Abstract
The ability of apolipoprotein E (apoE) to bind to cell-surface glycosaminoglycans (GAG) is important for lipoprotein remnant catabolism. Using surface plasmon resonance, we previously showed that the binding of apoE to heparin is a two-step process; the initial binding involves fast electrostatic interaction, followed by a slower hydrophobic interaction. Here we examined the contributions of the N- and C-terminal domains to each step of the binding of apoE isoforms to heparan sulfate (HS) and dermatan sulfate (DS). ApoE3 bound to less sulfated HS and DS with a decreased favorable free energy of binding in the first step compared to heparin, indicating that the degree of sulfation has a major effect on the electrostatic interaction of GAG with apoE. Mutation of a key Lys residue in the N-terminal heparin binding site of apoE significantly affected this electrostatic interaction. Progressive truncation of the C-terminal α-helical regions which favors the monomeric form of apoE3 greatly reduced the binding ability of apoE3 to HS, with much reduced favorable free energy of binding of the first step, suggesting that the C-terminal domain contributes to the GAG binding of apoE by the oligomerization effect. Supporting this, dimerization of the apoE3 N-terminal fragment via disulfide linkage restored the electrostatic interaction of apoE with HS. Significantly, apoE4 showed much greater binding to HS and DS than apoE2 or apoE3 in both lipid-free and lipidated states, perhaps resulting from enhanced electrostatic interaction through the N-terminal domain. This isoform difference in GAG binding of apoE may be physiologically significant such as in the retention of apoE-containing lipoproteins in the arterial wall.
Heparan sulfate proteoglycan (HSPG) is a common constituent of cell surfaces and the extracellular matrix, and is involved in a wide range of biological functions (1). Normal uptake and catabolism of atherogenic lipoproteins is mediated by the interaction of lipoproteins with heparan sulfate (HS) in the liver (2). The substantial reduction of hepatic HS sulfation associated with diabetes (3) is known to cause impaired hepatic uptake of lipoproteins, leading to increased atherosclerosis (4). Apolipoprotein E (apoE) is a critical ligand for high-affinity binding of remnant lipoproteins to members of the low density lipoprotein receptor (LDLR) family and HSPG (5–7). Binding of apoE to HSPG is an initial step in the localization of apoE-enriched remnant lipoproteins to the cell surface, followed by the internalization into hepatocytes through processes mediated by receptors, including the LDLR and the LDLR-related protein, or through interaction with HSPG alone (8). Binding of apoE to HSPG is also involved in the inhibition of platelet-derived growth factor-stimulated smooth muscle cell proliferation (9) and the differential effects of the apoE isoforms on neurite growth, repair, and consequently, the progression of late onset familiar Alzheimer’s disease (10–12).
Besides the apoE-HSPG interaction, the binding of apoE to cell-surface glycosaminoglycans (GAG) such as chondroitin sulfate (CS) and dermatan sulfate (DS) proteoglycans has been demonstrated to play an important role in lipoprotein catabolism. A major pool of apoE on the surface of human liver cells is associated with CS proteoglycans (13). The interaction of apoE with CS and/or DS proteoglycans is responsible for the retention of high density lipoproteins (HDL) on the extracellular matrix of arterial smooth muscle cells (14), and the uptake of β-very low density lipoprotein in the brain (15). In addition, apoE-mediated cholesterol efflux from macrophages is modulated by cell surface proteoglycans (16, 17).
ApoE contains two independently folded functional domains; these are a 22-kDa N-terminal domain (residues 1–191) and a 10-kDa C-terminal domain (residues 216–299) linked by a hinge region (18, 19). The N-terminal domain is folded into a four-helix bundle of amphipathic α-helices and contains the LDLR binding region (around residues 136–150 in helix 4) (20), whereas the C-terminal domain contains amphipathic α-helices that are involved in binding to lipoproteins with high affinity (21, 22). Both the N- and C-terminal domains contain a heparin binding site (23, 24): the N-terminal domain site is located between residues 136–147, overlapping with the LDLR binding region (25, 26), whereas the C-terminal site involves basic residues around lysine 233 (27). Although both sites are functional in the separated fragments, only the N-terminal site is available for interaction in both the lipid-free and lipidated states of the intact apoE molecule (27, 28). The C-terminal site may be involved in the binding of apoE to the protein core of biglycan of the vascular extracellular matrix (29, 30).
Given the physiological importance of apoE-GAG interactions, we have previously characterized the kinetics and affinity of the binding of apoE isoforms to heparin using surface plasmon resonance (SPR) measurements (28). Although the variations in the distribution of acidic groups in different types of GAG (31) are likely to influence the interaction with apoE (32, 33), quantitative information on the interaction with various GAG molecules is limited (26, 34). In the present study, we extended the SPR approach to explore the contributions of the N- and C-terminal domains of apoE to its interaction with more physiological GAG, such as HS and DS.
EXPERIMENTAL PROCEDURES
Materials
Porcine intestinal mucosa HS (average molecular weight of 13,655; sulfur content 5.51%), DS (average molecular weight of 41,400; sulfur content 6.85%), and heparin (average molecular weight of 13,000; sulfur content <38%) were purchased from Celsus Laboratories (Cincinnati, OH). Degrees of sulfation determined by the turbidimetry method (35) were 1.6, 0.7, and 0.7 sulfate groups per disaccharide for heparin, HS, and DS, respectively. 1,2-Dimyristoyl phosphatidylcholine (DMPC) was purchased from Avanti Polar Lipids (Pelham, AL), and stock solutions were stored in chloroform/methanol (2/1) under nitrogen at −20 °C. Ultrapure guanidine HCl was from ICN Pharmaceuticals (Costa Mesa, CA).
Protein Preparations
The full-length human apoE2, apoE3 and apoE4 and their 22-kDa (residues 1–191) fragments were expressed and purified as described (22, 36). The mutations in apoE to create K146E and K233E, and the truncated forms (Δ251–299, Δ261–299 and Δ273–299) were made using PCR methods as described (27, 37). The apoE preparations were at least 95% pure as assessed by SDS-PAGE. In all experiments, the apoE sample was freshly dialyzed from 6 M GdnHCl and 1% β-mercaptoethanol (or 10 mM DTT) solution into buffer solution before use. To prepare dimerized form of apoE3 22-kDa, the apoE3 22-kDa fragment at 10 mg/ml in oxygenated Tris buffer (10 mM Tris-HCl. 150 mM NaCl, 0.02% NaN3, 1mM EDTA, pH 7.4) in the presence 6 M urea was incubated for 2 weeks at 4 °C. Residual monomer form was removed by passage through a Thiopropyl Sepharose 6B column (GE Healthcare). DMPC discoidal complexes with the apoE3 variants were prepared as described (38).
Biotinylation of GAG
Heparin, HS, and DS were biotinylated at their reducing end (39, 40). Heparin, HS, or DS (4.5 mg/ml) was oxidized with 3.3 mM sodium metaperiodate (Wako Pure Chemicals, Osaka, Japan) in 0.1 M sodium acetate (pH 5.5) for 1 h at room temperature. The oxidation was stopped by adding an excess of sodium sulfite. The oxidized GAG was then incubated with a six-fold molar excess of 6-(biotinylamino) hexanoylhydrazine (Dojindo Laboratories, Kumamoto, Japan) for 2 h. The excess biotin was removed by dialysis.
SPR Experiments
SPR measurements were performed on a Biacore X instrument (Biacore, Inc., Uppsala, Sweden) (28). For immobilization of GAG on a SA chip, an injection of biotinylated GAG (10 μg/ml) in Tris buffer was made at a flow rate of 5 μl/min followed by a 10-μl injection of 2 M NaCl. Typically, 100–500 resonance units (RU) of GAG were immobilized and the effects of mass transport were not significant due to the low surface density of ligand (41). An untreated flow cell was used as a control. For kinetic measurement of apoE interaction with GAG, a 30 μl injection of the apoE sample was passed over the sensor surface at a flow rate of 20 μl/min. At the end of the sample plug, the same buffer was passed over the sensor surface to facilitate dissociation, and then the sensor surface was regenerated for the next sample using a 10-μl pulse of 2 M NaCl and 0.1 % CHAPS. The resultant sensorgrams were analyzed using BIA evaluation software (version 4.1). The response curves of various analyte concentrations were globally fitted to the two-state binding model described by the following equation (42, 43):
where the equilibrium constants of the each binding steps are K1 = ka1/kd1 and K2 = ka2/kd2 and the overall equilibrium binding constant is calculated as KA = K1(1 + K2) and Kd = 1/KA. In this model, the analyte (A) binds to the ligand (B) to form an initial complex (AB) and then undergoes subsequent binding or conformational change to form a more stable complex (ABx). Gibbs free energies of binding of each binding steps are calculated according to ΔG1 = −RT ln (55.5 K1) and ΔG2 = −RT ln K2, respectively. The maximum binding response (Rmax) value which reflects the saturated amount of apoE bound to GAG was obtained from the global fitting.
Circular Dichroism (CD) Spectroscopy
Far-UV CD spectra were recorded from 185 to 260 nm at 25 °C using an Aviv 62DS spectropolarimeter. After dialysis, the apoE sample was diluted to 25–50 μg/ml in 10 mM phosphate buffer (pH 7.4) for obtaining the CD spectrum. The results were corrected by subtracting the buffer baseline. The α-helix content was derived from the molar ellipticity at 222 nm, [θ]222, according to the following equation (44):
Fluorescence measurements
Fluorescence measurements were carried out with a Hitachi F-7000 fluorescence spectrophotometer at 25 °C in Tris buffer (pH 7.4). Trp emission fluorescence of proteins at a concentration of 25 μg/ml was recorded from 300 to 420 nm using a 295 nm excitation wavelength to avoid tyrosine fluorescence. For chemical denaturation experiments, the change in the wavelength of maximum fluorescence was monitored at various urea concentrations in the absence or presence of HS.
Statistical analysis
Results were compared statistically by t-test using GraphPad Prism 5.0.
RESULTS
Two-step Binding of ApoE to GAG
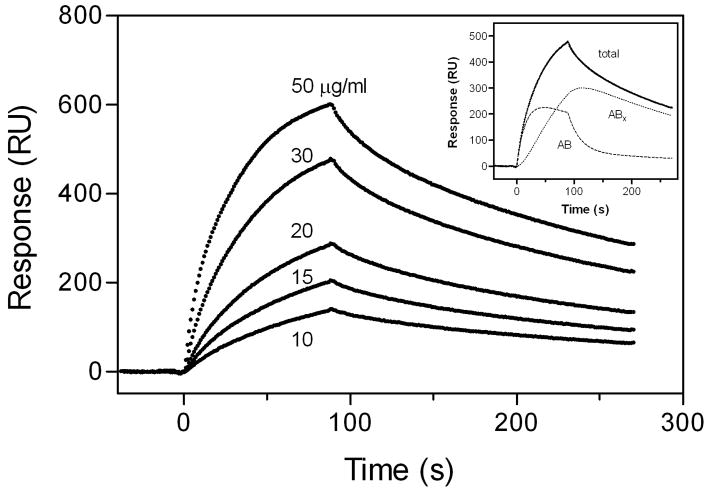
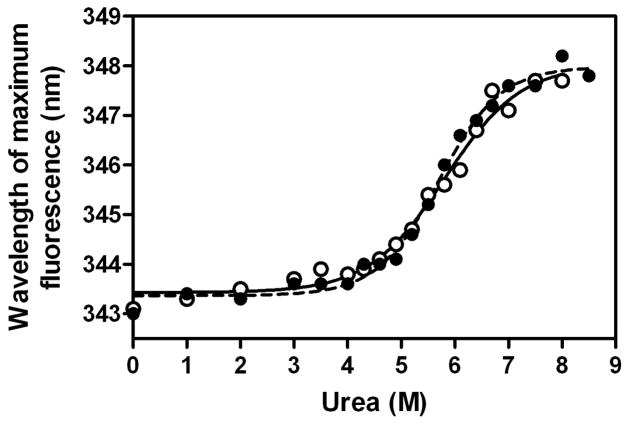
We previously demonstrated that the binding of apoE to heparin is a two-step process; the initial binding involves fast electrostatic interaction, followed by a slower hydrophobic interaction (28). Fig. 1 shows typical sensorgrams for the binding of lipid-free apoE3 to HS. The response curves were well fitted by a two-state binding model where binding of the analyte to the ligand is followed by either another subsequent binding event or a conformational change (Fig. 1, inset). In far-UV CD measurements, no change in the spectra of apoE3 was observed in the absence or presence of HS (data not shown). In addition, HS had no effect on the urea-induced denaturation behavior of the apoE3 22-kDa fragment (Fig. 2). These results suggest that there is no major conformational change in apoE upon HS binding. Similar two-step binding was also observed in the binding of lipid-free apoE3 to DS (data not shown).
Fig. 1. SPR sensorgrams of binding of lipid-free apoE3 to immobilized HS.
The inset shows the fitting of experimental data (●) with the two-state binding model, where A + B ⇔ AB ⇔ ABx (see “Experimental Procedures”). Simulated curves displaying the initial binding (AB) and subsequent binding (ABx) are the additive components from the fitted curve (solid line). RU, resonance units.
Fig. 2. Urea denaturation of apoE3 22-kDa fragment in the absence or presence of HS.
Protein concentration was 25 μg/ml. apoE3 22-kDa alone (○) or incubated with 50 μg/ml of HS (●).
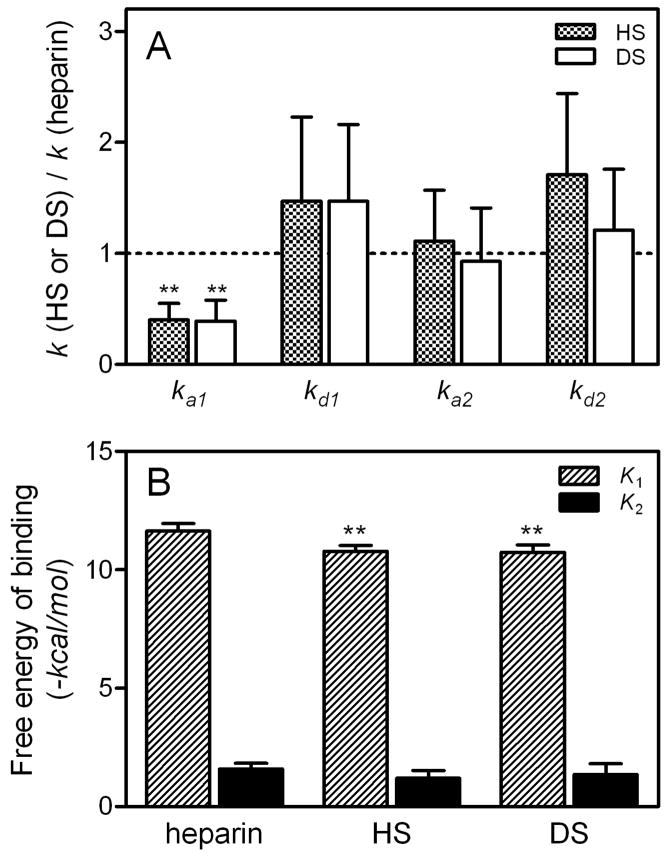
Fig. 3 summarizes the relative changes in the rate constants for each step of lipid-free apoE3 binding to GAG (Fig. 3A) and the relative contributions of each step to the overall free energy of binding estimated from the affinity constants for the two individual steps (Fig. 3B). Compared to heparin, the less sulfated HS and DS showed a slower first association rate and, consequently, a significant decrease in the favorable free energy of binding for the first step. This indicates that the degree of sulfation in GAG has a major effect on the electrostatic interaction with apoE (26).
Fig. 3. SPR rate constants and free energy of binding for the interaction of lipid-free apoE3 with GAG.
A, Relative change in SPR rate constants for each binding steps of apoE3 to GAG. B, Free energy for each binding steps of apoE3 to GAG. Free energy was calculated according to ΔG = −RT ln K using binding constants for each steps, K1 and K2. *, p < 0.05, **, p < 0.01 compared to heparin.
Effects of Lysine Mutations in the Heparin-Binding Sites of ApoE on GAG Binding
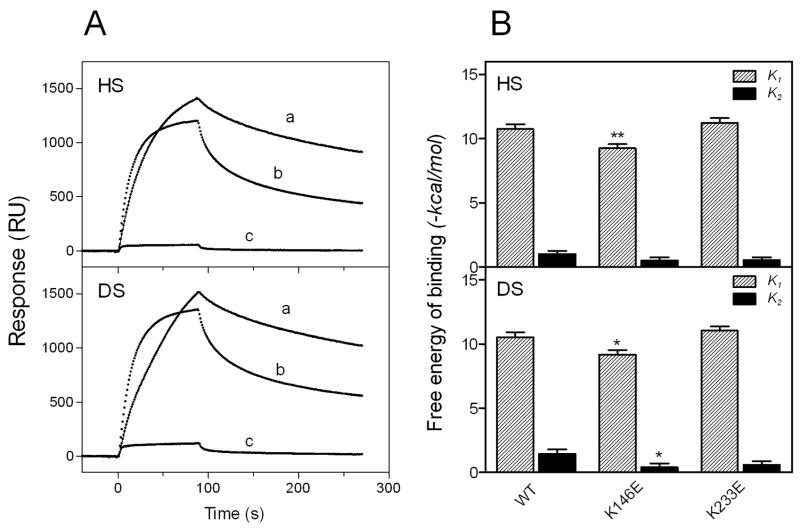
To confirm the importance of an ionic interaction in the apoE-GAG interaction, we examined the effects of substitutions of lysine residues 146 and 233 in apoE on the GAG binding; these residues are located in the N- and C-terminal heparin binding sites, respectively, and contribute to an ionic interaction with heparin (26, 27). As shown in Fig. 4A, large decreases in binding to HS and DS were observed with the K146E mutant whereas the K233E mutant exhibited responses similar to the wild-type, consistent with the previous finding that only the N-terminal site is available for the interaction with heparin even in the lipid-free state (27, 28). Comparison of the free energy of binding for each step among lysine mutants (Fig. 4B) clearly demonstrates that such decreases in GAG binding for the K146E mutant is due to the reduction of the electrostatic interaction between apoE and GAG (28). In addition, such a disruption of the electrostatic interaction resulted in a decrease in the saturated amount of apoE bound to GAG: for example, Rmax values for the HS binding of apoE3 WT, K146E, and K233E were 1300, 180, and 1020 RU respectively.
Fig. 4. SPR sensorgrams (A) and free energy of binding (B) for the interaction of Lys mutants of apoE3 with HS and DS.
A, WT apoE3 (a), apoE3 (K233E) (b), and apoE3 (K146E) (c). The protein concentration was 30 μg/ml. B, Free energy for each binding steps of apoE3 to HS and DS.*, p < 0.05, **, p < 0.01 compared to WT.
Effects of Oligomerization of ApoE on GAG binding
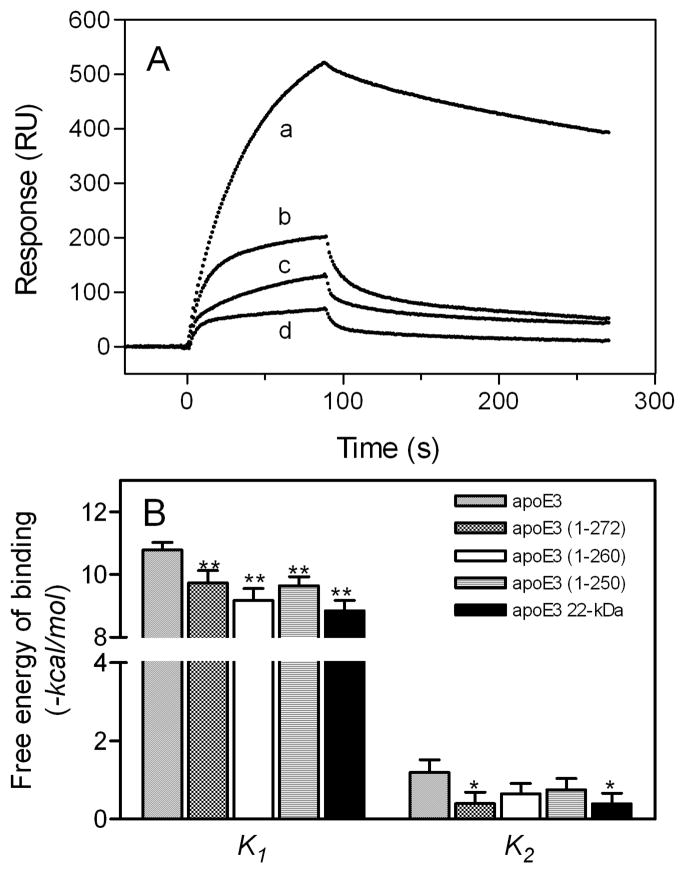
ApoE is known to self-associate through the C-terminal domain in solution and the C-terminal α-helical regions are responsible for the self-association (21, 45–47). To examine the role of the self-association of apoE in GAG binding, we compared the HS binding of progressive C-terminally truncated mutants of apoE3. Using gel filtration chromatography, we showed previously that, even at a very low protein concentration (5 μg/ml), full-length apoE3 predominantly forms tetramer and that more than half of apoE3 (1–272) forms tetramer whereas apoE3 (1–260) exists completely as a monomer (47). Although the C-terminal heparin binding site is conserved in both apoE3 (1–272) and apoE3 (1–260), significant decreases in binding response (Fig. 5A) and favorable free energy of binding in the first step (Fig. 5B) were observed for both mutants. This indicates that the oligomerization through the C-terminal α-helical regions has a major influence on the electrostatic interaction between apoE and GAG. However, Rmax values were not significantly different among all mutants (880, 840, 1200, and 710 RU for full-length, 1–272, 1–260, and 1–250 apoE3, respectively), suggesting that the oligomerization does not affect the level of apoE binding to GAG.
Fig. 5. Effect of C-terminal truncation on the interaction of lipid-free apoE3 with HS.
A, SPR sensorgrams of binding of apoE3 C-terminal-truncated mutants to HS. apoE3 (a), apoE3 (1–272) (b), apoE3 (1–260) (c), and apoE3 22-kDa (d). The protein concentration was 30 μg/ml. B, Free energy for each binding steps of apoE3 C-terminal-truncated mutants.
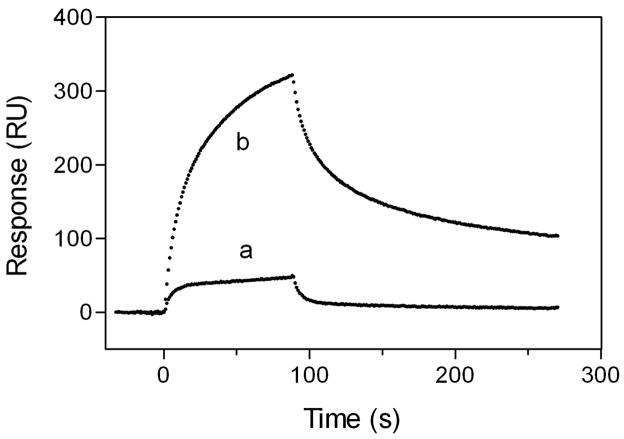
It is possible that the better binding of oligomeric apoE compared to the monomeric form is due to cooperative molecular interaction of apoE with GAG. To confirm this point, we compared the binding behaviors of the apoE3 22-kDa fragment to HS in the monomeric and dimeric states. As shown in Fig. 6, a greatly increased response in HS binding of apoE3 22-kDa dimer was observed compared to the monomeric form, and the kinetic parameters for the apoE3 22-kDa dimer are comparable to those for full-length apoE3 (Table 1). In contrast, Rmax values for monomer and dimer were similar (1050 and 1250 RU, respectively). These results suggest that the cooperative binding through multiple copies of the heparin binding sites of apoE molecules is important for the high affinity binding to GAG (27).
Fig. 6. SPR sensorgrams of binding of apoE3 22-kDa variants to HS.
apoE3 22-kDa monomer (a) and apoE3 22-kDa dimer (b). The protein concentration was 20 μg/ml.
Table 1.
Kinetic parameters for binding of lipid-free apoE3 variants to HS
| apoE | ka1 | kd1 | ka2 | kd2 | K1 | K2 | Kd |
|---|---|---|---|---|---|---|---|
| 104 M−1s−1 | 10−2 s−1 | 10−2 s−1 | 10−3 s−1 | 105 M−1 | μM | ||
| apoE3 | 3.6 ± 0.6 | 2.5 ± 0.8 | 3.1 ± 1.0 | 4.2 ± 0.8 | 15 | 7.4 | 0.18 ± 0.03 |
| apoE3 22-kDa | 1.0 ± 0.6 | 20 ± 5 | 0.8 ± 0.3 | 4.2 ± 1.1 | 0.6 | 1.9 | 6.2 ± 2.9 |
| apoE3 22-kDa dimer | 2.7 ± 0.8 | 6.8 ± 1.8 | 1.4 ± 0.4 | 4.4 ± 0.5 | 4.1 | 3.1 | 0.61 ± 0.12 |
Effects of Lipidation on GAG Binding of ApoE
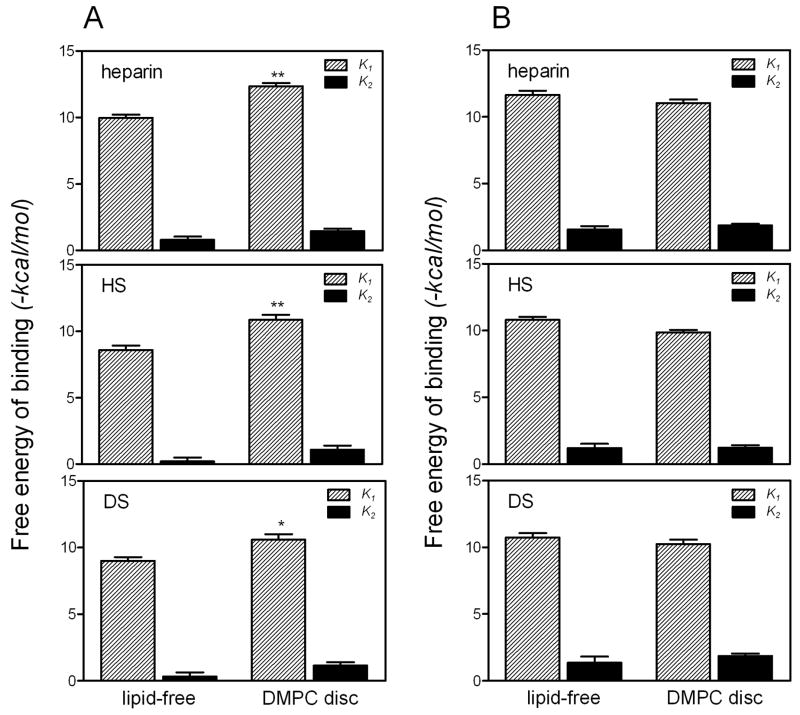
Although lipid-free apoE can bind effectively to GAG, we demonstrated previously that a discoidal complex of apoE 22-kDa fragments with DMPC exhibited much higher affinity in binding to heparin compared to the lipid-free form, mainly due to the increased free energy of binding in the first step (28). As shown in Fig. 7A, similar effects of lipidation on the binding of the apoE3 22-kDa fragment to HS and DS were observed. In contrast, there was no significant increase but rather a tendency to a decrease in the free energy of the first binding step between the lipid-free and lipidated forms of full-length apoE3 (Fig. 7B), suggesting that the electrostatic interactions of full-length apoE with GAG are similar in the lipid-free and lipidated forms.
Fig. 7. Comparison of free energy for GAG binding of apoE3 in the lipid-free form or complexed with DMPC.
A, apoE3 22-kDa. B, full-length apoE3. *, p < 0.05, **, p < 0.01 compared to lipid-free form.
Effects of ApoE Isoforms on GAG Binding
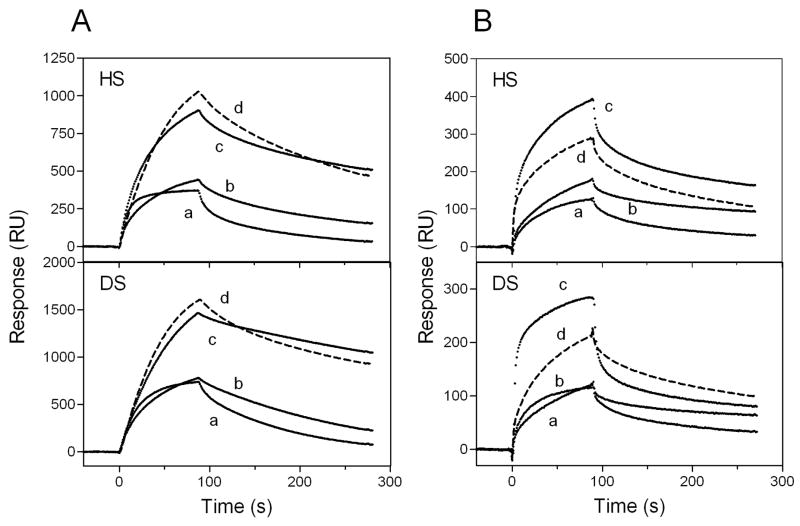
We further compared the association and dissociation kinetics for the interaction of apoE isoforms with GAG. As shown in Fig. 8, apoE4 displayed two- or three-fold higher binding to HS and DS compared to apoE2 or apoE3 both in the lipid-free and lipidated states. In fact, Rmax values for lipid-free apoE2, apoE3, and apoE4 were 670, 810, and 1210, respectively and, those for the DMPC discs were 360, 320, and 635, respectively. Such higher binding of apoE4 does not appear to result from the interaction between the N- and C-terminal domains in apoE4 because the apoE4 E255A mutant in which the domain interaction is disrupted (48) still exhibited much higher binding than apoE3 (Figs. 8A and 8B). As shown in Table 2, there was not much difference in association and dissociation rate constants and equilibrium affinity among apoE isoforms complexed with DMPC.
Fig. 8. SPR sensorgrams of binding of apoE isoforms to HS and DS.
apoE2 (a), apoE3 (b), apoE4 (c), and apoE4 E255A (d) in the lipid-free form (A) or DMPC discoidal complexes (B). The protein concentration was 30 μg/ml.
Table 2.
Kinetic parameters for binding of DMPC discoidal complexes of apoE isoforms to HS
| apoE | ka1 | kd1 | ka2 | kd2 | K1 | K2 | Kd |
|---|---|---|---|---|---|---|---|
| 105 M−1s−1 | 10−2 s−1 | 10−2 s−1 | 10−3 s−1 | 105 M−1 | μM | ||
| apoE2/DMPC | 0.4 ± 0.1 | 9.2 ± 2.7 | 1.5 ± 0.1 | 6.6 ± 2.0 | 4.4 | 2.3 | 0.68 ± 0.21 |
| apoE3/DMPC | 0.4 ± 0.1 | 13 ± 2 | 2.7 ± 0.1 | 3.4 ± 1.0 | 3.1 | 8.0 | 0.35 ± 0.15 |
| apoE4/DMPC | 1.1 ± 0.2 | 15 ± 3 | 1.6 ± 0.1 | 4.2 ± 1.0 | 7.5 | 4.0 | 0.27 ± 0.08 |
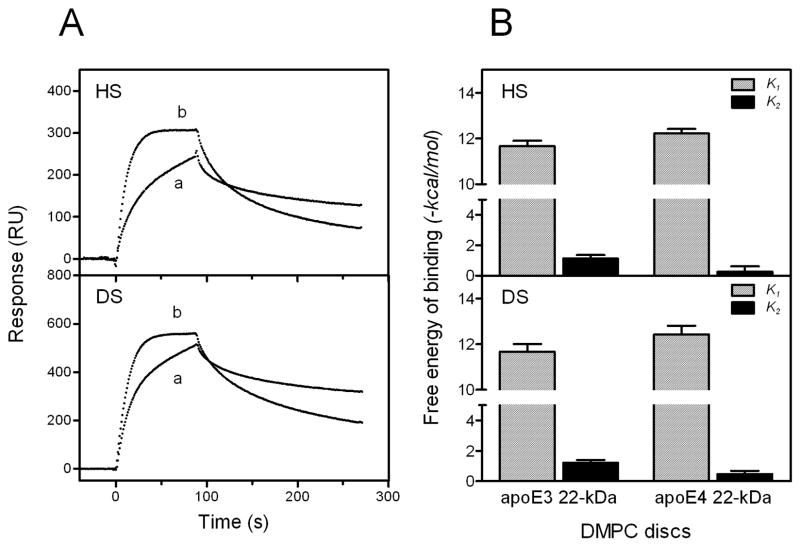
To explore why apoE4 binds better to GAG than other isoforms, the association and dissociation kinetics for the GAG binding of the 22-kDa fragments of apoE3 and apoE4 complexed with DMPC were compared. Although the binding level of apoE4 was similar to that of apoE3, apoE4 exhibited a faster first association, followed by a slower second step compared to apoE3 (Fig. 9A). Consequently, the free energy of binding for the first step tended to increase and decrease for the second step for apoE4 compared to apoE3 (Fig. 9B). This suggests that the increased binding of full-length apoE4 to GAG comes from enhanced electrostatic interaction through the N-terminal binding site (49).
Fig. 9. SPR sensorgrams (A) and free energy of binding (B) for the interaction of DMPC discoidal complexes of 22-kDa apoE3 and apoE4 with HS and DS.
A, DMPC discoidal complex of apoE3 22-kDa (a) and apoE4 22-kDa (b). The protein concentration was 3 μg/ml.
DISCUSSION
The various anti-atherogenic functions of apoE, such as facilitated remnant lipoprotein uptake by the liver (8, 50) and cholesterol efflux from macrophages (16, 17), are achieved through its interaction with cell-surface GAG. Because receptor-ligand interactions at the cell surface do not occur at equilibrium, it is important to establish the kinetics of apoE-GAG association and dissociation. Based on SPR measurements to characterize the kinetics and affinity of binding of apoE to heparin, we previously proposed a two-step mechanism for the apoE-heparin interaction; the first step involving a fast electrostatic interaction, followed by a slower hydrophobic interaction (28). In the present study, we extended this approach to examine the role of the N- and C-terminal domains of apoE isoforms in their interactions with more physiological GAG, HS and DS.
The current SPR data demonstrate that the binding of apoE to HS and DS involves a two-state binding process, similar to the situation with heparin. However, the large decreases in the association rate and the favorable free energy of binding in the first step observed for HS and DS compared to heparin (Fig. 3) indicate that the contribution of the initial electrostatic interaction to the overall binding is lower for HS and DS than for heparin (26). In addition, the mutation in lysine 146 which disrupts direct ionic interaction between basic residues on apoE and sulfate groups on the GAG chain (26) greatly reduced the favorable free energy for the first binding step to HS and DS (Fig. 4B). The reduction of this free energy from the wild-type (1.5 and 1.3 kcal/mol for HS and DS, respectively) was much lower than that in heparin (2.7 kcal/mol) (Fig. 6 in ref 28). These results are consistent with the fact that heparin has an average of 2–3 sulfate groups per disaccharide whereas HS and DS contain less than 1 sulfate per disaccharide (31). Comparison of the free energies of binding in the two processes (Fig. 3) indicates that even in the less sulfated HS and DS, the electrostatic interaction still contributes greatly (over 90%) to the overall free energy of binding of apoE to GAG. In fact, analysis of the dependence of Kd and Rmax values on sodium chloride concentration for binding of the apoE3 22-kDa fragment to various GAG showed that an increase in ionic strength greatly reduces both the affinity and saturated level of apoE binding to GAG (data not shown).
In contrast to the dominant role of the N-terminal site in apoE to GAG binding, the C-terminal site (basic residues around lysine 233) is not available for the interaction of full-length apoE with GAG (Fig. 4) (27, 28). Rather, the C-terminal domain appears to contribute to the high-affinity binding of apoE to GAG through an oligomerization effect (27). Using C-terminally truncated mutants of apoE, we showed previously that the C-terminal helical residues, 261–299 or 272–299 modulate the tetramerization of apoE in solution (37, 47). As shown in Fig. 5A, progressive truncation of the C-terminal helical segments which leads to the monomeric form greatly reduced binding of apoE3 to HS. The results that the C-terminal truncation mainly reduced the favorable free energy for the first binding step (Fig. 5B) indicate that oligomerization through the C-terminal α-helical region has a major influence on the electrostatic interaction between apoE and GAG. This notion that multiple binding sites of apoE molecules increase the GAG binding is consistent with the previous study of peptides including heparin binding consensus sequences showing that multiple copies of the heparin binding site enhance the binding affinity through cooperativity effects (51). The findings that dimerization of apoE3 22-kDa fragment via disulfide linkage markedly increased the binding to HS compared to the monomeric form (Fig, 6) and restored the kinetic rate constants and equilibrium affinity in the first step to the level comparable to full-length protein (Table 1) further support this idea. We found that the dimerization of the apoE3 22-kDa fragment destabilized the N-terminal helix bundle and exposed hydrophobic surface (data not shown). However, such a hydrophobic exposure contributes to the second step of GAG binding (28) whereas binding of the apoE3 22-kDa dimer to the GAG is associated with an increased contribution of the first step. This indicates that the dimerization enhances the electrostatic interaction similarly to the case of the C-terminal truncated mutants. In this regard, the similar electrostatic interaction in GAG binding of full-length apoE3 in the lipid-free state and in a discoidal complex (Fig. 7) may result from the fact that apoE is a tetramer in the lipid-free state and that there are about four apoE molecules bound to DMPC discs (52).
An important finding in this study is that apoE4 binds better to HS and DS than does apoE2 or apoE3 in both the lipid-free and lipidated forms (Fig. 8). The fact that the E255A mutation did not significantly reduce the binding of apoE4 indicates that the domain interaction in apoE4 is not mainly responsible for the higher binding of apoE4. Since apoE4 has a lesser propensity to self-associate than apoE3 in solution (47) and the number of apoE molecules on DMPC discs is similar among apoE isoforms (52), it is unlikely that the higher ability of binding of apoE4 to GAG is due to the cooperative effects of the heparin binding sites. Rather, it is likely that the greater electrostatic interaction through the N-terminal binding site of apoE4 leads to its higher binding ability to GAG (Fig. 9). It is possible that the positively charged residues involved in the heparin binding in apoE4 have a different electrostatic potential compared to apoE3 (49), leading to the enhanced electrostatic interaction of apoE4 with GAG. Indeed, analysis of the dependence of affinity constants on sodium chloride concentration (53, 54) indicated that the numbers of purely ionic interactions involved in the HS binding of apoE4 is about 3-fold larger than that for apoE3 (data not shown).
Such an increased binding ability of apoE4 to GAG than other apoE isoforms gives important insights into the atherogenic or anti-atherogenic properties of apoE isoforms. The retention of lipoproteins in the arterial extracellular matrix is a critical step in atherogenesis in which lipoproteins with higher affinity for arterial proteoglycans are more atherogenic (55, 56). It was reported that apoE3 and apoE4 have different effects on HDL metabolism (57), and the interaction of apoE with CS and/or DS proteoglycans is responsible for the retention of HDL on the extracellular matrix of arterial smooth muscle cells (14). Since apoE4 appears to bind better to cell-surface GAG than apoE3 (17), apoE4-containing lipoproteins could be retained more than apoE3-containing lipoproteins by vascular proteoglycans, resulting in oxidative and other potential atherogenic modifications like serum amyloid A-containing HDL (58). Thus, besides being a risk factor for Alzheimer’s disease, apoE4 is associated with an increased risk for coronary heart disease (7).
In summary, we have characterized the role of the N- and C-terminal domains in the interaction of apoE isoforms with GAG using an SPR approach. The N-terminal binding site dominantly contributes to the electrostatic interaction of apoE with GAG, whereas the C-terminal domain facilitates this electrostatic interaction through cooperative effects. The enhanced electrostatic interaction with apoE4 compared to apoE3 appears to underlie the greater binding of apoE4 to GAG, probably explaining the different retentions of apoE isoform-containing lipoproteins in the arterial wall.
Footnotes
This work was supported by NIH grant HL56083, Grants-in-Aid for Scientific Research from JSPS (Grants 17390011 and 19590048), the Naito Foundation, and NOVARTIS Foundation (Japan) for the Promotion of Science.
The abbreviations used are: apoE, apolipoprotein E; CD, circular dichroism; CS, chondroitin sulfate; DMPC, dimyristoylphosphatidylcholine; DS, dermatan sulfate; GAG, glycosaminoglycan; GdnHCl, guanidine hydrochloride; HS, heparan sulfate; HSPG, heparan sulfate proteoglycan; HDL, high density lipoprotein; LDLR, low density lipoprotein receptor; SPR, surface plasmon resonance; RU, resonance unit.
References
- 1.Salmivirta M, Lidholt K, Lindahl U. Heparan sulfate: a piece of information. Faseb J. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 2.Mahley RW, Huang Y. Atherogenic remnant lipoproteins: role for proteoglycans in trapping, transferring, and internalizing. J Clin Invest. 2007;117:94–98. doi: 10.1172/JCI30889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams KJ, Liu ML, Zhu Y, Xu X, Davidson WR, McCue P, Sharma K. Loss of heparan N-sulfotransferase in diabetic liver: role of angiotensin II. Diabetes. 2005;54:1116–1122. doi: 10.2337/diabetes.54.4.1116. [DOI] [PubMed] [Google Scholar]
- 4.Ebara T, Conde K, Kako Y, Liu Y, Xu Y, Ramakrishnan R, Goldberg IJ, Shachter NS. Delayed catabolism of apoB-48 lipoproteins due to decreased heparan sulfate proteoglycan production in diabetic mice. J Clin Invest. 2000;105:1807–1818. doi: 10.1172/JCI8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 6.Cooper AD. Hepatic uptake of chylomicron remnants. J Lipid Res. 1997;38:2173–2192. [PubMed] [Google Scholar]
- 7.Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 8.Mahley RW, Ji ZS. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- 9.Swertfeger DK, Hui DY. Apolipoprotein E: Receptor versus heparan sulfate proteoglycan binding in its regulation of smooth muscle cell migration and proliferation. J Biol Chem. 2001;276:25043–25048. doi: 10.1074/jbc.M102357200. [DOI] [PubMed] [Google Scholar]
- 10.Holtzman DM, Pitas RE, Kilbridge J, Nathan B, Mahley RW, Bu G, Schwartz AL. Low density lipoprotein receptor-related protein mediates apolipoprotein E-dependent neurite outgrowth in a central nervous system-derived neuronal cell line. Proc Natl Acad Sci U S A. 1995;92:9480–9484. doi: 10.1073/pnas.92.21.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji ZS, Pitas RE, Mahley RW. Differential cellular accumulation/retention of apolipoprotein E mediated by cell surface heparan sulfate proteoglycans. Apolipoproteins E3 and E2 greater than E4. J Biol Chem. 1998;273:13452–13460. doi: 10.1074/jbc.273.22.13452. [DOI] [PubMed] [Google Scholar]
- 12.Bazin HG, Marques MA, Owens AP, 3rd, Linhardt RJ, Crutcher KA. Inhibition of apolipoprotein E-related neurotoxicity by glycosaminoglycans and their oligosaccharides. Biochemistry. 2002;41:8203–8211. doi: 10.1021/bi025817e. [DOI] [PubMed] [Google Scholar]
- 13.Burgess JW, Liang P, Vaidyanath C, Marcel YL. ApoE of the HepG2 cell surface includes a major pool associated with chondroitin sulfate proteoglycans. Biochemistry. 1999;38:524–531. doi: 10.1021/bi9819778. [DOI] [PubMed] [Google Scholar]
- 14.Olin-Lewis K, Benton JL, Rutledge JC, Baskin DG, Wight TN, Chait A. Apolipoprotein E mediates the retention of high-density lipoproteins by mouse carotid arteries and cultured arterial smooth muscle cell extracellular matrices. Circ Res. 2002;90:1333–1339. doi: 10.1161/01.res.0000024691.82864.f0. [DOI] [PubMed] [Google Scholar]
- 15.Rapp A, Huttinger M. Role of chondroitin sulphate in the uptake of beta-VLDL by brain cells. Eur J Neurosci. 2005;22:1400–1408. doi: 10.1111/j.1460-9568.2005.04313.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin C-Y, Huang ZH, Mazzone T. Interaction with proteoglycans enhances the sterol efflux produced by endogenous expression of macrophage apoE. J Lipid Res. 2001;42:1125–1133. [PubMed] [Google Scholar]
- 17.Hara M, Matsushima T, Satoh H, Iso-o N, Noto H, Togo M, Kimura S, Hashimoto Y, Tsukamoto K. Isoform-dependent cholesterol efflux from macrophages by apolipoprotein E is modulated by cell surface proteoglycans. Arterioscler Thromb Vasc Biol. 2003;23:269–274. doi: 10.1161/01.atv.0000054199.78458.4b. [DOI] [PubMed] [Google Scholar]
- 18.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 19.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 21.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J Biol Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 22.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Lund-Katz S, Phillips MC. Lipid binding-induced conformational change in human apolipoprotein E. Evidence for two lipid-bound states on spherical particles. J Biol Chem. 2001;276:40949–40954. doi: 10.1074/jbc.M106337200. [DOI] [PubMed] [Google Scholar]
- 23.Cardin AD, Hirose N, Blankenship DT, Jackson RL, Harmony JA, Sparrow DA, Sparrow JT. Binding of a high reactive heparin to human apolipoprotein E: identification of two heparin-binding domains. Biochem Biophys Res Commun. 1986;134:783–789. doi: 10.1016/s0006-291x(86)80489-2. [DOI] [PubMed] [Google Scholar]
- 24.Weisgraber KH, Rall SC, Jr, Mahley RW, Milne RW, Marcel YL, Sparrow JT. Human apolipoprotein E. Determination of the heparin binding sites of apolipoprotein E3. J Biol Chem. 1986;261:2068–2076. [PubMed] [Google Scholar]
- 25.Dong J, Peters-Libeu CA, Weisgraber KH, Segelke BW, Rupp B, Capila I, Hernaiz MJ, LeBrun LA, Linhardt RJ. Interaction of the N-terminal domain of Apolipoprotein E4 with heparin. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 26.Libeu CP, Lund-Katz S, Phillips MC, Wehrli S, Hernaiz MJ, Capila I, Linhardt RJ, Raffai RL, Newhouse YM, Zhou F, Weisgraber KH. New insights into heparan sulfate proteoglycan-binding activity of apolipoprotein E. J Biol Chem. 2001;276:39138–39144. doi: 10.1074/jbc.M104746200. [DOI] [PubMed] [Google Scholar]
- 27.Saito H, Dhanasekaran P, Nguyen D, Baldwin F, Weisgraber KH, Wehrli S, Phillips MC, Lund-Katz S. Characterization of the heparin binding sites in human apolipoprotein E. J Biol Chem. 2003;278:14782–14787. doi: 10.1074/jbc.M213207200. [DOI] [PubMed] [Google Scholar]
- 28.Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, Saito H. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J Biol Chem. 2005;280:5414–5422. doi: 10.1074/jbc.M411719200. [DOI] [PubMed] [Google Scholar]
- 29.Klezovitch O, Formato M, Cherchi GM, Weisgraber KH, Scanu AM. Structural determinants in the C-terminal domain of apolipoprotein E mediating binding to the protein core of human aortic biglycan. J Biol Chem. 2000;275:18913–18918. doi: 10.1074/jbc.M909644199. [DOI] [PubMed] [Google Scholar]
- 30.Klezovitch O, Scanu AM. Domains of apolipoprotein E involved in the binding to the protein core of biglycan of the vascular extracellular matrix: potential relationship between retention and anti-atherogenic properties of this apolipoprotein. Trends Cardiovasc Med. 2001;11:263–268. doi: 10.1016/s1050-1738(01)00121-9. [DOI] [PubMed] [Google Scholar]
- 31.Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays. 1998;20:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 32.Olsson U, Ostergren-Lunden G, Moses J. Glycosaminoglycan-lipoprotein interaction. Glycoconj J. 2001;18:789–797. doi: 10.1023/a:1021155518464. [DOI] [PubMed] [Google Scholar]
- 33.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 34.Shuvaev VV, Laffont I, Siest G. Kinetics of apolipoprotein E isoforms-binding to the major glycosaminoglycans of the extracellular matrix. FEBS Lett. 1999;459:353–357. doi: 10.1016/s0014-5793(99)01285-5. [DOI] [PubMed] [Google Scholar]
- 35.Lundquist P, Martensson J, Sorbo B, Ohman S. Turbidimetry of inorganic sulfate, ester sulfate, and total sulfur in urine. Clin Chem. 1980;26:1178–1181. [PubMed] [Google Scholar]
- 36.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC, Lund-Katz S. Effects of polymorphism on the lipid interaction of human apolipoprotein E. J Biol Chem. 2003;278:40723–40729. doi: 10.1074/jbc.M304814200. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka M, Vedhachalam C, Sakamoto T, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry. 2006;45:4240–4247. doi: 10.1021/bi060023b. [DOI] [PubMed] [Google Scholar]
- 38.Lund-Katz S, Zaiou M, Wehrli S, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC. Effects of lipid interaction on the lysine microenvironments in apolipoprotein E. J Biol Chem. 2000;275:34459–34464. doi: 10.1074/jbc.M005265200. [DOI] [PubMed] [Google Scholar]
- 39.Raut S, Gaffney PJ. Interaction of heparin with fibrinogen using surface plasmon resonance technology: investigation of heparin binding site on fibrinogen. Thromb Res. 1996;81:503–509. doi: 10.1016/0049-3848(96)00024-2. [DOI] [PubMed] [Google Scholar]
- 40.Lin Y, Pixley RA, Colman RW. Kinetic analysis of the role of zinc in the interaction of domain 5 of high-molecular weight kininogen (HK) with heparin. Biochemistry. 2000;39:5104–5110. doi: 10.1021/bi992048z. [DOI] [PubMed] [Google Scholar]
- 41.Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 42.Karlsson R, Falt A. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J Immunol Methods. 1997;200:121–133. doi: 10.1016/s0022-1759(96)00195-0. [DOI] [PubMed] [Google Scholar]
- 43.Lipschultz CA, Li Y, Smith-Gill S. Experimental design for analysis of complex kinetics using surface plasmon resonance. Methods. 2000;20:310–318. doi: 10.1006/meth.1999.0924. [DOI] [PubMed] [Google Scholar]
- 44.Morrisett JD, David JS, Pownall HJ, Gotto AM., Jr Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry. 1973;12:1290–1299. doi: 10.1021/bi00731a008. [DOI] [PubMed] [Google Scholar]
- 45.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II Properties of the amino- and carboxyl-terminal domains. J Biol Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 46.Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A monomeric, biologically active, full-Length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto T, Tanaka M, Vedhachalam C, Nickel M, Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Contributions of the carboxyl-terminal helical segment to the self-association and lipoprotein preferences of human apolipoprotein E3 and E4 isoforms. Biochemistry. 2008;47:2968–2977. doi: 10.1021/bi701923h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 49.Lund-Katz S, Wehrli S, Zaiou M, Newhouse Y, Weisgraber KH, Phillips MC. Effects of polymorphism on the microenvironment of the LDL receptor-binding region of human apoE. J Lipid Res. 2001;42:894–901. [PubMed] [Google Scholar]
- 50.MacArthur JM, Bishop JR, Stanford KI, Wang L, Bensadoun A, Witztum JL, Esko JD. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J Clin Invest. 2007;117:153–164. doi: 10.1172/JCI29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verrecchio A, Germann MW, Schick BP, Kung B, Twardowski T, San Antonio JD. Design of peptides with high affinities for heparin and endothelial cell proteoglycans. J Biol Chem. 2000;275:7701–7707. doi: 10.1074/jbc.275.11.7701. [DOI] [PubMed] [Google Scholar]
- 52.Schneeweis LA, Koppaka V, Lund-Katz S, Phillips MC, Axelsen PH. Structural analysis of lipoprotein E particles. Biochemistry. 2005;44:12525–12534. doi: 10.1021/bi050872j. [DOI] [PubMed] [Google Scholar]
- 53.Goncalves E, Kitas E, Seelig J. Structural and thermodynamic aspects of the interaction between heparan sulfate and analogues of melittin. Biochemistry. 2006;45:3086–3094. doi: 10.1021/bi052221t. [DOI] [PubMed] [Google Scholar]
- 54.Klocek G, Seelig J. Melittin interaction with sulfated cell surface sugars. Biochemistry. 2008;47:2841–2849. doi: 10.1021/bi702258z. [DOI] [PubMed] [Google Scholar]
- 55.Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler Thromb Vasc Biol. 1995;15:551–561. doi: 10.1161/01.atv.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gustafsson M, Boren J. Mechanism of lipoprotein retention by the extracellular matrix. Curr Opin Lipidol. 2004;15:505–514. doi: 10.1097/00041433-200410000-00003. [DOI] [PubMed] [Google Scholar]
- 57.Hopkins PC, Huang Y, McGuire JG, Pitas RE. Evidence for differential effects of apoE3 and apoE4 on HDL metabolism. J Lipid Res. 2002;43:1881–1889. doi: 10.1194/jlr.m200172-jlr200. [DOI] [PubMed] [Google Scholar]
- 58.Lewis KE, Kirk EA, McDonald TO, Wang S, Wight TN, O’Brien KD, Chait A. Increase in serum amyloid a evoked by dietary cholesterol is associated with increased atherosclerosis in mice. Circulation. 2004;110:540–545. doi: 10.1161/01.CIR.0000136819.93989.E1. [DOI] [PubMed] [Google Scholar]