Abstract
Objective
To examine the presence of microRNAs within exosomes isolated from human saliva and to optimize and test methods for successful downstream applications.
Design
Exosomes isolated from fresh and frozen glandular and whole human saliva were used as a source of microRNAs. The presence of microRNAs was validated with TaqMan Real Time PCR and microRNA microarrays.
Results
We successfully isolated exosomes from human saliva from healthy controls and a patient with Sjögren’s syndrome. MicroRNAs extracted from the exosomal fraction were sufficient for quantitative PCR and microarray profiling.
Conclusions
The isolation of microRNAs from easily and non-invasively obtained salivary exosomes with subsequent characterization of the microRNA expression patterns is promising for the development of future biomarkers of the diagnosis and prognosis of various salivary gland pathologies.
Keywords: salivary exosomes, microRNA, biomarkers
Introduction
Exosomes are small, right-side out cell-secreted vesicles of about 30–100 nm, derived from fusion of multivesicular bodies to plasma membranes(Lakkaraju & Rodriguez-Boulan, 2008). They are morphologically distinct from secreted microvesicles, which are larger (~1 micron), and are instead derived from pinching off of the plasma membrane(Smalheiser, 2007). Both microvesicles and exosomes retain cytoplasmic contents, but exosomes have characteristic surface markers, such as CD63, CD9, CD81, and TSG101(Wang et al., 2008), not found on other secreted vesicle populations. They are derived from a wide range of cells, primarily hematopoetic cells such as reticulocytes, platelets, dendritic cells, B & T lymphocytes, and macrophages(Denzer et al., 2000). However, exosomes are also secreted by various epithelial and tumor cells(Valadi et al., 2007). Examples of epithelial cells that produce exosomes include alevolar lung tissue(Denzer et al., 2000), tubule cells and podocytes from nephrons(Zhou et al., 2008), and intestinal cells(Bunning et al., 2008). Exosome secretion occurs both in vitro from cell lines, ex vivo primary cells, and in vivo in animal models and humans(Valadi et al., 2007).
Beyond their characteristic repertoire of surface markers, exosomes feature a wide range of surface and internal proteins specific to their source (Lakkaraju & Rodriguez-Boulan, 2008), and recent studies found that they can also transport mRNA and microRNA(Valadi et al., 2007). Given the diversity of cargo transported by exosomes, it should come as no surprise that exosomes have already been implicated in the development of polarized epithelial cells, neuronal development, and tumor growth(Lakkaraju & Rodriguez-Boulan, 2008).
In the clinical setting, exosomes are present in a variety of bodily fluids, including blood, plasma, urine, amniotic fluid, and tumor malignant effusions (Lakkaraju & Rodriguez-Boulan, 2008). Given the relative ease and non-invasive nature of isolating exosomes from patient samples, and their distinctive protein and nucleotide contents, several studies have suggested using exosomal biomarkers for disease diagnostic purposes(Skog et al., 2008, Taylor & Gercel-Taylor, 2008, Zhou et al., 2008). The majority of these studies investigated exosomes isolated from serum, although several papers have focused on proteomic exosomal biomarkers in urine for renal disease(Gonzales et al., 2009, Zhou et al., 2008) prostate cancer(Mitchell et al., 2009) and saliva(Kapsogeorgou et al., 2005) (Gonzalez-Begne et al., 2009).
Beyond diagnostics, exosomes have also emerged as an exciting potential candidate for immunotherapy and vaccination modalities(De La Peña et al., Schorey & Bhatnagar, 2008), as well as a novel vector for gene therapy(Seow & Wood, 2009).
MicroRNAs are a group of small RNAs, 19–25 nucleotides in length, involved in the regulation of development and cell differentiation, proliferation and survival(Guarnieri & DiLeone, 2008, Lodish et al., 2008, Stefani & Slack, 2008). They exert their effects by two mechanisms: messenger RNA degradation and inhibition of translation. A single mRNA is usually translated into a single protein; however, a single miRNA is capable of regulating the translation of a multitude of genes by targeting specific regions in the 3′-UTR of their mRNA transcripts. Changes in mRNA levels can be ultimately controlled or cancelled out by post-transcriptional regulation; hence, miRNA expression levels may provide a better indication of a cell’s physiological state than mRNA expression.
Since a single microRNA can regulate hundreds of genes and may act as a master regulator of processes, select subsets of miRNAs can be used as biomarkers of physiologic and pathologic states. A recent study showed that the expression of as few as two miRNAs could accurately discriminate acute lymphoid from acute myeloid leukemia(Mi et al., 2007). Another feature that makes microRNAs excellent candidates for biomarker studies is their remarkable stability and resistance to degradation, especially compared to mRNA. We have been able to isolate miRNA from archived clinical specimens, including urine, saliva and formalin-fixed paraffin embedded tissues.
Relatively few studies, however, have investigated exosomal microRNAs (miRNAs) as potential diagnostic biomarkers: Hunter et al (Hunter et al., 2008) identified the presence of various miRNAs in human serum exosomes, while Skog et al (Skog et al., 2008) suggested that glioblastoma tumor-derived exosomes in patient serum carry a distinctive miRNA payload that can be used diagnostically.
Here, we report for the first time the successful isolation and initial characterization of miRNA-carrying exosomes from saliva. The purpose of this paper is to present our method for isolating and characterizing exosomal microRNAs from glandular and whole saliva.
Methods
Research subjects
Subjects were enrolled in a protocol for healthy volunteers or in a study of the natural history of Sjögren’s syndrome. Saliva was collected from 4 normal volunteers and 4 Sjögren’s syndrome patients. The Institutional Review Board of the National Institute of Dental and Craniofacial Research approved the study and all participants signed an informed consent.
Saliva Collection
To stimulate glandular salivary flow, subjects received a 2% citric acid solution to the posterior lateral surfaces of the tongue, applied bilaterally with a cotton swab for 5 seconds every 30 seconds. The citric acid stimulation continued for 30-second intervals during the entire collection procedure.
We collected parotid saliva as follows: Carlson Crittenden parotid collectors were placed bilaterally on the opening of Stenson’s duct orifice on the buccal mucosa opposite the upper second molar tooth. The parotid collectors were positioned on the mucosa so that the inner ring surrounded the duct orifice. Suction from the outer ring held the collector on the mucosa, with a vacuum created by squeezing and holding the deflated bulb during placement over the duct orifice and subsequent release of the bulb when the cup was in place.
Submandibular/sublingual saliva was collected as follows: With the orifices of the parotid ducts covered by the collectors, after applying 2% citric acid on the tongue for at least 5 times, the floor of the mouth was dried and saliva was collected with gentle suction into a tube on ice for twenty seconds. The collection was then stopped, a 2×2 gauze was placed over the orifice of the submandibular ducts and 2% citric acid was applied on the tongue. Saliva was collected in the same tube with gentle suction and the collection was stopped again with gauze. The whole process was repeated up to 8 times.
Salivary Exosome Isolation
The protocol for salivary exosome isolation was adapted and modified from a previous method for urinary exosome isolation(Zhou et al., 2008). Immediately after collection saliva was placed on ice, transferred to the laboratory and centrifuged at 1500 g for 10 minutes at 4 °C. The supernatant was then removed, placed in another tube and centrifuged at 17,000 g for 15 minutes at 4 °C to further remove unwanted organelles and cell fragments. Following initial centrifugation steps, the supernatant was transferred to sterile tubes for ultracentrifugation at 160,000 g for 1 hour at 4 °C. Following ultracentrifugation, the aqueous layer, which is viscous in whole saliva samples, was removed and the pellet containing the exosomes was washed with PBS and ultracentrifuged again at 160,000 g for 1 hour at 4 °C.
After the end of the second ultracentrifugation, the supernatant was removed and the pellet was briefly allowed to dry. The samples were then ready for protein or RNA isolation.
Protein Isolation and Western Blotting
Prior to exosome protein analysis, a stock solution of isolation buffer was made by mixing 10mM triethanolamine, 250 mM sucrose and deionized water. The isolation buffer pH was then adjusted to pH 7.6 with 1N sodium hydroxide. Deionized water was added to bring the total volume of the isolation buffer stock solution to 50 mL. Solution was stored at −20 °C. Protease inhibitors were added to 1 mL of isolation buffer just prior to use (50 microliters of phenylmethylsulphonyl fluoride [2mg/ml] and 10 microliters of leupeptin [1 mg/ml], both stored at −20 °C). Following exosome isolation, the pellet was resuspended in 100 microliters of isolation buffer containing the protease inhibitors. An equal volume of 2X Laemmli buffer (Biorad, Hercules, CA, USA) was added and the sample was denatured at 60 °C for 10 minutes.
Presence of TSG101 was determined with Western blotting. The samples were subjected to NuPAGE Novex 4–12% Bis-Tris Gel (Invitrogen, Carlsbadm CA, USA). The protein was transferred onto membranes using a semidry transfer unit. Western blotting was performed with TSG101 antibody (Abcam, (ab83), Cambridge, MA, USA) diluted 1:7500.
RNA Isolation and Analysis
Following exosome isolation, the pellet was treated with RNase A to degrade any residual cellular RNAs in order to ensure that all detected RNA was exosomal in origin. Some samples were treated with RNase A (Puregene-Gentra Systems, Valencia, CA, USA), 4 mg/ml solution, working concentration of 0.4 mg/ml in deionized water for 10 minutes at 37 °C. The sample exosomes were then lysed with 600 microliters of miRNeasy lysis buffer (Qiagen, Valencia, CA, USA) and stored at −80°C for later use or immediately processed using Qiagen’s miRNeasy Kit according to the manufacturer’s protocol. All RNA samples were eluted in 50 microliters of RNase free water.
To aid in the concentration and precipitation of exosomal RNA, Novagen’s pellet paint was used according to the manufacturer’s protocol with minor modifications; two microliters of pellet paint was added to the RNA samples. Following pellet paint addition, 0.1 volumes of 3M sodium acetate was added to the sample and the sample was mixed for 10 seconds. After mixing, 2.5 volumes of 100% ethanol were added to sample and vortexed briefly. The sample was then incubated at room temperature for two minutes and centrifuged for five minutes at 4 °C. Following centrifugation, the pellet containing exosomal RNA was washed with 200 microliters of 70% ethanol and allowed to air dry prior to resuspension in RNase free water. RNA was then quantitated using a UV-Vis spectrophotometer (Nanodrop 8000) and quality was assessed using the Agilent 2100 Bioanalyzer, where the presence of small RNAs was verified in both RNase treated and untreated samples.
After the isolation and quantitation of the exosomal RNA, five nanograms of input RNA were used for a reverse transcription reaction with the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). Gene specific primers to hsa-mir-203, hsa-mir-768-3p and hsa-mir-574-3p were used in separate reactions. A positive control reverse transcription reaction with the small nucleolar RNA U48 was performed using specific primers. Negative controls using 5 microliters of water in place of the RNA were performed alongside each reaction. cDNA obtained from the reverse transcription reactions were stored at −20 °C or immediately used for real-time quantitative PCR.
Real-time quantitative PCR was used to detect and quantify micro RNAs of interest. All samples were run in triplicate using 5 ng of cDNA for each reaction as described by the manufacturer’s protocol.
Microarray Studies
Microarray hybridization was performed using the Exiqon miRNA microarray system (miRCURY LNA™ microRNA Array, v.10.0) on exosomal miRNAs isolated from parotid and submandibular gland saliva, as well as from parotid salivary exosomal miRNAs from Sjögren’s syndrome patient. Sample labeling and hybridization were performed as described in the manufacturer’s protocol with the exception that starting material used was on the lower limit than the array manufacturer recommends. Briefly, miRNA spike-in controls were added to 250 ng of salivary exosomal microRNAs and were treated with calf intestinal phosphatase. The samples were then labeled with either Hy3 or Hy5, denatured, hybridized on the array at 56°C for 16 hours, washed and scanned on an Agilent scanner (Model G2505B). Data were processed with the Feature Extraction algorithm of Agilent.
Results
Isolation of exosomes
Saliva samples ranging from 200ul up to 5mL volume yielded an adequate amount of exosomal RNA for quantitative PCR. We isolated microRNAs from even smaller volumes of saliva, but the RNA yield was sufficient for only for a small number of quantitative PCR reactions. We were also able to isolate exosomes from saliva that was frozen at −20 °C for 7 days (Figure 1). Although we could isolate exosomes from both glandular and whole saliva, the viscosity and cellular contamination of whole saliva make it a less than ideal medium for exosomal isolation. Therefore, we primarily focused this study on glandular saliva only.
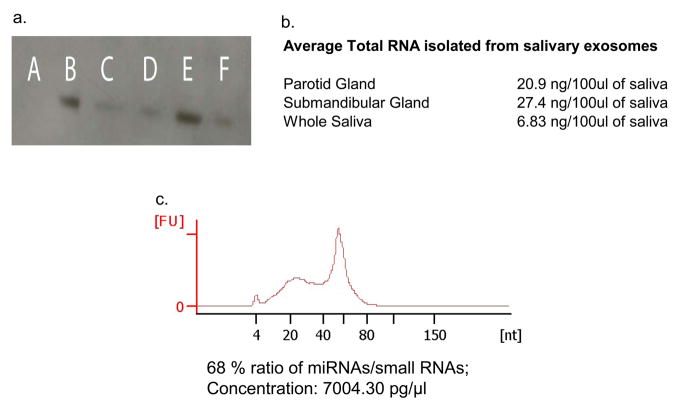
Figure 1.
(a) Western blot analysis of TSG101, a classic exosomal marker, of the exosomal lysates isolated from parotid saliva (40mg loaded sample) (B), from parotid saliva (20mg loaded sample) (C), submandibular saliva (D), frozen parotid saliva stored at −20°C for 7 days (E) and from frozen submandibular saliva stored at −20°C for 7 days (F). Negative control w as run in (A).
(b) Average total RNA concentration per 100ul of saliva collected. Technical difficulties with the mucin content of saliva precludes a higher RNA concentration to be obtained.
(c) A bioanalyzer profile of parotid saliva derived exosomal microRNAs. The enrichment in RNAs of the sizes of microRNAs is evident; 68% of the RNAs of size between 0 and 233 nucleotides falls within the microRNA range of 10–40 nucleotides and has an average size of 25 nucleotides.
To verify the presence of exosomes in the pellet after the series of centrifugations, we lysed pellets from both submandibular and parotid saliva, and confirmed the presence of TSG101, a standard exosomal marker by Western blot (Figure 1).
Assessment of exosomal microRNA
In order to assess the microRNA content of exosomes, and to ensure that isolated miRNAs originated from within the exosomes, we treated the exosomal pellets with RNaseA, as described in the methods section, and then isolated miRNA from exosomal lysates with a kit that also preserved mRNA. The total concentration of RNA that we isolated varied among individuals, with an average of 20.9ng per 100ul for parotid and 27.4ng per 100ul of submandibular saliva collected (Figure 1). Increased collection times did not increase the RNA concentration linearly; in continuous saliva collection, the first 100ul collected consistently had a greater RNA concentration than the subsequent 100ul. We suggest that the exosomes present in the cell are released in the saliva promptly upon stimulation and are collected quickly by this method. Once this extant supply is exhausted, the de novo synthesis of exosomes requires longer periods than our saliva collection times.
To confirm the presence of microRNAs within the exosomes, we performed TaqMan microRNA quantitative PCR amplification for three microRNAs (hsa-miR-203, hsa-miR-768-3p and hsa-miR-574-3p) that we have previously identified as present in minor salivary glands, as well as whole saliva. PCR reactions with negative and positive controls demonstrated the presence of microRNAs within the exosomes (Figure 1). For a more comprehensive assessment of exosomal miRNAs we ran two miRNA microarrays: one microarray was hybridized with microRNAs from parotid saliva against microRNAs from submandibular saliva from the same normal volunteer, and the second microarray was hybridized with miRNAs from parotid saliva from a normal volunteer against miRNAs from a Sjögren’s syndrome patient saliva sample (Table 1).
Table 1.
List of the most highly expressed human microRNAs in parotid exosomes from:
| Sjögren’ Syndrome Patient | Normal Volunteer |
|---|---|
| Gene Name | Gene Name |
| hsa-let-7b | hsa-let-7b |
| hsa-miR-150* | hsa-let-7c* |
| hsa-miR-23a* | hsa-miR-128 |
| hsa-miR-27b* | hsa-miR-150* |
| hsa-miR-29b | hsa-miR-17 |
| hsa-miR-29c | hsa-miR-1908 |
| hsa-miR-335 | hsa-miR-212 |
| hsa-miR-379* | hsa-miR-27b* |
| hsa-miR-433 | hsa-miR-29b |
| hsa-miR-454 | hsa-miR-29c |
| hsa-miR-483-3p | hsa-miR-335 |
| hsa-miR-584 | hsa-miR-379* |
| hsa-miR-621 | hsa-miR-433 |
| hsa-miR-652 | hsa-miR-454 |
| hsa-miR-760 | hsa-miR-483-3p |
| hsa-miR-888* | hsa-miR-584 |
| miRPlus_17824 | hsa-miR-621 |
| miRPlus_17841 | hsa-miR-652 |
| miRPlus_17848 | hsa-miR-760 |
| miRPlus_17858 | hsa-miR-888* |
| miRPlus_17824 | |
| miRPlus_17841 | |
| miRPlus_17848 | |
| miRPlus_17858 | |
| miRPlus_42487 | |
| miRPlus_42526 |
The human microRNAs were selected as having a normalized average expression level of 100 among replicates, after backrgound subtraction and dye normalization. miRPlus probes represent Exiqon’s proprietary sequences. The asterisk on some of the microRNAs is part of the microRNA name. The different microRNA patterns are shown only as a proof of concept and are not intended to draw any disease specific conclusions.
Discussion
In this report we show that exosomes can be readily isolated from saliva, and that these exosomes contain microRNAs in quantities adequate for both qPCR and microarray hybridization. To the best of our knowledge, this is the first report describing such a process. We have successfully amplified exosomal microRNAs from both parotid and submandibular gland saliva samples of a healthy volunteer, and from the parotid saliva of Sjögren’s patients. The different microRNA patterns are shown only as a proof of concept and are not intended to draw any disease specific conclusions. However, we believe that this report opens the door to reliable and reproducible salivary nucleic acid biomarker discovery. Previous reports investigating saliva for nucleic acid diagnostics analyzed mRNA expression in whole saliva. Although whole saliva is relatively easy to obtain, it has significant disadvantages as a medium for the isolation of mRNA. Whole saliva contains hundreds of thousands of cells of different origin, as well as contaminants such as commensal bacteria, that can easily alter levels of targeted mRNAs just by the differential presence of one cell type over another, even between different saliva collections of the same donor. For example, the periodontal status of a donor can easily alter the relative expression level of nucleic acids, by “contaminating” whole saliva with numerous inflammatory cells from the crevicular fluid. Second, nucleases are numerous in saliva, and what some groups may be describing as “free” circulating nucleic acids are typically degraded quickly, making identification and quantitation difficult. Many of these disadvantages are greatly reduced by the use of glandular saliva.
Exosomes isolated from individual salivary glands are derived from cells within that specific gland and may reflect the physiologic state of the gland not only at the protein level as previously examined ex vivo in human salivary gland epithelial cell lines (Kapsogeorgou et al., 2005), but also at the regulatory level. Thus, salivary exosomal miRNAs may be valuable not only as a diagnostic tool, but may also provide an insight in the role microRNAs play in the underlying pathophysiologic processes of various salivary gland diseases. Among others they may help understanding if specific miRNAs are involved abnormalities in saliva production or regulation of the peripheral inflammatory response in the salivary gland and oral characteristic of Sjögren’s syndome. Exosomal miRNA analysis may also be valuable in understanding the pathogenesis of salivary gland tumors since a number of studies have identified miRNA dysregulation as a characteristic marker in cancer cell proliferation in vivo(Hiyoshi et al., 2009, Noonan et al., 2009), and have found distinctive exosomal miRNA profiles in blood plasma, urine, and other fluids.
Isolation of exosomal microRNAs from the salivary gland holds the promise of focused biomarker discovery for pathologies that directly or indirectly affect the salivary glands. We have developed a method that allows the isolation of exosomal microRNA from saliva in quantities sufficient for microRNA microarrays. We have also provided pilot data suggesting that exosomal microRNA patterns between healthy controls and patients with salivary gland disease can be studied using micorarrays. More studies are needed to further characterize these differences and to better assess the value of salivary exosomal microRNAs in the diagnosis and prognosis of salivary gland diseases.
Acknowledgments
We would like to thank Stefanie Alexander and Dr Oscar Cheng for their technical assistance and suggestions. This research was supported by the Intramural Research Program of the NIH, NIDCR.
References
- Bunning, Smolinski Dv, Tafazzoli K, Zimmer K-P, Strobel S, Apostolaki M, Kollias G, Heath JK, Ludwig D, Gebert A. Multivesicular bodies in intestinal epithelial cells: responsible for MHC class II-restricted antigen processing and origin of exosomes. Immunology. 2008;125:510–521. doi: 10.1111/j.1365-2567.2008.02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Peña H, Madrigal JA, Rusakiewicz S, Bencsik M, Cave GWV, Selman A, Rees RC, Travers PJ, Dodi IA. Artificial exosomes as tools for basic and clinical immunology. Journal of Immunological Methods. doi: 10.1016/j.jim.2009.03.011. In Press, Accepted Manuscript. [DOI] [PubMed] [Google Scholar]
- Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. Journal of Cell Science 113 Pt. 2000;19:3365–74. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, Yates JR. Proteomic Analysis of Human Parotid Gland Exosomes by Multidimensional Protein Identification Technology (MudPIT) 2009:1304–1314. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Annals of Medicine. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M, Baba H. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clinical Cancer Research. 2009;15:1915–22. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee M-LT, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PLoS ONE. 2008;3:e3694–e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapsogeorgou EK, Abu-Helu RF, Moutsopoulos HM, Manoussakis MN. Salivary gland epithelial cell exosomes: A source of autoantigenic ribonucleoproteins. Arthritis & Rheumatism. 2005;52:1517–21. doi: 10.1002/art.21005. [DOI] [PubMed] [Google Scholar]
- Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends in Cell Biology. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nature Reviews Immunology. 2008;8:120–30. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19971–6. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Welton J, Staffurth J, Court J, Mason MD, Tabi Z, Clayton A. Can urinary exosomes act as treatment response markers in prostate cancer? Journal of Translational Medicine. 2009;7:4–4. doi: 10.1186/1479-5876-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–24. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- Schorey JS, Bhatnagar S. Exosome Function: From Tumor Immunology to Pathogen Biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow Y, Wood MJ. Biological Gene Delivery Vehicles: Beyond Viral Vectors. Mol Ther. 2009 doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser N. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biology Direct. 2007;2:35–35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature Reviews Molecular Cell Biology. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic Oncology. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Wang G-J, Liu Y, Qin A, Shah SV, Deng Z-b, Xiang X, Cheng Z, Liu C, Wang J, Zhang L, Grizzle WE, Zhang H-G. Thymus Exosomes-Like Particles Induce Regulatory T Cells. J Immunol. 2008;181:5242–5248. doi: 10.4049/jimmunol.181.8.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cheruvanky A, Hu X, Matsumoto T, Hiramatsu N, Cho ME, Berger A, Leelahavanichkul A, Doi K, Chawla LS, Illei GG, Kopp JB, Balow JE, Austin HA, Yuen PST, Star RA. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008;74:613–621. doi: 10.1038/ki.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]