Abstract
Members of the CHD protein family play key roles in gene regulation through ATP-dependent chromatin remodeling. This is facilitated by chromodomains that bind histone tails, and by the SWI2/SNF2-like ATPase/helicase domain that remodels chromatin by moving histones. Chd6 is ubiquitously expressed in both mouse and human, with the highest levels of expression in the brain. The Chd6 gene contains 37 exons, of which exons 12-19 encode the highly conserved ATPase domain. To determine the biological role of Chd6, we generated mouse lines with a deletion of exon 12. Chd6 without exon 12 is expressed at normal levels in mice, and Chd6 Exon 12 −/− mice are viable, fertile, and exhibit no obvious morphological or pathological phenotype. Chd6 Exon 12 −/− mice lack coordination as revealed by sensorimotor analysis. Further behavioral testing revealed that the coordination impairment was not due to muscle weakness or bradykinesia. Histological analysis of brain morphology revealed no differences between Chd6 Exon 12 −/− mice and wild-type (WT) controls. The location of CHD6 on human chromosome 20q12 is overlapped by the linkage map regions of several human ataxias, including autosomal recessive infantile cerebellar ataxia (SCAR6), a nonprogressive cerebrospinal ataxia. The genomic location, expression pattern, and ataxic phenotype of Chd6 Exon 12 −/− mice indicate that mutations within CHD6 may be responsible for one of these ataxias.
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-010-9248-8) contains supplementary material, which is available to authorized users.
Introduction
Dynamic remodeling of the chromatin structure during DNA replication, repair, recombination, and transcription is a vital facet of eukaryotic DNA metabolism and involves changes and modifications of DNA-protein interactions. The position of a nucleosome relative to a given DNA sequence directly influences the plasticity of chromatin and transcription of nearby genes (de la Serna et al. 2006). The processes of nucleosome sliding and nucleosome displacement are energy dependent and carried out by members of the sucrose nonfermenting 2 (SNF2) superfamily of DNA-dependent ATPase proteins (reviewed in Brown et al. 2007), which function in large multiprotein chromatin-remodeling complexes.
Chromodomain-Helicase-DNA-binding (CHD) proteins make up one family of SNF2-like chromatin-remodeling enzymes. The family name is based upon the distinct combination of structural domains common to all CHD enzymes; tandem N-terminal Chromodomains, followed by a SWI2/SNF2-like Helicase/ATPase domain, and a C-terminal portion with less well characterized DNA-binding capability (Delmas et al. 1993; reviewed in Hall and Georgel 2007; Marfella and Imbalzano 2007). The catalytic SWI2/SNF2-like helicase/ATPase chromatin-remodeling domain shares sequence similarities with bacterial RecA-like helicase domains.
The SWI2/SNF2-like ATPase domain contains seven motifs that are conserved throughout evolution (Eisen et al. 1995; Singleton and Wigley 2002; Thoma et al. 2005). Motif I was first characterized by Walker et al. (1982) and plays a critical role in the catalytic function of the ATP binding pocket. Within motif I there are 15 amino acids that are very highly conserved, 9 of which are 100% conserved among the 16 SNF2 family members compared here (Supplementary Fig. 1). Point mutation of key conserved residues in any of the seven motifs has been shown to remove function of the domain (Richmond and Peterson 1996). Mutation of the lysine within the 100% conserved GLGKT sequence of motif I has repeatedly been shown to result in complete loss of ATPase function of SNF2 family members (Citterio et al. 1998, 2000; Khavari et al. 1993; Laurent et al. 1993), including the recent characterization of Chd8 (Thompson et al. 2008). Several human diseases are associated with mutations within the SWI2/SNF2-like ATPase domain, indicating the importance of the ATPase domain in protein function. Mutations within the ATPase domain of Cockayne syndrome B protein result in ultraviolet sensitivity and neurodevelopmental abnormalities (Citterio et al. 2000), while mutations in the ATRX ATPase domain cause α-thalassemia/mental retardation syndrome, X-linked (ATR-X syndrome) (Wada et al. 2000).
There are nine CHD proteins in both human and mouse genomes [NCBI UaWE, Consensus CDS (CCDS) project in build for 36.3/37.1, 2008] and they are divided into three subfamilies based on the presence of other protein domains (Schuster and Stöger 2002). Mutations in subfamilies I and II are associated with developmental defects (Kulkarni et al. 2008; Williams et al. 2004; Yoshida et al. 2008) and perinatal lethality in mice (Marfella et al. 2006). Genes CHD5–CHD9 encode the proteins that make up CHD subfamily III. Currently, mutations within two CHD genes from subfamily III are associated with pathology. CHD5 is a tumor suppressor gene, mutations of which contribute to neuroblastoma (Bagchi et al. 2007; Fujita et al. 2008; Thompson et al. 2003; White et al. 2005), while mutations of CHD7 account for over 60% of individuals affected by CHARGE syndrome, a complex congenital disorder characterized by developmental malformations of the eye, ear, heart, and genitals as well as cognitive impairment (Aramaki et al. 2006; Pagon et al. 1981; Vissers et al. 2004). These and related findings highlight the essential role of CHD family members in normal neural development and brain function (Bultman et al. 2000; Picketts et al. 1996; reviewed in Brown et al. 2007; Hsieh and Gage 2005).
CHD6 (formerly called CHD5) is the founding member of CHD subfamily III (Schuster and Stöger 2002). First identified based on the highly conserved ATPase domain, the additional presence of ill-defined domains that make up the DNA-binding domain distinguished CHD6 from subfamilies I and II (Fig. 1a). CHD6 is a ubiquitously expressed gene (Lutz et al. 2006; Schuster and Stöger 2002) and is particularly abundant in regions of the adult mouse brain, such as the cerebellum, medulla, globus pallidus, and the retrohippocampal area (Lein et al. 2007; Su et al. 2004). CHD6 is a large (2711 amino acids) nuclear protein that colocalizes with RNA polymerase II during preinitiation and elongation stages of RNA synthesis, suggesting that this chromatin-remodeling enzyme plays a role in modulating and facilitating the transcription of target genes, which have yet to be identified (Lutz et al. 2006). CHD6 precipitates as part of a large complex of proteins and has been shown to function in the phase-two stress response pathway by interaction with the NFE2 related factor 2 (NRF2) transcription factor to regulate expression of NQO1 (Nioi et al. 2005). We previously demonstrated that CHD6 is a DNA-dependent ATPase with a functional SNF2/SWI2-like helicase/ATPase domain (Lutz et al. 2006).
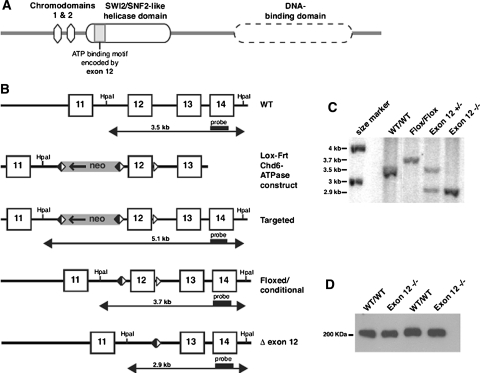
Fig. 1.
Generation of a Chd6 Exon 12 −/− mouse line. a Domain architecture of the Chd6 protein. The SWI2/SNF2 helicase domain contains seven conserved motifs, including motif I (ATP-binding region) and Ia (DNA binding), which are encoded by exon 12 of mChd6. b Gene targeting strategy: A portion of the mChd6 gene structure is shown, including exons 11-14 (boxes), and the HpaI restriction enzyme sites used for Southern blot analysis. The targeting vector “lox-frt-Chd6-ATPase” was designed to enable the deletion of mChd6 exon 12 by loxP site-specific recombination. The loxP sites (white triangles) and direction of transcription of the Neomycin-resistance gene (Neo) are indicated (arrow). The Frt-flanked Neo-cassette was removed by flp-mediated, site-specific recombination (Frt recombination elements are black triangles). The structure of the desired allele (Δ-exon 12) was obtained after Cre-mediated excision of the floxed exon 12. The Southern probe and the sizes of HpaI restriction fragments detected by this probe in wild-type (WT) and targeted DNA are shown. c Southern analysis of tail clip genomic DNA: The WT Chd6 allele (3.5 kb band) is distinguishable from the floxed conditional Chd6 allele (3.7 kb) and the Chd6 Exon 12 −/− allele (2.9 kb). d Western analysis: Detection of WT Chd6 (305 kDa) and mutant Δ479-558Chd6 (296 kDa) in lung extracts from WT and Chd6 Exon 12 −/− animals, respectively. The small difference in molecular weights between WT and mutant Chd6 could not be detected by this assay
The biological and physiological importance of CHD6 is not known. A recent case study of a chromosomal translocation between CHD6 and the basic helix-loop-helix transcription factor 4 (TCF4) gene identified a phenotype of mental retardation without Pitt-Hopkins syndrome characteristics that was attributed to the truncation of TCF4, but the resulting effect on CHD6 expression and function was not determined (Kalscheuer et al. 2008). Several human ataxias have linkage map regions that encompass the position of CHD6 on chromosome 20q12 (Bennetts et al. 2007; Hertz et al. 2004; Tranebjaerg et al. 2003).
To assess the functional role of Chd6 we generated a Chd6 Exon 12 −/− mouse. Because exon 12 encodes a highly conserved portion of the ATPase domain, it is expected that this deletion inactivates the ATPase domain, but this expectation has not been experimentally addressed. A splice from exon 11–13 is in-frame and the mutant protein is expressed at normal levels. The homozygous mutant mice are viable and fertile and have no obvious phenotype. Behavioral testing revealed that homozygous Chd6 Exon 12 −/− animals exhibit coordination defects most consistent with a cerebellar neuron disorder.
Materials and methods
Generation of the mutant ES cells and mice
The conditional deletion of exon 12 of the mouse Chd6 gene utilized a targeting construct that contains an Frt/loxP flanked neomycin cassette positioned 5′ of exon 12, with a third loxP site inserted 3′ to the exon (Fig. 1b). The integrity of the lox-frt-Chd6-ATPase construct was confirmed by restriction digestion and sequencing of Chd6 exons 11, 12, and 13, including the intron-exon boundaries.
Using standard techniques (Nagy 2003), targeted 129 SvIMJ embryonic stem (ES) cells were generated and C57BL/6/129 SvIMJ mouse lines were developed from one correctly targeted ES cell clone. The presence of the targeted allele was identified by PCR and Southern blot. Mixed WT mice were derived from homozygous wild-type littermates of mutant mice and were maintained with breeding strategies identical to those for the mutant mice by the intercross of multiple-mating pairs with the appropriate genotype. For genotyping of animals, genomic DNA was isolated from mouse tails using the Gentra Puregene kit (Qiagen, Valencia, CA, USA) followed by phenol-chloroform extraction. The derived animals were mated to mice that express either Cre in the germline (Jax® Mice, stock number 003465) to generate Exon 12 −/− alleles (Chd6 Exon 12 −/− mice), or flp in the germline (Jax® Mice, stock number 003800) to delete the selectable marker but leave the ATPase domain intact (mixed WT). Deletion of exon 12 was confirmed in genomic DNA by PCR and in the expressed mRNA by RT-PCR and sequencing of the exon 11-13 junction (data not shown) (see Table 1 for primer sequences). Mice were either maintained in a mixed genetic background or backcrossed into the 129SvImJ parental strain (Jax® Mice, stock number 002448) until they were 97% 129 genetically (N5), at which point heterozygotes were crossed to produce the homozygous 129 Exon 12 −/− line. All mouse work was performed under protocols approved by the Dartmouth IACUC.
Table 1.
Primers used in the phenotype assessment of Chd6 Exon 12 −/− mice
| Name | Primer sequence 5′-3′ | Annealing Tm/product size |
|---|---|---|
| Exon 12 −/− foward | GACATACACAATGGCATGTATGCT | 60°C |
| Exon 12 −/− reverse | GTGTACCATCATGCCTGACTCAC | 345 bp (−/−), 900 bp (WT) |
| WT forward | ACCCATACTTCTTACCCTCTGCAG | 60°C |
| WT reverse | GTGTACCATCATGCCTGACTCAC | 605 bp (WT) |
| Exon 11 forward | GAAACTGGAGACATCCCGTG | 61°C |
| Exon 13 reverse | GCAGTTCCTATTCTTCAGTCT | 238 bp (−/−), 483 bp (WT) |
| Nqo1 forward | ACAGGTGAGCTGAAGGACTC | 57°C |
| Nqo1 reverse | CTGCTACGAGCACTCTCTCA | 202 bp |
| β-actin forward | GTGGGCCGCTCTAGGCACCA | 57°C |
| β-actin reverse | CGGTTGGCCTTAGGGTTCAGG | 245 bp |
Physiological analysis of mutant mice
Physiological analysis of the mice included observations of appearance and behavior in the home cage, weight from 3 to 28 weeks of age, the effect of consuming high- and low-fat diets, and histological analysis of every major tissue and organ. Samples of each tissue were embedded in paraffin for sectioning and histological analysis of morphology and cellular structure (see Supplementary Methods for detailed descriptions). Mice were sacrificed by avertin overdose and whole blood was obtained by cardiac puncture. Blood was collected either without anticoagulant or into K2 EDTA for analysis of serum chemistry or complete blood count, respectively; analysis was performed by Charles River Laboratories (Wilmington, MA, USA). Protein was isolated from individual tissues and Chd6 Western analysis was performed as previously described in Lutz et al. (2006).
Behavioral analysis
For the ledge test, mice were placed on the 0.75 cm edge of a cage and allowed to either progress along the ledge, climb down head first in a controlled manner, fall off hind feet first in an uncontrolled manner, or sit stationary for up to 3 min. Collected data were distance traveled if they moved, or if they did not, the type of descent or whether they were stationary for the 3 min trial.
Behavioral analysis of age/gender-matched mice began at 8 weeks of age using mice with known genotypes. Male and female animals performed equally in all tests so we interpreted the results based solely on genotype. The data are represented as the average for all mice of each genotype used (Crawley 2007). The mice were put through a battery of tests in the following order: open field maze, Barnes maze, pole test, and rotorod. Later tests were added, including wire-hang, beam-walking, and olfactory tests, which were performed at age 18 weeks or at age 10 weeks. Age-specific differences were not observed in the wire-hang or beam-walking tests between the early and later test groups so the data were pooled for genotype analysis.
Mice were trained on the accelerating rotorod (model 775, IITC, Woodale, CA, USA) for 2 days and tested for 3 days. The mouse is placed on the rod as it slowly accelerates and the rotational speed at which the mouse falls off is recorded. Each trial consisted of the rotorod accelerating from 1 to 40 rpm over 120 s, with up to 10 s at 40 rpm. Three trials were performed each day for three consecutive days with a 6 min rest between trials. Mice that accomplish motor learning will improve in performance (higher rpm before falling off) with each trial. Mice will often regress partially for the first trial of the following day, resulting in a stepped pattern. Trials in which mice fell off the rod when rotating slower than 2 rpm were not included in the data and the trial was repeated. Detailed methods for behavioral analysis are provided as supplementary methods; a detailed chart of the behavioral analysis data is included as Supplementary Table 1. Statistical analysis was performed with R software (version 2.5.0; http://www.r-project.org). Data were analyzed by Welch’s t test and significance was determined as P < 0.05.
Immunohistochemistry
Brains from Chd6 Exon 12 −/− mice and WT controls were prepared at approximately 17 weeks of age by perfusion with 4% paraformaldehyde. Immunohistochemistry was carried out as previously described using standard procedures and with antibody dilutions recommended by the manufacturers (Chakrabarti et al. 2006). Counterstain of nuclei was achieved using a DAPI type reagent (Hoechst-H stain). Histology with Cresyl violet was performed on dewaxed sections.
Oxidative stress response analysis
Induction of the stress response pathway followed by reactive oxygen species challenge was measured by flow cytometry. Mouse embryonic fibroblast (MEF) cells from mixed WT and mixed Exon 12 −/− embryos were cultured and then treated with vehicle control or 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO) for 18 h. Cells were harvested and treated with 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA) or vehicle for 15 min before addition of tert-butyl hydroperoxide (tBHP) or vehicle. FACScan (Becton–Dickinson, San Jose, CA) analysis measured the change in fluorescence intensity. Stress response stimulation of the Nrf2 pathway was determined by qPCR analysis of Nqo1 gene expression. Mixed WT and mixed Exon 12 −/− MEFs cells were treated with DMSO vehicle control, 5 μM sulforaphane, or 100 nM CDDO for 20 h. RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA, USA) and reverse transcribed. Nqo1 and β-actin expression levels were assayed by iQ SYBER Green PCR (Bio-Rad, Hercules, CA, USA), with Nqo1 normalized to β-actin expression (see Table 1 for primer sequences).
Results
Exon 12 −/− mice do not show obvious anatomic or pathologic phenotype
To test the biological importance of Chd6 we generated a conditional knockout mouse line that enables the deletion of Chd6 exon 12 (Fig. 1). To avoid interference of the selection marker with expression of Chd6, the Neomycin resistance gene was removed by flp-mediated, site-specific recombination. Mice with a correctly targeted allele (Targeted) were crossed with C57BL/6 mice expressing flp in the germline to generate the conditional (floxed) allele mice (mixed WT), or with Cre-expressing BALB/c mice to generate the Exon 12 −/− mice (mixed Exon 12 −/−) (Fig. 1b). Heterozygous and homozygous targeted mice were identified through Southern blotting (Fig. 1c). Deletion of exon 12 removes 81 amino acids of the SWI2/SNF2-like domain which encodes motif I, a key ATP-binding motif, and motif Ia, which has DNA-binding activity (Durr et al. 2005). Exon 11 is spliced in-frame with exon 13 and the Exon 12 −/− protein is synthesized at WT levels (Fig. 1d).
Exon 12 −/− mice on a mixed C57BL/6/129 Sv/BALB/c genetic background (mixed Exon 12 −/−) are viable and fertile and born in normal Mendelian ratios. Mixed Exon 12 −/− mice were compared to mixed lineage mice that had a floxed conditional exon 12 allele but did not have the deletion (mixed WT). Pathological analysis of mixed Exon 12 −/− mice included histological analysis of tissues (including lung, bone marrow, skin/fat, liver, gastrointestinal tract, spleen/lymph nodes, kidney, pancreas, brain, heart, thymus, skeletal muscle, and testis or uterus) at two developmental time points, 26 days or 90 days of age; there were no differences in any tissue (data not shown). Whole blood and serum analysis revealed no changes in complete blood counts or serum chemistry (data not shown). Analysis of mouse body weight while consuming both high-fat and low-fat diets identified no difference between mixed Exon 12 −/− and mixed WT animals in body mass and did not indicate any metabolic deficiencies (Supplementary Fig. 2). Mouse embryonic fibroblast (MEF) cell lines of mixed Exon 12 −/− mice and mixed WT mice were used to determine that there was no difference in the DNA repair pathways of the Chd6 Exon 12 −/− mice, and analysis of splenocytes by flow cytometry (FACScan analysis) indicated that there was no difference in the cell cycle dynamics (data not shown).
To further determine potential strain-specific phenotypes, the Exon 12 −/− line was backcrossed into the 129 SvImJ parental strain until it was 97% pure 129 Sv statistically (N5 generation), at which point heterozygotes were crossed to produce homozygous 129 SvImJ Chd6 Exon 12 −/− mice (129 Exon 12 −/−), which were then compared to pure 129 SvImJ strain (129 WT) from The Jackson Laboratory (Bar Harbor, ME, USA). 129 Exon 12 −/− mice also exhibit no developmental problems and are born in expected Mendelian ratios.
Behavioral studies reveal Exon 12 −/− mice have impaired coordination and balance
The analysis of potential phenotypes included behavioral studies, as Chd6 is highly expressed in the brain and particularly in the cerebellum (Lein et al. 2007; Su et al. 2004). Mice from both mixed and 129 Sv genetic backgrounds displayed no difference in behavior when observed while in the home cage. However, a preliminary test of neurological function, the ledge test (Chen et al. 2005), indicated a coordination deficiency.
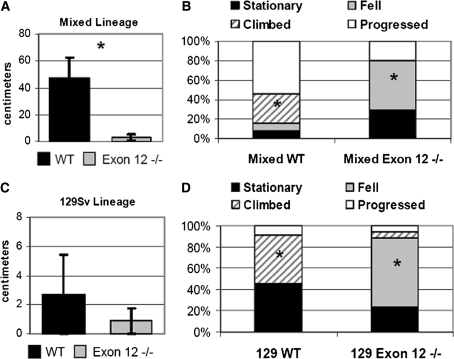
The ledge test measures the behavior of a mouse placed along the 0.75 cm-wide ledge of a mouse cage over a specified period of time. The mice were scored for movement along the ledge and if they did not progress, they were scored on whether they sat still for a full 3 min until removed, fell off the ledge, or made a controlled descent. Most mixed WT mice progressed (n = 13) with an average progression of 47 cm compared with the average distance of 3 cm per mouse observed in mixed Exon 12 −/− mice (n = 10), most of whom did not progress (Fig. 2a). Of the mixed WT that did not progress, most climbed into the cage in a controlled manner head first, and a small percentage either did not move or fell off the ledge (Fig. 2b). 129 WT (n = 11) and 129 Exon 12 −/− mice (n = 17) were equally unlikely to progress and there was no difference between the progression distances in the 129 strains (Fig. 2c). Most of the 129 Exon 12 −/− fell into the cage and rarely climbed down, while 129 WT generally either climbed down or sat still and never fell (Fig. 2d). The ledge test shows that there is a clear difference in the ability of Exon 12 −/− mice to move on the ledge or descend in a controlled manner, regardless of the genetic background. The inability to progress along the ledge or descend off the ledge in a controlled, head-first manner suggests a deficiency in motor coordination in the Exon 12 −/− mice.
Fig. 2.
Exon 12 −/− mice are impaired in the ledge test compared to WT mice. Mice were placed on the 0.75 cm-wide ledge of a mouse cage and observed for 3 min. Mice were scored as having performed one of four behaviors: remaining stationary on the ledge, falling off the ledge, climbing into the cage in a controlled manner, or progressing along the ledge. a Average distance progressed by mixed lineage WT and Exon 12 −/− mice. b Percent of mixed lineage mice to perform each behavior. c Average distance crossed by 129 Sv lineage Exon 12 −/− and WT mice. d Percent of 129 Sv lineage mice to perform each behavior. Data are represented as a percentage of the total number of mice tested. Error bars are ±SEM, significant difference is indicated by * (P < 0.05 for the same behavior compared between WT and Exon 12 −/− of the same lineage)
Several conditions may result in loss of motor coordination, including muscle weakness and ataxia (impaired coordination or balance) (Gowen and Miall 2007). Following the indication of impaired motor coordination, Exon 12 −/− mice from both mixed lineage and 129 Sv backgrounds were analyzed with a battery of behavioral tests, including sensorimotor assays.
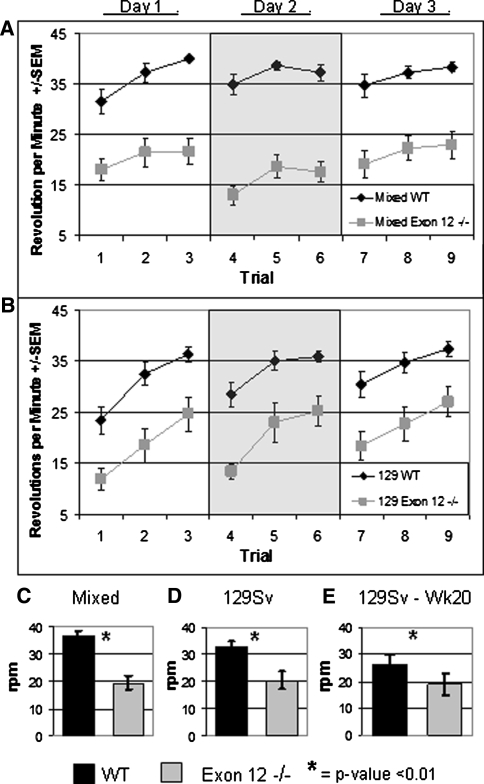
The rotorod assay measures motor coordination and, with repeated trials is able to assay motor learning (Crawley 2007). The impaired ability of the Exon 12 −/− mice to run on the rotorod at greater than moderate speeds is the most obvious and highly significant phenotype of the behavioral testing (P < 0.0001). Mixed Exon 12 −/− mice (n = 21) fell off significantly earlier than the mixed WT controls (n = 21) in every trial (Fig. 3a). This was true for all three trials run on a given day and for all 3 days of testing. It was common for the mixed WT mice to reach the top speed tested on the rotorod (40 rpm) and maintain that pace, while this was highly unusual for mixed Exon 12 −/− mice. The 129 Exon 12 −/− mice (n = 13) also fell off significantly earlier than the 129 WT mice (n = 17) for every trial (Fig. 3b). Interestingly, motor learning was not affected by the deletion of the Chd6 exon 12. All groups of mice generally improve the average trial rpm over the three trials of a given day (Fig. 3a, b). This manifestation of motor learning is particularly clear for the 129 lineage. This learning is not fully retained overnight as the first trial of a new day of testing has a lower average than the last trial of the previous day, but there was some retained motor learning because often a given day’s averages are higher than those of previous days. For all trials, the average maximum rpm that the mixed Exon 12 −/− mice reached before falling was 19.4 rpm (±2.46 SEM). This is significantly slower than the mixed WT mice, which averaged 36.6 rpm (±1.68) before falling (Fig. 3c). For all trials, the average maximum rpm that the 129 Exon 12 −/− mice reached before falling was 20.51 rpm (±3.19), and the average for the 129 WT control group was 32.6 rpm (±2.21) (Fig. 3d).
Fig. 3.
Exon 12 −/− mice have impaired rotorod performance. Mice were tested for 3 days, with three trials per day for a total of nine trials. a Average rpm reached for individual trials with mixed lineage mice. b Average rpm reached for individual trials with 129 Sv lineage mice. For both lineages, in each trial the WT results are significantly different from the Exon 12 −/− results (P < 0.01). c Average rpm for all trials with mixed lineage mice. d Average rpm for all trials with 129 Sv lineage mice. e Average rpm for all trials with 129 Sv lineage mice tested at week 20. Error bars are ±SEM, Significance of P < 0.01 is indicated by *
The lack of coordination seen in both the ledge test and the rotorod and the high levels of Chd6 present in the cerebellum (Lein et al. 2007) are indicative of an ataxia phenotype caused by neural impairment. To determine whether the impaired coordination of the Exon 12 −/− mice was due to a progressive ataxia, 129 Exon 12 −/− mice and 129 WT mice that had previously run the rotorod apparatus at 10 weeks of age were tested again at 20 weeks of age. The ability of aged 129 WT mice (n = 9) to run on the rotorod was diminished (129 WT, 32.6 rpm; aged 129 WT, 26.2 rpm average), and the aged 129 Exon 12 −/− mice (n = 6) had a slightly reduced performance (129 Exon 12 −/−, 20.5 rpm; aged 129 Exon 12 −/−, 18.9 rpm). The difference in aged 129 WT performance and aged 129 Exon 12 −/− performance continued to be significant, but it did not increase appreciably (Fig. 3e), indicating that the ataxia is not strongly progressive between 10 and 20 weeks of age.
Mice may display impaired ability during the rotorod assay for several reasons such as poor coordination or balance, bradykinesis (slowed movement) (Ogawa et al. 1985), increased weight (Cook et al. 2002; McFadyen et al. 2003), or muscular deficiencies (Koch et al. 2008). Previous studies had determined no difference in the weights of the mice (Supplementary Fig. 2). To investigate the impaired movement of Chd6 Exon 12 −/− mice, further behavioral tests were performed.
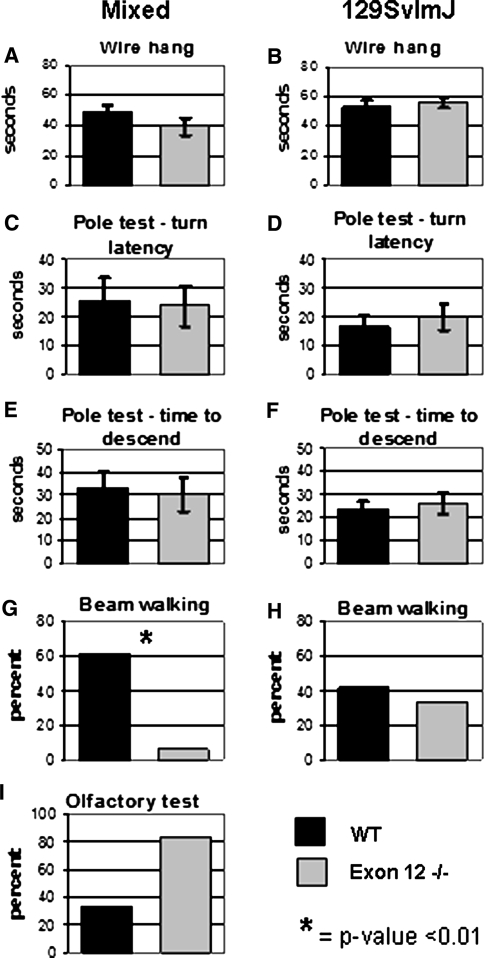
To determine whether muscle weakness was the cause of the poor rotorod performance, the wire-hang test was performed on all lineages (mixed WT, n = 18; mixed Exon 12 −/−, n = 14; 129 WT, n = 12; 129 Exon 12 −/−, n = 9). The wire-hang test directly measures the ability of the mouse to hang from a wire grid, reflecting muscle strength (Crawley 2007). Exon 12 −/− mice in both lineages performed as well as their WT counterparts in the wire-hang test (Fig. 4a, b).
Fig. 4.
Behavior analysis of Exon 12 −/− mice demonstrates further evidence that the sensorimotor defect is not due to muscle weakness or bradykinesis. The average for all trials is indicated for mixed lineage and 129 Sv lineage mice were tested on the wire hang (a, b), pole test (c-f), beam walking (g, h), and olfaction (i). Black bars indicate WT mice values; gray bars indicate Exon 12 −/− mice values. Error bars are ±SEM. For beam-walking and olfaction test data, the number of mice to accomplish the task is represented as a percent of the total number tested. Significance of P < 0.01 is indicated by *
The pole test is used to identify bradykinesis and also is indicative of muscle strength (Berardelli et al. 2001; Bouet et al. 2007) and proper dopaminergic neuronal signaling (Iwamoto et al. 2003; Matsuura et al. 1997). The pole test determines whether mice placed head up on a thin upright pole can both successfully invert and travel down the pole. Mixed Exon 12 −/− mice (n = 20) and 129 Exon 12 −/− mice (n = 9) were able to invert (Fig. 4c, d) and descend (Fig. 4e, f) the pole in times similar to those of the mixed WT (n = 21) and 129 WT mice (n = 12). Results from the wire-hang and pole tests suggest that the difficulty in the rotorod test is not likely due to poor muscle development or bradykinesis.
The beam-walking test measures both coordination and balance by assessing the ability to walk across a tape-covered cylindrical beam (1.5 cm wide and 61 cm long) and enter the home cage (Crawley 2007). The mixed Exon 12 −/− mice have impaired beam-walking ability compared with controls. Thirteen of 21 mixed WT mice walked the beam and successfully entered the home cage, while 1 of 17 mixed Exon 12 −/− mice walked the beam and entered the home cage (Fig. 4g). Both 129 WT (n = 12) and 129 Exon 12 −/− (n = 9) mice performed reasonably well on the beam-walking test (Fig. 4h), though a lower percentage of 129 mice performed the task compared to mixed lineage mice.
The majority of the mixed Exon 12 −/− mice did not fall off while attempting to cross the beam; they jumped off into the test area from the start location. This could be because they lack the coordination and balance to cross the beam or because of an impaired olfactory capacity. The mixed Exon 12 −/− mice may have been unable to smell their home cage and thus had no desire to travel along the beam. However, an olfactory test determined that mixed Exon 12 −/− mice are able to smell. After an 18 h fast the mice were given 3 min to find a piece of food hidden under bedding in a clean cage. Five of six mixed Exon 12 −/− mice found the food, while only two of six of the mixed WT mice accomplished this (Fig. 4i).
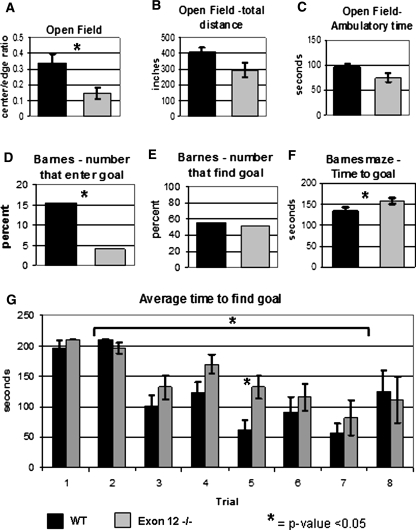
The open-field test directly measures the level of anxiety in the mouse. The test measures the amount of time a mouse spends in the center of an open field compared to time spent along the wall. Higher anxiety levels are demonstrated by increased time spent near the wall. The mixed Exon 12 −/− mice (n = 17) displayed slightly increased anxiety compared with mixed WT (n = 20), with a ratio of center to wall of 0.144 (±0.035) for the mixed Exon 12 −/− and 0.342 (±0.057) for the mixed WT indicating less anxiety (Fig. 5a). The mice showed no difference in other parameters measured by this test, including distance traveled (Fig. 5b) or ambulatory time (Fig. 5c). No difference was observed between 129 WT and 129 Exon 12 −/− mice in the open-field test (data not shown). An increased level of anxiety is characteristic of the 129SvImJ mouse strain (Hagenbuch et al. 2006), and 129 WT and 129 Exon 12 −/− mice exhibited similarly high levels of anxiety in this test.
Fig. 5.
Behavioral analysis of mixed-lineage Exon 12 −/− mice indicates normal spatial learning and mild anxiety. Mixed-lineage mice were tested for anxiety in the open-field maze and spatial learning and anxiety in the Barnes maze. Open-field measurements included (a) center-to-wall time ratio, (b) total distance traveled in centimeters, (c) the amount of ambulatory time. The Barnes maze measurements included (d) the percentage of mice that entered the goal box, (e) the percentage of mice finding and investigating the goal, (f) the average time to find and investigate the goal for all trials, and (g) the average time to find the goal for each trial. For Barnes maze data, the number of mice to accomplish the task is represented as a percent of the total number tested. Error bars are ±SEM. Significance of P < 0.05 is indicated by *
To further explore the differences in anxiety levels in Exon 12 −/− mice, the Barnes maze was used to study spatial learning ability as well as anxiety levels. The Barnes maze consists of a bright open circular platform with 40 holes along the edge, with a dark goal box beneath one hole into which the mouse can climb down and hide. Mice with higher anxiety levels will use spatial cues to learn where the goal box is located and will enter more quickly to escape the bright platform than less anxious control mice (Crawley 2008). The results of the Barnes maze studies indicated that mixed Exon 12 −/− mice (n = 21) had no deficiency in spatial learning, no increase in anxiety, and there is no impairment in vision compared to mixed WT mice (n = 21). Of 122 trials, 19 mixed WT mice entered the goal box compared with only 5 mixed Exon 12 −/− mice that entered the goal box in the same number of trials (Fig. 5d). This difference in entering the goal box was not due to difficulty locating the goal; mixed WT mice located the goal in 68 of 122 total trials, and mixed Exon 12 −/− mice located the goal in 63 of 122 trials (Fig. 5e). Repeated trials in the Barnes maze measures spatial learning ability, as mice use visual cues within the testing room and learn to find the goal box faster on consecutive days. Mice were tested twice each day for 4 days. While the average time it took the mice to first find and investigate the goal box was statistically greater in mixed Exon 12 −/− mice when measured as a function of the total number of trials (Fig. 5f), mixed Exon 12 −/− mice and mixed WT mice decreased the amount of time to find the goal over the eight trials (see the change in average time between trials 1 and 2 compared to 7 and 8, P < 0.01 for both WT and Exon 12 −/− mice, Fig. 5g). The mixed Exon 12 −/− mice were significantly slower than mixed WT mice only during trial 5; this shows that the slow phenotype is not consistent (Fig. 5g). These data suggest that the mixed Exon 12 −/− mice were not increasingly anxious and spatial learning is not impaired. While the Exon 12 −/− mice were able to find the goal as well as their WT counterparts, they did not enter it as often; this may be due to the impaired coordination phenotype. The high level of anxiety observed in 129 Sv lineage mice, both WT and Exon 12 −/−, during the open-field test was also observed in the Barnes maze test and made it an unproductive test for 129 strain mice (data not shown).
A variety of neurodegenerative diseases cause ataxia. Histological analysis of whole brain sections did not reveal any obvious differences in overall morphology between WT and Exon 12 −/− animals in either genetic background (data not shown). Cerebellar defects are frequently associated with ataxias, so to identify potential structural changes we histologically analyzed the cerebellum. Nissl staining was performed to visualize the overall lobe structure and neuronal arrangement; this appeared to be normal in all analyzed Exon 12 −/− animals (Supplementary Fig. 3a). By immunofluorescence we further studied the integrity of Purkinje cells and looked for evidence of axonal damage. We did not detect any histological abnormalities in Exon 12 −/− cerebella and no signs of neuronal degeneration were apparent (Supplementary Fig. 3b, c).
Gene regulation by Chd6 is not completely dependent on exon 12
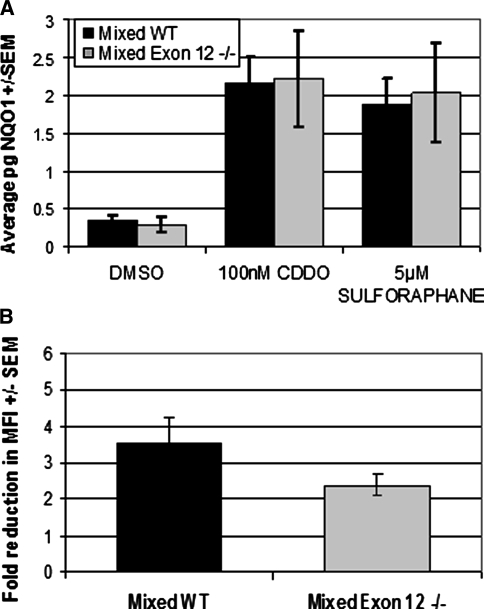
Chd6 has been shown to play a role in the induction of the phase-two stress response pathway (Nioi et al. 2005). Loss of exon 12 is expected to remove the ATPase function of Chd6, which should inhibit transcription of Chd6 target genes. To determine if exon 12 of Chd6 plays a critical role in the phase-two stress response, we stimulated the Nrf2 pathway in mouse embryonic fibroblasts from mixed Exon 12 −/− and mixed WT mice. Treatment of MEF cells with compounds that stimulate stress response pathways increased expression of Nqo1 in like manner in Exon 12 −/− mice and WT mice, irrespective of the stimulating compound used (Fig. 6a). The increased expression of Nrf2 pathway protective genes generated by the pretreatment with CDDO resulted in more efficient response upon exposure to reactive oxygen species. The level of protection, measured as the reduction in fluorescence intensity of DCFDA upon cell exposure to tBHQ, did not identify differences in the protective response of Exon 12 −/− MEFs compared to that of WT (Fig. 6b). These results indicate that the Chd6 exon 12 is not critical for Chd6 function in the response to oxidative stress.
Fig. 6.
Stimulation of the Nrf2 stress response pathway does not require the Chd6 exon 12. a Treatment of MEF cells with Nrf2 pathway stimulants CDDO or Sulforaphane increased the expression of Nqo1 in Exon 12 −/− MEF cells and WT MEFs to similar levels. qPCR analysis of the level of Nqo1 normalized to β-actin expression levels. b Increased expression of Nrf2 target genes results in protection from ROS challenge at similar levels in Exon 12 −/− MEF cells and WT MEF cells. MEFs pretreated with control or CDDO were labeled with DCFDA and challenged with tBHQ. The level of DCFDA fluorescence was measured by FACScan, and the change in mean fluorescence intensity was measured as treated over untreated levels. The average of six experiments is shown
Discussion
Chd6 Exon 12 −/− mice are ataxic
The studies reported here demonstrate that deletion of the Chd6 exon 12 causes impaired performances in a variety of sensorimotor tests, particularly on the rotorod, ledge test, and balance beam. Several different pathological problems can contribute to sensorimotor impairment, such as lack of coordination, poor balance, bradykinesis, or poor muscle strength. Further behavioral testing indicated that only coordination and balance are impaired in Chd6 Exon 12 −/− mice. The sensorimotor deficiency can be attributed to a lack of coordination; therefore, it can be defined as an ataxia (Gowen and Miall 2007). The ataxia phenotype can be classified, based on the affected region of the brain, as a cerebellar, sensory, vestibular, or optic ataxia. The lack of coordination is further distinguished as progressive or nonprogressive by whether the ataxia gets more severe with age. Many mutations that cause cerebellum degeneration result in an ataxic phenotype (Chakrabarti et al. 2008; Cheron et al. 2008; Dusart et al. 2006; Harkins and Fox 2002; Millen and Gleeson 2008). The ataxia of Chd6 Exon 12 −/− mice is not noticeably progressive and does not change significantly with age. Combined, our results demonstrate a deficiency in the coordination and balance of Chd6 Exon 12 −/− mice that most likely represents cerebellar ataxia.
While the cerebellum is responsible for many functions in the brain, including sensory perception, the olfactory test demonstrated that mixed Exon 12 −/− mice detect odor with normal efficiency, and the improved spatial learning demonstrated in the Barnes maze suggests that vision is not impaired. Morphological analysis of the brain determined that the apparent cerebellar impairment is not due to visible malformation or cell structure deficiencies, indicating that the ATPase domain of Chd6 is not a dominant factor in the regulation of genes necessary for cerebellar development and survival. Despite a lack of immunohistochemical evidence for cerebellar dysfunction in our current study, there is a distinct possibility that subcellular changes may affect cerebellar signaling and function as a result of exon 12 deletion altering Chd6 target gene expression levels.
CHD6 is a candidate gene for human recessive ataxias
In recent years two human ataxia mutations were mapped to regions that contain 20q12, the site of CHD6. Autosomal recessive infantile cerebellar ataxia (SCAR6 or CL3) is a nonprogressive ataxia that has been mapped to a 19.5-Mb region on chromosome 20q11-13 (Bennetts et al. 2007; Tranebjaerg et al. 2003), a region that contains many genes, including CHD6. A balanced translocation with resulting ataxia has breakage points that map proximal to CHD6 (Hertz et al. 2004). A translocation between CHD6 and TCF4 interrupts CHD6 between exon 1 and exon 2, and the resulting phenotype has been attributed to the truncation of the TCF4 gene (Kalscheuer et al. 2008). The phenotype observed in the Chd6 Exon 12 −/− mice makes the human CHD6 gene a promising candidate gene for the mutations that cause one or both of the ataxias, and suggests that the phenotype of the translocation patient may have multiple causes.
What little is known about Chd6 suggests its role as a chromatin-remodeling protein that interacts with several partners (Lutz et al. 2006). These partners include transcription factor NRF2 of the stress response pathway, and its target gene NQO1.(Nioi et al. 2005). Loss of exon 12 is expected to remove ATPase activity from Chd6, which in turn may affect transcriptional regulation of Chd6 target genes. To determine whether loss of exon 12 affects the Chd6/Nrf2 interaction, we assayed the stress response signaling pathway in mouse embryonic fibroblasts using two different techniques for activating the stress response pathway; however, there was no significant difference in either test suggesting that exon 12 is not necessary for the role Chd6 plays in regulation of the Nrf2 stress response pathway. The difference between our results and those of Nioi et al. (2005) could have a number of explanations. The functional interaction of CHD6 and NRF2 in regulating stress response genes in human cells was acting on the pathway in a dominant negative manner. It is possible that the ATPase domain is not required for the functional involvement of Chd6 in the pathway. And finally, it is possible that the pathways do not function identically in a specific human cancer cell line (Hela), as studied by Nioi et al., and in primary mouse cells, as studied here.
Although Chd6 is expressed ubiquitously, the only consistent phenotype appears to be the impairment in sensorimotor performance. The hypothesis that initiated these studies was that chromatin remodeling by the ATPase domain was the central function of Chd6. Based on the compiled biochemical analyses of mutations within the highly conserved motif I of the ATPase domain (Richmond and Peterson 1996), the deletion of exon 12 eliminates the ability of Chd6 to utilize ATP in the process of chromatin remodeling. The phenotype of the Chd6 Exon 12 −/− mice may not include all the potential phenotypes of a full Chd6 null mutation. CHD proteins are large proteins that work as part of multisubunit complexes. There are several other domains within Chd6 that may have ATPase-independent functional activity, in particular, the DNA-binding domain; the BRK domain (also called TCH), the function of which is not clearly understood (Allen et al. 2007; Doerks et al. 2002); and the SANT domain, which binds to histone N-terminal domains (Aasland et al. 1996; Boyer et al. 2002, 2004). Chd6 may act as a “scaffold” protein in complexes containing other ATPase proteins. The Drosophila CHD6 homolog, kismet-L, colocalizes with chd1 at sites of transcription initiation but does not colocalize with chd1 after promoter clearance by Pol II (which is indicated by the phosphorylation of the Pol II C-terminal domain serine 5) when active transcription is occurring (Srinivasan et al. 2005). The potential for CHD6 to bind to other ATPases suggests that the ATPase activity of its binding partners could compensate for the lack of ATPase activity in the exon 12 deletion mutant described here, while the remaining domains of CHD6 contribute their usual activity. A third possible explanation for the subtle phenotype observed is the potential for redundancy in the chromatin-remodeling pathways. Finally, it is possible, though unlikely, that the deletion of exon 12 has in fact destroyed all the functionality of Chd6. A role in improving coordination has obvious selective advantage and could be the primary contribution of the Chd6 protein.
Electronic supplementary material
Below is the link to the electronic supplementary material.
The Chd6 ATPase domain shares a high level of homology with SNF2 family members. The amino acid sequence of mouse Chd6. Exon 12 amino acids are boxed. Percentage of homology between the compared sequences is indicated above the sequences by height of the red bar. The highly conserved motif I (ATPase) is indicated in dark blue, the conserved motif Ia is shown in green, and motif II (DEXH helicase) is indicated in magenta. Genome indication is m, mouse; h, human; d, Drosophila; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe (TIFF 181 kb)
Exon 12 −/− mice on either a high-fat or a low-fat diet show no differences in body weight compared to control mice. Adult mixed WT mice (triangles) and mixed Exon 12 −/− mice (open circles) were fed a high-fat (45% kcal fat) or low-fat (10% kcal fat) diet for 6 months and weighed weekly. No significant difference in body weight was measured when mice were fed either the (A) high-fat diet or (B) low-fat diet. WT data are shown ± SEM; Exon 12 −/− SEM values were similar in size, but are not shown (TIFF 70 kb)
Immunohistochemistry analysis of Exon 12 −/− cerebellum revealed no differences in cellular morphology. Cerebella from WT and Chd6 Exon 12 −/− mice were compared at around 17 weeks of age; examples from the 129Sv lineage are shown. A Nissl-stained midline sagittal sections were compared and folia were counted (examples are shown at 2× magnification). The general cellular architecture was found to be similar in mutants compared with WT controls (n > 6). B Calbindin antibody (Swant, Switzerland) was used to detect potential changes within the Purkinje cell layer; consistent differences indicating Purkinje cell loss were not noted in mutant animals and the data were similar in all respects with that of age-matched WT controls (n > 3). (Calbindin - red fluorescence, nuclei are stained blue - H stain, Hoechst). C Antibody for Neurofilament protein (Abcam) was applied to cerebellar sections (Neurofilament protein – green fluorescence, blue – nuclear stain as before). Cerebellum was examined carefully throughout, from deep cerebellar nuclei to the folia (shown in C). The antibody against Neurofilament protein did not reveal any axonal damage in either mutant or WT samples (n > 3). Scale bars (white lines) in B and C represent 100 μm (TIFF 555 kb)
Acknowledgments
This work was supported by NIH grant T32 AI07363. The authors acknowledge the assistance provided by Laurie Horne; the Dartmouth Transgenic and Genetic Construct Shared Resource; Val Galton and Cheryl Withrow for advice on behavioral analysis; Gwenn Garden for immunohistochemistry analysis; Brent Harris for the use of equipment; and Diane Genereux for help with statistical analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Reinhard Stöger, Email: Reinhard.Stoger@nottingham.ac.uk.
Steven Fiering, Email: Fiering@dartmouth.edu.
References
- Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Allen MD, Religa TL, Freund SM, Bycroft M. Solution structure of the BRK domains from CHD7. J Mol Biol. 2007;371:1135–1140. doi: 10.1016/j.jmb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Aramaki M, Udaka T, Kosaki R, Makita Y, Okamoto N, et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J Pediatr. 2006;148:410–414. doi: 10.1016/j.jpeds.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Bennetts JS, Rendtorff ND, Simpson F, Tranebjaerg L, Wicking C. The coding region of TP53INP2, a gene expressed in the developing nervous system, is not altered in a family with autosomal recessive non-progressive infantile ataxia on chromosome 20q11–q13. Dev Dyn. 2007;236:843–852. doi: 10.1002/dvdy.21064. [DOI] [PubMed] [Google Scholar]
- Berardelli A, Rothwell JC, Thompson PD, Hallett M. Pathophysiology of bradykinesia in Parkinson’s disease. Brain. 2001;124:2131–2146. doi: 10.1093/brain/124.11.2131. [DOI] [PubMed] [Google Scholar]
- Bouet V, Freret T, Toutain J, Divoux D, Boulouard M, et al. Sensorimotor and cognitive deficits after transient middle cerebral artery occlusion in the mouse. Exp Neurol. 2007;203:555–567. doi: 10.1016/j.expneurol.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002;10:935–942. doi: 10.1016/S1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Brown E, Malakar S, Krebs JE. How many remodelers does it take to make a brain? Diverse and cooperative roles of ATP-dependent chromatin-remodeling complexes in development. Biochem Cell Biol. 2007;85:444–462. doi: 10.1139/O07-059. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/S1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, Neal JT, Miles M, Martinez RA, Smith AC, et al. The Purkinje cell degeneration 5J mutation is a single amino acid insertion that destabilizes Nna1 protein. Mamm Genome. 2006;17:103–110. doi: 10.1007/s00335-005-0096-x. [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, Eng J, Martinez RA, Jackson S, Huang J, et al. The zinc-binding domain of Nna1 is required to prevent retinal photoreceptor loss and cerebellar ataxia in Purkinje cell degeneration (pcd) mice. Vision Res. 2008;48:1999–2005. doi: 10.1016/j.visres.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Collins LL, Uno H, Chang C. Deficits in motor coordination with aberrant cerebellar development in mice lacking testicular orphan nuclear receptor 4. Mol Cell Biol. 2005;25:2722–2732. doi: 10.1128/MCB.25.7.2722-2732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheron G, Servais L, Dan B. Cerebellar network plasticity: from genes to fast oscillation. Neuroscience. 2008;153:1–19. doi: 10.1016/j.neuroscience.2008.01.074. [DOI] [PubMed] [Google Scholar]
- Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, et al. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/MCB.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MN, Bolivar VJ, McFadyen MP, Flaherty L. Behavioral differences among 129 substrains: implications for knockout and transgenic mice. Behav Neurosci. 2002;116:600–611. doi: 10.1037/0735-7044.116.4.600. [DOI] [PubMed] [Google Scholar]
- Crawley JN. What’s wrong with my mouse? Behavioral phenotyping of transgenic and knockout mice. 2. Hoboken, NJ: Wiley-Interscience; 2007. [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Dusart I, Guenet JL, Sotelo C. Purkinje cell death: differences between developmental cell death and neurodegenerative death in mutant mice. Cerebellum. 2006;5:163–173. doi: 10.1080/14734220600699373. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen E, Miall RC. The cerebellum and motor dysfunction in neuropsychiatric disorders. Cerebellum. 2007;6:268–279. doi: 10.1080/14734220601184821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch N, Feldon J, Yee BK. Use of the elevated plus-maze test with opaque or transparent walls in the detection of mouse strain differences and the anxiolytic effects of diazepam. Behav Pharmacol. 2006;17:31–41. doi: 10.1097/01.fbp.0000189811.77049.3e. [DOI] [PubMed] [Google Scholar]
- Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- Harkins AB, Fox AP. Cell death in weaver mouse cerebellum. Cerebellum. 2002;1:201–206. doi: 10.1080/14734220260418420. [DOI] [PubMed] [Google Scholar]
- Hertz JM, Sivertsen B, Silahtaroglu A, Bugge M, Kalscheuer V, et al. Early onset, non-progressive, mild cerebellar ataxia co-segregating with a familial balanced translocation t(8;20)(p22;q13) J Med Genet. 2004;41:e25. doi: 10.1136/jmg.2003.011510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Okumura S, Iwatsubo K, Kawabe J, Ohtsu K, et al. Motor dysfunction in type 5 adenylyl cyclase-null mice. J Biol Chem. 2003;278:16936–16940. doi: 10.1074/jbc.C300075200. [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Feenstra I, Van Ravenswaaij-Arts CM, Smeets DF, Menzel C, et al. Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am J Med Genet A. 2008;146A:2053–2059. doi: 10.1002/ajmg.a.32419. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Koch JD, Miles DK, Gilley JA, Yang CP, Kernie SG. Brief exposure to hyperoxia depletes the glial progenitor pool and impairs functional recovery after hypoxic-ischemic brain injury. J Cereb Blood Flow Metab. 2008;28:1294–1306. doi: 10.1038/jcbfm.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Nagarajan P, Wall J, Donovan DJ, Donell RL, et al. Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am J Med Genet A. 2008;146A:1117–1127. doi: 10.1002/ajmg.a.32178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lutz T, Stöger R, Nieto A. CHD6 is a DNA-dependent ATPase and localizes at nuclear sites of mRNA synthesis. FEBS Lett. 2006;580:5851–5857. doi: 10.1016/j.febslet.2006.09.049. [DOI] [PubMed] [Google Scholar]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CG, Ohkawa Y, Coles AH, Garlick DS, Jones SN, et al. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol. 2006;209:162–171. doi: 10.1002/jcp.20718. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Kabuto H, Makino H, Ogawa N. Pole test is a useful method for evaluating the mouse movement disorder caused by striatal dopamine depletion. J Neurosci Methods. 1997;73:45–48. doi: 10.1016/S0165-0270(96)02211-X. [DOI] [PubMed] [Google Scholar]
- McFadyen MP, Kusek G, Bolivar VJ, Flaherty L. Differences among eight inbred strains of mice in motor ability and motor learning on a rotorod. Genes Brain Behav. 2003;2:214–219. doi: 10.1034/j.1601-183X.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Gleeson JG. Cerebellar development and disease. Curr Opin Neurobiol. 2008;18:12–19. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo: a laboratory manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, Hirose Y, Ohara S, Ono T, Watanabe Y. A simple quantitative bradykinesia test in MPTP-treated mice. Res Commun Chem Pathol Pharmacol. 1985;50:435–441. [PubMed] [Google Scholar]
- Pagon RA, Graham JM, Jr, Zonana J, Yong SL. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J Pediatr. 1981;99:223–227. doi: 10.1016/S0022-3476(81)80454-4. [DOI] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, et al. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster EF, Stöger R. CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm Genome. 2002;13:117–119. doi: 10.1007/s00335-001-3042-6. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wigley DB. Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, et al. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranebjaerg L, Teslovich TM, Jones M, Barmada MM, Fagerheim T, et al. Genome-wide homozygosity mapping localizes a gene for autosomal recessive non-progressive infantile ataxia to 20q11–q13. Hum Genet. 2003;113:293–295. doi: 10.1007/s00439-003-0967-8. [DOI] [PubMed] [Google Scholar]
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Wada T, Kubota T, Fukushima Y, Saitoh S. Molecular genetic study of Japanese patients with X-linked alpha-thalassemia/mental retardation syndrome (ATR-X) Am J Med Genet. 2000;94:242–248. doi: 10.1002/1096-8628(20000918)94:3<242::AID-AJMG11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Chd6 ATPase domain shares a high level of homology with SNF2 family members. The amino acid sequence of mouse Chd6. Exon 12 amino acids are boxed. Percentage of homology between the compared sequences is indicated above the sequences by height of the red bar. The highly conserved motif I (ATPase) is indicated in dark blue, the conserved motif Ia is shown in green, and motif II (DEXH helicase) is indicated in magenta. Genome indication is m, mouse; h, human; d, Drosophila; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe (TIFF 181 kb)
Exon 12 −/− mice on either a high-fat or a low-fat diet show no differences in body weight compared to control mice. Adult mixed WT mice (triangles) and mixed Exon 12 −/− mice (open circles) were fed a high-fat (45% kcal fat) or low-fat (10% kcal fat) diet for 6 months and weighed weekly. No significant difference in body weight was measured when mice were fed either the (A) high-fat diet or (B) low-fat diet. WT data are shown ± SEM; Exon 12 −/− SEM values were similar in size, but are not shown (TIFF 70 kb)
Immunohistochemistry analysis of Exon 12 −/− cerebellum revealed no differences in cellular morphology. Cerebella from WT and Chd6 Exon 12 −/− mice were compared at around 17 weeks of age; examples from the 129Sv lineage are shown. A Nissl-stained midline sagittal sections were compared and folia were counted (examples are shown at 2× magnification). The general cellular architecture was found to be similar in mutants compared with WT controls (n > 6). B Calbindin antibody (Swant, Switzerland) was used to detect potential changes within the Purkinje cell layer; consistent differences indicating Purkinje cell loss were not noted in mutant animals and the data were similar in all respects with that of age-matched WT controls (n > 3). (Calbindin - red fluorescence, nuclei are stained blue - H stain, Hoechst). C Antibody for Neurofilament protein (Abcam) was applied to cerebellar sections (Neurofilament protein – green fluorescence, blue – nuclear stain as before). Cerebellum was examined carefully throughout, from deep cerebellar nuclei to the folia (shown in C). The antibody against Neurofilament protein did not reveal any axonal damage in either mutant or WT samples (n > 3). Scale bars (white lines) in B and C represent 100 μm (TIFF 555 kb)