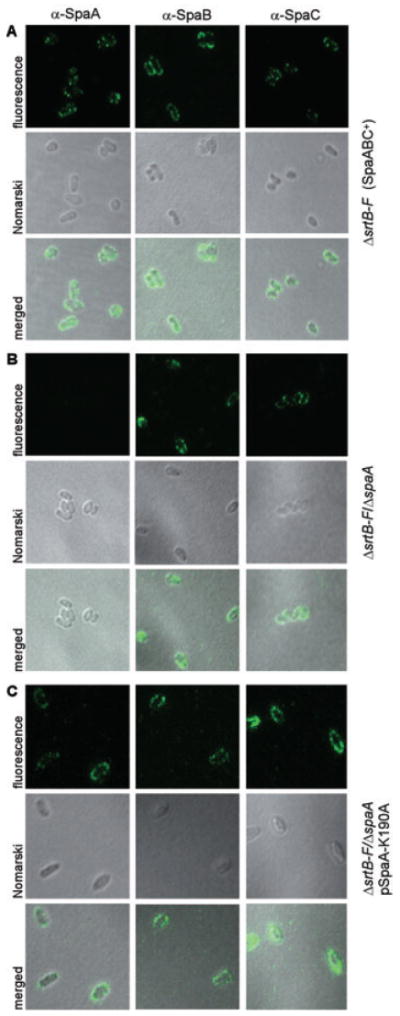
Immunofluorescent detection of SpaB and SpaC pilins on the bacterial surface. Corynebacteria were stained with a specific antibody against SpaA (α-SpaA), SpaB (α-SpaB) or SpaC (α-SpaC) and AlexaFluor 488 chicken anti-rabbit IgG. Shown are the fluorescent, the Nomarski DIC and the merged images of strain ΔsrtB–F (A), its isogenic derivative ΔspaA (B) or its isogenic derivative ΔspaA transformed with a plasmid expressing the SpaA protein mutated at the K190 in the pilin motif (C). The samples were observed on a Zeiss LSM 510 confocal microscope.
The above experiments showed that SpaB and SpaC pilins are displayed on the bacterial surface in the absence of SpaA pilins, i.e. when SpaA is not synthesized. We wondered whether the two minor pilins are surface-displayed when SpaA pilin is synthesized but it fails to form the pilus structures. Previous studies showed that the substitution of K190 of the SpaA pilin motif by alanine abrogates pilus assembly, although the synthesis of SpaABC pilins is not affected (Ton-That and Schneewind, 2003). The indirect immunofluorescence experiment described above showed that this SpaA-K190A mutant was surface-displayed along with SpaB and SpaC (Fig. 8C). This pilus-independent display of pilins on the cell surface is a general phenomenon as corynebacteria that express either SpaD-K107A or SpaH-K202A displayed respective minor pilins on cell surface in the absence of the pilus shaft (A. Swierczynski and H. Ton-That, unpublished data).
Finally, if the surface display of minor pilins involves the normal sortase-mediated cross-linking to the cell wall, this process should be dependent on the conserved LPXTG motif (Ton-That and Schneewind, 2003). Therefore, by site-directed mutagenesis, we replaced the LAFTG sorting signal of SpaB or LPLTG sorting signal of SpaC with scrambled motifs (GTALF or GTPLL respectively). Cell wall anchoring of SpaB and SpaC pilins was examined by immunofluorescence experiments. In a strain that expressed SpaA-GTPLL, SpaA signals were no longer detected on the cell surface (Fig. 9, second column). Consistent with above results, SpaB and SpaC labelling were still observed on the bacterial surface in this strain (Fig. 9, second column). When the LPXTG motif of SpaB/SpaC was mutated, however, neither SpaB-GTALF nor SpaC-GTPLL pilin was observed on the surface (Fig. 9, fourth and sixth columns). Immunoblotting showed that these mutations did not affect the synthesis of the respective proteins (data not shown). We conclude that SpaB and SpaC are indeed cross-linked to the cell wall and surface displayed, and that this process is dependent on both sortase and the LPXTG motif as is true for the pilus assembly.
Fig. 9.
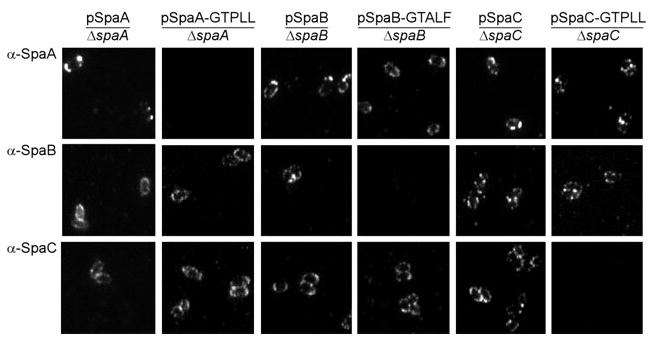
Anchoring of minor pilins SpaB and SpaC required the conserved LPXTG motif. Strain ΔsrtB–F with a deletion of spaA, spaB or spaC gene was transformed with a plasmid expressing the wild-type or mutated SpaA, SpaB or SpaC respectively. Corynebacteria were stained with a specific antibody against SpaA (α-SpaA), SpaB (α-SpaB) or SpaC (α-SpaC) and AlexaFluor 488 chicken anti-rabbit IgG. The samples were observed on a Zeiss LSM 510 confocal microscope. Only fluorescent images are shown.
Discussion
Adherence to specific organs and tissues is a key step in the successful colonization of humans by many bacterial pathogens (Pizarro-Cerda and Cossart, 2006). This host–bacterial interaction is mediated by a variety of cell surface pili (or fimbriae) in the case of several model Gram-negative pathogens (Olsen et al., 1989; Freitag et al., 1995; Kirn et al., 2000; Martinez et al., 2000; Craig et al., 2003; Kikuchi et al., 2005). To date, very little has been reported in this regard for Gram-positive pathogens (see recent review by Telford et al., 2006). In this study, we began to characterize host cell adhesion by C. diphtheriae, the toxigenic strain of diphtheria that assembles three distinct types of pili on the cell surface (Ton-That and Schneewind, 2003; Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). Employing a large battery of C. diphtheriae mutant strains, we assessed systematically the importance of various pili as well as their individual pilin subunits in corynebacterial adherence to a panel of three epithelial cell lines. We demonstrate for the first time that pili are indeed involved in corynebacterial attachment to specific epithelial cells, and that it is the minor pilins that act as adhesins. Significantly, our results reveal that the minor pilins not only decorate the pilus shaft, they are also displayed in clusters throughout the cell surface. In fact, we show that the pilus fibre is dispensable for corynebacterial adherence. Therefore, the minor pilins that are anchored on the cell wall are sufficient to mediate the specific host cell attachment. These findings, in conjunction with those recently reported on streptococci (Dramsi et al., 2006), suggest that the adhesive function of minor pilins is a conserved feature of Gram-positive pathogens.
To investigate the adhesive function of corynebacterial pili, we chose three different human cell lines that are all epithelial in nature, but they are derived from distinct tissues: pharynx (D562), larynx (HEp2) and lung (A549). Wild-type C. diphtheriae bound avidly to each cell line with little discrimination (Fig. 3A). However, when these cells were exposed to a C. diphtheriae mutant that is devoid of all six sortases, bacterial binding to the pharyngeal cell was dramatically reduced, while bacterial binding to the lung and the laryngeal epithelial cells suffered only a modest decrease (approximately two- to threefold). This sortase-less mutant bacterium fails to assemble each of the three pili (Swierczynski and Ton-That, 2006); in addition, the mutant must be defective in displaying many other surface-anchored proteins predicted from the genome sequence (Cerdeno-Tarraga et al., 2003). Thus, the gross defect in pharyngeal cell binding might not be ascribed to the loss of pili. In fact, the residual binding observed with the lung and the laryngeal cells must be mediated by some bacterial surface molecules whose assembly is sortase independent.
That the interaction between corynebacteria and the pharyngeal cells rely upon a specific pilus was hinted by our analysis of another C. diphtheriae mutant in which we deleted the gene-encoding SrtA, the sortase that specifically catalyses assembly of the SpaA-type pilus. This ΔsrtA mutant bound well to both lung and laryngeal cells but not the pharyngeal cells (Fig. 3A). In contrast, the C. diphtheriae mutant ΔsrtB–F, which encodes SrtA but lacks all other sortases, bound pharyngeal cells avidly (Fig. 3B). These two pieces of data are consistent with an adhesive role of the SpaA-type pili in pharyngeal cell binding. However, because we cannot be certain that SpaA-pilus assembly is the only sorting function of SrtA, these experiments did not provide the conclusive evidence for the nature of the surface molecules that mediate corynebacterial adherence.
The direct evidence for the role of SpaA-type pili in corynebacterial adherence to the pharyngeal cells came from our analysis of in-frame deletion mutations that knock out the individual pilins. Recall that the ΔsrtB–F mutant assembles only the SpaA-type pilus (Fig. 2) and that it adheres well to the pharyngeal cells. To determine whether this binding is mediated by the SpaA pili or not, we introduced into this strain individual pilin subunit deletion mutations. SpaA is the major pilin that is essential for assembly of the pilus fibre. Its deletion abolished pilus assembly (Ton-That and Schneewind, 2003; Fig. 3), but surprisingly there was little consequence of the loss of pilus fibres to pharyngeal cell binding (Fig. 4A). In sharp contrast, the pharyngeal cell adherence was greatly compromised when we deleted either of the two minor pilins SpaB and SpaC (Fig. 4A). That each minor pilin contributes to specific corynebacterial adherence was established by several lines of further evidence. First, the deletion of both SpaB and SpaC in the ΔsrtB–F parental strain minimized pharyngeal cell binding (Fig. 5A). Second, the defective binding is complemented by plasmids that encode either SpaB or SpaC (Fig. 5A). Third, antibodies directed against either SpaB or SpaC block the binding of the ΔsrtB–F mutant to pharyngeal cells (Fig. 5C). Finally, latex beads with SpaB- or SpaC-bound protein bind readily to pharyngeal epithelial cells in a specific manner (Fig. 6), but bind poorly to lung epithelial cells (data not shown).
The fact that the minor pilins could mediate corynebacterial adherence in the absence of a pilus fibre was an unexpected development. To allow adhesion in the absence of a pilus shaft, the minor pilins of corynebacteria must somehow be anchored to the cell surface, a logical scenario we did not envision until now. Like the major pilin subunits, each minor pilin contains the LPXTG motif, which permits the sortase-catalysed cross-linking of the carrier protein to the cell wall. It is therefore reasonable to suspect that the SpaBC pilins are components of the cell wall as well as the pilus. We obtained several lines of complementary evidence that establish this important fact. First, the presence of SpaB and SpaC on bacterial surface is detected by indirect immunofluorescence microscopy in the absence of SpaA, irrespective of whether the spaA deletion mutant is generated in the ΔsrtB–F strain (Fig. 7) or the wild-type strain (Ton-That and Schneewind, 2003, and data not shown). Second, the surface display of SpaB and SpaC occurs in a strain that produces abundantly a mutant form of SpaA (SpaA-K190A), which cannot form a pilus shaft due to a defect in homopolymer formation (Fig. 8). Finally, the surface display of minor pilins is sortase catalysed as the surface assembly is abolished when the respective LPXTG motifs are scrambled (Fig. 9).
Our findings provoke two basic questions on the cell surface assembly of Gram-positive bacteria and its role in colonization. First, how does the secretion-sorting machinery of Gram-positive bacteria orchestrate the assembly of pilins as components of both pilus fibres and/or the cell wall? Second, how do pili and surface-associated minor pilins facilitate bacterial adhesion to the host tissue?
The pilus proteins contain signal sequence at their N-termini and they are likely exported by the general secretory apparatus. Unlike other secreted proteins, however, the pilus proteins within the exoplasm are recognized and processed by a sortase, which can catalyse the covalent cross-linking of a pilin either to another protein substrate or to the lipid II precursor of the cell wall (Ton-That and Schneewind, 2004). According to current models for this cross-linking reaction (Ton-That and Schneewind, 2004; Marraffini et al., 2006; Telford et al., 2006), sortase cleaves the LPXTG motif of a pilin forming a covalent intermediate. In assembling the pilus fibre, this intermediate acts as a donor, transferring and joining the cleaved pilin subunit to another pilin located nearby, which acts as the acceptor by providing a suitable amino group. The alternative acceptor for this transfer reaction is the membrane-bound lipid II precursor, in which the diaminopimelic acid (DAP) moiety provides a reactive amine group. When the sortase-pilin intermediate catalyses this latter reaction, the pilin monomer becomes linked to DAP and eventually incorporated into the growing cell wall. Thus, during the sorting process, the sortase-pilin cross-linked intermediate has two choices that are mutually exclusive. One choice permits extension of the pilus fibre while the other terminates this polymerization by cell wall anchoring. Currently, we have no information about the factors that govern this choice; however, it is tempting to speculate that the cell wall anchoring and/or pilus incorporation of pilins may involve a regulatory switch.
Our finding that the pilus fibre itself is not essential for pharyngeal cell adhesion by corynebacteria shows that the clusters of SpaB/SpaC minor pilins anchored on the cell wall can act as specific adhesive patches to bind the host cell. Consistent with its role as an adhesin (Figs 5 and 6), homology searches revealed that the N-terminus of SpaC harbours a VWF domain, which is known to be important in interacting with extracellular matrix components and host cell membrane receptors (Ruggeri, 2001). The VWF domain also contributes to the generation of fibrin (Ruggeri, 2001), a critical component of the pseudomembrane produced by diphtheria (Love and Murphy, 2006). We have not identified the host cell molecules involved in corynebacterial adherence. However, it is clear that the adhesion involves two separate interactions, as each of the minor pilins alone suffices for strong host cell adhesion and the two proteins are disparate in their primary sequence. We have yet to determine whether SpaB and SpaC cross-link to each other or not. In either case, the two pilins may bind two separate receptors or two separate regions of a single receptor.
The evidence that the minor pilins are components of both the cell wall and pili provides an attractive mechanism for the intimate adherence of corynebacteria to the host cell (Telford et al., 2006). During the initial stages of infection, the pilus-associated minor pilins would secure the piliated bacteria onto pharyngeal epithelial cells through distant contacts. Once anchored to the target cell, the patches of pilins on the bacterial surface would bind to additional host membrane receptors, thereby creating a zone of tight adhesion. Conceivably, this intimate attachment serves important functions in pathogenesis. First, the tight adherence would not only prevent bacterial dissociation, it would strengthen host cell signalling. Second, the delivery of diphtheria toxin to the host cell in contact would be greatly facilitated. The secreted toxin would readily bind to the receptor displayed on the host cell surface and be internalized, ultimately paralysing the host cell through the inhibition of protein synthesis. This would create a safe niche for the adherent bacteria to grow and divide, and throw off progenies for colonization of new host cells.
In conclusion, specific pilus proteins displayed on the bacterial cell surface govern the host cell tropism. The resulting intimate attachment is pivotal to colonization by both Gram-negative and Gram-positive pathogens. So far, while the assembly and adhesive functions of various types of pili in Gram-negative bacteria have been dissected in detail (Sauer et al., 2000), our knowledge of many aspects of the sortase-catalysed pilus assembly in Gram-positive bacteria still remains rudimentary. Our demonstration that pilins can be deposited on the cell wall in addition to their directed assembly as fibrilar polymers opens up a number of intriguing mechanistic and biological questions. Is the secretion of pilins coupled to their sorting? If so, how is this achieved? Is pilus assembly compartmentalized? If so, how? Are there factors that regulate pilus polymerization and the surface display of pilins? Does this regulation involve cues from the host? Future challenge is to answer some of these basic questions and utilize the information to develop novel antimicrobial therapies.
Experimental procedures
Bacterial strains, plasmids and media
Corynebacterium diphtheriae NCTC13129 was obtained from the American Type Culture Collection. C. diphtheriae strains (Table S1) were grown on heart infusion broth (HIB), heart infusion agar (HIA) or trypticase soy agar supplemented with 5% sheep blood (Hemostat) (TSASB). E. coli strains were grown on Luria broth. Kanamycin was added at 50 μg ml-1 as needed. Polyclonal antibodies raised against recombinant pilins were obtained as previously described (Ton-That and Schneewind, 2003; Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). Reagents were purchased from Sigma unless otherwise indicated.
Generation of C. diphtheriae mutants
Corynebacterium diphtheriae in-frame, non-polar deletion mutants were generated by homologous recombination by a previously described procedure (Ton-That and Schneewind, 2003). The deletion mutants were verified by PCR, Western and Southern blotting techniques. Briefly, gene deletions were constructed by cross-over PCR using appropriate primer sets (Table S2) (Ton-That and Schneewind, 2003) and cloned between suitable restriction sites of pK18mobsacB (Schafer et al., 1994). Recombinant plasmid was purified, characterized and transformed into E. coli S17-1 (de Lorenzo et al., 1993). Overnight cultures of E. coli S17-1 and C. diphtheriae were mixed in equal volumes and spread on agar plates at 30°C for 16 h. Corynebacterial co-integrates were isolated by plating conjugal bacteria on HIA plates supplemented with 35 μg ml-1 nalidixic acid and 50 μg ml-1 kanamycin. The deletion mutants were then selected by plating co-integrates on HIA plates supplemented with 10% sucrose and 35 μg ml-1 nalidixic acid. For complementation analysis (Table S2), DNA fragments containing the promoter and open reading frames of gene of interest were cloned into the E. coli/Corynebacterium shuttle vector pCGL0243 (Ankri et al., 1996), and the recombinant plasmids were electroporated into C. diphtheriae (Ton-That and Schneewind, 2003).
Site-directed mutagenesis of recombinant plasmids
A PCR-based site-directed mutagenesis of double-stranded DNA was used in this study (Ton-That et al., 2004). Recombinant plasmid DNA was used as a template for PCR amplification with Pfu DNA polymerase using primer sets (5′and 3′) flanking six codons on both sides of the desired site of mutation (Table S2). After PCR, the amplified plasmids were digested overnight at 37°C with DpnI to select against parental DNA molecules. Digested plasmids were transformed into E. coli. Mutant plasmids were verified by sequencing and then transformed into C. diphtheriae by electroporation (Ton-That and Schneewind, 2003).
Expression and purification of recombinant pilus proteins
Appropriate primer sets (R-Spa primers, Table S2) were used to PCR amplify the spaA, spaB or spaC sequence from the chromosome of C. diphtheriae strain NCTC13129 that encoded the mature SpaA, SpaB lacking the cell wall sorting domain or the E19-N344 region of SpaC respectively. The DNA fragments were digested with BamHI, inserted into pQE30 (Qiagen, Chatsworth, CA)-cut BamHI. The resulting plasmids were verified by sequencing, transformed into E. coli XL1-Blue and selected on Luria agar with ampicillin (100 μg ml-1). The recombinant proteins were purified according to a published protocol (Ton-That et al., 1999).
Covalent coupling of proteins to fluorescent latex beads
The coupling process was performed according to a published protocol with modifications (Braun et al., 1998). Briefly, approximately 1 × 109 carboxylate-modified, fluorescent latex beads (1 μm in diameter, Yellow Green fluorescent Fluo-Spheres, Molecular Probes) were incubated with 300 μg of purified protein or with 1 mg of BSA (Sigma) in a MES buffer at pH 6.0 at room temperature for 15 min. EDAC [1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, hydrochloride] (Molecular Probes) was added to a final concentration of 1.6 mg ml-1 to activate surface carboxyl groups and the pH was adjusted to 6.5 with 0.2 M NaOH. The reaction mixture was then incubated with end-to-end shaking for 24 h in the dark. Glycine was added to a final concentration of 100 mM to quench unreacted groups, and the solution was incubated for 30 min at room temperature. The protein coated beads were then washed and blocked with 2 mg ml-1 BSA in phosphate-buffered saline (PBS) for 15 h in the dark at 4°C. The beads were then washed and stored in PBS with 1% BSA in the dark at 4°C. To determine the coupling efficiency, the protein concentration of the supernatant from the first wash (before addition of BSA) was measured using the BCA system (Pierce) and subtracted from the protein concentration in the mixture at the start of the coupling. Similar coupling efficiency was observed for different recombinant pilus proteins.
Cell cultures
HEp-2 cells (human larynx carcinoma CCL-23, ATCC Manassas, VA) and Detroit 562 cells (human pharynx carcinoma CCL-138, ATCC) were cultured in Minimum Essential Medium (MEM Eagle, ATCC) supplemented with non-essential amino acids, 1 mM sodium pyruvate, 2 mM l-glutamine, 1500 mg l-1 sodium bicarbonate, 100 U penicillin, 100 μg ml-1 streptomycin (Penicillin-streptomycin solution, ATCC) and 10% fetal bovine serum (Atlanta Biologicals). A549 cells (lung carcinoma CCL-185, ATCC) were cultured in RPMI Medium 1640 (Gibco) supplemented with 2 mM glutamine (Gibco), 100 U penicillin, 100 μg ml-1 streptomycin and 10% fetal bovine serum. Cell cultures were maintained in a 5% CO2/95% air atmosphere at 37°C and were passaged in a 1:5 ratio every 3–5 days.
Adherence assays
Epithelial cells were grown to confluency in 12-well plates, washed with sterile PBS, and fresh media (not containing antibiotic and serum) was added. C. diphtheriae cultures were grown to mid-exponential phase and used to infect epithelial cells with a multiplicity of infection (moi) of 10 [~107 colony-forming units (cfu) ml-1] and centrifugation at low speed (2000 r.p.m.). The assay was carried out for 1 h at 37°C in a 5% CO2/95% air atmosphere. Washed cells were detached and lysed by treating with diluted Trypsin-versene mixture (BioWhittaker) and 0.025% Triton X-100. The appropriate dilutions were plated on TSASB to enumerate adherent bacteria. The titre of adherent bacteria for each strain was compared with input titre and the percentage of adherent bacteria was determined. Each assay was performed in quadruplicates and repeated at least three times. Statistical analysis was performed by using the Student’s t-test.
In blocking experiments with antibodies, mid-log phase corynebacteria were incubated with a specific antiserum at a dilution of 1:100 for 1 h at 37°C with slight shaking. Pharyngeal cell monolayers were infected with antibody-bound bacteria at an moi of 10 (~107 cfu ml-1). The adhesion assay was then carried out as described above.
To test the adhesive properties of SpaA, SpaB or SpaC and BSA-coated latex beads, epithelial cells were grown on glass coverslips. Prior to infection, the semi-confluent monolayers were washed with sterile PBS and blocked with 0.5% BSA for 1 h at 37°C. Fluorescent latex beads with bound protein were added to cells at an moi of 100. After 1 h of incubation and five washes with PBS, the cells were fixed in 3% paraformaldehyde in PBS for 10 min at room temperature, rinsed twice in PBS. The cells were then permeabilized with PBS plus 0.1% Triton X-100 (Sigma) for 3–5 min, rinsed twice with PBS and incubated with Texas-Red-X phalloidin (Molecular Probes) for 20 min. After two final rinses with water, the coverslips were examined with an Olympus BX60 microscope.
Biochemical analysis of C. diphtheriae pili
Extractions of C. diphtheriae pili were carried out as previously described with modifications (Ton-That and Schneewind, 2003). Briefly, C. diphtheriae strains were scraped from TSASB plates after overnight growth and washed in SMM buffer (0.5 M sucrose, 10 mM MgCl2 and 10 mM maleate, pH 6.8). Cell pellets were re-suspended in the same buffer, treated with mutanolysin (300 U ml-1) at 37°C for 6 h. Solubilized pili isolated by centrifugation were precipitated with trichloroacetic acid, acetone-washed and dried under vacuum. Samples were boiled in SDS containing sample buffer, separated by 4–12% Tris-Glycine gradient gels (Invitrogen), subjected to immunoblotting with rabbit antisera (1:20 000 for α-SpaA, 1:5000 for α-SpaB and 1:10 000 for α-SpaC) and detected with chemiluminescence.
Immunofluorescence microscopy
Bacterial strains grown on TSASB plates were washed with PBS. The cells were then blocked for 1 h in PBS with 1% BSA. Pilins were stained with primary antibody diluted 1:100 in PBS with 0.1% BSA for 1 h followed by washing and blocking for 30 min. AlexaFluor 488 chicken anti-rabbit IgG (Molecular Probes, Invitrogen) diluted 1:100 in PBS with 0.1% BSA was added as secondary antibody for 1 h, followed by washing and final suspension in PBS. All incubations were carried out at 4°C on a rotisserie. The suspensions were then mounted on agarose-coated slides, covered with coverslip and sealed. The slides were observed on a Zeiss LSM 510 confocal microscope using the 100 × 1.4 planapochromat objective.
Immunoelectron microscopy
Electron microscopic experiments were carried out as previously described (Ton-That and Schneewind, 2003). Briefly, bacterial strains were grown on TSASB plates, washed in 0.1 M NaCl. A drop of bacterial suspension was placed on carbon grids, washed three times with PBS containing 2% BSA and blocked for 1 h in PBS with 0.1% gelatin. Pili were stained with primary antibody diluted 1:100 in PBS with 2% BSA for 1 h, followed by washing and blocking. Pili were stained with 12 nm gold-goat anti-rabbit IgG (Jackson ImmunoResearch) diluted 1:20 in PBS with 2% BSA for 1 h followed by washing in PBS with 2% BSA. The grids were washed five times with water before staining with 1% uranyl acetate. Samples were viewed in a Jeol 100CX electron microscope.
Supplementary Material
Acknowledgments
We thank Ann Cowan (Center for Cell Analysis and Modeling, University of Connecticut Health Center) for advice on imaging and Andrew Gaspar for advice and help in electron microscopy and bead binding assays. We are grateful to Sarkis Mazmanian (Harvard Medical School), Olaf Schneewind (University of Chicago) and members of our laboratory for valuable discussions and critical review of the manuscript. This work was supported by the US Public Health Service Grants AI061381 from the National Institute of Allergy and Infectious Diseases to H.T.-T.
Footnotes
This material is available as part of the online article from http://www.blackwell-synergy.com
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ankri S, Reyes O, Leblon G. Electrotransformation of highly DNA-restrictive corynebacteria with synthetic DNA. Plasmid. 1996;35:62–66. doi: 10.1006/plas.1996.0007. [DOI] [PubMed] [Google Scholar]
- Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun L, Ohayon H, Cossart P. The InIB protein of Listeria monocytogenes is sufficient to promote entry into mammalian cells. Mol Microbiol. 1998;27:1077–1087. doi: 10.1046/j.1365-2958.1998.00750.x. [DOI] [PubMed] [Google Scholar]
- Cerdeno-Tarraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, et al. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 2003;31:6516–6523. doi: 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar JO, Vatter AE, Clark WB, Curl SH, Hurst-Calderone S, Sandberg AL. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect Immun. 1988;56:2984–2989. doi: 10.1128/iai.56.11.2984-2989.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AV, Hirata R, Jr, de Souza CM, Monteiro-Leal LH, Previato JO, Formiga LC, et al. Corynebacterium diphtheriae surface proteins as adhesins to human erythrocytes. FEMS Microbiol Lett. 2001;197:235–239. doi: 10.1111/j.1574-6968.2001.tb10609.x. [DOI] [PubMed] [Google Scholar]
- Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- Freitag NE, Seifert HS, Koomey M. Characterization of the pilF–pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata R, Napoleao F, Monteiro-Leal LH, Andrade AF, Nagao PE, Formiga LC, et al. Intracellular viability of toxigenic Corynebacterium diphtheriae strains in HEp-2 cells. FEMS Microbiol Lett. 2002;215:115–119. doi: 10.1111/j.1574-6968.2002.tb11379.x. [DOI] [PubMed] [Google Scholar]
- Hirata R, Jr, Souza SM, Rocha-de-Souza CM, Andrade AF, Monteiro-Leal LH, Formiga LC, Mattos-Guaraldi AL. Patterns of adherence to HEp-2 cells and actin polymerisation by toxigenic Corynebacterium diphtheriae strains. Microb Pathog. 2004;36:125–130. doi: 10.1016/j.micpath.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Holmes RK. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis. 2000;181(Suppl 1):S156–S167. doi: 10.1086/315554. [DOI] [PubMed] [Google Scholar]
- Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci USA. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Eltis L, Kessler B, Timmis KN. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- Love JF, Murphy JR. Corynebacterium diphtheriae: iron-mediated activation of DtxR and regulation of diphtheria toxin expression. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive Pathogens. Washing, DC: American Society for Microbiology Press; 2006. pp. 726–737. [Google Scholar]
- Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, et al. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira Lde O, Andrade AF, Vale MD, Souza SM, Hirata R, Jr, Asad LM, et al. Effects of iron limitation on adherence and cell surface carbohydrates of Corynebacterium diphtheriae strains. Appl Environ Microbiol. 2003;69:5907–5913. doi: 10.1128/AEM.69.10.5907-5913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of Gram-positive bacteria and the mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell. 2006;124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Ruggeri ZM. Structure of von Willebrand factor and its function in platelet adhesion and thrombus formation. Best Pract Res Clin Haematol. 2001;14:257–279. doi: 10.1053/beha.2001.0133. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JR, Zahner D. Pili with strong attachments: gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- Swierczynski A, Ton-That H. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- Yeung MK. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10:120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- Yeung MK, Ragsdale PA. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun. 1997;65:2629–2639. doi: 10.1128/iai.65.7.2629-2639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung MK, Donkersloot JA, Cisar JO, Ragsdale PA. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. J Bacteriol. 1998;66:1482–1491. doi: 10.1128/iai.66.4.1482-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


